Abstract
Incubation of resting cells of Sphingobium indicum B90A, Sphingobium japonicum UT26, and Sphingobium francense Sp+ showed that they were able to transform β- and δ-hexachlorocyclohexane (β- and δ-HCH, respectively), the most recalcitrant hexachlorocyclohexane isomers, to pentachlorocyclohexanols, but only resting cells of strain B90A could further transform the pentachlorocyclohexanol intermediates to the corresponding tetrachlorocyclohexanediols. Moreover, experiments with resting cells of Escherichia coli expressing the LinB proteins of strains B90A, UT26, and Sp+ indicated that LinB was responsible for these transformations. Purified LinB proteins from all three strains also effected the formation of the respective pentachlorocyclohexanols. Although the three LinB enzymes differ only marginally with respect to amino acid sequence, they showed interesting differences with respect to substrate specificity. When LinB from strain B90A was incubated with β- and δ-HCH, the pentachlorocyclohexanol products were further transformed and eventually disappeared from the incubation mixtures. In contrast, the LinB proteins from strains UT26 and Sp+ could not catalyze transformation of the pentachlorocyclohexanols, and these products accumulated in the incubation mixture. A mutant of strain Sp+ lacking linA and linB did not degrade any of the HCH isomers, including β-HCH, and complementation of this mutant by linB from strain B90A restored the ability to degrade β- and δ-HCH.
Hexachlorocyclohexane (HCH), a broad-spectrum insecticide, was one of the most extensively used organochlorine pesticides for the control of agricultural pests and the control of mosquitoes in malaria health programs during the 1940s. Technical HCH is prepared by chlorination of benzene in the presence of UV, resulting in the formation of a mixture primarily containing the isomers γ-HCH (10 to 12%), α-HCH (60 to 70%), β-HCH (5 to 12%), and δ-HCH (6 to 10%) (20). Of these isomers, only γ-HCH (also known as lindane) has insecticidal properties. For purely economic reasons, technical HCH was used indiscriminately instead of lindane in many countries, and its use continued unabated until the 1990s, when the persistent and toxic nature of the components of technical mixtures was realized. Today, the use of technical HCH is banned in most countries, and the use of lindane is either banned or severely restricted. The extensive and widespread use, unregulated disposal, and persistent nature of HCH isomers have created the following two types of contamination problems: (i) low levels of contamination of agricultural soils and groundwater (3, 14, 25, 32, 39, 45), mainly caused by the intended usage, and (ii) high levels of contamination at the production sites caused by inappropriate disposal of the noninsecticidal isomers α-, β-, and δ-HCH. A large number of open or sealed dumping sites exist in The Netherlands (53), Brazil (36), Spain (24), Germany (13), India (39), and Eastern and Central Europe (53), which still pose serious risks for soils and groundwater.
α- and γ-HCH are degraded faster than β- and δ-HCH (2), which are quite persistent. The relative persistence of each isomer is mainly controlled by its chemical structure, i.e., the positions of the chlorine atoms on the cyclohexane ring, which also influence solubility and volatility. The compact spatial structure of β-HCH, with all chlorines in the equatorial position, seems to confer physical (β-HCH has a low vapor pressure and a high melting point) and metabolic stability (12). β-HCH residues have been reported predominantly from soil (9, 46), water (3, 46, 54), and food commodities (1, 43), and very high levels persist along dumping sites (35, 39). Since β-HCH is highly toxic to mammals, is a known endocrine disrupter (15), is suspected to cause breast cancer (55), bioaccumulates strongly (4, 35), and is quite resistant to microbial degradation (23, 52), it is the most problematic of the HCH isomers.
Although several organisms degrade γ-HCH, reports of strains degrading β-HCH are scarce (21). Three HCH-degrading bacterial strains, Sphingomonas paucimobilis B90A (41), Sphingomonas paucimobilis UT26 (44), and Sphingomonas paucimobilis Sp+ (6), were isolated from HCH-contaminated soils in India, Japan, and France, respectively (Table 1). The taxonomic positions of B90A, UT26, and Sp+ were ascertained recently, and they are now classified as three distinct species, i.e., Sphingobium indicum B90A, Sphingobium japonicum UT26, and Sphingobium francense Sp+, respectively (38). Studies of lin gene expression in these strains suggested that degradation of β- and δ-HCH proceeds by a different pathway from that of α- and γ-HCH (21). While Sphingobium indicum B90A reportedly degrades β- and δ-HCH (8, 12, 19, 41, 42), the primary gene(s) associated with their degradation is not established. Recently, Nagata et al. (31) reported that LinB (haloalkane dehalogenase) is responsible for the initial transformation of β-HCH to pentachlorocyclohexanol in strain UT26. Pentachlorocyclohexanol was not degraded further, even after incubation for 2 days. Although LinB of strain UT26 seems to be able to transform β-HCH, whole-cell incubation with strain UT26 did not effect any β-HCH transformation (31). This is in contrast to the case with strain B90A, which is able to transform β-HCH significantly (8, 12, 19, 41). These discrepancies led us to investigate the role of LinB in the degradation of β-HCH in strain B90A. Here we report that LinB is responsible not only for the transformation of β-HCH but also for that of δ-HCH. LinB of strain B90A also acted on pentachlorocyclohexanol as a substrate, yielding a tetrachlorocyclohexanediol as the final product. Furthermore, differences in the activities of LinB proteins originating from different strains were evident.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or referencea |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 phoA supE44 thi-1 gyrA96 relA1 | Amersham Pharmacia Biotech, Piscataway, N.J. |
| E. coli BL21 | F−ompT hsdSB(rB−mB−) gal dcm | Amersham Pharmacia Biotech, Piscataway, N.J. |
| Sphingobium indicum B90A | Yellow colonies, produces brown pigment, fast degrader of HCH isomers | N. Sethunathan, CRRI, Cuttack, India (41) |
| Sphingobium japonicum UT26 | Yellow colonies, degrades HCH isomers, albeit slowly | Y. Nagata, University of Tokyo, Tokyo, Japan (44) |
| Sphingobium francense Sp+ | Yellow colonies, degrades HCH isomers, albeit slowly | Tim Vogel, University of Lyon, Lyon, France (6) |
| Plasmids | ||
| pUC18 | 2.7 kb; Ampr; multiple cloning site internal to lacZ gene | Fermentas Inc. |
| pUC4K | 3.9 kb; Ampr Kanr pBR322 ori; restriction site mobilizing element | Amersham Pharmacia Biotech, Piscataway, N.J. |
| pUCIS | pUC18 containing 1-kb IS6100 element (amplified from pLINA57) | This study |
| pUCIS1K | pUCIS containing 1.2-kb Kanr fragment from pUC4K | This study |
| pUCIS1KB | pUCIS1K containing linB of S. indicum B90A cloned at KpnI site | This study |
| pGEX-5X-3 | 4.9 kb; GST fusion vector with tac promoter, lacIq, and factor Xa protease recognition site | Amersham Pharmacia Biotech, Piscataway, N.J. |
| pLINB35 | pWE15 carrying DNA fragment of B90A containing linB and IS6100 | 8 |
| pLINA57 | pWE15 carrying 41-kb DNA fragment of B90A containing linA1, linC, linX, and IS6100 | 8 |
| pLINSB | pUC18 containing HindIII-digested 2.16-kb fragment of S. francense Sp+ containing linB ORF and IS6100 | This study |
| pLINEBB | linB ORF of S. indicum B90A cloned into pGEX-5X-3 at BamHI and XhoI sites | This study |
| pLINEUB | linB ORF of S. japonicum UT26 cloned into pGEX-5X-3 at BamHI and XhoI sites | This study |
| pLINESB | linB ORF of S. francense Sp+ cloned into pGEX-5X-3 at EcoRI and XhoI sites | This study |
| Mutants | ||
| Sp+mt | Sp+ mutant lacking linA and linB, degrades none of the four HCH isomers | 8 |
| Sp+mtC1-9 | Sp+mt complemented with linB of B90A, degrades only β- and δ-HCH isomers | This study |
CRRI, Central Rice Research Institute.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 shows the bacterial strains and plasmids used in this study. All strains were generally grown in Luria broth (LB) (19). Escherichia coli strains were grown in LB at 37°C. Antibiotics, when required, were added to a final concentration of 150 μg/ml (ampicillin) or 50 μg/ml (kanamycin).
