Abstract
Vibrio vulnificus causes rare but frequently fatal septicemia associated with raw oyster consumption by persons with underlying hepatic or immune system dysfunction. The virulence potential of environmental reservoirs appears widely distributed, because most strains are virulent in animal models; however, several investigations recently demonstrated genetic divergence among strains from clinical versus environmental origin at independent genetic loci. The present study used PCR to screen DNA polymorphisms in strains from environmental (n = 35) or clinical (n = 33) sources, and genomic relationships were determined by repetitive extragenic palindromic DNA PCR (rep-PCR) typing. Significant (P < 0.01) association was observed for typical “clinical” or “environmental” polymorphism profiles based on strain origin. Most oyster isolates (88%), including all of those with the “environmental” profile, also formed a single rep-PCR genogroup. Clinical isolates within this group did not have the typical “clinical” profile. On the other hand, clinical isolates with the typical polymorphism profile were distributed among multiple rep-PCR genogroups, demonstrating greater genetic diversity than was evident by profiling genetic polymorphisms. Wound isolates were genetically distinct from typical blood isolates by all assays. Strains from an outbreak of wound infections in Israel (biotype 3) were closely related to several U.S. strains by rep-PCR, indicating potential reservoirs of emerging disease. Strains genetically related to blood isolates appeared to be relatively rare in oysters, as only one had the “clinical” polymorphism profile or clustered by rep-PCR. However, this study was not an extensive survey, and more sampling using rep-PCR for sensitive genetic discrimination is needed to determine the virulence potential of environmental reservoirs.
Vibrio vulnificus is associated with serious wound infections or frequently fatal (mortality rates are generally >50%) septicemia related to consumption of raw shellfish, particularly oysters (5, 37). The bacterium is indigenous to temperate estuaries, and prevalence in oysters and seawater approaches 100% during warmer months (25, 45). Most strains isolated from environmental reservoirs appear to be as virulent as clinical strains in animal models (13, 35, 36, 39). Also, multiple virulence factors have been proposed for V. vulnificus but are generally present in most strains and do not provide predictive value (16, 36, 39, 50). Virulent strains are distinguished by opaque colony morphology (34, 49), which reflects expression of a protective capsular polysaccharide (CPS); however, both clinical and environmental strains are generally encapsulated (45). Thus, appropriate markers to screen the virulence potential of V. vulnificus in environmental reservoirs are not available.
Recently, DNA sequence polymorphisms at individual loci discriminated isolates from clinical versus oyster origin in several independent studies. Polymorphic variants generally included two genotypes, such as types A and B of the 16S rRNA gene, whose distribution significantly correlated with either environmental or clinical origin, respectively (2, 20, 26, 42). Similar genetic distinctions were reported for types E and C derived from sequences of random amplification of polymorphic DNA (RAPD) typing (32, 43) and for CPS alleles 1 and 2 from the group 1 CPS operon (8). In addition, clinical strains were more likely than environmental strains to be positive for the viuB gene encoding siderophore biosynthesis (28). These studies disagreed with multiple analyses of V. vulnificus that reported no correlation of genotype (6, 11, 17, 18, 19, 38, 51), biotype (1, 40), or serotype (1, 18, 52) with clinical origin or virulence potential. Furthermore, 16S RNA genotypes showed equivalent virulence potential in animal and cell culture models of infection (13); therefore, inferences that allelic markers equate to virulence potential are still unsubstantiated.
