Abstract
In plants, reproductive development requires normal meiosis, which involved several highly coordinated events. Such meiotic events are regulated in a number of ways in yeast and animal systems, including transcriptional and checkpoint control mechanisms. Although a number of mutations that affect different aspects of meiosis have been characterized in plants, very little is known about the regulation of plant meiosis at the molecular level. In particular, no meiosis-specific transcriptional regulators have been identified in plants, and checkpoint control has not been observed during plant meiosis. We report here the isolation and characterization of a new Arabidopsis male-sterile mutant that exhibits meiotic defects. Meiocytes from mutant plants appeared normal up to diakinesis, when they exhibited signs of apoptosis, including defects in chromosome behavior, cytoplasmic shrinkage, and chromatin fragmentation, followed by cell death before cytokinesis. Therefore, the mutant was named male meiocyte death1 (mmd1). The MMD1 gene was cloned using Dissociation transposon tagging and encodes a plant homeo domain domain–containing protein. MMD1 is expressed preferentially during male meiosis. Our results suggest that MMD1 may be involved in the regulation of gene expression during meiosis and that the mmd1 mutation triggers cell death in male meiocytes.
INTRODUCTION
Meiosis has been characterized extensively in a number of organisms, including plants. It consists of a highly coordinated series of events, all of which are essential for the proper segregation of chromosomes. The establishment of cohesion between sister chromatids, which is essential for the correct attachment of chromosomes to the spindles, occurs during meiotic S-phase. Homologous chromosome pairing and synaptonemal complex formation and recombination occur during zygotene and pachytene, respectively. During diplonema and diakinesis, chromosomes desynapse and condense. The reorganization of chromosomes from long thin fibers into highly condensed, compact structures, which begins during leptonema and is completed during diakinesis, is one of the most striking but least understood events of meiosis. The condensin complex and topoisomerase II participate in chromosome condensation during mitosis and likely are involved in meiosis (Hirano, 2000; Cuvier and Hirano, 2003); however, how this process is controlled is unknown at present. Once condensed, the chromosomes align and attach to the meiotic spindle during metaphase, followed by segregation of homologous copies of each chromosome pair at anaphase I. Finally, during meiosis II, the sister chromosomes segregate in an equational division similar to that observed during mitosis.
Considerable progress has been made in understanding meiosis and elucidating factors associated with a number of meiotic events. Many meiotic processes are highly conserved among organisms (reviewed by Ashley and Plug, 1998; Dawe, 1998; Shaw and Moore, 1998; Zickler and Kleckner, 1999; Bhatt et al., 2001). For instance, it is clear that the mismatch repair system plays an important role during meiotic recombination in systems ranging from yeast to humans (Baker et al., 1995; Roeder, 1995; Cummings and Zolan, 1998). Likewise, characteristics of the cohesion and condensin complexes also are well conserved across a wide range of organisms (reviewed by Gillies, 1984; Dawe, 1998; Orr-Weaver, 1999; Hirano, 2000).
However, one aspect of meiosis that may vary significantly between organisms concerns how the cell controls the progression through the meiotic cell cycle. Regulation of the meiotic cell cycle is best understood in yeast (reviewed by Lee and Amon, 2001), in which numerous mutations have been identified that block chromosome synapsis and/or recombination and induce pachytene arrest (Roeder, 1997; Roeder and Bailis, 2000). The characterization of secondary mutations that bypass pachytene arrest in the presence of recombination and synapsis defects has led to the identification of a number of proteins that participate in meiotic checkpoint control mechanisms (Lydall et al., 1996; Leu and Roeder, 1999; SanSegundo and Roeder, 1999; Lindgren et al., 2001). The pachytene check-point also has been shown to operate in several other organisms, such as Drosophila, Caenorhabditis elegans, and mouse (Edelmann et al., 1996, 1999; Pittman et al., 1998; Ghabrial and Schupbach, 1999; Gartner et al., 2000); however, it has not yet been found in plants.
In Arabidopsis, maize, and other plants, genetic studies have identified a number of mutants that affect either meiocyte formation or the process of meiosis (Scott et al., 1991; McCormick, 1993; Chaudhury et al., 1994; Neuffer et al., 1997; Taylor et al., 1998; Sanders et al., 1999). Mutational analyses have been used to identify genes required for the formation or differentiation of anther cells at stages before meiosis. For example, the Arabidopsis SPOROCYTELESS/NOZZLE gene, which encodes a nuclear protein with similarity to transcription factors, is critical for the formation of male and female meiocytes (Schiefthaler et al., 1999; Yang et al., 1999b). Another gene that is required for anther cell differentiation is the EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS gene, which is required for the formation of the tapetum and encodes a putative receptor protein kinase (Canales et al., 2002; Zhao et al., 2002). Furthermore, a number of meiotic mutants have been characterized that exhibit defects in one or more critical events during meiosis, including chromosome cohesion and recombination (Sato et al., 1995; Klimyuk and Jones, 1997; Doutriaux et al., 1998; Bai et al., 1999; Bhatt et al., 1999; Couteau et al., 1999; Franklin et al., 1999; Caryl et al., 2000; Grelon et al., 2001; Mercier et al., 2001; Azumi et al., 2002).
In contrast to meiotic mutants in yeast and animals that cause meiotic cell arrest and/or cell death, most plant meiotic mutants complete meiosis and cytokinesis and produce abnormal microspores. The abnormal microspores produced in these plant meiotic mutants typically degenerate during pollen development, resulting in male sterility or reduced male fertility, suggesting that plants may not use meiotic checkpoint control. However, a number of maize male-sterile mutants have been isolated that show defects in male meiocytes, including ab-normal morphology and cell degeneration before cytokinesis (Albertsen and Phillips, 1981; West and Albertsen, 1985). Therefore, it is not clear if plant meiosis is under a checkpoint control system similar to that in yeast and animal meiosis.
As part of studies to better understand meiosis and its control in plants, we have characterized male-sterile mutants in Arabidopsis. Here, we describe the isolation and detailed characterization of one mutant, male meiocyte death1 (mmd1), that exhibited alterations in meiosis that resulted in male meiocyte arrest and cell death. We show that MMD1 encodes a plant homeo domain (PHD) domain–containing protein and that it is expressed preferentially in male meiocytes. The mutant phenotype and molecular characteristics of MMD1 suggest that it may participate in chromatin remodeling and/or transcriptional events required for successful progression through meiosis.
RESULTS
Isolation and Characterization of a Male Meiotic Mutant
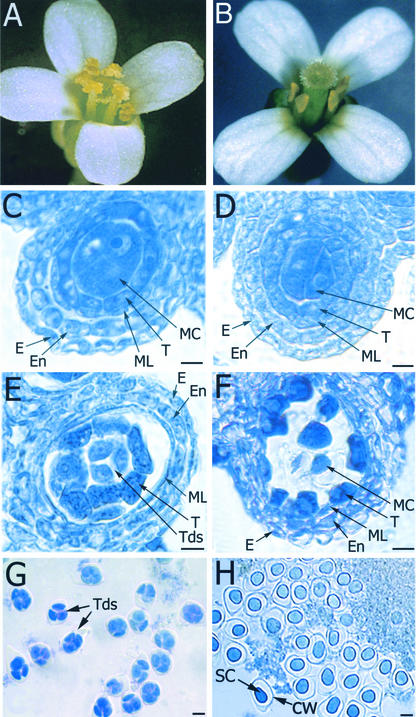
Arabidopsis lines carrying Dissociation (Ds) insertions were generated and screened for mutant plants that exhibit defects in fertility and meiosis. Progeny of one line (Y9287) segregated for fertile and sterile plants with a ratio of ∼3:1, suggesting that the parental plant was heterozygous for a recessive mutation. Mutant flowers failed to produce pollen but produced apparently normal seeds when cross-pollinated (data not shown), suggesting that the mutation affects male meiosis and/or pollen development but not female reproduction. Although most flowers resembled those of the wild type (Figure 1A), a fraction of flowers (30%) contained four or five stamens (Figure 1B) instead of the normal six stamens. In addition, the filaments of mutant flowers were consistently shorter than those of wild-type plants.
Figure 1.
Flower and Pollen Development in mmd1 and Wild-Type Plants.
(A) Wild-type flower with six stamens and normal pollen.
(B) mmd1 mutant flower containing four pollenless stamens.
(C) and (E) Sections of wild-type anthers from stage-9 and -10 anthers, respectively, showing epidermis (E), endothecium (En), middle layer (ML), tapetum (T), meiotic cells (MC), and tetrads (Tds).
(D) and (F) Sections of mmd1 anthers from stage-9 and -10 anthers, respectively, showing epidermis (E), endothecium (En), middle layer (ML), tapetum (T), and meiotic cells (MC).
(G) Contents of a stage-10 wild-type anther. Tetrads (Tds) of four immature microspores are observed.