Degradation of β-HCH and δ-HCH in Sphingobium indicum B90A, Sphingobium japonicum UT26, and Sphingobium francense Sp+.
Degradation of β- and δ-HCH (D-86199; Ehrenstorfer GmbH, Augsburg, Germany) with strains B90A, UT26, and Sp+ and the generated mutants was assessed with resting-cell assays. For this purpose, a cell pellet (∼200 mg) of each strain obtained from 500 ml culture in LB (optical density at 600 nm, 0.5) was washed twice with 0.1 M sodium phosphate buffer (pH 7) and suspended in 10 ml phosphate buffer. To this suspension, β- or δ-HCH (5 μg/ml) was added separately. An aliquot of 0.2 ml of the reaction mixture was withdrawn from each flask periodically, extracted twice with 0.5 ml of hexane, pooled, and analyzed by gas chromatography (GC; Shimadzu GC-17A gas chromatograph fitted with an electron-capture 63Ni detector) as described previously (19).
Cloning of linB into pGEX-5X-3 and expression in E. coli BL21.
In order to clone the linB genes into an expression vector, the linB open reading frames (ORFs) of B90A and Sp+ were amplified from a cosmid clone, pLINB35 (8), and a plasmid, pLINSB (Table 1), respectively. However, the linB gene from UT26 was amplified by using genomic DNA. The primers used for this purpose are listed in Table 2. PCR amplification was performed with a Robocycler (Stratagene), and amplified products were cloned into the E. coli BL21 expression vector pGEX-5X-3 (Stratagene). The clones were confirmed to carry the linB ORFs by restriction digestion and DNA sequencing (Avant 3100 genetic analyzer; Applied Biosystems). pGEX-5X-3 plasmids containing the linB ORFs of B90A, Sp+, and UT26 in E. coli BL21 were named pLINEBB, pLINESB, and pLINEUB, respectively.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′)a | Restriction site | Designation | Source or accession no. |
|---|---|---|---|---|
| 1 | GCGGATCCGCATGAGCCTCGGCGCAAAGCCA | BamHI | linB-sense | D14594 |
| 2 | GCCTCGAGTTATGCTGGGCGCAATCGCCGGAC | XhoI | linB-antisense | D14594 |
| 3 | GCGAATTCCATGAGCCTCGGCGCAAAGCCA | EcoRI | linB-senseb | This study |
| 4 | GCGGTACCAAAATGAGCCGGTTC | KpnI | linB-sense with promoter and SD sequences | This study |
| 5 | GCGGTACCCGATTCCTCGATTGA | KpnI | linB-antisense | This study |
| 6 | AAGAATTCTAAGCTCAACGGATGC | EcoRI | IS6100-sense | AY331258 |
| 7 | ATGAATTCCCTTGCTGCCCACGGA | EcoRI | IS6100-antisense | AY331258 |
Restriction sites are underlined.
The sense primer contained an EcoRI site for linB amplification from Sp+ because linB of Sp+ contains a BamHI site within the ORF.
The degradation of β- or δ-HCH (5 μg/ml) in E. coli BL21 containing pLINEBB, pLINESB, or pLINEUB was studied using a previously described protocol (19).
Purification and selectivity of LinB from E. coli BL21.
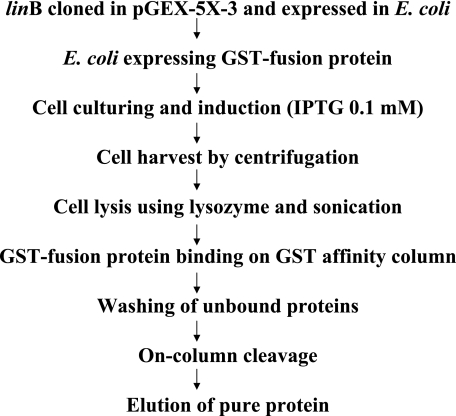
The overexpressed LinB enzymes from strains B90A, Sp+, and UT26 were purified from E. coli harboring pLINEBB, pLINESB, and pLINEUB, respectively, using a glutathione S-transferase (GST)-glutathione affinity column chromatography kit (Amersham Pharmacia). The steps involved in purification are depicted in Fig. 1. The amount of protein was determined by using the Bradford reagent (Bio-Rad), with bovine serum albumin (Amersham Pharmacia) as a standard. To remove the GST tag from LinB proteins, 1 mg of purified fusion protein was incubated at 25°C with factor Xa for 12 h and passed through the GST column to get the desired cleaved product. The GST fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The initial degradation experiments revealed that GST fused with LinB does not affect the activities of LinB proteins. In order to determine the specific activities (U/mg) of the enzymes, the purified LinB proteins were used without cleavage for β- and δ-HCH degradation. For this purpose, two reactions (separate reactions for β- and δ-HCH) were initiated by the addition of the respective LinB enzyme (final concentration, 20 mg/liter) to reaction mixtures containing 50 mM Tris buffer (pH 8) and β- or δ-HCH at a final concentration of 1.25 μg/ml (4 μM) at 37°C. At regular time intervals, samples of 0.2 ml were withdrawn, extracted with 0.5 ml of hexane, and analyzed as described above.