In the present study, clinical and environmental strains of V. vulnificus were screened for DNA polymorphisms at multiple loci in order to establish their distribution among the same group of strains and validate their application as markers for strain discrimination. Repetitive extragenic palindromic PCR (rep-PCR) was used to determine the similarity of these strains at the genomic level. rep-PCR targets conserved repetitive elements that are distributed throughout the genome for discrimination of interstrain variation based on amplicon size and intensity. rep-PCR greatly enhances assay reproducibility and strain discrimination compared to other PCR-based platforms (14, 24, 41) and has proven to be highly effective for molecular typing of V. cholerae (10, 15, 31, 33), V. parahaemolyticus (44), and V. vulnificus (30). The present study confirmed genetic distinctions of V. vulnificus strains from clinical versus environmental sources and showed that better discrimination was obtained by rep-PCR molecular typing than by other genetic markers at multiple loci.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
V. vulnificus strains included 68 isolates that were divided between clinical (n = 33) and environmental (n = 35) origins. Most of the clinical (n = 23) and environmental (n = 24) isolates were provided by P. Gulig and were selected to provide a comparison to previous genetic typing (26) and virulence analyses (13). The two strains for which genomic sequences are available, CMCP6 (21) and YJ018 (9), were also included, along with others that have been commonly used in multiple investigations: V. vulnificus strains 345 (34, 48, 50), MO6-24 (8, 45, 46, 47, 48, 49, 50), C7184 (29, 34, 48, 50), and E4125 (8, 22, 34, 48, 50), dating back 20 years or more. Additional strains included isolates (n = 10) from more recent environmental sampling in Florida (2004; unpublished data) and strains provided by A. DePaola from wound infections in Denmark (n = 2; biotype 1) and Israel (n = 2; biotype 3), as well as a biotype 3 strain from an unspecified environmental source. All strains from clinical cases of septicemia or from oysters were biotype 1. All strains were confirmed as V. vulnificus by the species-specific vvhA PCR and DNA probe, as previously described (7, 45). Pure cultures were stored in Luria-Bertani (LB) broth with 50% glycerol at −70°C and observed for opaque or translucent colony morphology on LB agar incubated at 30°C. Unless otherwise noted, reagents and media were purchased from Difco.
Multilocus PCR analysis of V. vulnificus.
Distribution of polymorphic DNA sequences was determined by PCR profiling using previously described or new primer sets that discriminated between two genotypes or alleles at each locus. Primer sets derived from the group 1 CPS operon (8) distinguished between divergent open reading frames (ORFs) for CPS alleles (1 versus 2) encoding proteins of unknown function. Primers from another ORF identified by RAPD typing (also of unknown function) determined C type versus E type (32). For these assays, DNA was extracted using an UltraClean Microbial DNA Isolation kit (MO BIO Laboratories, Inc., Solana Beach, CA), and templates (100 ng in a 25-μl reaction) were amplified with Pfu DNA Polymerase (Stratagene) on a GeneAmp PCR System 2400 (Perkin Elmer) or Mastercycler gradient (Eppendorf) thermocyclers under the previously described conditions.
A real-time PCR method was developed for discriminating between 16S rRNA variants that were previously described as 16S type A and 16S type B (26) based on DNA sequence polymorphisms (2). Primer sets are shown in Table 1. Real-time PCRs of 20 μl contained 2 mM MgCl2, 5 μM of each primer, 2 μl of LightCycler FastStart DNA Master SYBR Green I reaction mix (Roche Diagnostics, Germany), and PCR template (whole cells or extracted DNA). Positive and negative controls were included for each set of reactions. For type A primers, an environmental V. vulnificus isolate (vPvMH1003-12) was used, and for type B primers, V. vulnificus ATCC 33814 was used. The assay was optimized as a two-step PCR amplification, which allowed the maximum sensitivity and specificity of the primer sets. The amplification protocol involved an initial 10-min denaturation program at 95°C, followed by an amplification program of between 30 and 50 cycles of 95°C for 15 s and 74°C for 35 s; a melting curve program involving 1 cycle of temperature increase to 95°C, 65°C for 15 s, a gradual temperature increase back to 95°C with a 0.1°C/s slope (the slope for all other steps was kept at 20°C/s); and finally a cooling program of holding at 40°C for 30 s. Multiple reactions were performed to confirm reproducibility and specificity of results. Real-time PCR analysis of the 16S rRNA gene indicated the presence of the two 16S alleles (16S type A or B) or a combination of both alleles (type A+B) resulting from multiple copies of the gene on the chromosome.
TABLE 1.