(H) Contents of a stage-10 mmd1 anther. Arrested microsporocytes with shrunken cytoplasm (SC) and callose wall (CW) are observed.
Bars = 10 μm.
Light microscopic analysis indicated that anther development was normal through stage 8 (as defined by Smyth et al., 1990), just before male meiosis. In particular, compared with wild-type anthers (Figure 1C), no alterations were observed in the epidermis, endothecium, middle layer, tapetum, or meiocytes of mutant plants before meiosis (Figure 1D). In anthers of stage-9 flowers, when normal male meiosis occurs to form tetrads (Figure 1E), the four outer layers of the anther again appeared normal in the mutant (Figure 1F). However, arrested and/or degenerating male meiocytes were observed in the locules of mutant plants (Figure 1F). In anthers of stage-10 flowers from wild-type plants, tetrads (Figure 1G) and developing microspores were observed. By contrast, only shrunken and collapsed male meiocytes were observed in stage-10 and later flowers from the mutant (Figure 1H). Tetrads were not observed in the anthers of mutant plants, suggesting that meiocytes fail to undergo cytokinesis. Because no alterations were observed in the remaining four layers of the anther before, during, or after defects were observed in meiocytes, other aspects of anther development seem normal in the mutant. However, we cannot exclude the possibility that subtle alterations in anther development were present and not detected. Because the main defects associated with the mutant were meiotic arrest and cell death in male meiocytes (see below for more details), we named the mutant mmd1 for male meiocyte death1.
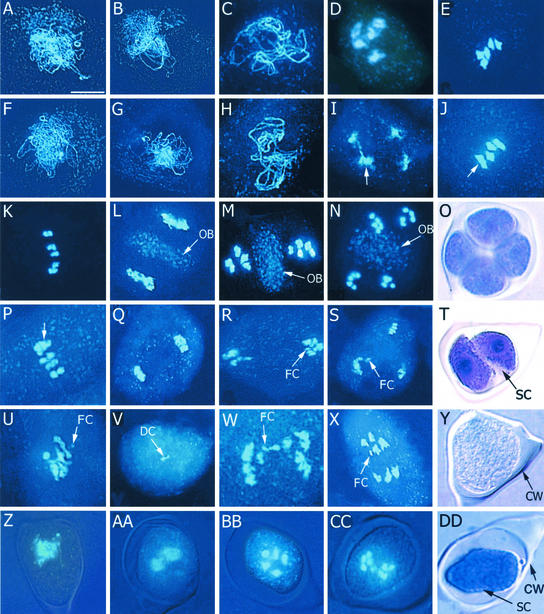
To further understand the major effect of the mmd1 mutation, male meiosis in mmd1 plants was analyzed using 4′,6-diamidino-2-phenylindole (DAPI) staining of spreads of meiotic chromosomes and was compared with wild-type meiosis (Ross et al., 1996; Bai et al., 1999; Azumi et al., 2002). Wild-type chromosomes displayed characteristic morphologies at leptotene (Figure 2A), zygotene (Figure 2B), pachytene (Figure 2C), diakinesis (Figure 2D), metaphase I (Figure 2E), early anaphase I (Figure 2K), and meiosis II (Figures 2L to 2N). We found that mmd1 male meiocytes underwent meiosis with normal chromosomal morphology during early meiosis, including leptotene (Figure 2F), zygotene (Figure 2G), and pachytene (Figure 2H), indicating that chromatid cohesion and homologous chromosome pairing is relatively normal in the mutant. At diakinesis, ∼30% of cells contained five relatively normal-looking bivalents. However, most cells at diakinesis were abnormal. They contained one to six groups of chromosomes, with four being the most numerous (∼50%) (Figure 2I), suggesting that two pairs of chromosomes often are associated abnormally with each other. Cells containing four groups of chromosomes also were observed commonly at metaphase I and anaphase I (Figures 2J and 2P).
Figure 2.
Meiotic Alterations in mmd1 Plants.
Meiotic spreads of wild type ([A] to [E] and [K] to [N]) and mmd1 plants ([F] to [J], [P] to [S], and [U] to [X]) were prepared and stained with DAPI. (Z) to (CC) show composite images of bright-field and DAPI staining of unspread mmd1 microsporocytes; (O) and (Y) (wild type) and (T) and (DD) (mmd1) show bright-field images of toluidine blue–stained, unspread cells. Bar in (A) = 10 μm for all panels.
(A) and (F) Leptotene.
(B) and (G) Zygotene.
(C) and (H) Pachytene.
(D) and (I) Diakinesis. Note the improperly condensed and resolved chromosomes in the mmd1 meiocyte (arrow in [I]).
(E) and (J) Metaphase I. Note the two bivalents associated in the mmd1 meiocyte.
(K) and (P) Metaphase I–to–anaphase I transition. Note the two associated heterochromatids associated in the mmd1 meiocyte (arrow in [P]).
(L) and (Q) Telophase I. Note the distinctive organelle band (OB) in the wild-type cell that is missing in mmd1 meiocytes.
(M) and (N) Wild-type meiocytes during meiosis II.
(O) Wild-type tetrad.
(R) and (S) Examples of abnormal mmd1 meiocytes observed during meiosis II. Note the fragmented chromosomes (FC).
(T) mmd1 “dyad” with a separated, shrunken cytoplasm (SC).
(U) to (X) Examples of defects observed in mmd1 meiocytes during meiosis I, including degrading chromosomes (DC).
(Y) Normal meiocyte with a narrow callose wall (CW).
(Z) to (CC) Composite images of a normal mmd1 meiocyte during pachytene (Z) and meiocytes with shrunken cytoplasms after diakinesis ([AA] to [CC]).
(DD) mmd1 meiocyte showing shrunken cytoplasm and callose wall.
Regardless of chromosomal morphology, all mmd1 cells observed at diakinesis exhibited signs of cytoplasmic shrinkage (Figures 2AA to 2DD), indicating that they are clearly defective by this stage. This finding is in contrast to the results seen in mmd1 meiocytes before diakinesis, which did not exhibit cytoplasmic shrinkage (Figure 2Z). Some mmd1 meiocytes appeared to arrest and undergo nuclear degradation at diakinesis (Figure 2U), whereas others progressed past diakinesis and exhibited several different alterations. Cells with fewer than five bivalents were observed at metaphase I and early anaphase I (Figures 2J and 2P). Uneven distribution of chromosomes, chromosome bridges, and chromosome fragmentation also was observed (Figures 2V to 2X). These defects in chromosome segregation may result from alterations in the resolution of nonhomologous chromosomes observed at diakinesis. Small numbers of meiocytes also were observed at metaphase II and at anaphase II/telophase II with two or three clusters of degrading chromosomes (Figures 2R and 2S), suggesting that in some cells chromosomes are able to complete meiosis I and progress into meiosis II. Finally, a considerable number of collapsing cells were observed with two to four DAPI-staining areas (data not shown). In these cells, which typically were surrounded by a thin callose wall, vesicles sometimes were observed partitioning the cytoplasm (Figure 2T), in contrast to the typical cell membrane and callose wall that surrounded wild-type cells at the tetrad stage (Figure 2O). No evidence of cytokinesis or the formation of the callose wall associated with cytokinesis was observed in mmd1 meiocytes. Ultimately, all mmd1 meiocytes underwent nuclear degradation and collapse before anthesis. In contrast to most meiotic mutants characterized in plants, microspores were not formed in mmd1 plants.
One of the more notable alterations during meiosis in mmd1 meiocytes was observed from telophase I to telophase II. In wild-type meiocytes, a distinct organelle band formed between the separated chromosomes along what would become the division plane (Figures 2L and 2M). This organelle band was not observed in meiocytes of mmd1 plants (Figures 2Q to 2S), although DAPI-staining material was detected in the cytoplasm. Therefore, it is not clear whether the organelles are degraded in the mutant or just dispersed throughout the cytoplasm. The absence of an organelle band in mmd1 meiocytes may be associated with the failure of meiocytes to form normal cell/callose walls during cytokinesis. It is worth noting that programmed cell death in animal cells often is associated with mitochondrial degeneration (Green and Reed, 1998).
