Abstract
While many anaerobic microbial communities are capable of reductively dechlorinating tetrachloroethene (PCE) and trichloroethene (TCE) to dichloroethene (DCE), vinyl chloride (VC), and finally ethene, the accumulation of the highly toxic intermediates, cis-DCE (cDCE) and VC, presents a challenge for bioremediation processes. Members of the genus Dehalococcoides are apparently solely responsible for dechlorination beyond DCE, but isolates of Dehalococcoides each metabolize only a subset of PCE dechlorination intermediates and the interactions among distinct Dehalococcoides strains that result in complete dechlorination are not well understood. Here we apply quantitative PCR to 16S rRNA and reductase gene sequences to discriminate and track Dehalococcoides strains in a TCE enrichment derived from soil taken from the Alameda Naval Air Station (ANAS) using a four-gene plasmid standard. This standard increased experimental accuracy such that 16S rRNA and summed reductase gene copy numbers matched to within 10%. The ANAS culture was found to contain only a single Dehalococcoides 16S rRNA gene sequence, matching that of D. ethenogenes 195, but both the vcrA and tceA reductive dehalogenase genes. Quantities of these two genes in the enrichment summed to the quantity of the Dehalococcoides 16S rRNA gene. Further, between ANAS subcultures enriched on TCE, cDCE, or VC, the relative copy number of the two dehalogenases shifted 14-fold, indicating that the genes are present in two different Dehalococcoides strains. Comparison of cell yields in VC-, cDCE-, and TCE-enriched subcultures suggests that the tceA-containing strain is responsible for nearly all of the TCE and cDCE metabolism in ANAS, whereas the vcrA-containing strain is responsible for all of the VC metabolism.
Bioremediation of tetrachloroethene (PCE) and trichloroethene (TCE) has become an attractive alternative to traditional remediation technologies (6, 21). While many microbial species have been shown to degrade chlorinated solvents both aerobically and anaerobically (7, 10, 24, 33), anaerobic bacteria are especially promising for in situ bioremediation applications since PCE and TCE groundwater plumes are often anaerobic. Of particular interest are members of the genus Dehalococcoides, the only organisms known to reductively dechlorinate PCE and TCE beyond dichloroethene (DCE) to vinyl chloride (VC) and ethene (5, 11, 21, 30). A number of Dehalococcoides isolates have been described with differing substrate specificities. The first identified isolate, Dehalococcoides ethenogenes strain 195, reduces PCE or TCE to cis-DCE (cDCE) and VC in energy-yielding reactions and VC to ethene in a cometabolic reaction (20). Similarly, strain FL2 metabolically dechlorinates TCE and cDCE to VC and then cometabolically to ethene, but it does not dechlorinate PCE (2, 13). Two Dehalococcoides strains that metabolically degrade DCEs and VC to ethene, Dehalococcoides strain BAV1 (11, 12) and Dehalococcoides strain VS (5, 23), have also been described. Strain BAV1, which contains the bvcA reductive dehalogenase gene, degrades PCE or TCE only cometabolically, whereas strain VS, which contains the vcrA reductive dehalogenase gene, is reported to degrade TCE metabolically (3, 11). Most recently, a strain that metabolically degrades TCE to ethene, strain GT, has been isolated and characterized (30). An additional Dehalococcoides strain, CBDB1, has the ability to dechlorinate chlorobenzenes and polychlorinated dibenzodioxins (1, 2).
Recent studies characterizing Dehalococcoides activity suggest that the rapid dechlorination of PCE to ethene observed in laboratory and field microbial communities may be a cooperative effort between multiple Dehalococcoides strains (9, 14, 17, 27, 31). Unfortunately, 16S rRNA gene sequences cannot be used to distinguish specific Dehalococcoides strains in a microbial community because these sequences are highly conserved among many identified Dehalococcoides strains (25). For example, the 16S rRNA gene sequences of two recently isolated strains, GT and KB-1/VC, were shown to be identical to those of two previously identified Dehalococcoides strains, FL2 and CBDB1 (The Ribosomal Database Project [http://rdp.cme.msu.edu/html/]; 9, 30, 32). The former two dechlorinate TCE and VC, respectively, to ethene, while neither of the latter two can efficiently dechlorinate VC. In addition, the 16S rRNA gene sequence of a recently described TCE-dechlorinating strain (KB-1/TCE) was found to be identical to that of the VC-dechlorinating strain BAV1 (9, 11).