FIG. 1.
Purification scheme for GST-LinB fusion proteins of Sphingobium indicum B90A, Sphingobium francense Sp+, and Sphingobium japonicum UT26. GST-LinB was also cleaved with Prescission protease (factor Xa).
Production and identification of metabolites.
For the production of B1 and D1, 100 ml E. coli culture (containing pLINEBB) was raised. The culture pellet (∼700 mg wet weight) was washed with 0.1 M sodium phosphate buffer (pH 7.0) and suspended in 80 ml of 0.1 M sodium phosphate buffer (pH 7.0), and β- or δ-HCH was added separately to a final concentration of 20 mg/liter. The samples were incubated at 37°C with constant shaking. After 36 h of incubation, the entire sample was harvested by centrifugation at 10,000 × g, and the supernatant was collected and filtered through Whatman filter paper (125-mm diameter) to remove residual impurities. Supernatants were extracted twice with equal volumes (80 ml) of hexane, pooled, and concentrated in a Rotavapour instrument (Buchi Rotavapour, Switzerland) to 1 ml.
The B1 and D1 intermediates were separated by thin-layer chromatography on silica gel 60 F254 plates (20 by 20 cm, with a thickness of 0.25 mm; Merck, Germany) by using the solvent acetone-hexane (5:95) and were visualized under UV light after being sprayed with 2% ortho-toluidine in acetone. Two spots appeared in each lane, presumably corresponding to β-HCH and B1 and to δ-HCH and D1. The spots were scraped off the plates and extracted with hexane; the silica particles were removed by centrifugation and passed through a column with glass wool. To determine the structure of the metabolites, GC-mass spectrometry (GC-MS) analysis was carried out using a Shimadzu GCMS-QP2010 system (Toshvin Analytical Pvt. Ltd., Mumbai, India). The gas chromatograph was equipped with a 30 m by 0.25 mm (internal diameter) by 0.25 μm DB-5 column. The oven temperature program was as follows: 100°C for 1 min and 270°C for 20 min. The flow rate of the carrier gas (He) was 1 ml/min. The temperature of the ion source was held at 200°C, and the temperature of the interface was held at 250°C.
To produce the metabolites B2 and D2, a preculture of S. indicum B90A grown overnight in LB was transferred to fresh medium (1% [vol/vol]) and incubated at 28°C until the optical density at 600 nm reached ∼1.0. Cells were harvested by centrifugation at 4,985 × g for 10 min and washed twice with sterile potassium phosphate buffer (10 mM, pH 7). Washed cells were resuspended in the same buffer to a cell density of 2.09 × 109 cells/ml and divided into batches of 10 ml, with each batch spiked with either β- or δ-HCH. The final concentrations were 5 mg/liter for β-HCH and 20 mg/liter for δ-HCH. After appropriate intervals, whole flasks were extracted with equal volumes of ethyl acetate. After phase separation, aqueous phases were acidified to pH 2.0 with 1 N HCl and reextracted with equal volumes of ethyl acetate. The acidic and neutral fractions were pooled and dried over anhydrous sodium sulfate. After evaporation to dryness in a rotary evaporator at 40°C, the residue was dissolved in hexane, and appropriate dilutions were subjected to GC-MS on a VG Tribrid double-focusing magnetic-sector hybrid mass spectrometer (VG Analytical, Manchester, England). Samples were injected at 50°C, and the column temperature was programmed as follows: 50°C for a 2-min isothermal hold, 20°C/min to 120°C, and then 5°C/min to 280°C, followed by an isothermal hold at this temperature.
Complementation of linB in deletion mutant.
In our earlier study (8), one mutant of Sphingobium francense Sp+ (designated Sp+mt) (Table 3) lacking linA as well as linB was obtained. This mutant (Sp+mt) was analyzed for the ability to degrade β- and δ-HCH isomers as described above. The mutant strain Sp+mt was complemented with linB of B90A (Table 3). For this purpose, plasmid pUCIS1KB, carrying the insertion element IS6100, the kanamycin resistance gene, and the linB gene of B90A (containing promoter and Shine-Dalgarno [SD] sequences) in pUC18, was constructed. Plasmid pUCIS1KB was transferred into Sp+mt by electroporation as described by Iwasaki et al. (10). Electroporation was performed in 2-mm cuvettes with a Bio-Rad gene pulser apparatus (Bio-Rad) by applying the following parameters: capacitance, 25 μF; voltage, 7.5 kV/cm; resistance, 200 Ω; pulse time, 3 to 4 ms. Among several transformants (transformation efficiency, 0.4 × 102) which appeared after 3 to 4 days, nine colonies (designated Sp+mtC1-9) were selected and analyzed for β- and δ-HCH degradation as described above. The presence of linB genes in these clones was also confirmed by hybridizing BamHI-digested DNAs of clones Sp+mtC1-9 with a [32P]ATP-labeled linB gene as a probe (8).
TABLE 3.
Degradation pattern of α-, γ-, β-, and δ-HCH isomersa
| Strain or mutant | Presence of geneb
|
Degradation of HCH isomerc
|
||||||
|---|---|---|---|---|---|---|---|---|
| linA | linB | linC | linDER | α | γ | β | δ | |
| Sphingobium indicum B90Ad | + | + | + | + | +++ | +++ | +++ | +++ |
| Sphingobium francense Sp+ | + | + | + | + | ++ | ++ | ++ | ++ |
| Sphingobium japonicum UT26 | + | + | + | + | ++ | ++ | + | ++ |
| Sp+mt | − | − | + | + | − | − | − | − |
| Sp+mtC1-9 (containing linB of B90A) | − | + | + | + | − | − | ++ | ++ |
| E. coli containing pLINEBB | − | + | − | − | − | − | +++ | +++ |
| E. coli containing pLINESB | − | + | − | − | − | − | + | + |
| E. coli containing pLINEUB | − | + | − | − | − | − | + | + |
RESULTS
Degradation of β- and δ-HCH by resting cells of Sphingobium indicum B90A, Sphingobium francense Sp+, and Sphingobium japonicum UT26.