Primer sequences developed for real-time PCR of type A and B V. vulnificus
rep-PCR molecular typing.
rep-PCR genomic typing was performed using a DiversiLab Microbial Typing System (Bacterial Barcodes, Inc., Houston, TX). Extracted DNA was amplified by PCR using the DiversiLab Salmonella kit (DL-SE01). In short, 2 μl of genomic DNA (100 ng) was added to a master mix that contained 18 μl of rep-PCR MM1 solution, 2.5 μl of GeneAmp 10× PCR Buffer, 2 μl of Primer Mix P, and 0.5 μl of AmpliTaq DNA Polymerase in a total volume of 25 μl. PCR was performed on an Eppendorf Mastercycler gradient thermocycler with the following conditions: 94°C for 2 min, 35 cycles of 94°C for 30 s, 50°C for 30 s, and 70°C for 90 s, with a final extension at 70°C for 3 min. PCR amplicons were applied to a Caliper LabChip for capillary separation by the Agilent 2100 instrument. Electrophoretograms were transmitted electronically and integrated for similarity analysis by DiversiLab software.
Statistics.
Significant differences in the distribution of genetic profiles were determined by the Student's t test or the chi-squared analysis.
RESULTS AND DISCUSSION
Distribution of V. vulnificus genotype profiles.
Previous reports correlated specific genotypes with clinical origin in V. vulnificus, but each of these studies examined single loci among different groups of strains. In the present study, clinical and environmental isolates were examined for distribution of genetic polymorphisms at multiple loci within the same strain collection. Although these studies are not meant to be extensive in terms of geographic and temporal distribution, this collection includes strains from 1979 through 2004 and provides the first examination of clinically associated genetic profiles in the same set of V. vulnificus strains and in the context of genomic rep-PCR typing.
Differential distribution of polymorphic sequences for strains from clinical versus environmental origin was confirmed by PCR screening. Significant differences (P < 0.01) were noted in these populations for all loci. In agreement with previous reports, strains of clinical origin were predominantly 16S type B, RAPD C type, and CPS allele 1 (Table 2), and this “clinical” profile was significantly (P < 0.001) associated with clinical origin (57.6%), while only one (3%) of the environmental strains exhibited this profile. Similarly, an “environmental” profile (16S type A, RAPD E type, and CPS allele 2) was more common among strains of environmental origin (60%), whereas only 27.6% of clinical strains showed this pattern. Results did vary somewhat from previous studies, as association of clinical or environmental populations with the typical genotype profile was somewhat less than that reported previously for individual loci (72 to 94%). These discrepancies probably reflect differences in strain selection, as wound isolates were not included in previous studies. It was also noted that some strains (23 of the 68) showed a strong PCR signal for one RAPD genotype but were also faintly positive for the second, despite PCR optimization efforts. These strains were scored for RAPD E or C type based on the most predominant band. Interestingly, PCR analysis of 16S genes detected some strains with strong signals for both 16S rRNA genotypes (A and B) within a single strain. This result was not detected by previous analysis (26), but it is consistent with the fact that bacteria have multiple copies of 16S rRNA genes, and dual signals have been noted by other researchers (42). Overall, results confirmed significant associations of distinct 16S, CPS, and RAPD genetic profiles with clinical origin but also suggested greater diversity within this group than previously reported.
TABLE 2.
Allelic distribution among clinical versus environmental V. vulnificus isolates
| Identifiera | Genotype distribution (%)b
|
|
|---|---|---|
| Clinical | Environmental | |
| 16S type | ||
| A | 27.3 | 80.0 |
| B | 66.7 | 8.6 |
| A+B | 6.0 | 11.4 |
| CPS allele | ||
| 1 | 63.6 | 20.0 |
| 2 | 36.4 | 71.4 |
| Allele absent | 0 | 8.6 |
| RAPD type | ||
| C | 66.7 | 8.6 |
| E | 33.3 | 91.4 |
| Profile | ||
| Clinical | 57.6 | 3.0 |
| Environmental | 27.3 | 60.0 |
| Atypical | 15.1 | 34.0 |
DNA polymorphisms were screened by PCR as described in Materials and Methods and are based on previously described assays for detection of alternate polymorphic sequences for DNA targets derived from 16S rRNA genes (27), the CPS operon (9), and RAPD (30) or for profiles of composite genotypes as described in the text.
