Abstract
This study characterizes the transcriptional expression of the reductive dehalogenase (RDase)-encoding tceA and vcrA genes and evaluates their applicability as potential biological markers of Dehalococcoides activity. When Dehalococcoides ethenogenes 195 was provided with trichloroethene (TCE) as the electron acceptor, the expression of the tceA gene increased by 90-fold relative to that in cells starved of chlorinated ethenes, demonstrating that tceA gene expression is indicative of the active physiological state of this strain. In a Dehalococcoides-containing enrichment culture that contains both the tceA and vcrA genes, the tceA gene was up-regulated in response to TCE and cis-1,2-dichloroethene (cDCE) exposure, while the vcrA gene was up-regulated in response to TCE, cDCE, and vinyl chloride (VC). When chlorinated ethenes were depleted, the RDase-encoding gene transcripts decayed exponentially, with a half-life between 4.8 and 6.1 h, until they reached a stable background level after 2 days. We found that while gene expression correlated generally to the presence of chlorinated ethenes, there was no apparent direct relationship between RDase-encoding transcript numbers and respective rates of TCE, cDCE, and VC dechlorination activities. However, elevated tceA and vcrA expression did correlate with chlorinated-ethene reduction beyond cDCE, suggesting that elevated RDase-encoding transcript numbers could serve as a biomarker for the physiological ability of Dehalococcoides spp. to dechlorinate beyond cDCE.
Trichloroethene (TCE) is a probable human carcinogen (42) and common groundwater contaminant that is endangering many groundwater supplies across the United States (5, 44). In recent years, in situ bioremediation has become a favorable alternative for the remediation of TCE-contaminated sites (25, 26, 31). In field applications, determining the presence or absence of the key organisms and the functional genes that catalyze the desired bioremediation reactions is a critical initial step in evaluating the approach that will most likely lead to successful bioremediation. Further, improved diagnostic data on the in situ physiological state of the key microorganisms catalyzing the desired biochemical reactions could provide information that will facilitate the manipulation of site conditions to optimize cellular activity for improved bioremediation processes.
One useful method for monitoring the physiological state of microorganisms is to analyze the mRNA transcript levels of key functional genes. This strategy has been applied to the analysis of both pure cultures (4, 10, 34, 41) and environmental samples (2, 12, 19, 21, 33) to demonstrate that up-regulated mRNA levels for particular genes correspond with specific observable metabolic functions. In some cases, mRNA levels have gone beyond qualitative agreement to be quantitatively correlated with microbial activity rates. For example, the number of nahA transcripts was shown to increase with naphthalene mineralization rates in sediments (12). A direct correlation has also been reported between the omcB gene, a gene coding for an outer membrane c-type cytochrome, and Fe(III) reduction rates in Geobacter sulfurreducens (4). In contrast, the expression of genes for the reduction of fumarate, dimethyl sulfoxide, and nitrate in Escherichia coli exhibited no correlation with cell growth rates (41), and a correlation was not consistently observed between Hg(II) volatilization rates and the expression of the indigenous mercuric reductase merA gene at a mercury-contaminated site (33). These results demonstrate that gene expression may sometimes, but not always, correlate with measured functional activity.
For TCE reductive dechlorination, only members of the genus Dehalococcoides have been reported to be capable of dechlorination beyond that of dichloroethenes (DCEs) (39). Recently, studies targeting the 16S rRNA gene for identification have shown that Dehalococcoides species are indigenous to many chlorinated-ethene-contaminated sites distributed globally (11, 18, 35). The presence of Dehalococcoides 16S rRNA gene sequences was also shown to qualitatively correlate with the generation of ethene from chlorinated ethenes, while the absence of Dehalococcoides at sites correlated with the accumulation of cis-1,2-dichloroethene (cDCE) (18).
Within the Dehalococcoides genus, four distinct strains, Dehalococcoides ethenogenes 195 (29, 30, 38), Dehalococcoides sp. strain FL2 (17), Dehalococcoides sp. strain VS (7), and Dehalococcoides sp. strain GT (40), have been demonstrated to metabolically reduce TCE by anaerobic dehalorespiration. The tceA gene is present in both strain 195 (38) and strain FL2 (17, 20) and has been shown to participate in the sequential metabolic transformation of TCE to cDCE and vinyl chloride (VC) and in the cometabolic reduction of VC to ethene (27, 28). Strains VS (32) and GT (40) also metabolize TCE, but the vcrA gene rather than the tceA gene is implicated in this reaction, while strain BAV1 metabolically dechlorinates VC via the bvcA gene (15, 16, 24). Another Dehalococcoides strain that possesses differing metabolic activities is strain CBDB1, which metabolically dechlorinates chlorobenzenes and some polychlorinated dibenzodioxin congeners but exhibits no activity on chlorinated ethenes (1, 3).