Although both β- and δ-HCH isomers were degraded by strains B90A, Sp+, and UT26 at a biomass concentration of 20 mg/ml, the rate at which degradation occurred was found to be strain dependent. Whereas β-HCH disappeared completely within 4 h during incubations with strain B90A, in the case of the Sp+ and UT26 strains, complete disappearance of β-HCH was observed only after 8 h and 24 h, respectively (data not shown). δ-HCH disappeared completely within 4 h during incubations with all strains. These results were further confirmed by degradation studies carried out with purified LinB proteins from all strains (see below).
Degradation of β- and δ-HCH by B90A, Sp+, and UT26 was accompanied by the appearance of the intermediates B1 and D1, with retention times of 12.7 and 13.7 min, respectively, when analyzed by GC. These intermediates disappeared eventually in B90A; however, they were not further degraded in Sp+ and UT26. In subsequent incubation experiments with B90A and β- and δ-HCH (as the substrate), further metabolites (B2 and D2) were detected.
β- and δ-HCH degradation in E. coli containing pLINEBB, pLINESB, and pLINEUB.
The linB genes from B90A, Sp+, and UT26 were cloned into pGEX-5X-3 and expressed in E. coli BL21. E. coli BL21 expressing LinB as a fusion protein with GST (GST-LinB) from pLINEBB (B90A) completely degraded not only β- but also δ-HCH within 12 h, whereas there was no degradation of α- and γ-HCH. As observed with intact cells of B90A, there was a concomitant appearance of the intermediates B1 and D1, which did not persist and eventually disappeared. There were noticeable differences in the activities with β- and δ-HCH when the corresponding linB genes from Sp+ and UT26 were expressed in E. coli. While E. coli containing pLINESB and pLINEUB degraded β-HCH (60% and 40%, respectively, within 72 h), the degradation was much slower than that in E. coli containing pLINEBB. While δ-HCH disappeared completely within 12 h of incubation with E. coli containing LinB of strain B90A, only 60% of the δ-HCH was degraded in E. coli containing LinB of Sp+. E. coli containing LinB of UT26 (pLINEUB) was able to degrade δ-HCH, but the degradation was quite slow compared to that in E. coli containing pLINEBB, and only 40% δ-HCH degradation was observed within 72 h (data not shown).
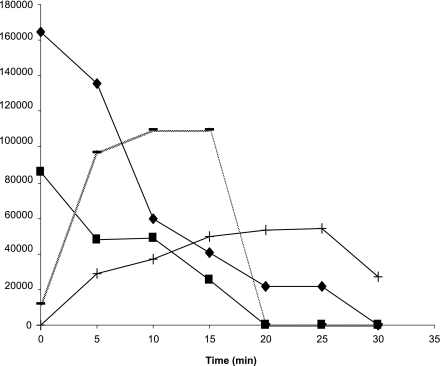
The LinB proteins from B90A, Sp+, and UT26 differ in their amino acid sequences at seven positions (1 to 2% differences). The enzymes from the three strains were purified as GST fusion proteins, and their substrate spectra were determined. The sizes of the GST-LinB fusion proteins were ∼58 kDa. Taking into account a size of 26 kDa for GST, this corresponds well to the value reported for LinB of strain UT26 (30). The purified GST-LinB proteins from strains B90A, Sp+, and UT26 degraded both β- and δ-HCH, with the concomitant appearance of intermediates. Similar results were obtained with GST-cleaved LinB (using factor Xa). Further degradation of the intermediates (B1 and D1) was observed only for LinB of strain B90A (Fig. 2). The other LinB proteins could not further degrade the intermediates, which accumulated in the reaction mixture.
FIG. 2.
Degradation of β- and δ-HCH by purified GST-LinB fusion protein of Sphingobium indicum B90A, with concomitant appearance of their respective intermediates and their disappearance. ▪, β-HCH; −, B1; ⧫, δ-HCH; +, D1. The degradation of β-HCH and production of the intermediate are depicted as peak areas instead of absolute concentrations of the compounds because a standard for intermediate B1 was not available.
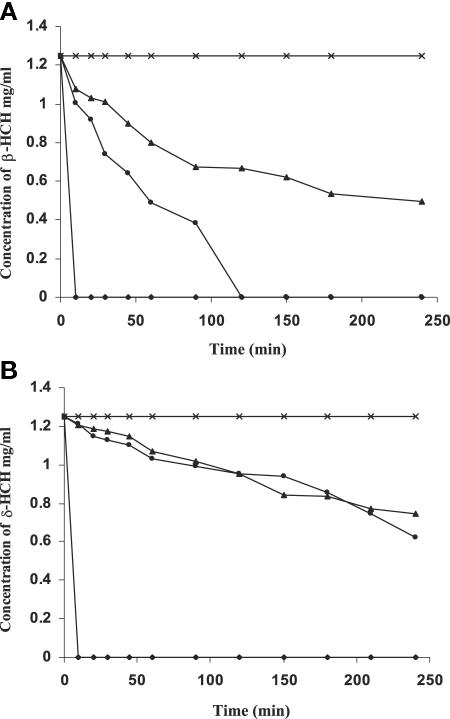
Both β- and δ-HCH were degraded after the addition of the LinB proteins of B90A, UT26, and Sp+, with the concomitant appearance of their intermediates (Fig. 3A and B). The specific activities of purified LinB proteins using β- and δ-HCH as substrates were found to be 100, 2.2, and 2 U/mg for B90A, Sp+, and UT26, respectively. This confirms the strong activity of LinB of B90A for degrading β-and δ-HCH, which is approximately 50 times higher than those of the LinB proteins of UT26 and Sp+.
FIG. 3.
(A) Progress curves of β-HCH conversion by the LinB proteins of Sphingobium indicum B90A (⧫), Sphingobium francense Sp+ (•), and Sphingobium japonicum UT26 (▴). (B) Progress curves of δ-HCH conversion by the LinB proteins of Sphingobium indicum B90A (⧫), Sphingobium francense Sp+ (•), and Sphingobium japonicum UT26 (▴).
Identification of β- and δ-HCH metabolites.