The percentage of each genotype is shown for clinical versus environmental strains.
rep-PCR analysis of clinical versus environmental strains.
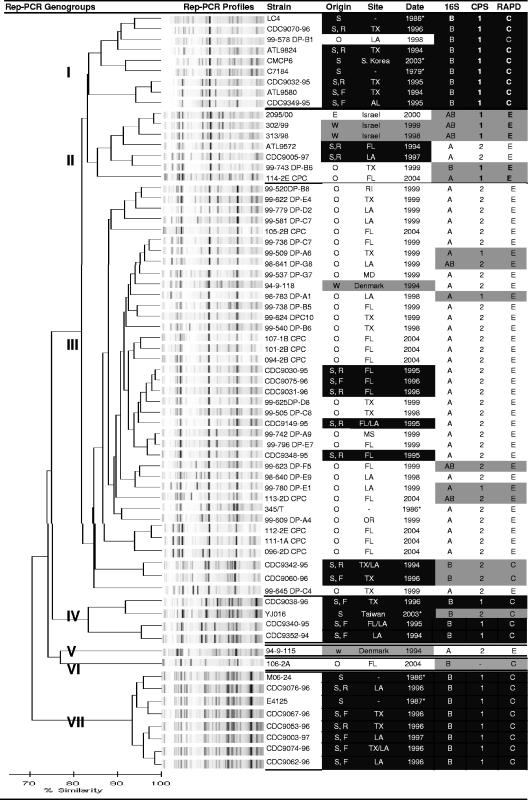
rep-PCR molecular typing was used to determine the relatedness of strains that shared similar genotype profiles. Reproducibility of the rep-PCR assay was confirmed by replicate reactions (n = 7) of individual strains (n = 3), which demonstrated >95% similarity within the same strain (data not shown). rep-PCR of all V. vulnificus strains showed at least 70% similarity, and an arbitrary value of ≥85% similarity segregated strains into seven distinct rep-PCR genogroups, although two strains (assigned to genogroups V and VI) did not cluster with any other strains (Fig. 1).
FIG. 1.
Comparison of rep-PCR analysis and genotype profiles for clinical and environmental strains of V. vulnificus. A scale for rep-PCR similarity is shown at the bottom of the figure. Strain sources are described in detail in the text, and a description of strains in the table includes the following: S, septicemia from oyster consumption; W, wound infection; O, oyster; E, environmental, not oyster; F, fatal outcome of infection; R, recovered from infection. Strains from septicemia origins are shaded in black, wound infection isolates are shaded in gray, and environmental isolates are not shaded. Where available, information on the site and date of isolation of strains is provided; alternatively, the date that the strains first appeared in publication is shown (asterisk). Specific genotype profiles were determined by PCR as described in the text and included the following: 16S rRNA gene (type A, B, or A+B), group 1 CPS genes (alleles 1 and 2; a dash is used to indicate where CPS primers did not amplify), and RAPD-associated locus (C type and E type). Typical “clinical” profiles (16S B, CPS allele 1, and RAPD C type) are shaded in black, “environmental” profiles (16S A or A+B, CPS allele 2 or CPS negative, and RAPD E type) are not shaded, and atypical combinations are shaded in gray.
Clinical strains were distributed among all genogroups, but the majority (61%) segregated into genogroups I, IV, and VII, which also accounted for 69% of strains from septicemia cases. Conversely, only one oyster isolate and none of the isolates from wound infections was found within these groups. Genogroups II and III, on the other hand, contained 94% of the oyster isolates but only 27% of clinical strains. Most (88%) environmental strains were in genogroup III. Thus, rep-PCR confirmed the differential distribution of clinical versus environmental strains but suggested that clinical strains may represent a more diverse population than environmental strains derived from oysters.