Despite the observed variety of metabolic functions, the above-mentioned Dehalococcoides strains have highly similar 16S rRNA gene sequences. For example, of the 1,421 and 1,420 base pairs of 16S rRNA gene sequences in strains CBDB1 and BAV1, respectively, 1,404 and 1,402 base pairs are the same as those in strain 195 (GenBank accession numbers AY165308, AF230641, and AF004928). Further, the 16S rRNA gene sequences of FL2 and CBDB1 are identical to that of KB-1/VC, a highly enriched TCE-to-ethene-dechlorinating strain (9). Because of the highly conserved nature of the 16S rRNA gene, its phylogeny alone is an inadequate predictor of the metabolic activities of Dehalococcoides (9, 26). Further, the presence or absence of a metabolic gene in a community may also be an inadequate predictor of activity since the presence of a gene alone does not guarantee expression (26). Gene expression data may overcome these limitations by verifying that the functionally important Dehalococcoides spp. are physiologically active and are carrying out specific metabolic functions of interest in a microbial community.
Previous findings (22, 23, 43) have provided some evidence that reductive dehalogenase (RDase)-encoding genes are expressed in the presence of a variety of chlorinated organics and under different environmental conditions but the relationship between activity and expression has not been examined in detail. In this study, we assess the uses of tceA and vcrA gene expression as biological markers of the physiological state and the TCE-to-ethene dechlorination activity of Dehalococcoides spp. The findings presented here represent an important step toward developing gene expression analysis as a diagnostic tool for the improved monitoring of in situ bioremediation processes.
MATERIALS AND METHODS
Culture growth and maintenance.
D. ethenogenes 195 was grown in batch cultures in defined medium (15; J. He et al., unpublished data). The 100-ml cultures were grown in 160-ml serum bottles with 5 mM acetate as a carbon source and a H2-CO2 (80:20) headspace as the electron donor source. Cultures were routinely transferred at a 1:50 dilution and amended with one to three doses of 56 μmol TCE.
The Dehalococcoides-containing TCE enrichment culture was derived from a contaminated site at Alameda Naval Air Station in California and has been designated ANAS. The culture conditions and maintenance procedures have been described previously (36). Briefly, 500 ml of culture is maintained in a 1.5-liter reactor at 25 to 28°C under 0.80 atm of positive pressure of N2-CO2 (90:10) in a semibatch feeding mode of 25 mM of lactate and 0.1 mM of TCE. When the amended TCE has been completely dechlorinated to ethene, typically after 4 to 7 days, the reactor is purged with N2 and 60 to 100 ml of culture is withdrawn and replaced with an equal volume of fresh anaerobic medium. This enrichment culture has been functionally stable in our laboratory for at least 6 years.
Chemical analyses.
Ethene and chlorinated ethenes (TCE, cDCE, and VC) were analyzed by injecting 100 μl of headspace samples into an HP 5890 gas chromatograph (GC) fitted with a flame ionization detector. The GC was programmed to hold at 45°C for 1 min, ramp to 180°C at 30°C/min, and then hold at 180°C for 5 min. The injector and detector temperatures were isothermal at 220°C and 250°C, respectively. Gas flow was set at 3.0 ml/min, and the column was a GC-Gas Pro (J&W Scientific, Folsom, CA). External calibration curves were generated using serum bottles containing anaerobic medium with known amounts of each compound. The concentrations of gas (CG) were converted to total mass of chlorinated ethenes (M) in vials by using Henry's constants (H) (13, 14), according to a material balance, as follows: M = CGVG + CG/H · VL, where VG and VL are the gas and liquid volumes in the vials, respectively.
Clone library and phylogenetic analysis.
A clone library of the ANAS enrichment culture was produced with DNA extracted from 1 ml of culture taken at the end of a feeding cycle. Universal primers 8F (5′-AGAGTTTGATCCTGGCTCAG) and 1492R [5′-GG(C/T)TACCTTGTTACGACTT] (8) were used to amplify the whole culture community 16S rRNA gene. The following PCR parameters were used for amplification: an initial denaturation step at 94°C (12 min), followed by 30 cycles of 94°C (60 s), 50°C (45 s), and 72°C (120 s), with a final extension at 72°C for 12 min. The clone library was constructed with a TOPO TA cloning kit (with pCR2.1-TOPO vector) (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A total of 92 plasmid inserts was later digested with the restriction enzyme HhaI (Promega, Madison, WI) at 34°C for 3 h, and the resulting fragments were analyzed with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) using DNA 7500 chips according to the manufacturer's protocol. Distinct and/or prominent clones (34 out of 92 clones) were selected for sequencing at Elim Biopharmaceuticals, Inc. (Hayward, CA) and aligned using Cap3 assembly software (http://bio.ifom-firc.it/ASSEMBLY/assemble.html). After alignment and visual checking, the nearly full-length 16S rRNA gene sequences (1,458 bp) were compared to sequences in GenBank with BLASTn (http://www.ncbi.nlm.nih.gov/), and the nearest cultured neighbors were identified.