Several publications have used quantitative real-time PCR (qPCR) to compare Dehalococcoides 16S rRNA gene copy numbers to total bacteria and/or the sum of reductive dehalogenase copy numbers (27, 29, 30). An initial study examined several technical limitations of comparing 16S rRNA gene copy numbers between genera and of accurately quantifying total numbers of bacterial 16S rRNA genes (29). Subsequently, this comparative qPCR method has been used to track the recent isolation of strain GT from other Dehalococcoides in a mixed community (30) and to effectively quantify the fraction of Dehalococcoides and the relative proportion of reductive dehalogenases therein in enrichment cultures, microcosms, and field samples (27). In these applications qPCR results were accurate over 8 orders of magnitude and precise to within a few fold (27).
The present study uses qPCR of the Dehalococcoides 16S rRNA gene and multiple chlorinated-solvent reductive dehalogenase genes to characterize the growth of two functionally distinct Dehalococcoides strains in a TCE enrichment culture and to determine their respective yields on specific solvents. By applying a unique qPCR standard plasmid assembled from fragments of all four targeted genes, hereafter referred to as the four-gene plasmid, precision was increased to the point where the sum of reductive dehalogenase gene copy numbers consistently matched the Dehalococcoides 16S rRNA gene copy number to within 10%. With this improved precision, conclusions could confidently be based on small differences in gene copy number.
MATERIALS AND METHODS
Culture and growth conditions. (i) ANAS culture.
The TCE-dechlorinating microbial enrichment was derived from soil taken from the Alameda Naval Air Station (ANAS). Procedures for culture maintenance have been described previously (16, 26). Briefly, cultures were maintained at 25 to 28°C in a 1.5-liter continuously stirred semi-batch reactor (400-ml liquid volume) pressurized to 1.8 atm with N2/CO2 (90:10 [vol/vol]) to prevent oxygen intrusion. After each complete dechlorination of 111 μmol of TCE to ethene (4 to 7 days), the reactor was purged with N2, 60 to 100 ml of culture was withdrawn, and an equal volume of fresh anaerobic minimal medium (26) (containing l-cysteine and sodium sulfide [0.2 mM each]) was added, along with 10 μl of pure TCE and 10 ml of sodium lactate (from sterile, anaerobic 1 M stock).
(ii) ANAS subcultures.
Subcultures of the ANAS culture were grown in 160-ml bottles containing 98 ml of minimal medium (26) and inoculated with 2 ml of ANAS culture taken from the semi-batch reactor after dechlorination was complete. The 100-ml cultures were fed chlorinated solvents as described in individual experiments and 2 mmol of sodium lactate (20 mM final). Bottles were shaken at 90 rpm at 25°C. Total solvent concentrations were measured by analyzing headspace samples on a gas chromatograph with a flame ionization detector and normalizing the values to a standard curve generated using bottles with the same gas to liquid ratio and maintained at the same temperature as the cultures.
(iii) Dehalococcoides isolates.
Dehalococcoides sp. strain BAV1 and D. ethenogenes 195 were grown in defined medium as described previously (11). Briefly, the 100-ml cultures were maintained in 160-ml serum bottles with 5 mM acetate as the carbon source and hydrogen (80:20 H2/CO2 headspace) as an electron donor. Strains BAV1 and 195 were allowed to dechlorinate 40 μmol (1 ml) of VC and 56 μmol of TCE, respectively, two or three times before being transferred. Cultures inoculated at a 1:50 dilution usually required a 14- to 21-day lag before significant dechlorination was observed.
Initial rate measurements.
Initial VC and TCE dechlorination rates were measured in subcultures after completion of dechlorination by purging bottles with nitrogen to remove ethene and transferring 13 ml of culture to each of four 58-ml anaerobic serum bottles containing a H2/CO2 (80:20) headspace and 5 μmol of either VC or TCE. Rates were calculated from linear (R2 > 0.98) TCE or VC disappearance during the first 31 h of incubation. Liquid samples were taken at the final time point for qPCR analysis.
DNA isolation.