Strains sharing multilocus genotype profiles described above were also related by rep-PCR analysis (Fig. 1). For example, clinically associated genogroups I, IV, and VII contained strains with the typical “clinical” genotype profile with only one exception (YJ016), and it differed at only one locus (CPS allele 2). Furthermore, the single oyster isolate among these groups was the only environmental strain with a “clinical” profile. Similarly, most strains (70%) in the predominantly oyster-associated genogroup III exhibited an “environmental” profile with 16S type A (87%), CPS allele 2 (82%), and RAPD E type (92%). Clinical strains in this group showed either the “environmental” profile (n = 5) or atypical profiles (n = 2). Genogroup II was diverse in terms of genotype profile, and strains were RAPD E type but with a mixture of 16S types A, B, or A+B, as well as both CPS alleles 1 and 2.
Overall, clinical strains were more diverse by rep-PCR analysis than was previously indicated by multilocus genotyping. For example, some strains with identical genotype profiles were only about 70% similar by rep-PCR. In fact, “clinical” genogroup I was more closely related to “environmental” genogroups II and III than to the other clinical genogroups, IV and VII, in the study. On the other hand, strains within clinical genogroup VII showed the greatest degree of similarity to each other compared to other groups and several appeared clonal by rep-PCR, with >95% similarity. Wound isolates were genetically distinct from most blood-derived strains and from each other, with the exception of biotype III strains from Israel, which appeared to be clonal as predicted from previous studies (3, 4). In summary, rep-PCR showed general agreement with multilocus genotype profile distributions but provided more sensitive discrimination for tracking genetic distinctions among strains from clinical and environmental sources.
rep-PCR analysis and virulence potential.
The colony morphology of V. vulnificus shows phase variation and alternates between opaque colonies, which are virulent in animal models of infection, and avirulent translucent colonies. We investigated whether or not opaque/translucent phase variation corresponded to differences in genotype profile or rep-PCR analysis. Translucent variants of originally opaque strains from both clinical (n = 6) or oyster (n = 4) origin were derived from standard subculture. Phase variants were examined by rep-PCR, 16S, and CPS alleles. All phase variants were >95% similar to each other by rep-PCR pattern (data not shown), indicating variants were clonal by this assay. CPS and 16S genotypes were also identical to parent strains. Thus, rep-PCR patterns and/or profile did not discriminate among phase variants.
Speculations (26, 32) that specific genetic markers may be indicative of virulence potential in V. vulnificus were not validated by experimental infections (13) or by genetic profiles of avirulent phase variants in the present study. However, as previously noted (13), most strains (82%) derived from fatal cases of infection share typical genetic profiles, while those for which patients recovered were more likely (58%) to have atypical profiles. Assuming a range of virulence potentials, strain differences may not be accurately reflected in available animal models. Only one oyster isolate exhibited the clinical profile and was also related to clinical strains by rep-PCR, suggesting that relatively few environmental isolates are clinically significant. However, Kim and Jeong (20) showed a preponderance (65%) of “clinical” 16S type B in environmental samples, and Lin and Schwarz (23) found that the prevalence of 16S type B was seasonal and more common in summer months when the disease incidence also increases. Discrepancies among data are likely the consequence of limited databases, but the extreme rarity of V. vulnificus food-borne disease compared to the relative abundance of the bacterium in oyster tissues supports the hypothesis that few environmental strains have the potential for disease.
Human infection appears to be a dead end for evolution of V. vulnificus, as person-to-person transmission is not known to occur. Divergence in genetic populations is more likely to be related to survival in estuarine habitats or alternate host species, i.e., fish or oysters, than to survival in humans. The contribution of fish to V. vulnificus populations is indicated by the large numbers (1010 CFU/g) that are associated with shellfish-feeding fish species compared to the typical V. vulnificus content (ca. 103 CFU/g) in oysters (12). Biotypes 2 and 3 are primarily fish pathogens but also produce human wound infections (4). Biotype 3 infections were derived from exposure to aquacultured fish, and the large numbers of cases (n = 63) derived from clonal strains led to speculation of an emergent and more virulent phenotype (3, 4). Interestingly, two clinical and two oyster isolates from the United States were 85 to 90% similar to biotype 3 by rep-PCR, suggesting potential for a public health risk in the United States. Concerns are underscored by the first outbreak of V. vulnificus wound infections in the United States at a fishing contest in Texas (27). rep-PCR molecular typing provides rapid and sensitive monitoring of divergent genetic populations of V. vulnificus, and future applications should include a broader survey of strains in the context of more detailed clinical history and more sensitive virulence models in order to determine the virulence potential for environmental reservoirs.