Quantification of mRNA and DNA.
Nucleic acids were extracted from 1- and 2-ml culture samples from the ANAS enrichment culture and strain 195 isolate, respectively. Cells were collected by centrifugation (12,000 × g at 4°C), the supernatants were discarded, and the cell pellets were stored at −80°C until processing. Genomic DNA was isolated from frozen cell pellets using an UltraClean microbial DNA kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's instructions. Total RNA was isolated from frozen cell pellets by the acid phenol method, described previously (22). For RNA isolation, 2 μl of 107 transcripts μl−1 of luciferase control RNA (Promega, Madison, WI) (ref mRNA) was added prior to cell lysis to serve as a sample-specific internal reference for mRNA losses during mRNA isolation, reverse transcription, and quantification (22).
To quantify tceA and vcrA genes, real-time quantitative PCR (qPCR) was applied to genomic DNA in conjunction with the absolute standard curve method as previously described (23). Primers and probes used for qPCR of tceA were 5′-ATCCAGATTATGACCCTGGTGAA (forward), 5′-GCGGCATATATTAGGGCATCTT (reverse), and 5′-FAM-TGGGCTATGGCGACCGCAGG-TAMRA (where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxymethylrhodamine) (probe) (22); and those used for vcrA were 5′-CTCGGCTACCGAACGGATT (forward), 5′-GGGCAGGAGGATTGACACAT (reverse), and 5′-FAM-CGCACTGGTTATGGCAACCACTC-TAMRA (probe) (19a).
To quantify tceA or vcrA expression, multiplex reverse transcription (RT) followed by multiplex qPCR was applied in conjunction with the absolute standard curve method to independently quantify tceA or vcrA mRNA and ref mRNA as previously described (22). RT was performed using the tceA or vcrA reverse primers listed above and the ref reverse primer (5′-GGAAGTTCACCGGCGTCAT) (22). Multiplex qPCR was performed using the tceA or vcrA primers listed above, the ref reverse primer listed above, the ref forward primer (5′-TACAACACCCCAACATCTTCGA), and the ref probe (5′-VIC-CGGGCGTGGCAGGTCTTCCC-TAMRA) (22). To control for sample-specific losses of mRNA during cell lysis, RNA isolation, and DNA removal steps, the quantity of tceA or vcrA mRNA was divided by the fractional recovery of ref mRNA, which typically ranged between 20 and 35% (22). The expression of tceA or vcrA was then calculated as the ref mRNA-normalized quantity of tceA or vcrA mRNA per ml of ANAS sample divided by the quantity of the tceA or vcrA gene per ml of ANAS sample, giving units of transcription as the number of tceA transcripts per tceA gene or number of vcrA transcripts per vcrA gene.
Expression and dechlorination activity studies.
For experiments conducted with strain 195, after the first dose of TCE was completely dechlorinated to VC and ethene in duplicate bottles, one of the cultures was amended with an additional 33 μmol of TCE, and the other was left without any chlorinated ethene. Dechlorination activity and tceA expression were tracked over 24 h.
Prior to the start of each experiment conducted with ANAS, a subculture was withdrawn from the source reactor after TCE had been completely dechlorinated to ethene and then stored in an anaerobic serum bottle filled with N2-CO2 (90:10) in the absence of chlorinated ethenes for either 2 or 6 days to allow the degradation of mRNA transcripts that remained from the previous feeding cycle. All experimental incubations were performed at 30°C with shaking at 150 rpm.
Measurements of tceA and vcrA expression over a feeding cycle in ANAS were conducted in 58-ml anaerobic serum bottles with a N2-CO2 (90:10) headspace, 30 ml of culture, 11 to 14 μmol of TCE, cDCE, or VC, and 42 mM lactate.
In the 3-day expression study, 50 ml of ANAS in a 70-ml serum bottle was amended with a H2-CO2 (80:20) headspace instead of lactate to ensure that the supply of electron donors was not limited. Twelve micromoles of TCE was added at the onset, and thereafter, TCE was reamended as needed throughout the 3 days to keep the culture active. Every 24 h, liquid samples were collected for DNA and RNA analyses, and the headspace was flushed with H2-CO2 (80:20) to replenish electron donors.
For the decay experiment, 65 ml of ANAS was initially provided with 13 μmol of TCE and 30 mM of lactate in a 160-ml serum bottle, and substrates were reamended as necessary to maintain activity. At the end of the 24 h, N2 gas was used to strip all chlorinated ethenes from the culture and liquid samples were immediately collected to analyze the decay of tceA mRNA with time.
Relating tceA transcript levels with initial dechlorination rates.
For experiments evaluating the relationship between tceA transcript levels and initial TCE dechlorination rates, five 16-ml cultures in 58-ml anaerobic serum bottles were maintained with different total concentrations of TCE and cDCE for 24 h to generate different transcript levels (23). After 24 h of chlorinated-ethene exposure, the cultures were purged with N2 to remove all chlorinated ethenes, and an aliquot of each culture was transferred to a clean 58-ml anaerobic serum bottle for the initial rate assay. At the onset of the initial rate assay, each culture was provided with a H2-CO2 (80:20) headspace along with chloramphenicol and 4 μmol of TCE.