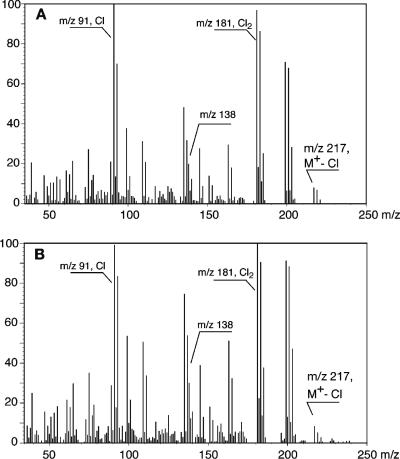
Although B1 and D1, when analyzed with GC, showed different retention times, i.e., 12.7 and 13.7 min, respectively, the mass spectra of peaks B1 and D1, formed by the action of LinB on β- and δ-HCH, respectively, were almost identical. This indicates that the corresponding intermediates of β- and δ-HCH have identical constitutions but are stereochemically different. The mass spectra showed different sets of chlorine-containing mass fragment ions, including set A, corresponding to a four-chlorine-atom-containing fragment at m/z 235 (monoisotopic); and set B, with ions m/z 198 to 203, likely corresponding to two different fragment ions (m/z 198 and 199 [monoisotopic]) containing three chlorine atoms. Presumably, the loss of HCl or Cl from the fragment ions of set B leads to the formation of the ions of set D, at m/z 163 (monoisotopic). The ion cluster C at m/z 170 could be formed from the loss of CO and CHO from the ion clusters at m/z 198 and 199, respectively. Although it could not be proven that the ions at m/z 135 are from those at m/z 170 or are formed by an alternate route, apparently the ions at m/z 170 to 175 lose one chlorine atom, leading to the formation of a cluster F, with m/z 135, 137, and 139 (100, 66.7, and 11.1 Da, respectively). The major peak G at m/z 109 is again formed by the loss of C2H2 (loss of 26 atomic mass units) from m/z 135. The ion at m/z 199 as well as that at m/z 156 (set E) contains three chlorine atoms, and the difference of m/z 43 between these two ions again can be accounted for by the loss of two carbon atoms and one oxygen atom in the molecules, although we do not have evidence that ions 156 and 199 are directly related. Although the mass spectral data alone do not prove the configurational structures of the two intermediates unequivocally, the data provided support the notion that LinB catalyzes the formation of the respective pentachlorocyclohexanols from β- and δ-HCH. The mass spectra of two other, later-eluting metabolites (B2 and D2) were very similar to each other (Fig. 4A and B), with both showing fragment ions (monoisotopic) at m/z 217 (Cl3; minor), 199 (Cl3; major), 181 (Cl2; major), etc., suggestive of tetrachlorocyclohexanediols (M+ = 252) and interpretation of the above-mentioned ions as M-Cl, M-Cl-H2O, and M-Cl-HCl, respectively. Identification of the metabolites B1 and D1 as well as B2 and D2 as pentachlorocyclohexanols and tetrachlorocyclohexanediols was confirmed by the formation of corresponding mono- and diacetates upon acetylation (shift of highest mass ions by 42 Da and 84 Da to m/z 277 and 301 [monoisotopic]), respectively. The mass spectra of the acetates were very similar to those published previously by Sahu et al. (42).
FIG. 4.
Mass spectra of β- and δ-HCH intermediates B2 and D2 from resting-cell incubations of Sphingobium indicum B90A.
Complementation of linB deletion mutant.
Complementation of the linB mutant (Sp+mt) was carried out with pUCIS1KB, which contained IS6100, the kanamycin resistance gene, and the linB ORF along with promoter and SD sequences of B90A, by electroporation. Several kanamycin-resistant transformants were obtained. On average, a transformation efficiency of 0.4 × 102 transformants/μg of plasmid DNA was obtained. Among several transformants, nine transformants (designated Sp+mtC1-9) were analyzed for the presence of the linB gene and the degradation of β- and δ-HCH. In contrast to Sp+mt, which lacked both linB and linA and did not degrade any of the HCH isomers (Table 3), all nine kanamycin-resistant transformants degraded β- and δ-HCH, confirming the functionality of the complemented linB gene of B90A in Sp+mt. All nine transformants also hybridized with an [α-32P]dATP-labeled internal fragment of IS6100. Although all of the selected transformants degraded β- and δ-HCH, degradation was not as fast as that with strain B90A (Table 3). Interestingly, LinB of B90A in the environment of Sp+ did not degrade the intermediates of β- and δ-HCH (B1 and D1, respectively).
DISCUSSION
The data presented in this study reconfirm the potential of Sphingobium indicum B90A for transforming β- and δ-HCH. Previous studies stated that only strain B90A could degrade β-HCH (12, 41), whereas strains Sp+ (8) and UT26 (29) could not. However, these studies were conducted with a relatively low biomass concentration (7 mg/ml). Recent incubations of strain UT26 with β-HCH at high biomass concentrations revealed that strain UT26 was also able to transform β-HCH, albeit at very low rates (1.5 pmol/min/mg of biomass) (31). Our results confirm these findings and clearly show that strain B90A is superior to the other strains with respect to transformation of β-HCH. Furthermore, our studies revealed that the LinB enzymes of strains B90A, Sp+, and UT26, when expressed in E. coli, not only degrade β-HCH but also degrade δ-HCH. In experiments with intact cells, δ-HCH disappeared much faster than it did in experiments with E. coli expressing LinB. These differences can be explained partly by the fact that in intact cells, δ-HCH is also a substrate of the HCH dehydrochlorinase (LinA) (19, 26). LinB of B90A transformed β- and δ-HCH to mono- and dihydroxylated metabolites that were identified as pentachlorocyclohexanols (B1 and D1) and probably transformed these to tetrachlorocyclohexanediols (B2 and D2). This is in agreement with a previous report showing that LinB of UT26 does not further degrade the pentachlorocyclohexanol intermediate formed during enzyme incubations (31). It is interesting that the three haloalkane dehalogenases from B90A, Sp+, and UT26 showed different degradation abilities with respect to β- and δ-HCH. It is unclear, however, why the β- and δ-HCH degradation rates differed among sphingomonads that degrade HCH isomers. Such differences in the ability to degrade β-HCH among sphingomonads isolated from different geographic locations appear to be widespread (5, 24). A close analysis revealed that the LinB enzymes of Sp+ and UT26 differ from each other by three amino acids and that LinB of B90A differs from those of Sp+ and UT26 by six and seven amino acids residues, respectively. These differences in amino acids are outside the putative catalytic domain (D-108, H-272, and E-132) (7, 11, 17, 18, 22, 27, 28, 33, 34, 37, 40, 47), but they seem to play an important role in the substrate specificity of the enzymes. Although the studies with purified LinB proteins from B90A and Sp+ are preliminary, they strongly indicate that strain B90A is best suited for biodegradation of β-HCH, the most recalcitrant isomer. The finding that strains Sp+mtC1-9 containing linB of B90A degraded β- and δ-HCH, whereas strain Sp+mt lacking linA and linB did not, further confirmed that LinB is important for the transformation of β- and δ-HCH. In addition, LinB of B90A did not further degrade B1 and D1 when integrated into strain Sp+, while it did degrade these intermediates when expressed in E. coli. The activity of LinB of B90A towards β- and δ-HCH degradation was slowed down in the environment of Sp+. Such examples of strain-dependent activities of identical catabolic enzymes exist in the literature. For instance, the biphenyl deoxygenases of Pseudomonas pseudoalcaligenes KF707 and Pseudomonas cepacia LB400 exhibit a distinct difference in substrate ranges of polychlorinated biphenyls, despite having nearly identical amino acid sequences (16). While it is difficult presently to point out the factors leading to the differences in activity of the LinB proteins from three different strains, we can only state that in addition to the differences in amino acids, the host environment might also play an important role.