Acknowledgments
We thank Ayana McCoy and Melissa Evans for assistance in strain collection. Angelo DePaola and Paul Gulig kindly provided strains.
This research was supported in part by Sea Grants R/LR-Q-26A and R/LR-Q-27.
REFERENCES
- 1.Amaro, C., E. G. Biosca, B. Fouz, and E. Garay. 1992. Electrophoretic analysis of heterogeneous lipopolysaccharides from various strains of Vibrio vulnificus biotypes 1 and 2 by silver staining and immunoblotting. Curr. Microbiol. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 2.Aznar, R., W. Ludwig, R. I. Amann, and K. H. Schleifer. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of V. vulnificus with rRNA-targeted oligonucleotide probes. Int. J. Syst. Bacteriol. 44:330-337. [DOI] [PubMed] [Google Scholar]
- 3.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, J. J. Farmer III, and the Israel Vibrio Study Group. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 4.Bisharat, N., D. I. Cohen, R. M. Harding, D. Falush, D. W. Crook, T. Peto, and M. C. Maiden. 2005. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 11:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake, P. A., M. H. Merson, R. E. Weaver, D. G. Hollis, and P. C. Heublein. 1979. Disease caused by a marine Vibrio. Clinical characteristics and epidemiology. N. Engl. J. Med. 300:1-5. [DOI] [PubMed] [Google Scholar]
- 6.Buchrieser, C., V. V. Gangar, R. L. Murphree, M. L. Tamplin, and C. W. Kaspar. 1995. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogenous electric field gel electrophoresis. Appl. Environ. Microbiol. 61:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, M. S., and A. C. Wright. 2003. Real-time PCR analysis of Vibrio vulnificus from oysters. Appl. Environ. Microbiol. 69:7137-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatzidaki-Livanis, M., M. K. Jones, and A. C. Wright. 2006. Genetic variation in the Vibrio vulnificus group 1 capsular polysaccharide operon. J. Bacteriol. 188:1987-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo, M. M., S. Mastrandrea, F. Leite, A. Santona, S. Uzzau, P. Rappelli, M. Pisano, S. Rubino, and P. Cappuccinelli. 1997. Tracking of clinical and environmental Vibrio cholerae O1 strains by combined analysis of the presence of toxin cassette, plasmid content and ERIC PCR. FEMS Immunol. Med. Microbiol. 19:33-45. [DOI] [PubMed] [Google Scholar]
- 11.Dalsgaard, A., N. Frimodt-Moller, B. Bruun, L. Hoi, and J. L. Larsen. 1996. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur. J. Clin. Microbiol. Infect. Dis. 15:227-232. [DOI] [PubMed] [Google Scholar]
- 12.DePaola, A., G. M. Capers, and D. Alexander. 1994. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast. Appl. Environ. Microbiol. 60:984-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePaola, A., J. L. Nordstrom, A. Dalsgaard, A. Forslund, J. Oliver, T. Bates, K. L. Bourdage, and P. A. Gulig. 2003. Analysis of Vibrio vulnificus from market oysters and septicemia cases for virulence markers. Appl. Environ. Microbiol. 69:4006-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folgosa, E., S. Mastrandrea, P. Cappuccinelli, S. Uzzau, P. Rappelli, M. J. Brian, and M. M. Colombo. 2001. Molecular identification of pathogenicity genes and ERIC types in Vibrio cholerae O1 epidemic strains from Mozambique. Epidemiol. Infect. 127:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gulig, P. A., K. L. Bourdage, and A. M. Starks. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43:118-131. [PubMed] [Google Scholar]