Chloramphenicol was added to cultures from an anaerobic stock solution to obtain a final concentration of 0.17 g/liter (37) to inhibit de novo protein synthesis. Chlorinated ethene measurements were taken over 10 h, and the slope of the dechlorination curve was used to calculate the initial dechlorination rate of TCE. Dechlorination rates were normalized to the number of copies of the tceA gene.
Dechlorination activities of constitutive and/or stable reductases.
The activities of constitutively expressed and/or stable reductases in ANAS were tested by transferring 90 ml of culture from the source reactor to a 160-ml anaerobic serum bottle with a N2-CO2 (90:10) headspace. The culture was starved of chlorinated ethenes and lactate following its removal from the reactor. After 0, 4, 9, 15, and 42 days, 13-ml aliquots of the starved culture were transferred into 58-ml anaerobic serum bottles where the initial TCE dechlorination rate was measured in the presence of chloramphenicol, H2-CO2 (80:20), and 4 μmol of TCE. Chlorinated ethene concentrations were measured for 32 h to analyze TCE dechlorination activity as well as VC formation activity.
RESULTS
tceA gene expression of Dehalococcoides ethenogenes 195.
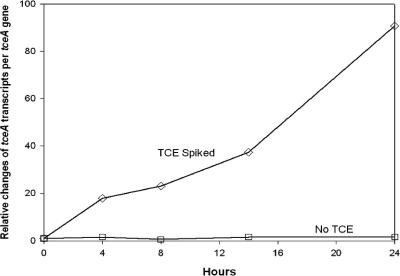
Changes in the expression level of the tceA gene after exposure to TCE were measured in a pure culture of D. ethenogenes 195. The expression of tceA was up-regulated 90-fold during the 24-h exposure, while a control culture that contained no chlorinated ethenes showed no significant change in expression (Fig. 1). These results demonstrate that tceA expression is induced by TCE exposure in previously starved cells of D. ethenogenes 195. During the 24-hour exposure experiment, 2% of the amended TCE was reduced to cDCE. The observed up-regulation of the tceA gene is similar to the behavior observed previously in the Dehalococcoides-containing ANAS enrichment culture (22, 23).
FIG. 1.
tceA gene expression profile of a pure culture of Dehalococcoides ethenogenes 195 in the presence and absence of TCE over 24 h. Measurements of tceA transcripts per tceA gene at each time point are normalized to the value at time zero to obtain the relative changes. Data are calculated from averages of triplicate qPCRs and RT-qPCRs, and error bars are smaller than the box symbol.
Phylogenetic characteristics of the TCE enrichment culture.
Prior to studying the activity and expression of RDase-encoding genes in the ANAS enrichment, a clone library was constructed to evaluate the species distribution and to determine whether the phylogenetic makeup of this enrichment had changed significantly since its previous characterization (36). Reassuringly, the species distributions in the two libraries showed strong similarity. The functionally important Dehalococcoides spp. comprised 10% of the clones, and Dehalobacter restrictus strain TEA was most similar to 2% of the clones. Fermenting organisms were also well represented in the clones, including Clostridium lactatifermentans (48%) and Desulfovibrio desulfuricans (4%). Sequence analysis of the 10 clones belonging to the genus Dehalococcoides showed that all 10 had at least 97% similarity to the isolated Dehalococcoides strains deposited in GenBank.
Dechlorination activity and expression of the tceA and vcrA genes in ANAS.
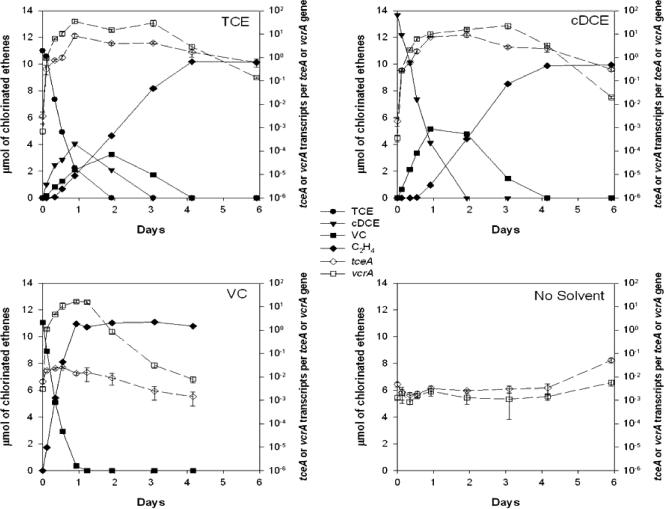
The expression dynamics of the two reductive dehalogenase genes were characterized in the ANAS enrichment culture. After the chlorinated-ethene-starved cells were exposed to TCE or cDCE, both the tceA and vcrA genes were up-regulated by 3 to 4 orders of magnitude (Fig. 2). However, while the vcrA gene was significantly up-regulated by 3 orders of magnitude during VC exposure, the tceA gene showed only a weak response, with an increase of less than 3.5-fold (Fig. 2). The culture amended with no electron acceptor showed no change in its expression profile throughout the experiment (Fig. 2).