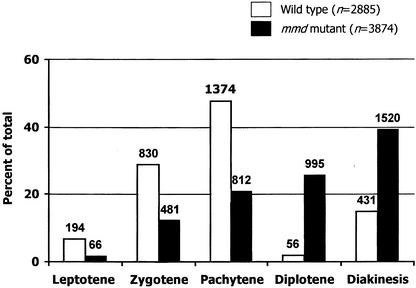
Our observation that mmd1 meiocytes exhibited cytoplasmic shrinkage and signs of chromosome degradation at diakinesis raised the possibility that the mutation may evoke a meiotic checkpoint control mechanism, which may arrest meiosis or cause a delay in the progression of meiosis. To investigate this possibility, we examined the distribution of meiocytes at various substages of prophase I in stage-9 flowers and compared these numbers with those observed for wild-type plants. When data from samples containing only or largely prophase I cells were analyzed, we found that in wild-type anthers (six flowers), nearly half of the meiocytes (1374 of 2885) were pachytene cells (Figure 3). Moderate numbers of cells at zygotene (29%) and diakinesis (13%) were observed, with considerably fewer cells at leptotene (7%) and diplotene (2%). A similar distribution of meiotic cells during prophase was reported previously for wild-type Arabidopsis (Azumi et al., 2002). By contrast, a dramatically different distribution of prophase I substages was observed in stage-9 flowers from mmd1 plants. Of the 3874 cells observed from nine flowers, ∼40% were at diakinesis, with progressively fewer cells observed at diplotene (25%), pachytene (21%), zygotene (12%), and leptotene (2%). These results suggest that in contrast to wild-type meiocytes, which move relatively quickly through diplotene and diakinesis, mmd1 male meiocytes are delayed starting at diplotene, consistent with the idea that the mutation may invoke a checkpoint control mechanism. However, at present, we cannot exclude the alternative possibility that mmd1 meiocytes move more quickly through early stages of prophase.
Figure 3.
Distribution of Prophase I Cells in Anthers of Stage-9 Flowers in the Wild Type and the mmd1 Mutant.
The number of cells at each stage is shown above the appropriate bar. The mmd1 stages were assigned based on chromosome condensation and overall chromosome morphology.
In contrast to the alterations in male fertility observed in mmd1 plants, female fertility appeared to be relatively normal. Nevertheless, we examined female meiosis in wild-type and mmd1 mutant plants. At least 150 female meiocytes ranging from early prophase I to telophase II were examined in mmd1 plants and compared with wild-type meiocytes. Female meiosis appeared completely normal in mmd1 plants (data not shown). Therefore, the mmd1 mutation disrupts male meiosis starting at approximately diakinesis in microsporocytes but does not affect female meiosis.
Male Meiocytes in mmd1 Plants Undergo Chromosome Fragmentation
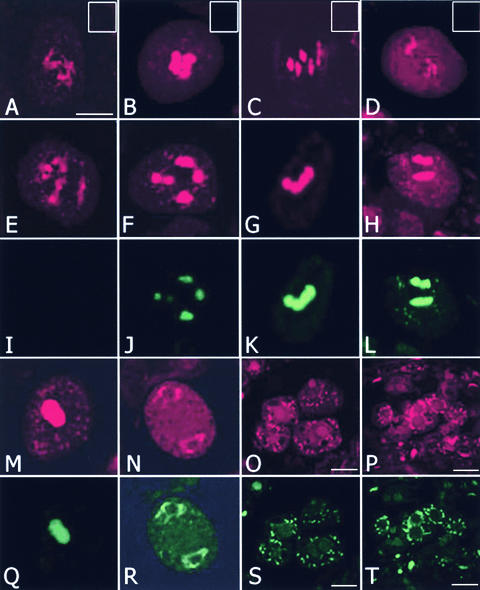
Several characteristics of meiocytes in mmd1 plants, including cytoplasmic shrinkage and chromosome degradation, are typical of cells undergoing apoptosis, a process used by both animals and plants for the selective elimination of damaged or otherwise unwanted cells (Ellis et al., 1991). One hallmark of cells undergoing apoptosis is the cleavage of nuclear DNA into fragments (Cohen et al., 1994). To determine whether mmd1 male meiocytes contain fragmented chromosomes, TdT-mediated dUTP nick-end labeling (TUNEL) assays were conducted on sections of developmentally staged wild-type and mmd1 anthers as well as isolated male meiocytes. TUNEL labeling was not observed in male meiocytes (n > 100) of wild-type plants at any stage (Figures 4A to 4D), indicating that the assay was not sensitive enough to detect the low level of double-strand breaks that occurs normally during recombination. In addition, labeling was not found in male meiocytes (n > 150) of mmd1 plants before early diakinesis (Figures 4E and 4I).
Figure 4.
Chromosomes Undergo DNA Fragmentation in mmd1 Microsporocytes.
TUNEL analysis was performed on wild-type and mmd1 microsporocytes. Nicked DNA was detected by incorporating fluorescein isothiocyanate–labeled dUTP (green) by 3′ end labeling. Chromosomes were stained with propidium iodide (red). Composite images of the red and green channels are shown for wild-type cells at diakinesis (A), prometaphase I (B), metaphase I (C), and anaphase I (D), with insets at top right showing only the green channel. Individual images of the red ([E] to [H]) and green ([I] to [L]) channels are shown for microsporocytes from mmd1 plants at early diakinesis ([E] and [I]), late diakinesis ([F] and [J]), metaphase I ([G] and [K]), and anaphase I ([H] and [L]). Chromosomes in mmd1 cells labeled TUNEL positive from diakinesis onward ([J] to [L]). Individual images of the red ([M] to [P]) and green ([Q] to [T]) channels are shown for arrested microsporocytes from mmd1 plants at different developmental stages. Bars = 10 μm.
By contrast, TUNEL labeling of DNA was first detected at approximately mid to late diakinesis in mmd1 male meiocytes (Figure 4J), which is consistent with when we first observed cytoplasmic shrinkage in mmd1 meiocytes. Strong labeling was observed in cells at metaphase I and anaphase I and in cells arrested at diakinesis and the dyad stage (Figures 4K, 4L, 4Q, and 4R). Labeling was observed typically throughout the chromosomes in these cells. Weak labeling also was detected in degenerating meiotic cells from anthers of stage-10 flowers (Figures 4S and 4T). The labeling patterns of these cells typically appeared as foci concentrated around the periphery of the nucleus. The foci also stained positive with DAPI and propidium iodide, confirming that they contained DNA. This finding is reminiscent of the apoptotic bodies—membrane-bound bodies that contain fragmented DNA—that are observed in animal cells undergoing apoptosis (Bursch et al., 1990). The TUNEL-positive results, coupled with the cytoplasmic shrinkage and the lack of an organelle band in degenerating mmd1 male meiocytes, are consistent with the hypothesis that they are apoptotic. However, we cannot exclude the possibility that TUNEL labeling is the result of cell death unrelated to a specific apoptosis-related pathway.
The mmd1 Mutation Was Caused by a Ds Insertion
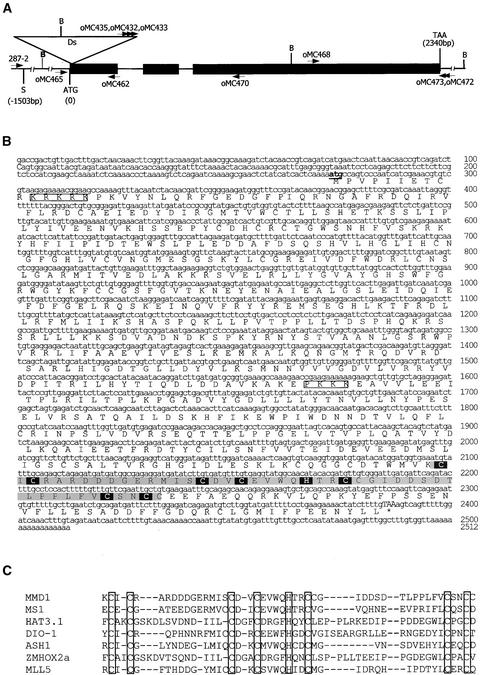
Genetic and DNA gel blot analyses indicated that the mmd1 mutant contained a single Ds insertion that was linked with the sterility phenotype (data not shown). For convenience, this Ds element is designated Ds-mmd1. A series of thermal asymmetric interlaced PCRs (Liu et al., 1995) was used to isolate the Ds-mmd1 insertion site. These experiments, along with DNA gel blot analyses, indicated that Ds-mmd1 has an unusual rearranged structure (our unpublished data). Searches of the Arabidopsis database with genomic sequences flanking Ds-mmd1 showed that the insertion was in a predicted gene, At1g66170 (Figure 5A). DNA gel blot hybridization with a wild-type probe confirmed that this gene is disrupted by the Ds insertion (data not shown).
Figure 5.
MMD1 Is Predicted to Be Nuclear Localized and to Contain a PHD Domain.
(A) MMD1 gene structure and the Ds insertion site. Closed rectangles represent exons. The Ds insertion site is indicated by the open inverted triangle. BamHI sites in the region are shown (B). The positions of primers used in RT-PCR for expression studies and cDNA cloning are shown above and below the map.
(B) MMD1 cDNA and predicted amino acid sequences. Putative nuclear localization signals are boxed. The PHD domain is highlighted in gray. Amino acids corresponding to the Cys4-His-Cys3 consensus motif of PHD domains are highlighted in black.