- 17.Gutacker, M., N. Conza, C. Benagli, A. Pedroli, M. V. Bernasconi, L. Permin, R. Aznar, and J. C. Piffaretti. 2003. Population genetics of Vibrio vulnificus: identification of two divisions and a distinct eel-pathogenic clone. Appl. Environ. Microbiol. 69:3203-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Høi, L., I. Dalsgaard, A. DePaola, R. J. Siebeling, and A. Dalsgaard. 1998. Heterogeneity among isolates of Vibrio vulnificus recovered from eels (Anguilla anguilla) in Denmark. Appl. Environ. Microbiol. 64:4676-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, J. K., R. L. Murphree, and M. L. Tamplin. 1997. Evidence that mortality from Vibrio vulnificus infection results from single strains among heterogeneous populations in shellfish. J. Clin. Microbiol. 35:2098-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, M. S., and H. D. Jeong. 2001. Development of 16S rRNA targeted PCR methods for the detection and differentiation of Vibrio vulnificus in marine environments. Aquaculture 193:199-211. [Google Scholar]
- 21.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreger, A., L. d. Gray, and J. Testa. 1981. Interaction of Vibrio vulnificus with human polymorphonuclear leukocytes: association of virulence with resistance to phagocytosis. J. Infect. Dis. 144:244-248. [DOI] [PubMed] [Google Scholar]
- 23.Lin, M., and J. R. Schwarz. 2003. Seasonal shifts in population structure of Vibrio vulnificus in an estuarine environment as revealed by partial 16S ribosomal DNA sequencing. FEMS Microbiol. Lett. 45:23-27. [DOI] [PubMed] [Google Scholar]
- 24.Merino, L. A., M. C. Ronconi, M. M. Navia, J. Ruiz, J. M. Sierra, N. B. Cech, N. S. Lodeiro, and J. Vila. 2003. Analysis of the clonal relationship among clinical isolates of Salmonella enterica serovar Infantis by different typing methods. Rev. Inst. Med. Trop. Sao Paulo 45:119-123. [DOI] [PubMed] [Google Scholar]
- 25.Motes, M. L., A. DePaola, D. W. Cook, J. E. Veazey, J. C. Hunsucker, W. E. Garthright, R. J. Blodgett, and S. J. Chirtel. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson, W. B., R. N. Paranjype, A. DePaola, and M. S. Strom. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, J. D. 2005. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol. Infect. 133:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panicker, G., M. C. Vickery, and A. K. Bej. 2004. Multiplex PCR detection of clinical and environmental strains of Vibrio vulnificus in shellfish. Can. J. Microbiol. 50:911-922. [DOI] [PubMed] [Google Scholar]
- 29.Poole, M. D., and J. D. Oliver. 1977. Experimental pathogenicity and mortality in ligated ileal loop studies of the newly reported halophilic lactose-positive Vibrio sp. Infect. Immun. 20:126-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radu, S., G. Yuherman, L. Rusul, L. K. Yeang, and M. Nishibuchi. 2000. Detection and molecular characterization of Vibrio vulnificus from coastal waters of Malaysia. Southeast Asian J. Trop. Med. Public Health 31:668-673. [PubMed] [Google Scholar]
- 31.Rivera, I. G., M. A. Chowdhury, A. Huq, D. Jacobs, M. T. Martins, and R. R. Colwell. 1995. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl. Environ. Microbiol. 61:2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosche, T. M., Y. Yano, and J. D. Oliver. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381-389. [DOI] [PubMed] [Google Scholar]
- 33.Shangkuan, Y. H., H. C. Lin, and T. M. Wang. 1997. Diversity of DNA sequences among Vibrio cholerae O1 and non-O1 isolates detected by whole-cell repetitive element sequence-based polymerase chain reaction. J. Appl. Microbiol. 82:335-344. [DOI] [PubMed] [Google Scholar]