FIG. 2.
Reductive dechlorination of chlorinated ethenes (solid lines) and expression profile of the tceA and vcrA genes (dashed lines) over a feeding cycle for ANAS. Expression measurements are calculated from averages of triplicate qPCRs and RT-qPCRs, and error bars represent 1 standard deviation.
Dechlorination of chlorinated ethenes coincided with the up-regulation of the tceA and/or vcrA gene. The rates of reduction of the initially amended chlorinated ethenes were constant during the first 24 h, resulting in linear regressions on TCE, cDCE, and VC reduction, with an r2 of >0.98 (Fig. 2). In all the actively dechlorinating cultures, near-stoichiometric conversion to ethene was achieved, with little accumulation of intermediates in the cases of TCE and cDCE.
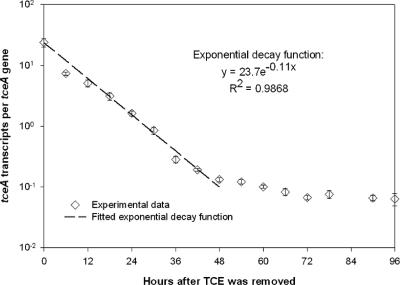
In general, the dynamics of the functional gene profiles showed an up-regulation in response to the chlorinated ethenes, reaching a steady-state level until the available electron acceptors were reduced, followed by a gradual decay (Fig. 2). The decay in transcripts occurred only after the chlorinated ethenes were depleted. With the data from the VC feeding (Fig. 2), the decay of the vcrA transcripts following the complete reduction of VC to ethene could be described by an exponential decay function (r2 = 0.98) with a rate constant of 0.14 h−1 and a half-life of 4.8 h. Because the tceA gene was induced by both TCE and cDCE and the endpoint of cDCE was less clear, the decay rate of the tceA transcripts was measured in a separate experiment following the abrupt stripping of TCE from the culture. In the absence of TCE, the tceA mRNAs began to decay exponentially and returned to a lower stable level within 2 days (Fig. 3). Similarly, an exponential decay function with a rate constant of 0.11 h−1 and a half-life of 6.1 h could be fitted to the tceA data for the first 2 days following TCE removal (r2 = 0.99).
FIG. 3.
Decay profile of tceA transcripts when chlorinated ethenes were stripped from the culture after 24 h of active dechlorination.
Relationship between transcript levels and dechlorination activity.
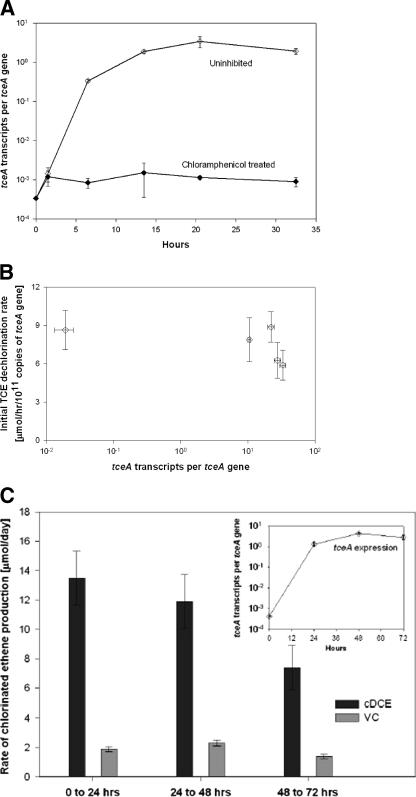
In order to determine whether a proportional correlation could be found between tceA transcript levels and TCE dechlorination activity, experiments were conducted in which TCE dechlorination rates were measured in cultures that expressed a wide range of tceA transcript levels. In order to prevent de novo protein synthesis at the conclusion of these experiments during the TCE dechlorination rate assay, chloramphenicol was applied during the assay. An experiment measuring the effects of chloramphenicol on tceA transcription (Fig. 4A) indicated that chloramphenicol inhibited tceA expression, in addition to presumably inhibiting translation. Hence, dechlorination activity measured in the presence of chloramphenicol is expected to be the result only of enzymes synthesized prior to chloramphenicol addition.
FIG. 4.