(C) Alignment of the PHD domain in MMD1 with PHD domains from several transcription factors. The sequences are derived from the following sources. MS1, an Arabidopsis PHD-finger protein involved in anther and pollen development (Wilson et al., 2001); HAT3.1, an Arabidopsis homeobox protein (Schindler et al., 1993); DIO-1, death-associated transcription factor (Garcia-Domingo et al., 1999); ASH1, a putative chromosome-remodeling factor (Nakamura et al., 2000); ZMHOX2a, a maize homeobox gene (Klinge et al., 1996); MLL5, human MIXED-LINEAGE LEUKEMIA5 protein (Emerling et al., 2002). Conserved PHD consensus sequences are shown as open boxes.
To determine whether Ds-mmd1 caused the meiotic defects observed in mmd1 plants, further linkage analysis was performed. PCR with Ds and plant-specific primers indicated that all 63 sterile plants analyzed were homozygous for the Ds-mmd1 insertion, demonstrating a linkage of <1 centimorgan between the sterility defect and the Ds insertion. Furthermore, revertant analysis was conducted to ascertain if the Ds-mmd1 insertion caused the sterility defect observed in mmd1 plants by searching for fertile flowers and elongated siliques on mutant plants carrying an active Activator (Ac). Of the 58 mmd1 mutant plants examined, 26 produced one or more elongated siliques, whereas mutant plants lacking Ac produced none. Flowers of one large revertant sector produced significant amounts of pollen, demonstrating that the fertility was caused by reversion and not by cross-pollination. The progeny of six independent revertant sectors were planted and segregated for both wild-type and mutant plants. DNA sequence analysis of PCR fragments flanking the Ds insertion site in plants from 10 independent reversion events identified only the wild-type sequence, suggesting that restoration of the wild-type MMD1 sequence might be required for functional reversion.
Complementation experiments also were performed. A 4.8-kb wild-type genomic DNA fragment containing MMD1 and 1.5 and 1.0 kb of upstream and downstream DNA, respectively, was introduced into a segregating population of mmd1 plants. Thirty-six transgenic plants were obtained and analyzed for the presence of the complementation clone and Ds-mmd1. PCR using Ds and MMD1 primers showed that of 36 transgenic plants analyzed, 9 were homozygous for Ds-mmd1, 10 were heterozygous, and the remaining 17 were wild type. All of the nine mmd1 homozygous lines were confirmed to contain the complementation clone. Of these nine lines, two lines were completely fertile, three lines were semifertile, and four lines were sterile (data not shown). Therefore, male sterility can be restored by the 4.8-kb fragment containing a wild-type copy of MMD1. At this time, it is not clear why several of the lines showed no or partial restoration of fertility; however, it is possible that the transgene may not be expressed properly in these lines.
MMD1 Encodes a Novel PHD-Containing Nuclear Protein
Sequence analysis of a cDNA clone isolated using reverse transcriptase–mediated (RT) PCR confirmed the predicted gene structure of MMD1 and indicated that the transcript encodes an 80.8-kD protein (Figure 5B) that contains a C-terminal PHD domain (Figure 5C). Basic Local Alignment Search Tool (BLAST) searches identified limited similarity between MMD1 and a number of proteins that contain PHD domains, including MALE STERILITY1 (MS1), a protein that was shown recently to be required for pollen development in Arabidopsis (Wilson et al., 2001), and two predicted gene products (At2g01810 and At1g33420) in the Arabidopsis genome. Similarity between MMD1, MS1, and the two predicted proteins spans the entire length of the proteins and ranges from 26 to 35% overall identity. Although the greatest similarity is found in the C-terminal PHD domain, two other highly conserved domains (>50% over ∼30 to 40 amino acids) are found in the middle of the proteins. After the three PHD domain–containing Arabidopsis proteins, MMD1 exhibited the greatest similarity (50% over 88 amino acids) with the PHD domain of the human MIXED-LINEAGE LEUKEMIA5 protein, which is a homolog of Drosophila TRITHORAX (Emerling et al., 2002). However, with the exception of the presence of a PHD domain, MMD1 does not contain any other motifs or similarities to provide insight into its function.
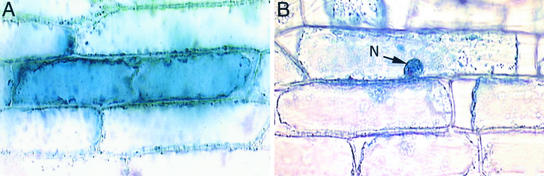
The predicted protein also contains two potential nuclear localization signals (amino acids 11 to 15 and 432 to 435), suggesting that the protein is targeted to the nucleus. To investigate this possibility, a translational fusion of MMD1 to β-gluc-uronidase (GUS) was generated for transient expression in plant cells driven by the 35S promoter. Introduction of the MMD1-GUS fusion into onion epidermal cells by particle bombardment resulted in GUS staining in the nucleus (Figure 6). By contrast, GUS was found throughout the cell in control bombardments with the vector alone (Figure 6). Nuclear targeting of MMD1 is consistent with the observations that it is essential for meiosis and that it contains a PHD domain, which often is found in proteins that participate in chromatin remodeling.
Figure 6.
MMD1 Is Targeted to the Nucleus.
Onion epidermal cells were bombarded with pBK16-MMD1-GUS and a control construct, pBK16-35S-GUS.
(A) The GUS protein is localized throughout the cytoplasm of the cell in a control bombardment.
(B) The MMD1-GUS fusion protein is detected only in the nucleus (N).
The MMD1 Transcript Accumulates Preferentially in Male Meiocytes
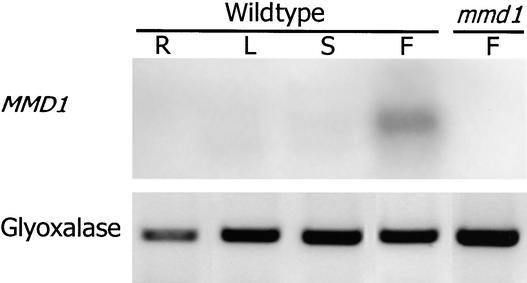
To obtain additional clues about the mechanism of MMD1 function, we analyzed its expression pattern. MMD1 transcripts were not detectable on RNA gel blots of poly(A+) RNA (1.5 μg) isolated from various Arabidopsis organs, including flower buds (data not shown), indicating that MMD1 is expressed at low levels. Using RT-PCR, MMD1 transcripts were detected only in RNA samples from young buds (24 cycles of PCR; Figure 7). However, 45 cycles of amplification also resulted in the detection of MMD1 transcripts in RNA samples from roots, leaves, and seedlings (data not shown). Therefore, MMD1 transcript levels were highest in floral buds. Amplification was not obtained with bud RNA isolated from mmd1 plants (Figure 7), indicating that the Ds element completely inactivated MMD1 expression.
Figure 7.
MMD1 Transcript Levels Are Highest in Flower Buds.
MMD1 gene expression was examined by RT-PCR analysis of total RNA from different plant organs: R, roots; L, leaves; S, stems; F, flower buds. Transcripts for the ubiquitously expressed GLYOXALASE2-2 gene (Maiti et al., 1997) were used as controls in RT-PCR.
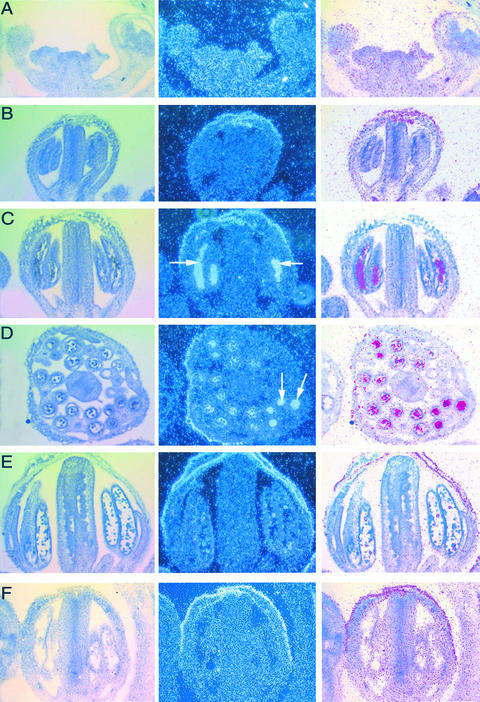
The temporal and spatial expression patterns of MMD1 in flowers were determined by RNA in situ hybridization experiments. Our results show that MMD1 transcripts were below detection levels in reproductive meristems, early floral primordia, and floral buds before the formation of meiocytes (Figures 8A and 8B). Approximately at the time of male meiosis, MMD1 transcripts clearly were present inside anther locules in areas corresponding to meiocytes but not in other anther tissues or other organs (Figures 8C and 8D) or in postmeiotic anthers (Figure 8E). Binding of both the antisense and sense (negative control) probes to the sepals also was observed. This nonspecific sticking of RNA probes to sepals has been reported previously (Yanofsky et al., 1990; Chen et al., 2002). These observations indicate that MMD1 transcripts are highest in male meiocytes at approximately the time of meiosis.