For molecular analyses, duplicate 1- and 10-ml culture samples were taken from ANAS subcultures and isolates, respectively, and replaced with an equal volume of medium at each sampling event. Samples were centrifuged to pellet cells, the supernatant was discarded, and the pellet was frozen at −80°C until DNA extraction and analysis. DNA was extracted from frozen cell pellets by using a MoBio microbial DNA extraction kit as recommended by the manufacturer, except that a second 50-μl volume of elution buffer was added to and spun through the affinity column, giving a 100-μl final volume.
Dehalococcoides-specific 16S rRNA gene library.
Primers 1492R and DHC16S9F (5′-CTAGCGGCGTGCCTTAT) were used in a PCR with genomic DNA from the ANAS culture (8, 25). The resulting 1.5-kb fragment was cloned by using the Invitrogen TopoTA kit. Clones were sequenced using the M13F and M13R primer sites flanking the insertion site on the plasmid and the 704R universal 16S rRNA gene primer (complement of 704F [15]). Sequences were manually corrected in overlapping regions and compared to known sequences in GenBank (http://www.ncbi.nlm.nih.gov) by using NCBI BLAST.
PCR for reductive dehalogenase genes.
Genomic DNA from pure strains or the ANAS culture was reacted with the published primers for tceA (797F and 2490R [18]) to yield a product of 1,732 bp, for vcrA (5′-CTATGAAGGCCCTCCAGATGC-3′ and 5′-GTAACAGCCCCAATATGCAAGTA-3′ [23]) to yield a product of 1,482 bp, or for bvcA (bvcAF and bvcAR [17]) to yield a fragment of 839 bp. All three reactions were run with an initial 94°C denaturation (12 min), followed by 30 cycles of 94°C (60 s), 50°C (45 s), and 72°C (120 s), with a final extension at 72°C for 12 min.
qPCR.
TaqMan primers and probe sets (PE Applied Biosystems, Foster City, CA) for qPCR were designed by using the Primer Express software package suite. The primers for the tceA gene (AF228507) were as follows: forward, 5′-ATCCAGATTATGACCCTGGTGAA; probe, 5′-TGGGCTATGGCGACCGCAGG; and reverse, 5′-GCGGCATATATTAGGGCATCTT (16). The primers for the vcrA gene (AY322364) were as follows: forward, 5′-CTCGGCTACCGAACGGATT; probe, 5′-CGCACTGGTTATGGCAACCACTC; and reverse, 5′-GGGCAGGAGGATTGACACAT. The primers for the bvcA gene (AY563562) were as follows: forward, 5′-GGTGCCGCGACTTCAGTT; probe, 5′-TGCCGAATTTTCACGACTTGGATGAAG; and reverse, 5′-TCGGCACTAGCAGCAGAAATT. The primers specific to the 16S rRNA gene of the genus Dehalococcoides, based on the published D. ethenogenes 195 genome (CP000027) (21, 28), were as follows: forward, 5′-GGTAATACGTAGGGAAGCAAGCG; probe, 5′-ACATCCAACTTGAAAGACCACCTACGCTCACT; and reverse, 5′-CCGGTTAAGCCGGGAAATT.
Assembly of the four-gene plasmid standard.
A plasmid containing fragments of three functional reductive dehalogenase genes along with the 16S rRNA gene of Dehalococcoides was derived from PCR2.1topo (Invitrogen). First, a 1,732-bp fragment of the vcrAB operon was cloned into the plasmid as directed. Next, a 450-bp fragment of the tceA gene was added between the vcrA fragment and the plasmid M13R sequence by using the unique BamHI restriction enzyme plasmid target site. The tceA gene has a BamHI site at position 1054, and a second one was engineered into a tceA PCR product using the primers 797F and 1483RBamHI (5′-CGTATGGATCCTTCAGGCGTACCCTCCCAC).
Third, a 1-kb fragment of the Dehalococcoides 16S rRNA gene was cloned into the plasmid's unique KpnI site, between the M13R sequence and the tceA fragment, using both a KpnI site at position 440 in the 16S rRNA gene and a second site engineered with a modified 1492R primer, 1492KpnI (5′-CGTATGGTACCTTACGGYTACCTTGTTACGACTT).