It is clear from the present study that LinB of B90A converts β- and δ-HCH through hydrolytic halogenation to pentachlorocyclohexanols in a first step and, in a second step, to tetrachlorocyclohexanediols. This indicates that a new pathway(s) for the degradation of β- and δ-HCH exists in B90A. This pathway appears to be independent of that being used for the degradation of α- and γ-HCH by this strain. Many previous reports (8, 19, 31, 42, 48, 49) agree with this view. Strain B90 (a mutant of B90A which lacks linDER) degraded β- and even δ-HCH without any evidence of accumulation or further degradation of chlorohydroquinone and hydroquinone (8), indicating that β- and δ-HCH degradation is not mediated through the lower pathway and the common central intermediate hydroquinone, as proposed for γ-HCH (29). In addition, in Sphingobium indicum B90A, linA1/A2, linB, and linC were constitutively expressed, whereas linDER could be induced by the addition of α- and γ-HCH but not by the addition of β- and δ-HCH (48).
Although metabolites B1 and D1 as well as B2 and D2 are represented with identical constitutional formulas, it needs to be pointed out that they are not identical but are stereoisomers. LinB acts on both β- and δ-HCH, and mono- as well as dihydroxylated intermediates are formed. Since δ-HCH is also a substrate of LinA (51), δ-pentachlorocyclohexene will also be formed during resting-cell incubations. However, whether this intermediate will be completely metabolized in strain B90A needs to be elucidated. Further elucidation of the degradation of B2 and D2 will also be crucial for depicting the ultimate fate of β- and δ-HCH in B90A as well as in bioremediation situations. At the moment, the environmental fate of the tetrachlorocyclohexanediols is not known, nor is it known whether there are any bacterial strains able to transform them.
Here we showed that LinB is responsible for the transformation of β-HCH to pentachlorocyclohexanol and tetrachlorocyclohexanediol in S. indicum B90A. Moreover, we were able to demonstrate that LinB also acts on δ-HCH, analogously forming the respective pentachlorocyclohexanol and tetrachlorocyclohexanediol intermediates.
Acknowledgments
We are grateful to R. Girijan, Toshvin Analytical Pvt. Ltd., Bombay, India, for providing the GC-MS facility. Thanks are also due to Frederic Gabriel for his help in interpreting the GC-MS data. We also thank Jan Roelof van der Meer, Rakesh Jain, and Banwari Lal for providing valuable suggestions.
Part of this work was supported by grants under the Indo-Swiss Collaboration in Biotechnology (ISCB) from the Swiss Agency for Development and Cooperation (SDC), Switzerland, and the Department of Biotechnology (DBT), India. P.S., S.M., C.D., and H.K. gratefully acknowledge CSIR-UGC, Government of India, for providing research fellowships.
REFERENCES
- 1.Ahuja, A. K., and M. D. Awasthi. 1993. Contamination of rice and wheat grains with residues of HCH and DDT. Pestic. Res. J. 5:83-86. [Google Scholar]
- 2.Bachmann, A., P. Walet, P. Wijnen, W. de Bruin, J. L. M. Huntjens, W. Roelofsen, and A. J. B. Zehnder. 1988. Biodegradation of alpha- and beta-hexachlorocyclohexane in soil slurry under different redox conditions. Appl. Environ. Microbiol. 54:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakore, N., P. J. John, and P. Bhatnagar. 2004. Organochlorine pesticide residues in wheat and drinking water samples from Jaipur, Rajasthan, India. Environ. Monit. Assess. 98:381-389. [DOI] [PubMed] [Google Scholar]
- 4.Ballschmiter, K. 1996. Persistent, ecotoxic and bioaccumulative compounds and their possible environmental effects. Pure Appl. Chem. 68:1771-1780. [Google Scholar]
- 5.Boltner, D., S. M. Morillas, and J. L. Ramos. 2005. 16S rDNA phylogeny and distribution of lin genes in novel hexachlorocyclohexane-degrading Sphingomonas strains. Environ. Microbiol. 7:1329-1338. [DOI] [PubMed] [Google Scholar]
- 6.Cérémonie, H., H. Boubakri, P. Mavingui, P. Simonet, and T. M. Vogel. 2006. Plasmid-encoded γ-hexachlorocyclohexane degradation genes and insertion sequences in Sphingobium francense (ex-Sphingomonas paucimobilis Sp+). FEMS Microbiol. Lett. 257:243-252. [DOI] [PubMed] [Google Scholar]
- 7.Damborsky, J., E. Rorije, A. Jesenska, Y. Nagata, G. Klopman, and W. J. G. M. Peijnenburg. 2001. Structure-specificity relationships for haloalkane dehalogenases. Environ. Toxicol. Chem. 20:2681-2689. [PubMed] [Google Scholar]
- 8.Dogra, C., V. Raina, R. Pal, M. Suar, S. Lal, K. H. Gartemann, C. Holliger, J. R. van der Meer, and R. Lal. 2004. Organization of genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J. Bacteriol. 186:2225-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong, Z. M., F. L. Xu, R. Dawson, J. Cao, W. X. Liu, B. G. Li, W. R. Shen, W. J. Zhang, B. P. Qin, R. Sun, and S. Tao. 2004. Residues of hexachlorocyclohexane isomers and their distribution characteristics in soils in the Tianjin area, China. Arch. Environ. Contam. Toxicol. 46:432-437. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki, K., K. Uchiyama, O. Yagi, T. Kurabayashi, K. Ishizuka, and Y. Takamura. 1994. Transformation of Pseudomonas putida by electroporation. Biosci. Biotechnol. Biochem. 58:851-854. [DOI] [PubMed] [Google Scholar]
- 11.Janssen, D. B., F. Pries, and J. R. van der Ploeg. 1994. Genetics and biochemistry of dehalogenating enzymes. Annu. Rev. Microbiol. 48:163-191. [DOI] [PubMed] [Google Scholar]
- 12.Johri, A. K., M. Dua, D. Tuteja, R. Saxena, D. M. Saxena, and R. Lal. 1998. Degradation of alpha, beta, gamma, and delta hexachlorocyclohexane by Sphingomonas paucimobilis. Biotechnol. Lett. 20:885-887. [Google Scholar]
- 13.Kalbitz, K., and P. Popp. 1999. Seasonal impacts on β-hexachlorocyclohexane concentration in soil solution. Environ. Pollut. 106:139-141. [DOI] [PubMed] [Google Scholar]
- 14.Kannan, K., S. Tanabe, J. P. Giesy, and R. Tatsukawa. 1997. Organochlorine pesticides and polychlorinated biphenyls in foodstuffs from Asian and oceanic countries. Rev. Environ. Contam. Toxicol. 152:1-55. [DOI] [PubMed] [Google Scholar]
- 15.Kendall, R., and R. Dickerson. 1998. Principles and processes for evaluating endocrine disruption in wildlife. SETAC technical publications series. Society of Environmental Toxicology and Chemistry, Pensacola, Fla.