Figure 8.
MMD1 Transcript Levels Are Highest in Male Meiocytes.
RNA in situ hybridization in wild-type inflorescences. The left column shows bright-field images of sections, the center column shows dark-field images of the emulsion grains, and the right column shows composite images from the left and center columns rendered by Photoshop processing (Adobe Systems, Mountain View, CA).
(A) Section of a wild-type inflorescence apex with the inflorescence meristem and early floral primordia.
(B) A young flower at approximately stage 8, before meiosis.
(C) A stage-9 flower containing microsporocytes in the anther. Dense grains (arrows) are observed in the locules, indicating accumulation of the MMD1 transcript.
(D) Cross-section of a stage-9 flower showing MMD1 transcript accumulation in the anther locules (arrows).
(E) A flower at approximately stage 10, with no detectable MMD1 transcripts.
(F) A sense probe control at stage 9, showing no signal in the anther locules.
DISCUSSION
The MMD1 Gene Is Required for Normal Male Meiosis
The mmd1 mutant is male sterile but female fertile. The mutation has its greatest effect on meiosis. Male meiosis appeared to proceed normally in mmd1 plants through pachytene. No alterations were observed in >1300 cells observed in leptotene, zygotene, and pachytene. In particular, sister chromosome cohesion and the synapsis of homologous chromosomes appeared normal in meiotic chromosome spreads of mmd1 meiocytes (Figure 2). Consistent with our conclusion that mmd1 meiocytes proceed normally through pachytene was our observation that the distribution of SYN1, the meiotic cohesin protein (Bai et al., 1999), was normal on mmd1 meiotic chromosomes up to diakinesis, when chromosomes began to exhibit signs of fragmentation (data not shown). At approximately diakinesis, male meiocytes began to exhibit a general breakdown in meiosis and signs of cell death, including cytoplasmic shrinkage and chromatin fragmentation. Numerous meiotic defects were observed in mmd1 male meiocytes beginning at diakinesis, including alterations in chromosome condensation and difficulties in resolving nonhomologous chromosomes, which resulted in missegregation of chromosomes, chromosome bridges, and fragmentation of chromosomes at anaphase I. In addition, mmd1 meiocytes lacked the distinctive organelle band found in normal meiocytes from telophase I to telophase II. Male meiocytes in mmd1 plants arrested at various stages of meiosis I and II, and none of them underwent cytokinesis. All of our analyses indicate that alterations associated with the mmd1 mutation are first manifest at diakinesis. However, we cannot exclude the possibility that minor alterations in an earlier stage of meiosis may be present in mmd1 meiocytes.
To our knowledge, mmd1 is the only example of a meiotic mutation in Arabidopsis that exhibits apoptosis-like phenotypes before cytokinesis. However, several maize male-sterile mutants have been isolated that show abnormal male meiocyte morphology and cell degeneration. For example, in ms8 mutants, meiotic defects are observed as early as leptotene; a few pollen mother cells complete meiosis, but microspores degenerate soon afterward (Albertsen and Phillips, 1981). In ms22 mutants, male meiocytes begin to degenerate before pachytene, and cells generally are not observed beyond meiosis I (West and Albertsen, 1985). The ms23 mutation also causes cellular arrest beginning in meiosis I, with some cells progressing into meiosis II; cells with cytoplasmic degeneration, similar to mmd1, also were observed (West and Albertsen, 1985). Further analyses are needed to better understand the phenotypes of these maize mutants and to determine if any of the mutations occur in genes related to MMD1.
In contrast to the meiotic defects observed during male meiosis, no alterations were detected during female meiosis, and female fertility appeared normal in mmd1 plants. In addition, no alterations were observed in the epidermis, endothecium, or middle layer of the anther. These observations, along with the fact that MMD1 transcripts were detected mainly in male meiocytes, suggest that MMD1 is essential only for male meiosis. However, very low levels of MMD1 transcript were detected in other tissues, raising the possibility that it may play an additional minor role outside of male meiosis. Consistent with this possibility is our observation that 30% of mmd1 flowers contained abnormal numbers of stamens and that filaments in mmd1 flowers were somewhat shorter than those in wild-type flowers. This finding suggests that mmd1 also could play a minor role in flower development.
Does the Absence of MMD1 Trigger Meiotic Cell Cycle Control and Associated Apoptosis?
The apoptosis-like phenotypes of mmd1 meiocytes raise the possibility that the mutation may activate a meiotic cell cycle checkpoint and trigger a cell death pathway. This is rare for a plant meiotic mutant. There are many examples of meiotic mutations that activate cell cycle control mechanisms in other organisms. Numerous meiotic mutations that block chromosome synapsis and/or recombination induce pachytene arrest in yeast (Roeder, 1997; Roeder and Bailis, 2000). The pachytene checkpoint also has been shown to operate in several other organisms, including Drosophila (Ghabrial and Schupbach, 1999), C. elegans (Gartner et al., 2000), and mouse (Edelmann et al., 1996, 1999; Pittman et al., 1998). However, mutations in genes that trigger pachytene arrest in yeast, worms, and mice do not appear to activate a typical meiotic checkpoint in plants. This finding suggests that plants do not use the same meiotic checkpoint control mechanisms. Several Arabidopsis mutants have been identified that appear to affect meiotic cell cycle progression (Yang et al., 1999a; Siddiqi et al., 2000; Magnard et al., 2001; Azumi et al., 2002); however, most studies of meiotic mutants have not compared the distribution of meiocytes at various stages of meiosis. Therefore, it is possible that alterations in chromosome synapsis and/or recombination may trigger a prophase I checkpoint in plants but that after a brief arrest the meiocytes proceed through prophase I to ultimately produce polyads. The observation that most plant meiotic mutants do not trigger meiotic arrest led us to speculate that the cell death associated with mmd1 meiocytes is not the result of a typical meiotic cell cycle control response, as observed in worms and mice (reviewed by Cohen and Pollard, 2001).
It is possible that the mmd1 mutation may trigger a checkpoint control mechanism other than the classic meiotic checkpoint. For example, mutations in mice that cause telomere dysfunction trigger a germ cell surveillance system that results in developmentally regulated germ cell apoptosis (Hemann et al., 2001). There also is a strong possibility that the mmd1 mutation does not trigger a checkpoint control mechanism of any kind. Rather, MMD1 may be responsible for repressing a male meiocyte cell death pathway. Likewise, the mutation may disrupt chromatin structure, cellular transcription patterns, and/or protein degradation in male meiocytes in a way that directly or indirectly triggers cell death.
What Is the Role of MMD1?
Insight into potential roles of MMD1 comes from its expression patterns, predicted protein sequence, and the phenotype of mmd1 plants. MMD1 is expressed mainly in male meiocytes. Consistent with this fact, mmd1 plants exhibit a general breakdown of male meiosis beginning at diakinesis that results in cell death before cytokinesis. Finally, a transiently expressed MMD1-GUS fusion protein is localized to the nucleus. These observations, coupled with the presence of a PHD domain in MMD1, raise the possibility that it may participate in chromatin-remodeling events associated with either chromosome condensation or possibly meiotic gene expression. PHD domains often are found in transcriptional regulatory proteins and proteins associated with chromatin-remodeling complexes. They have been proposed to mediate protein–protein, protein–DNA, and protein–RNA interactions (Aasland et al., 1995). In animals, PHD domains are found in a number of transcriptional regulators, including members of the Polycomb and Trithorax groups of proteins (reviewed by Kennison, 1995). They also have been identified as components of the RNA polymerase II TFIID complex (Gangloff et al., 2001) and chromatin-remodeling complexes (Bochar et al., 2000; Halback et al., 2000; Loewith et al., 2000; Schultz et al., 2001). In plants, PHD domains have been identified in a regulator of defense genes (Korfhage et al., 1994), a putative transcriptional regulatory protein required for proper development and fertility (Mussig et al., 2000), and a chromatin-remodeling protein that regulates the transition from embryonic to vegetative development (Ogas et al., 1999).
Recently, PHD domains were found to be associated with E3 ubiquitin ligase activity. Several PHD-containing viral proteins that are targeted to cellular membranes and the cytosol have been shown to function as E3 ubiquitin ligases (Cosoy and Ganem, 2003). In addition, MEKK1, a cellular mitogen-activated protein kinase kinase kinase that phosphorylates several different mitogen-activated kinases contains a PHD domain and has E3 activity (Lu et al., 2002). To date, all PHD domain–containing proteins that exhibit E3 ligase activity are membrane or cytosol localized and contain a conserved Trp residue between Cys-6 and Cys-7 of the PHD domain. MMD1 lacks the conserved Trp and is localized to the nucleus; therefore, it more closely resembles proteins that are associated with transcriptional regulation and/or chromatin-remodeling events.