Finally, a 250-bp fragment of the bvcA gene was added to the three-gene plasmid. A pCR2.1-topo plasmid that had the bvcA gene cloned into it as directed was used as the template for a PCR using the standard M13F primer and the bvcA254RXba primer (5′-TCTAGACGCGCCGTCATTACGCGCAAG). The resulting PCR fragment was cut and ligated into the XbaI-cut three-gene plasmid.
The sequences and positions of the four gene fragments were verified by DNA sequencing.
RESULTS
16S rRNA gene sequence of Dehalococcoides in the ANAS culture.
The ANAS culture, the lactate and TCE enrichment culture used in the present study, effectively degrades TCE to ethene with a brief accumulation of VC but does not degrade PCE. This culture has been functionally stable for approximately 6 years. A previous study characterizing the microbial composition of the ANAS culture suggested that 20 to 30% of the microbial cells were Dehalococcoides (26).
Because the dechlorination abilities of the ANAS culture consortium (no PCE dechlorination, VC degraded rapidly and completely to ethene) are different from those reported for strain 195 (PCE dechlorination, VC converted slowly and incompletely to ethene), it seemed likely that dechlorinating strains other than strain 195 were present in the ANAS culture. In a genus-specific clone library generated by using 16S rRNA gene primers specifically targeting Dehalococcoides, 48 analyzed clones generated identical RFLP patterns. A subsample of 21 generated the same sequence (GenBank accession number DQ855129), matching the D. ethenogenes 195 genome sequence (28) to >99% identity.
Presence of reductive dehalogenase genes in the ANAS culture.
Because none of the Dehalococcoides reductive dehalogenases characterized to date could account solely for the observed dechlorination activity of the ANAS culture, experiments were performed to evaluate whether multiple reductive dehalogenases were present at significant levels. First, genomic DNA from the ANAS culture was queried by PCR with primer sets targeting three previously identified Dehalococcoides reductive dehalogenase genes: tceA, vcrA, and bvcA. Pure cultures of strains 195 and BAV1 were used as positive controls for tceA and bvcA, respectively.
The ANAS culture consortium produced strong PCR products (Fig. 1) for both tceA and vcrA which, after cloning and sequencing, showed >95% nucleotide identity to the published homologs. The bvcA gene was not found in the culture. These findings suggest that the overall dechlorination ability of the ANAS culture could be due to the combined activities of the tceA and vcrA gene products.
FIG. 1.
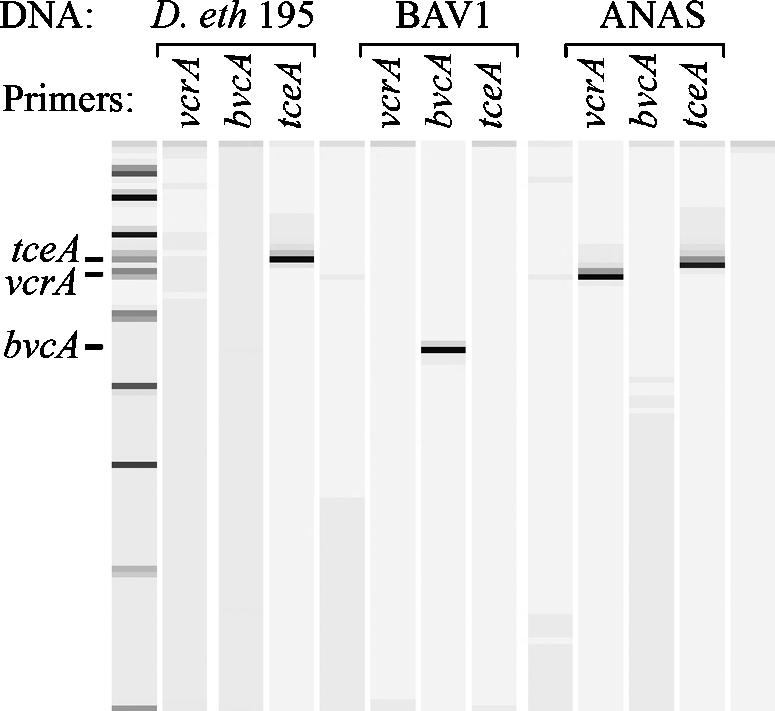
PCR with primers specific to three known chlorinated-solvent reductive dehalogenase genes—vcrA, bvcA, and tceA—applied to DNA from Dehalococcoides sp. strain 195, Dehalococcoides sp. strain BAV1, and the ANAS enrichment culture. The expected mobility of each PCR product is indicated on the left. The analysis was performed by capillary electrophoresis but is presented as a gel image for clarity.