- 16.Kimura, N., A. Nishi, M. Goto, and K. Furukawa. 1997. Functional analysis of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J. Bacteriol. 179:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kmunicek, J., K. Hynkova, T. Jedlicka, Y. Nagata, A. Negri, F. Gago, R. C. Wade, and J. Damborsky. 2005. Quantitative analysis of substrate specificity of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. Biochemistry 44:3390-3401. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, M., P. Chaudhary, M. Dwivedi, R. Kumar, D. Paul, R. K. Jain, S. K. Garg, and A. Kumar. 2005. Enhanced biodegradation of β- and δ- hexachlorocyclohexane in the presence of α- and γ-HCH in contaminated soils. Environ. Sci. Technol. 39:4005-4011. [DOI] [PubMed] [Google Scholar]
- 19.Kumari, R., S. Subudhi, M. Suar, G. Dhingra, V. Raina, C. Dogra, S. Lal, J. R. van der Meer, C. Holliger, and R. Lal. 2002. Cloning and characterization of genes responsible for the degradation of hexachlorocyclohexane isomers in Sphingomonas paucimobilis strain B90. Appl. Environ. Microbiol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutz, F. W., P. H. Wood, and D. P. Bottimore. 1991. Organochlorine pesticides and polychlorinated biphenyls in human adipose tissue. Rev. Environ. Contam. Toxicol. 120:1-82. [DOI] [PubMed] [Google Scholar]
- 21.Lal, R., C. Dogra, S. Malhotra, P. Sharma, and R. Pal. 2006. Diversity, distribution and divergence of lin genes in hexachlorocyclohexane-degrading sphingomonads. Trends Biotechnol. 24:121-130. [DOI] [PubMed] [Google Scholar]
- 22.Marek, J., J. Vevodova, I. K. Smatanova, Y. Nagata, L. A. Svenson, J. Newman, M. Takagi, and J. Damborsky. 2000. Crystal structure of haloalkane dehalogenase of Sphingomonas paucimobilis UT26. Biochemistry 39:14082-14086. [DOI] [PubMed] [Google Scholar]
- 23.Middledorp, P. J. M., M. Jaspers, A. J. B. Zehnder, and G. Schraa. 1996. Biotransformation of α-, β-, γ- and δ-hexachlorocyclohexane under methanogenic conditions. Environ. Sci. Technol. 30:2345-2349. [Google Scholar]
- 24.Mohn, W. W., B. Mertens, J. D. Neufeld, W. Verstraete, and V. de Lorenzo. 2006. Distribution and phylogeny of hexachlorocyclohexane-degrading bacteria in soils from Spain. Environ. Microbiol. 8:60-68. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee, I., and M. Gopal. 2002. Organochlorine insecticide residues in drinking and ground water in and around Delhi. J. Environ. Monit. Assess. 76:185-193. [DOI] [PubMed] [Google Scholar]
- 26.Nagata, Y., T. Hatta, R. Imai, K. Kimbara, M. Fukuda, K. Yano, and M. Takagi. 1993. Purification and characterization of γ-hexachlorocyclohexane (γ-HCH) dehydrochlorinase (LinA) from Pseudomonas paucimobilis. Biosci. Biotechnol. Biochem. 57:1582-1583. [DOI] [PubMed] [Google Scholar]
- 27.Nagata, Y., Z. Prokop, S. Marvanova, J. Sykorova, M. Monincova, M. Tsuda, and J. Damborsky. 2003. Reconstruction of mycobacterial dehalogenase Rv2579 by cumulative mutagenesis of haloalkane dehalogenase LinB. Appl. Environ. Microbiol. 69:2349-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagata, Y., K. Hynkova, J. Damborsky, and M. Takagi. 1999. Construction and characterization of histidine tagged haloalkane dehalogenase (LinB) of a new substrate class from a γ-hexachlorocyclohexane degrading bacterium Sphingomonas paucimobilis UT26. Protein Expr. Purif. 17:299-304. [DOI] [PubMed] [Google Scholar]
- 29.Nagata, Y., K. Miyauchi, and M. Takagi. 1999. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. Biotechnol. 23:380-390. [DOI] [PubMed] [Google Scholar]
- 30.Nagata, Y., K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova, and M. Takagi. 1997. Purification and characterization of a haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63:3707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagata, Y., Z. Prokop, Y. Sato, P. Jerabek, A. Kumar, Y. Ohtsubo, M. Tsuda, and J. Damborsky. 2005. Degradation of β-hexachlorocyclohexane by haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 71:2183-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nawab, A., A. Aleem, and A. Malik. 2003. Determination of organochlorine pesticides in agricultural soil with special reference to γ-HCH degradation by Pseudomonas strains. Biores. Technol. 88:41-46. [DOI] [PubMed] [Google Scholar]
- 33.Oakley, A. J., M. Klvana, M. Otyepka, Y. Nagata, M. C. J. Wilce, and J. Damborsky. 2004. Crystal structure of haloalkane dehalogenase LinB from Sphingomomas paucimobilis UT26 at 0.95A resolution: dynamics of catalytic residues. Biochemistry 43:870-878. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira, R. M., L. H. Bastos, A. E. Dias, S. A. Silva, and J. C. Moreira. 2003. Residual concentration of hexachlorocyclohexane in a contaminated site in Cidade dos Meninos, Duque de Caxias, Rio de Janeiro, Brazil, after calcium oxide treatment. Cad. Saude Publica 19:447-453. [DOI] [PubMed] [Google Scholar]
- 35.Ortiz, J. B., M. L. Gonzalez de Canales, and C. Sarasquete. 2001. The impact of persistent organochlorine contaminant (lindane, γ-HCH): histopathological alterations in fish tissues. Ecotoxicol. Environ. Restor. 4:45-52. [Google Scholar]