To date, PHD domain–containing proteins have not been linked with chromatin-remodeling events associated with chromosome condensation during either mitosis or meiosis. However, it should be noted that although it is clear that topoisomerase II and the condensin complex are required for chromosome condensation (Hirano, 2000; Cuvier and Hirano, 2003), very little is known about how the extended chromatin fibers ultimately are folded into condensed chromosomes during mitosis or meiosis in any organism. If MMD1 is required for chromosome-remodeling events during meiotic chromosome condensation, this would be the first example of a PHD-containing protein that functions in this manner.
As discussed above, Arabidopsis contains four MMD1-related genes, including MS1, which is required for microspore development (Wilson et al., 2001). In ms1 plants, microsporogenesis apparently is normal until microspore release from tetrads, when the microspores and tapetal cells exhibit signs of cytoplasmic degeneration followed by cell death. TUNEL experiments were not conducted with the ms1 mutant; therefore, it is not clear whether chromosome fragmentation occurs in ms1 microspores or tapetal cells. Like mmd1, the ms1 mutation does not appear to affect female fertility. The presence of two additional MMD1/MS1-related genes in the Arabidopsis genome raises the possibility that they represent a family of proteins that are required to control gene expression during microsporogenesis and microgametogenesis. Alternatively, these proteins may play a role in repressing cell death in specific cell types. Currently, we are characterizing mmd1 plants in more detail and examining plants with mutations in other MMD1/MS1-like genes to investigate these possibilities.
METHODS
Generation of Dissociation Insertional Lines, Isolation of the mmd1 Mutant, and Reversion Analysis
Plants used in this study were of the Arabidopsis thaliana ecotype Landsberg erecta. They were grown in the greenhouse or growth chambers with a 16-h-light/8-h-dark cycle at 22 to 24°C. Seeds were plated onto Murashige and Skoog (1962) agar medium with or without appropriate antibiotics under continuous light at 22°C.
Arabidopsis Dissociation insertional lines were generated according to Sundaresan et al. (1995). Approximately 2000 F1 seeds from crosses between Ds- and Activator (Ac)-carrying lines were planted and allowed to self-pollinate. F2 seedlings from individual families were screened on agar plates containing 0.4 mg/L naphthalene acetamide and 50 mg/L kanamycin. Double-resistant seedlings were transferred to soil, grown to maturity, and allowed to self-pollinate. F3 plants of one line, Y9287, segregated for sterile plants and were investigated further.
Putative revertant sectors were identified as fertile siliques on otherwise sterile F2 plants carrying both the mmd1 mutation and a recombinant Ac element, Ac1 (Sundaresan et al., 1995). Their seeds were harvested separately for further analysis. Genomic DNA from individual revertant progeny was isolated, and the sequence spanning the Ds insertion site was determined from PCR fragments with gene-specific primers (oMC465 and oMC462) that flanked the Ds insertion site.
PCR was conducted on DNA isolated from 63 sterile plants segregating from a cross between mmd1 and wild-type Columbia plants to test linkage between the Ds and male sterility. Two gene-specific primers flanking the Ds insertion site, oMC462 and oMC465, and a Ds-specific primer, oMC432, were used in separate PCR procedures to determine if the plants contained the mutant mmd1 or the wild-type MMD1 allele.
Phenotypic Characterization of Meiocytes
The chromosome spreads of male meiocytes were prepared essentially according to Ross et al. (1996) and stained with 10 μL of 4′,6-diamidino-2-phenylindole (1 μg/mL). Female meiosis was analyzed essentially as described (Armstrong and Jones, 2001) using floral buds at stages 10 to 11 (Smyth et al., 1990). Sections (7 μm) of wild-type and mmd1 inflorescences were prepared, stained with 1% toluidine blue, and visualized essentially as described (Peirson et al., 1996). Fragmentation of chromosomes in mmd1 male meiocytes was investigated using the TdT-mediated dUTP nick end-labeling apoptosis detection system (Promega) according to the manufacturer's instructions essentially as described (Balk and Leaver, 2001). DNA was counterstained with propidium iodide (Peirson et al., 1997). Fluorescent signals were viewed using a PMC200 confocal microscopy system (Nikon, Melville, NY).
Isolation and Characterization of MMD1 Genomic and cDNA Clones
Genomic DNA was isolated from wild-type and individual heterozygous and homozygous mmd1 plants using the cetyl-trimethyl-ammonium bromide method (Stewart and Via, 1993). Thermal asymmetric interlaced PCR (Liu et al., 1995) was used to amplify the genomic DNA adjacent to the Ds element using the degenerate primer AD4 (Liu et al., 1995) and the nested Ds-specific primers oMC435, oMC432, and oMC433 near the 3′ end of the Ds-mmd1 insertion.
The MMD1 cDNA was amplified in successive rounds of PCR using the primer pairs oMC465/oMC473 and oMC465/oMC472 on first-strand cDNA generated by reverse transcription with an oligo(dT) adaptor primer (oCM162) on poly(A+) RNAs isolated from wild-type buds (Figure 5A). The 3′ end of the mmd1 cDNA was amplified using primer oMC468 and the adaptor primer (oCM176). Amplification products were cloned and sequenced. The Arabidopsis BAC clone F15E21 was obtained from the Arabidopsis Stock Center (Columbus, OH). Primers used in this study are described in Table 1.
Table 1.
Primers Used in This Study
| Primer | Sequence | Application |
|---|---|---|
| Ds3-1 | 5′-CGATTACCGTATTTATCCCGTTCG-3′ | Amplifying Ds junction in mmd1 plants with Ds5-1 |
| Ds5-1 | 5′-CCGTTTACCGTTTTGTATATCCCG-3′ | Amplifying Ds junction in mmd1 plants with Ds3-1 |
| oMC435 | 5′-CCCTTCAGTGAACGTTATTAGTTC-3′ | 3′ end thermal asymmetric interlaced PCR primer for amplifying genomic DNA flanking the Ds |
| oMC432 | 5′-GTGTCGTAGATACTAGCCCCT-3′ | Nested primer for oMC435 |
| oMC433 | 5′-GCGCTCTATCATAGATGTCGC-3′ | Nested primer for oMC435 and oMC432 |
| oMC465 | 5′-CGTCTCCATCGAAGCTAAAA-3′ | Forward primer for cDNA synthesis and gene expression |
| oMC462 | 5′-ACAAACCTGTGCAACGACAG-3′ | Revertant analysis and transgenic plant confirmation |
| oMC470 | 5′-CTCTGAAAGTCTTCAAGTGTC-3′ | Reverse primer for gene expression by RT-PCR |
| oMC473 | 5′-GACTTTACAAAAGATAGTTTTCTTCAG-3′ | Reverse primer for cDNA isolation |
| oMC472 | 5′-GGCAAACAAATCACATATACAATTTGG-3′ | Nested primer for oCM473 |
| oMC468 | 5′-CCATTGAAACCTGGAGCTGAC-3′ | Transgenic plant analysis in complementation experiment |
| oCM017 | 5′-GATAATGGTGACAAGCTGAC-3′ | Forward primer for transcript detection of GLYOXALASE2-2 by RT-PCR |
| oCM162 | 5′-CGAGGATCCTCGAGTCGACGCT(dT)17-3′ | Adaptor-oligo(dT) primer for cDNA synthesis |
| oCM176 | 5′-CGAGGATCCTCGAGTCGACGCT-3′ | Adaptor primer for cDNA cloning |
| oCM267 | 5′-ATCAATTGGCGACTTGCAACCG-3′ | Reverse primer for transcript detection of GLYOXALASE2-2 by RT-PCR |
| oCM441 | 5′-ATCGCAATGATGGCATTTG-3′ | Left border primer of plant transformation vector pCAMBIA1390 |
| 287-2 | 5′-GCCGTGGATCGATTGATACGA-3′ | Transgenic plant analysis in complementation experiment |
| 287-10 | 5′-TGCTCTAGAGAACTCTATCATCACTCA-3′ | Forward primer for translational construct of MMD1 with GUS |
| 287-11 | 5′-CCGCTCGAGTCCAAAAGATAGTTTTCTTCAGG-3′ | Reverse primer for translational construct of MMD1 with GUS |
| AD4 | 5′-NGTA(G/C)A(G/C)(A/T)GTNA(A/T)CAA-3′ | Degenerate primer for thermal asymmetric interlaced PCR |
The Ds insertion site was analyzed by DNA gel blot hybridization with α-32P-dCTP–labeled probes for either the Ds element (1.8-kb EcoRI fragment) or the MMD1 genomic locus (2.4-kb PCR-derived genomic clone). Hybridized membranes were exposed to x-ray film.