(A) tceA gene expression profile with and without the influence of chloramphenicol. (B) Initial dechlorination rate of TCE in the presence of chloramphenicol after the establishment of different transcript levels in five respective cultures. Rate measurements are normalized to the tceA gene copy number. The error bars for the rate measurement represent the 95% confidence interval of the slope of TCE dechlorination over 10 h. (C) Dechlorination activity of the ANAS enrichment culture over three consecutive days as represented by the rate of production of the TCE daughter products cDCE and VC every 24 h. Error bars represent the 95% confidence interval. The inset graph shows tceA gene expression during the 3 days of active dechlorination.
Surprisingly, when TCE dechlorination rates were measured for cultures expressing tceA transcript levels that spanned 3 orders of magnitude, no significant relationship between TCE dechlorination rates and transcript levels was observed (Fig. 4B).
In an experiment that monitored tceA gene expression and TCE dechlorination rates over a 3-day period, a similar lack of relationship between expression and dechlorination activity was observed (Fig. 4C). Despite the occurrence of active dechlorination during each of the three 24-h intervals, the rate of daughter product generation did not correlate with the increase in transcript numbers, and in fact, the rate decreased slightly as time proceeded.
Measuring constitutive and/or stable dechlorination activity in ANAS.
In order to evaluate whether the lack of correlation between tceA transcript levels and TCE dechlorination activity was caused by other constitutively expressed and/or highly stable TCE reductases in the ANAS enrichment culture, the dechlorination activities of cultures that were starved of chlorinated ethenes for periods ranging from 0 to 42 days were measured (Table 1).
TABLE 1.
Dechlorination activity
| No. of days without chlorinated ethenes | Mass amt (μmol) ofa:
|
|
|---|---|---|
| TCE dechlorinated | VC produced | |
| 0 | 3.0 | 0.7 |
| 4 | 2.1 | 0.3 |
| 9 | 1.7 | 0.2 |
| 15 | 1.5 | 0 |
| 42 | 1.0 | 0 |
Data represent mass amounts of TCE dechlorinated and VC produced in ANAS enrichment culture that had been starved of chlorinated ethenes for various durations. Data were measured after 14 h. cDCE was produced during each day of sampling but was not included in this table.
Surprisingly, TCE dechlorination activity was detected in ANAS even after 42 days of starvation, when an aliquot of the starved culture was assayed in the presence of fresh TCE with chloramphenicol added to inhibit de novo protein synthesis. Dechlorination activity did decline with increasing starvation time, as reflected in the mass of TCE dechlorinated and VC generated (Table 1). This result indicates that ANAS contains constitutively expressed and/or highly stable TCE reductases that contribute to the overall measured TCE dechlorination activity. While TCE-to-cDCE dechlorination activity persisted for 42 days, the formation of VC ceased in cultures that had been starved for 9 or more days, indicating that the constitutively expressed and/or stable reductases in ANAS do not contribute significantly to dechlorination beyond DCE.
DISCUSSION
Dehalococcoides strains that contain the tceA or the vcrA gene are important players in the bioremediation of TCE-contaminated sites, since these strains can catalyze the complete dechlorination of TCE to ethene (17, 20, 27, 28, 32, 38, 40). Analytical methods that can effectively measure the presence and physiological activities of these strains would be extremely beneficial for optimizing the dechlorination of TCE. One direct method for monitoring the physiological state of these Dehalococcoides strains is to quantify their RDase-encoding gene expression.
To assess the applicability of RDase-encoding gene expression as a biomarker, we examined the response of Dehalococcoides cells to chlorinated-ethene exposure. When both a pure culture of D. ethenogenes 195 and the ANAS enrichment culture were actively dechlorinating TCE, the expression of the tceA and/or the vcrA gene increased by orders of magnitude compared to that of the chlorinated ethene-starved cells (Fig. 1 and 2), demonstrating that functional gene expression correlates with an active physiological state of Dehalococcoides strains. These results are similar to those found in previous studies of RDase gene expression dynamics in Dehalococcoides-containing enrichments (22, 23, 43). Together, the results presented here of the tceA and vcrA genes, along with previous findings, confirm that the expression of RDase-encoding genes can serve to indicate that Dehalococcoides strains are physiologically active. Such identification is especially valuable in microbial communities that contain multiple dechlorinating organisms, such as with the ANAS culture, where tracking the transformation of chlorinated ethenes alone is insufficient to determine the role or activity of Dehalococcoides strains.
Because the tceA and vcrA genes respond differently to VC, monitoring RDase-encoding gene expression can also indicate the likelihood of a complete reduction of TCE to ethene and assign activity to the appropriate strain. In the ANAS community, based on the expression response, we can conclude that both the tceA- and vcrA-carrying strains are active on TCE and cDCE reduction but that the vcrA strain is responsible primarily for the reduction of VC. Having two strains in ANAS that are active on TCE and cDCE reduction provides a redundancy in function and ensures the reduction of the higher-chlorinated ethenes to VC. From the expression profile, both strains in the ANAS culture responded simultaneously and independently to chlorinated-ethene exposure. This is demonstrated in the cDCE-amended experiment (Fig. 2), where the tceA gene starts to decay when cDCE is depleted, while the vcrA gene is maintained at a steady level until VC is transformed, and in the differential induction patterns caused by VC exposure.