- 36.Österreicher-Cunha, P., T. Langenbach, J. P. M. Torres, A. L. C. Lima, T. de Campos, E. A. Vargas, Jr., and A. R. Wagener. 2003. HCH distribution and microbial parameters after liming of a heavily contaminated soil in Rio de Janeiro. Environ. Res. 93:316-327. [DOI] [PubMed] [Google Scholar]
- 37.Otyepka, M., and J. Damborsky. 2002. Functionally relevant motions of haloalkane dehalogenases occur in the specificity-modulating cap domains. Protein Sci. 11:1206-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal, R., S. Bala, M. Dadhwal, M. Kumar, G. Dhingra, O. Prakash, S. R. Prabagaran, S. Shivaji, J. Cullum, C. Holliger, and R. Lal. 2005. The hexachlorocyclohexane-degrading bacterial strains Sphingomonas paucimobilis B90A, UT26 and Sp+, having similar lin genes, are three distinct species, Sphingobium indicum sp. nov., Sphingobium japonicum sp. nov. and Sphingobium francense sp. nov., and reclassification of (Sphingomonas) chungbukensis as Sphingobium chungbukense comb. nov. Int. J. Syst. Evol. Microbiol. 55:1965-1972. [DOI] [PubMed] [Google Scholar]
- 39.Prakash, O., M. Suar, V. Raina, C. Dogra, R. Pal, and R. Lal. 2004. Residues of hexachlorocyclohexane isomers in soil and water samples from Delhi and adjoining areas. Curr. Sci. 87:73-77. [Google Scholar]
- 40.Prokop, Z., M. Monincova, R. Chaloupkova, M. Klvana, Y. Nagata, D. B. Janssen, and J. Damborsky. 2003. Catalytic mechanism of the haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. J. Biol. Chem. 278:45094-45100. [DOI] [PubMed] [Google Scholar]
- 41.Sahu, S. K., K. K. Patnaik, M. Sharmila, and N. Sethunathan. 1990. Degradation of alpha-, beta-, and gamma- hexachlorocyclohexane by a soil bacterium under aerobic conditions. Appl. Environ. Microbiol. 56:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahu, S. K., K. K. Patnaik, S. Bhuyan, B. Sreedharn, N. Kurihara, T. K. Adhya, and N. Sethunathan. 1995. Mineralization of α-, γ- and β- isomers of hexachlorocyclohexane by a soil bacterium under aerobic conditions. J. Agric. Food Chem. 43:833-837. [Google Scholar]
- 43.Sanghi, R., and V. Tewari. 2001. Monitoring of pesticide residues in summer fruits and vegetables from Kanpur, India. Bull. Environ. Contam. Toxicol. 67:587-593. [DOI] [PubMed] [Google Scholar]
- 44.Senoo, K., and H. Wada. 1989. Isolation and identification of an aerobic γ-HCH-decomposing bacterium from soil. Soil Sci. Plant Nutr. 35:79-87. [Google Scholar]
- 45.Simonich, S. L., and R. A. Hites. 1995. Global distribution of persistent organochlorine compounds. Science 269:1851-1854. [DOI] [PubMed] [Google Scholar]
- 46.Singh, R. P. 2001. Comparison of organochlorine pesticide levels in soil and groundwater of Agra, India. Bull. Environ. Contam. Toxicol. 67:126-132. [DOI] [PubMed] [Google Scholar]
- 47.Streltsov, V. A., Z. Prokop, J. Damborsky, Y. Nagata, and M. C. J. Wilce. 2003. Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates. Biochemistry 42:10104-10112. [DOI] [PubMed] [Google Scholar]
- 48.Suar, M., J. R. van der Meer, K. Lawlor, C. Holliger, and R. Lal. 2004. Dynamics of multiple lin gene expression in Sphingomonas paucimobilis B90A in response to different hexachlorocyclohexane isomers. Appl. Environ. Microbiol. 70:6650-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suar, M., A. Hauser, T. Poiger, H. R. Buser, M. D. Müller, C. Dogra, V. Raina, C. Holliger, J. R. van der Meer, R. Lal, and H. P. E. Kohler. 2005. Enantioselective transformation of α-hexachlorocyclohexane by the dehydrochlorinases LinA1 and LinA2 from the soil bacterium Sphingomonas paucimobilis B90A. Appl. Environ. Microbiol. 71:8514-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Trantirek, L., K. Hynkova, Y. Nagata, A. Murzin, A. Ansorgova, V. Sklenar, and J. Damborsky. 2001. Reaction mechanism and stereochemistry of γ-hexachlorocyclohexane dehydrochlorinase LinA. J. Biol. Chem. 276:7734-7740. [DOI] [PubMed] [Google Scholar]
- 52.Van Eckert, M. H. A., N. J. P. van Ras, G. H. Mentink, H. H. M. Rijnaarts, A. J. M. Stams, J. A. Field, and G. Schraa. 1998. Anaerobic transformation of β-HCH by methanogenic granular sludge and soil microflora. Environ. Sci. Technol. 32:3299-3304. [Google Scholar]
- 53.Willem, J. K., and I. Wollent. 2005. Inventories of obsolete pesticide stocks in central and Eastern Europe, p. 37-39. In E. Elbestawy, L. Moklyachuk, V. Pidlisnyuk, N. Schulz, T. Stefanovska, and J. Vijgen (ed.), Proceedings of the 7th International HCH and Pesticides Forum, Kiev, Ukraine. International HCH and Pesticides Association, Holte, Denmark.
- 54.Zhulidov, A. V., R. D. Robarts, J. V. Headley, K. Liber, D. A. Zhulidov, O. V. Zhulidov, and D. F. Pavlov. 2002. Levels of DDT and hexachlorocyclohexane in burbot (Lota lota l.) from Russian Arctic rivers. Sci. Total Environ. 292:231-246. [DOI] [PubMed] [Google Scholar]
- 55.Zou, E., and F. Matsumura. 2005. Long-term exposure to beta-hexachlorocyclohexane (beta-HCH) promotes transformation and invasiveness of MCF-7 human breast cancer cells. Biochem. Pharmacol. 66:831-840. [DOI] [PubMed] [Google Scholar]