A 4.8-kb wild-type genomic DNA fragment containing MMD1 and 1.5 and 1.0 kb of upstream and downstream DNAs, respectively, was cloned into the plant transformation vector pCAMBIA1390 (Hajdukiewicz et al., 1994). The resulting plasmid, pCAM-MMD4.8, was introduced into the Agrobacterium tumefaciens line GV3101 and transformed into a segregating population of wild-type and mmd1 plants using the floral-dip method (Clough and Bent, 1998). Seeds were harvested and germinated on Murashige and Skoog (1962) medium containing either 35 mg/L hygromycin only or both hygromycin and 50 mg/L kanamycin. Thirty-six independent transgenic plants were obtained. PCR with two Ds-specific primers (Ds3-1 and Ds5-1) was used to detect the presence of the Ds, whereas PCR with the gene-specific primers 287-2 and oMC462 flanking the Ds was used to determine if the plants were heterozygous or homozygous for the Ds. The presence of the MMD1 transgene was verified using PCR with a vector primer (oMC441) and oMC468.
Expression Studies and Protein Localization
MMD1 expression was analyzed using the One-Step reverse transcriptase–mediated PCR kit (Promega). Equal amounts of poly(A+) RNA (150 ng) from unopened buds of mmd1 plants and from roots, leaves, stems, and buds of wild-type plants were subjected to reverse transcriptase–mediated PCR with the gene-specific primers oMC465 and oMC470. Control experiments were conducted with a primer pair (oCM017 and oCM267) that amplified a similar-sized fragment of GLYOXALASE2-2, a gene that is expressed ubiquitously in plants (Maiti et al., 1997). Amplification products were examined by DNA gel blot analysis, and radioactivity was detected using a Molecular Dynamics PhosphorImager (Sunnyvale, CA).
In situ RNA hybridization experiments were performed as described previously (Drews et al., 1991). Sense and antisense RNA probes were synthesized using full-length MMD1 cDNA cloned into pGEM-T vector (pGEMT-MMD1). Sense RNA probe was transcribed by T7 RNA polymerase incorporating 35S-UTP with NdeI-linearized pGEMT-MMD1 as a template. An antisense RNA probe was made with SP6 RNA polymerase using NcoI-linearized pGEMT-MMD1.
Nuclear localization of the MMD1 protein was tested by transient expression of the MMD1–β-glucuronidase (GUS) fusion protein. PCR using primers 287-10 and 287-11, which are located immediately upstream of the start codon and downstream of the stop codon, respectively, were used to amplify the full-length cDNA. The MMD1 cDNA was cloned downstream of the 35S promotor of Cauliflower mosaic virus and in frame with the GUS gene in the transient expression vector pBK16. The pBK16-MMD1-GUS plasmid was delivered into onion epidermal cells using a Biolistic PDS-1000/He gene gun system (Bio-Rad). The bombarded samples were kept overnight at 24°C and stained for GUS expression with 2 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid at 37°C until blue spots appeared.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The accession number for the MMD1 cDNA is AY158082.
Acknowledgments
We thank Y. Hu for generating F2 families for the screening of Ds insertional lines and for performing the RNA in situ hybridization experiment, A. Richardson for help with mutant screening, A. Omeis for plant care, W. Li for assistance in phenotypic analysis, and B. Bliss, W. Li, and D. Zhao for comments and critical reading of the manuscript. The Miami University Electron Microscope Facility provided resources for this work. This work was supported by grants from the National Science Foundation (MCB-9896340) and the National Institutes of Health (R01 GM63871-01) to H.M., by funds from the Department of Biology and the Huck Institute of Life Sciences at Pennsylvania State University, and by Grant R15 GM55956-02 from the National Institutes of Health to C.A.M.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010447.
References
- Aasland, R., Gibson, T., and Stewart, F. (1995). The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20, 50–59. [DOI] [PubMed] [Google Scholar]
- Albertsen, M.C., and Phillips, R.L. (1981). Developmental cytology of 13 genetic male sterile loci in maize. Can. J. Genet. Cytol. 23, 195–208. [Google Scholar]
- Armstrong, S.J., and Jones, G.H. (2001). Female meiosis in wild-type Arabidopsis thaliana and in two meiotic mutants. Sex. Plant Reprod. 13, 177–183. [Google Scholar]
- Ashley, T., and Plug, A. (1998). Caught in the act: Deducing meiotic function from protein immunolocalization. In Meiosis and Gametogenesis, M.A. Handel, ed (San Diego, CA: Academic Press), pp. 201–239. [DOI] [PubMed]
- Azumi, Y., Liu, D., Zhao, D., Li, W., Wang, G., Hu, Y., and Ma, H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21, 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X., Peirsion, B., Dong, F., Cai, X., and Makaroff, C. (1999). Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S.M., Bronner, C.E., Zhang, L., Plug, A.W., Robatzek, M., Warren, G., Elliott, E.A., Yu, J.A., Ashley, T., Arnheim, N., Flavell, R.A., and Liskay, R.M. (1995). Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 82, 309–319. [DOI] [PubMed] [Google Scholar]
- Balk, J., and Leaver, C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Canales, C., and Dickinson, H.G. (2001). Plant meiosis: The means to 1N. Trends Plant Sci. 6, 114–121. [DOI] [PubMed] [Google Scholar]
- Bhatt, A.M., Lister, C., Page, T., Fransz, P., Findlay, K., Jones, G.H., Dickinson, H.G., and Dean, C. (1999). The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 19, 463–472. [DOI] [PubMed] [Google Scholar]
- Bochar, D., Savard, J., Wang, W., Lafleur, D., Moore, P., Côté, J., and Shiekhattar, R. (2000). A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl. Acad. Sci. USA 97, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch, W., Paffe, S., Putz, B., Barthel, G., and Schultehermann, R. (1990). Determination of the length of the histological stages of apoptosis in normal liver and in altered hepatic foci of rats. Carcinogenesis 11, 847–853. [DOI] [PubMed] [Google Scholar]
- Canales, C., Bhatt, A., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12, 1718–1727. [DOI] [PubMed] [Google Scholar]
- Caryl, A.P., Armstrong, S.J., Jones, G.H., and Franklin, F.C.H. (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109, 62–71. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Lavithis, M., Taylor, P.E., Craig, S., Singh, M.B., Signer, E.R., Knox, R.B., and Dennis, E.S. (1994). Genetic control of male fertility in Arabidopsis thaliana: Structural analysis of premeiotic developmental mutants. Sex. Plant Reprod. 7, 17–28. [DOI] [PubMed] [Google Scholar]
- Chen, C., Marcus, A., Li, W., Hu, Y., Vielle Calzada, J.-P., Grossniklaus, U., Cyr, R., and Ma, H. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129, 2401–2409. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, G., Sun, X., Fearnhead, H., Macfarlane, M., Brown, D., Snowden, R., and Dinsdale, D. (1994). Formation of large molecular-weight fragments of DNA is a key committed step of apoptosis in thymocytes. J. Immunol. 15, 507–516. [PubMed] [Google Scholar]
- Cohen, P., and Pollard, J. (2001). Regulation of meiotic recombination and prophase I progression in mammals. Bioessays 23, 996–1009. [DOI] [PubMed] [Google Scholar]
- Cosoy, L., and Ganem, D. (2003). PHD domains and E3 ubiquitin ligases: Viruses make the connection. Trends Cell Biol. 13, 7–12. [DOI] [PubMed] [Google Scholar]
- Couteau, F., Belzile, F., Horlow, C., Grandjean, O., Vezon, D., and Doutriaux, M.P. (1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11, 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, W.J., and Zolan, M.E. (1998). Functions of DNA repair genes during meiosis. In Meiosis and Gametogenesis, M.A. Handel, ed (San Diego, CA: Academic Press), pp. 117–140. [DOI] [PubMed]
- Cuvier, O., and Hirano, T. (2003). A role of topoisomerase II in linking DNA replication and chromosome condensation. J. Cell Biol. 160, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K. (1998). Meiotic chromosome organization and segregation in plants. Annu. Rev. Plant Physiol. 49, 371–395. [DOI] [PubMed] [Google Scholar]
- Doutriaux, M.P., Couteau, F., Bergounioux, C., and White, C. (1998). Isolation and characterization of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257, 283–291. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002. [DOI] [PubMed] [Google Scholar]
- Edelmann, W., Cohen, P., Kneitz, B., Winand, N., Lia, M., Heyer, J., Kolodner, R., Pollard, J., and Kucherlapati, R. (1999). Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21, 123–127. [DOI] [PubMed] [Google Scholar]
- Edelmann, W., et al. (1996). Meiotic pachytene arrest in MLH1-deficient mice. Cell 85, 1125–1134. [DOI] [PubMed] [Google Scholar]
- Ellis, R.E., Yuan, J., and Horvitz, H.R. (1991). Mechanisms and functions of cell death. Annu. Rev. Cell Biol. 7, 663–698. [DOI] [PubMed] [Google Scholar]
- Emerling, B., Bonifas, J., Kratz, C., Donovan, S., Taylor, B., Green, E., Le Beau, M., and Shannon, K. (2002). MLL5, a homolog of Drosophila Trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene 21, 4849–4854. [DOI] [PubMed] [Google Scholar]