Quantification of reductive dehalogenase and 16S rRNA genes.
To determine whether the tceA and vcrA genes within the ANAS culture were present within one Dehalococcoides strain or whether the genes were present in two or more strains with identical 16S rRNA gene sequences, qPCR was used to quantify the copy number of the tceA, vcrA, and Dehalococcoides 16S rRNA genes.
Initially, inaccuracies associated with individually quantifying the concentration of a different standard plasmid for each target gene hindered attempts to simultaneously quantify multiple genes. For example, experiments conducted with pure cultures of strain 195, known to contain a 1:1 ratio of tceA to 16S rRNA gene (28), resulted in ratios ranging from 0.6 to 1.2, whereas those conducted with the ANAS culture resulted in ratios of summed reductive dehalogenase genes to 16S rRNA genes ranging from 0.9 to 1.6. Despite the reproducibility of qPCR, the summed errors associated with the application of individual plasmid standards for multiple genes were too large to effectively distinguish the number of individual Dehalococcoides strains using reductive dehalogenase gene copy numbers.
To increase the comparative accuracy of qPCR, a single plasmid standard was constructed that contained fragments of the three identified reductive dehalogenase genes—tceA, vcrA, and bvcA—and the Dehalococcoides 16S rRNA gene (Fig. 2A). Comparison of qPCR results using the individual plasmid standards with those using the four-gene plasmid standard on the ANAS culture (Fig. 2B and C) shows that this methodological improvement greatly decreased data variability (R2 increased from 0.79 to 0.99), resulting in a 1:1 ratio of the sum of reductive dehalogenase gene copy numbers to the 16S rRNA gene copy number.
FIG. 2.
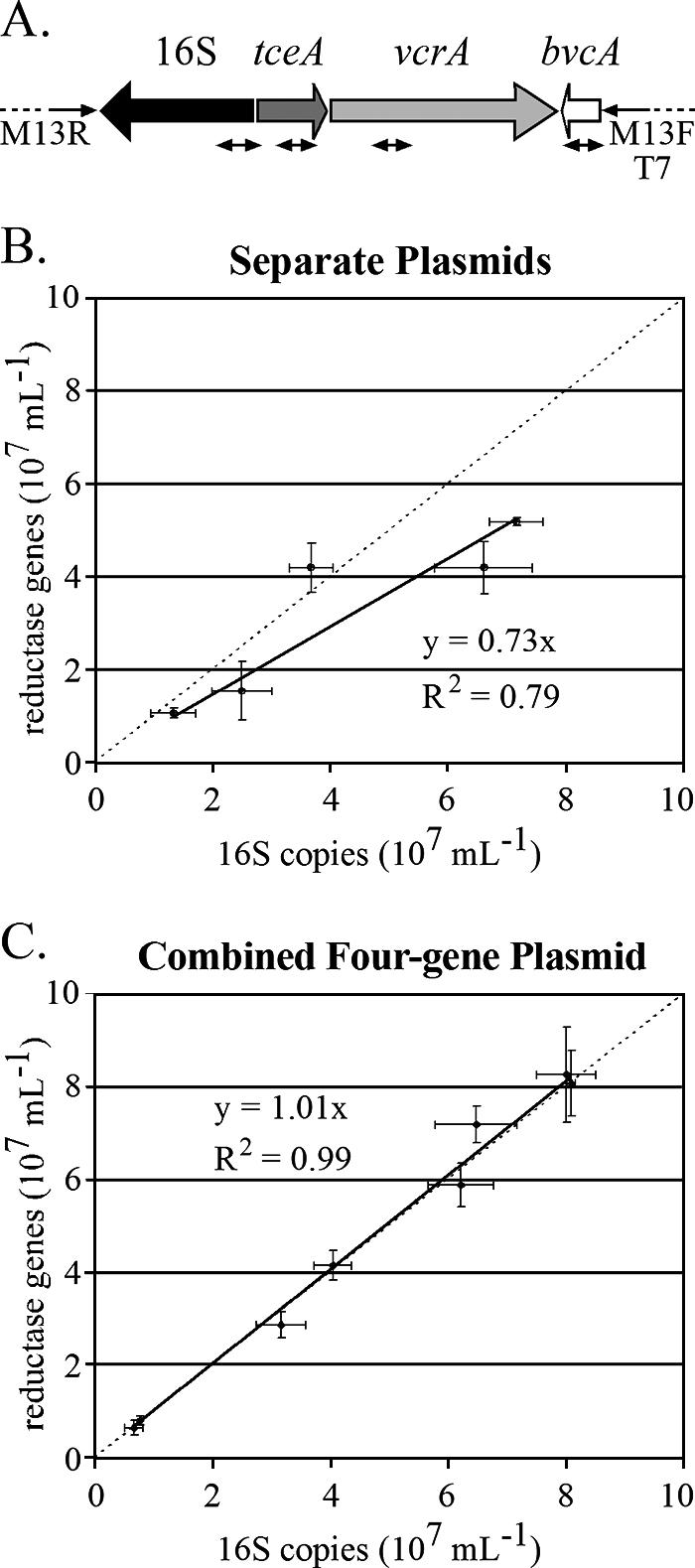