- 34.Simpson, L. M., V. K. White, S. F. Zane, and J. D. Oliver. 1987. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect. Immun. 55:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starks, A. M., T. R. Schoeb, M. L. Tamplin, S. Parveen, T. J. Doyle, P. E. Bomeisl, G. M. Escudero, and P. A. Gulig. 2000. Pathogenesis of infection by clinical and environmental strains of Vibrio vulnificus in iron-dextran-treated mice. Infect. Immun. 68:5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stelma, G. N., Jr., A. L. Reyes, J. T. Peeler, C. H. Johnson, and P. L. Spaulding. 1992. Virulence characteristics of clinical and environmental isolates of Vibrio vulnificus. Appl. Environ. Microbiol. 58:2776-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microb. Infect. 2:177-188. [DOI] [PubMed] [Google Scholar]
- 38.Tamplin, M. L., J. K. Jackson, C. Buchrieser, R. L. Murphree, K. M. Portier, V. Gangar, L. G. Miller, and C. W. Kaspar. 1996. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl. Environ. Microbiol. 62:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tison, D. L., and M. T. Kelly. 1986. Virulence of Vibrio vulnificus strains from marine environments. Appl. Environ. Microbiol. 51:1004-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tison, D. L., M. Nishibuchi, J. D. Greenwood, and R. J. Seidler. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Versalovic, J., F. de Bruijn, and J. Lupski. 1998. Repetitive sequence-based PCR (rep-PCR) DNA fingerprinting of bacterial genomes. In F. J. de Bruijn, J. R. Lupski, and G. M. Weinstock (ed.), Bacterial genomes: physical structure and analysis. Chapman & Hall, New York, N.Y.
- 42.Vickery, M. C. L., W. B. Nilsson, M. S. Strom, J. L. Nordstrom, and A. DePaola. Personal communication.
- 43.Warner, J. M., and J. D. Oliver. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other Vibrio species. Appl. Environ. Microbiol. 65:1141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong, H. C., and C. H. Lin. 2001. Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J. Clin. Microbiol. 39:4233-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright, A. C., R. T. Hill, J. A. Johnson, M. C. Roghman, R. R. Colwell, and J. G. Morris, Jr. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright, A. C., J. G. Morris, Jr., D. R. Maneval, Jr., K. Richardson, and J. B. Kaper. 1985. Cloning of the cytotoxin-hemolysin gene of Vibrio vulnificus. Infect. Immun. 50:922-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright, A. C., J. L. Powell, J. B. Kaper, and J. G. Morris, Jr. 2001. Identification of a group 1-like capsular polysaccharide operon for Vibrio vulnificus. Infect. Immun. 69:6893-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright, A. C., J. L. Powell, M. K. Tanner, L. A. Ensor, A. B. Karpas, J. G. Morris, Jr., and M. B. Sztein. 1999. Differential expression of Vibrio vulnificus capsular polysaccharide. Infect. Immun. 67:2250-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright, A. C., L. M. Simpson, J. D. Oliver, and J. G. Morris, Jr. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright, A. C., L. M. Simpson, K. Richardson, D. R. Maneval, Jr., J. D. Oliver, and J. G. Morris, Jr. 1986. Siderophore production and outer membrane proteins of selected Vibrio vulnificus strains under conditions of iron limitation. FEMS Microbiol. Lett. 35:255-260. [Google Scholar]
- 51.Wu, J. J., L. I. Hor, and S. L. Shiau. 1995. Differentiation of Vibrio vulnificus strains by an arbitrarily primed polymerase chain reaction. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi 28:70-78. [PubMed] [Google Scholar]
- 52.Zuppardo, A. B., A. DePaola, J. C. Bowers, K. L. Schully, J. A. Gooch, and R. J. Siebeling. 2001. Heterogeneity of environmental, retail, and clinical isolates of Vibrio vulnificus as determined by lipopolysaccharide-specific monoclonal antibodies. J. Food Prot. 64:1172-1177. [DOI] [PubMed] [Google Scholar]