- Franklin, A.E., McElver, J., Sunjevaric, I., Rothstein, R., Brown, B., and Cande, W.Z. (1999). Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11, 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff, Y., Pointud, J., Thuault, S., Carre, L., Romier, C., Muratoglu, S., Brand, M., Tora, L., Couderc, J., and Davidson, I. (2001). The TFIID components human TAF(II)140 and Drosophila BIP2 (TAF(II)155) are novel metazoan homologues of yeast TAF(II)47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21, 5109–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Domingo, D., Leonardo, E., Grandien, A., Martinez, P., Albar, J., Izpisua-Belmonte, J., and Martinez, A. (1999). DIO-1 is a gene involved in onset of apoptosis in vitro, whose misexpression disrupts limb development. Proc. Natl. Acad. Sci. USA 96, 7992–7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner, A., Milstein, S., Ahmed, S., Hodgkin, J., and Hengartner, M. (2000). A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5, 435–443. [DOI] [PubMed] [Google Scholar]
- Ghabrial, A., and Schupbach, T. (1999). Activation of meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1, 354–357. [DOI] [PubMed] [Google Scholar]
- Gillies, C. (1984). The synaptonemal complex in higher plants. CRC Crit. Rev. Plant Sci. 2, 81–116. [Google Scholar]
- Green, D., and Reed, J. (1998). Mitochondria and apoptosis. Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Halback, T., Scheer, N., and Werr, W. (2000). Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res. 28, 3542–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann, M., Rudolph, K., Strong, M., DePinho, R., Chin, L., and Greider, C. (2001). Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell 12, 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T. (2000). Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69, 115–144. [DOI] [PubMed] [Google Scholar]
- Kennison, J. (1995). The Polycomb and Trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu. Rev. Genet. 29, 289–303. [DOI] [PubMed] [Google Scholar]
- Klimyuk, V.I., and Jones, J.D.G. (1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Klinge, B., Uberlacker, B., Korfhage, C., and Werr, W. (1996). ZmHox: A novel class of maize homeobox genes. Plant Mol. Biol. 30, 439–453. [DOI] [PubMed] [Google Scholar]
- Korfhage, U., Trezzini, G., Meier, I., Hahlbrock, K., and Somssich, I. (1994). Plant homeodomain protein involved in transcriptional regulation of a pathogen defense-related gene. Plant Cell 6, 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B., and Amon, A. (2001). Meiosis: How to create a specialized cell cycle. Curr. Opin. Cell Biol. 13, 770–777. [DOI] [PubMed] [Google Scholar]
- Leu, J., and Roeder, G. (1999). The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell 4, 805–814. [DOI] [PubMed] [Google Scholar]
- Lindgren, A., Bungard, D., Pierce, M., Xie, J., Vershon, A., and Winter, E. (2001). The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19, 6489–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Loewith, R., Meijer, M., Lees-Miller, S., Riabowol, K., and Young, D. (2000). Three yeast proteins related to the human candidate tumor suppressor p33ING1 are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20, 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z., Xu, S., Joazeiro, C., Cobb, M., and Hunter, T. (2002). The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9, 945–956. [DOI] [PubMed] [Google Scholar]
- Lydall, D., Nikolsky, Y., Bishop, D.K., and Weinert, T. (1996). A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383, 840–843. [DOI] [PubMed] [Google Scholar]
- Magnard, J., Yang, M., Chen, Y., Leary, M., and McCormick, S. (2001). The Arabidopsis gene tardy asynchronous meiosis is required for the normal pace and synchrony of cell division during male meiosis. Plant Physiol. 127, 1157–1166. [PMC free article] [PubMed] [Google Scholar]
- Maiti, M.K., Krishnasamy, S., Owen, H.A., and Makaroff, C.A. (1997). Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Mol. Biol. 35, 471–481. [DOI] [PubMed] [Google Scholar]
- McCormick, S. (1993). Male gametophyte development. Plant Cell 5, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, R., Vezon, D., Bullier, E., Motamayor, J.C., Sellier, A., Lefevre, F., Pelletier, G., and Horlow, C. (2001). SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15, 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Mussig, C., Kauschmann, A., Clouse, S., and Altmann, T. (2000). The Arabidopsis PHD-finger protein SHL is required for proper development and fertility. Mol. Gen. Genet. 264, 363–370. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Blechman, J., Tada, S., Rozovskaia, T., Itoyama, T., Bullrich, F., Mazo, A., Croce, C., Geiger, B., and Canaani, E. (2000). huASH1 protein, a putative transcription factor encoded by a human homologue of the Drosophila ash1 gene, localizes to both nuclei and cell-cell tight junctions. Proc. Natl. Acad. Sci. USA 13, 7284–7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M., Coe, E., and Wessler, S. (1997). Mutants of Maize. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 263–266.
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T.L. (1999). The ties that bind: Localization of the sister-chromatid cohesin complex on yeast chromosomes. Cell 99, 1–4. [DOI] [PubMed] [Google Scholar]
- Peirson, B.N., Bowling, S.E., and Makaroff, C.A. (1997). A defect in synapsis causes male sterility in a T-DNA-tagged Arabidopsis thaliana mutant. Plant J. 11, 659–669. [DOI] [PubMed] [Google Scholar]
- Peirson, B.N., Owen, H.A., Feldmann, K.A., and Makaroff, C.A. (1996). Characterization of three male-sterile mutants of Arabidopsis thaliana exhibiting alterations in meiosis. Sex. Plant Reprod. 9, 1–16. [Google Scholar]
- Pittman, D.L., Cobb, J., Schimenti, K.J., Wilson, L.A., Cooper, D.M., Brignull, E., Handel, M.A., and Schimenti, J.C. (1998). Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1, 697–705. [DOI] [PubMed] [Google Scholar]
- Roeder, G. (1997). Meiotic chromosomes: It takes two to tangle. Genes Dev. 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roeder, G.S. (1995). Sex and the single cell: Meiosis in yeast. Proc. Natl. Acad. Sci. USA 92, 10450–10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G.S., and Bailis, J.M. (2000). The pachytene checkpoint. Trends Genet. 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Ross, K.J., Fransz, P., and Jones, G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4, 507–516. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.C., Lee, P.Y., Truong, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- SanSegundo, P.A., and Roeder, G.S. (1999). Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97, 313–324. [DOI] [PubMed] [Google Scholar]
- Sato, S., Hotta, Y., and Tabata, S. (1995). Structural analysis of a rec-A like gene in the genome of Arabidopsis thaliana. Chromosome Res. 2, 89–93. [DOI] [PubMed] [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, U., Beckmann, H., and Cashmore, A.R. (1993). HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4, 137–150. [DOI] [PubMed] [Google Scholar]
- Schultz, D., Friedman, J., and Rauscher, F. (2001). Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2 alpha subunit of NuRD. Genes Dev. 15, 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R., Hodge, R., Paul, W., and Draper, J. (1991). The molecular biology of anther differentiation. Plant Sci. 80, 167–191. [Google Scholar]
- Shaw, P., and Moore, C. (1998). Meiosis: Vive la difference! Curr. Opin. Plant Biol. 1, 458–462. [DOI] [PubMed] [Google Scholar]
- Siddiqi, I., Ganesh, G., Grossniklaus, U., and Subbiah, V. (2000). The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127, 197–207. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, C., and Via, L. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14, 748–749. [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene-action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Taylor, P.E., Glover, J.A., Lavithis, M., Craig, S., Singh, M.B., Knox, R.B., Dennis, E.S., and Chaudhury, A.M. (1998). Genetic control of male fertility in Arabidopsis thaliana: Structural analyses of post-meiotic developmental mutants. Planta 205, 492–505. [DOI] [PubMed] [Google Scholar]
- West, D.P., and Albertsen, M.C. (1985). Three new male sterile mutants of maize. Maize Genet. Coop. Newsl. 59, 87. [Google Scholar]
- Wilson, Z., Morroll, S., Dawson, J., Swarup, R., and Tighe, P. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28, 27–39. [DOI] [PubMed] [Google Scholar]
- Yang, M., Hu, Y., Lodhi, M., McCombie, W.R., and Ma, H. (1999. a). The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. USA 96, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.-C., Ye, D., Xu, J., and Sundaresan, V. (1999. b). The sporocyteless gene of Arabidopsis is required for the initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene Agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Zhao, D.-Z., Wang, G.-F., Speal, B., and Ma, H. (2002). The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., and Kleckner, N. (1999). Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754. [DOI] [PubMed] [Google Scholar]