(A) Map of the four-gene qPCR plasmid standard generated in the present study. The small double-headed arrows indicate the qPCR amplicons. (B) Dehalococcoides 16S rRNA copy number versus reductive dehalogenase copy number in qPCR experiments using DNA from the ANAS culture and D. ethenogenes 195 and separate plasmid standards for 16S, tceA, and vcrA, and bvcA. (C) Same as in panel B, but using the four-gene Dehalococcoides plasmid standard. The dashed lines in panels B and C show a 1:1 ratio for reductive dehalogenase and 16S rRNA genes; the solid lines indicate a best fit to the data.
Application of the four-gene plasmid with qPCR to the genomic DNA of strain 195, strain BAV1, and the ANAS culture showed that the sum of reductive dehalogenase gene copy numbers matched the 16S rRNA gene copy number to within 5% (0.96 ± 0.05, 0.96 ± 0.04, and 1.02 ± 0.1, respectively) (Fig. 3). This result demonstrates that each Dehalococcoides cell in the ANAS culture contains a single copy of either the vcrA or tceA gene.
FIG. 3.
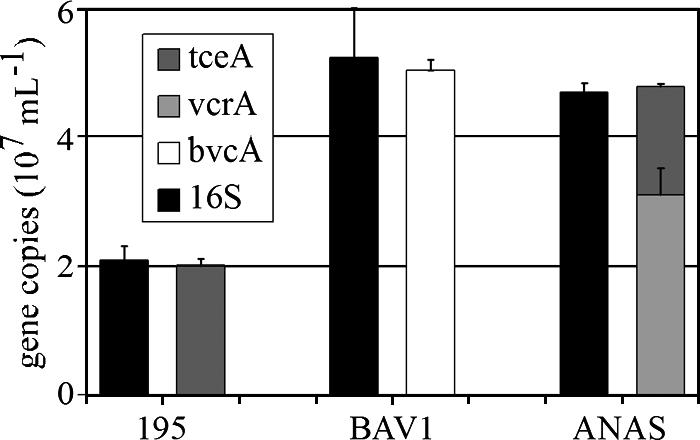
Comparison of copy numbers determined by qPCR for the Dehalococcoides 16S rRNA, vcrA, tceA, and bvcA reductive dehalogenase genes in strain 195, strain BAV1, and the ANAS culture consortium.
Perturbing the relative amounts of tceA and vcrA in ANAS enrichments.
A ratio of two vcrA to one tceA was consistently measured in samples of the batch-fed ANAS culture that spanned the TCE to ethene dechlorination and the feeding cycle. To confirm that the two reductive dehalogenase genes belonged to individual Dehalococcoides strains, an experiment was performed over a 2-month period to specifically enrich each of the two strains. Subcultures meant to enrich for vcrA were allowed to dechlorinate VC to ethene with repeated feedings of VC. In contrast, subcultures meant to enrich for tceA were allowed to dechlorinate TCE to cDCE and, whenever VC was detected, were flushed and refed with TCE. A third subculture, serving as a control, was starved of solvents during the experiment. During the 2-month incubation, the VC enrichment dechlorinated three VC amendments of 40 μmol, while the TCE enrichment dechlorinated to DCE roughly five TCE amendments of 44 μmol each.