In all of the expression profiles that showed a positive response to chlorinated ethenes (Fig. 2), the dynamics of the changes in expression could be categorized into three stages: an initial up-regulation followed by a steady state and finally a decay, when the inducing chlorinated ethenes have been transformed. In the absence of inducers, tceA and vcrA transcripts decayed exponentially, with half-lives on the order of 6.1 to 4.8 h and decay constants of 0.11 h−1 and 0.14 h−1, respectively (Fig. 2 and 3). Compared to that of aerobic organisms, such as E. coli, whose typical mRNA half-lives last for minutes (6), the decay of the reductive dehalogenase transcripts is relatively low. However, low decay rates mirror the relatively low growth rates reported for the genus Dehalococcoides, with a doubling time of about 19 h for D. ethenogenes 195 (30). Despite the relative stability of these reductive dehalogenase transcripts, they could still serve as effective biomarkers for the physiological state of Dehalococcoides cells in bioremediation projects, which typically occur over time scales of months to years. Further, considering that Dehalococcoides cells can decouple growth from dechlorination activity (30), reductive dehalogenase expression levels can be diagnostic of cell activity when cell numbers are not increasing.
Predictive quantitative correlations between transcript levels and dechlorination activity have been reported for some bacteria (4, 12), while a lack of correlation has been demonstrated for others (33, 41). In this study, the constant rate measured in the initial dechlorination of the respective electron acceptors in the feeding cycle (Fig. 2) indicates that the activity of the ANAS had no direct correlation with the magnitude of the increase in transcript levels. This observation held whether the chlorinated ethene was being catalyzed by vcrA alone for VC reduction or by both the vcrA and tceA genes for TCE and cDCE. In the follow-up experiments that focused on the tceA gene, the lack of a proportional correlation was also consistently observed (Fig. 4).
In the ANAS culture, a significant fraction of the TCE dechlorination activity appears to come from non-tceA reductases (Table 1). One possible source, D. restrictus strain TEA, an organism that dechlorinates TCE to cDCE (45), was detected by the clone library. It is possible that there are a variety of unidentified organisms with multiple TCE reductases in ANAS. However, despite the long-lived activity exhibited by the unidentified TCE reductases, dechlorination sustained beyond that of DCE to VC or ethene was not observed during TCE starvation (Table 1), suggesting the lack of involvement of Dehalococcoides cells in these reactions.
Besides reductases from non-Dehalococcoides spp., it is possible that multiple Dehalococcoides reductases other than tceA and vcrA are participating in chlorinated-ethene reduction. Annotation of the strain 195 genome revealed 17 intact putative reductive dehalogenases (38), and the actual role and importance of each of these reductases in TCE reduction have yet to be understood fully. A recent transcription analysis of the Dehalococcoides-containing enrichment culture KB-1 provided evidence that multiple reductive dehalogenase homolog genes were indeed transcribed in the presence of chlorinated organics (43). Therefore, TCE reduction to ethene is likely catalyzed by multiple reductases which might exhibit a range of expression responses, and to fully understand the reactions would require employing a more global transcription analysis of the genome. Surprisingly, though, despite the possibility that exposure to chlorinated ethenes induced multiple active RDases in this study, the lack of a subsequent increase in chlorinated-ethene dechlorination activity demonstrates that transcript numbers and activity indeed were not correlated. Furthermore, since transcription of the RDase-encoding genes is only the first step toward activity, with translation and proper protein placement potentially playing roles in the actual enzyme function, an improved understanding of function within a cellular context will aid in developing the most effective biomarkers for monitoring activity.
In summary, this study has shown that measurements of the expressions of the tceA and vcrA genes can be useful biomarkers for gauging the overall physiological activities of Dehalococcoides spp. but cannot be used quantitatively for predicting TCE-to-ethene dechlorination rates. The ability to confirm the physiological state of this key organism within microbial communities and its activity in the generation of VC and ethene will be useful in the optimization of bioremediation strategies for TCE.
Acknowledgments
We thank Steve Zinder of Cornell University for providing a pure culture of D. ethenogenes 195.
This work was supported by National Science Foundation grant no. BES-05-04244, by NIEHS Superfund Basic Research Project grant ES04705, and by the University of California Toxic Substances Research and Training Program.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Alfreider, A., C. Vogt, and W. Babel. 2003. Expression of chlorocatechol 1,2-dioxygenase and chlorocatechol 2,3-dioxygenase genes in chlorobenzene-contaminated subsurface samples. Appl. Environ. Microbiol. 69:1372-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 4.Chin, K.-J., A. Esteve-Núñez, C. Leang, and D. R. Lovley. 2004. Direct correlation between rates of anaerobic respiration and levels of mRNA for key respiratory genes in Geobacter sulfurreducens. Appl. Environ. Microbiol. 70:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Committee on Ground Water Cleanup Alternatives, National Research Council. 1994. Alternatives for groundwater cleanup. National Academy Press, Washington, D.C.
- 6.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 7.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2004. Comparative evaluation of chloroethene dechlorination to ethene by Dehalococcoides-like microorganisms. Environ. Sci. Technol. 38:4768-4774. [DOI] [PubMed] [Google Scholar]
- 8.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duhamel, M., K. Mo, and E. A. Edwards. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellington, M. J. K., G. Sawers, H. J. Sears, S. Spiro, D. J. Richardson, and S. J. Ferguson. 2003. Characterization of the expression and activity of the periplasmic nitrate reductase of Paracoccus pantotrophus in chemostat cultures. Microbiology 149:1533-1540. [DOI] [PubMed] [Google Scholar]
- 11.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 12.Fleming, J. T., J. Sanseverino, and G. S. Sayler. 1993. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ. Sci. Technol. 27:1068-1074. [Google Scholar]
- 13.Gossett, J. M. 1987. Measurement of Henry's law constants for C1 and C2 chlorinated hydrocarbons. Environ. Sci. Technol. 21:202-208. [Google Scholar]
- 14.Harvey, A. H. 1996. Semiempirical correlation for Henry's constants over large temperature ranges. AIChE J. 42:1491-1494. [Google Scholar]
- 15.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, J., K. M. Ritalahti, K. L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 17.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. E. Löffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 70:7251-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Holmes, V. F., J. He, P. K. H. Lee, and L. Alvarez-Cohen. 2006. Discrimination of functionally distinct Dehalococcoides strains in a trichloroethene enrichment by quantification of their functional reductases. Appl. Environ. Microbiol. 72:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. von Wintzingerode, H. Görisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffrey, W. H., S. Nazaret, and T. Barkay. 1996. Detection of the merA gene and its expression in the environment. Microb. Ecol. 32:293-303. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. R., P. K. H. Lee, V. F. Holmes, and L. Alvarez-Cohen. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, D. R., P. K. H. Lee, V. F. Holmes, A. C. Fortin, and L. Alvarez-Cohen. 2005. Transcriptional expression of the tcea gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 71:7145-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krajmalnik-Brown, R., T. Hölscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. E. Löffler. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovley, D. R. 2001. Bioremediation—anaerobes to the rescue. Science 293:1444-1446. [DOI] [PubMed] [Google Scholar]
- 26.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1:35-44. [DOI] [PubMed] [Google Scholar]
- 27.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maymó-Gatell, X., Y. T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 31.McCarty, P. L. 1997. Microbiology—breathing with chlorinated solvents. Science 276:1521-1522. [DOI] [PubMed] [Google Scholar]
- 32.Müller, J. A., B. M. Rosner, G. von Abendroth, G. Meshulam-Simon, P. L. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazaret, S., W. H. Jeffrey, E. Saouter, R. Von Haven, and T. Barkay. 1994. merA gene expression in aquatic environments measured by mRNA production and Hg(II) volatilization. Appl. Environ. Microbiol. 60:4059-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neretin, L. N., A. Schippers, A. Pernthaler, K. Hamann, R. Amann, and B. B. Jørgensen. 2003. Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environ. Microbiol. 5:660-671. [DOI] [PubMed] [Google Scholar]
- 35.Rahm, B. G., S. Chauhan, V. F. Holmes, T. W. Macbeth, K. S. Sorenson, Jr., and L. Alvarez-Cohen. 2006. Molecular characterization of microbial populations at two sites with differing reductive dechlorination abilities. Biodegradation 14:1-12. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 36.Richardson, R. E., V. K. Bhupathiraju, D. L. Song, T. A. Goulet, and L. Alvarez-Cohen. 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 36:2652-2662. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 38.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 39.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 40.Sung, Y., K. M. Ritalahti, R. P. Apkarian, and F. E. Löffler. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tseng, C.-P., A. K. Hansen, P. Cotter, and R. P. Gunsalus. 1994. Effect of cell growth rate on expression of the anaerobic respiratory pathway operons frdABCD, dmsABC, and narGHJI of Escherichia coli. J. Bacteriol. 176:6599-6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Department of Health and Human Services. 2005. Report on carcinogens, 11th ed. U.S. Department of Health and Human Services, Research Triangle Park, N.C. [Online.] http://ntp.niehs.nih.gov/ntp/roc/toc11.html.
- 43.Waller, A. S., R. Krajmalnik-Brown, F. E. Löffler, and E. A. Edwards. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westrick, J. J., J. W. Mello, and R. F. Thomas. 1984. The groundwater supply survey. J. Am. Water Works Assoc. 76:52-59. [Google Scholar]
- 45.Wild, A., R. Hermann, and T. Leisinger. 1997. Isolation of an anaerobic bacterium which reductively dechlorinates tetrachloroethene and trichloroethene. Biodegradation 7:507-511. [DOI] [PubMed] [Google Scholar]