After 2 months, whereas the starved subculture had shown only a 1.4-fold increase in 16S rRNA gene numbers, the fed subcultures showed a 10- to 20-fold increase in Dehalococcoides 16S rRNA genes and dramatic shifts in the vcrA to tceA ratio (Fig. 4). The VC subculture ended the experiment with a 14:1 ratio of vcrA genes to tceA genes; the sum of quantified reductive dehalogenase genes corresponded to 103% of the Dehalococcoides 16S rRNA genes. As expected, the number of tceA genes in the VC subculture was similar to that in the starved subculture. Surprisingly, in the TCE-to-DCE enrichment, the number of vcrA genes increased as much as the tceA genes, resulting in a near 1:1 final ratio of vcrA to tceA. The sum of reductive dehalogenase genes in this enrichment corresponded to 99% of the Dehalococcoides 16S rRNA genes. The excellent agreement between the sum of reductive dehalogenase genes and the 16S rRNA gene, regardless of the reductive dehalogenase gene ratio, supports the hypothesis that the two reductive dehalogenase genes belong to individual Dehalococcoides strains.
FIG. 4.

Copy numbers determined by qPCR of vcrA, tceA, and Dehalococcoides 16S rRNA genes in parallel 50-fold-diluted ANAS subcultures. Inoculum, dilution culture at time zero; Starved, culture incubated for 2 months in the absence of solvent; VC to Eth, the culture was fed VC and allowed to dechlorinate to ethene; TCE to cDCE, the culture was fed TCE and limited to dechlorination to DCE by flushing and refeeding as soon as VC was detected. The ratio of vcrA to tceA in each sample is shown above each set of columns. Note that a linear scale is used for copy numbers.
The increase in Dehalococcoides 16S rRNA genes in the fed cultures appeared proportional to the total moles of Cl− generated by each subculture. The TCE-to-DCE enrichment reached a density of 8 × 107 Dehalococcoides cells/ml and consumed a total of 220 μmol of Cl− equivalents, whereas the VC enrichment reached a density of 4 × 107 Dehalococcoides cells/ml and consumed a total of 120 μmol of Cl− equivalents.
Growth of each strain during dechlorination of VC, cDCE, or TCE.
An experiment was conducted to determine the contribution of individual ANAS Dehalococcoides strains to each dechlorination step between TCE and ethene. The growth of Dehalococcoides was measured in parallel 100-ml ANAS subcultures diluted 50-fold and, over a period of 3 months, allowed to completely dechlorinate either 88 μmol of TCE, 132 μmol of cDCE, or 264 μmol of VC, each corresponding to 264 μmol of Cl− (Table 1) .
TABLE 1.
Increases in Dehalococcoides gene copy number after dechlorination of equal molar equivalents of Cl− from VC, cDCE, or TCE
| Substrate added (amt [μmol]) | Mean no. of copies (1010)/liter ± SD (% of 16S)
|
||
|---|---|---|---|
| 16S gene | vcrA | tceA | |
| VC (264) | 2.7 ± 0.3 | 2.8 ± 0.2 (103) | <0 |
| cDCE (132) | 2.8 ± 0.4 | 1.5 ± 0.2 (54) | 1.6 ± 0.1 (56) |
| TCE (88) | 3.7 ± 0.3 | 1.1 ± 0.5 (31) | 2.8 ± 0.2 (74) |
In all three enrichments, the increases in the numbers of Dehalococcoides 16S rRNA genes corresponded, as expected, to equivalent increases in the summed reductive dehalogenase genes. In the VC culture, the vcrA-containing strain was responsible for the entire increase of 16S rRNA genes, whereas this strain accounted for only 54 and 31% of the increase in 16S rRNA genes in the cDCE- and TCE-fed cultures, respectively. The tceA-containing strain was responsible for the other 56 and 74% of increases in 16S rRNA genes for the DCE- and TCE-fed subcultures.
In order to determine contributions of individual strains to individual dechlorination steps, yields were calculated for 16S rRNA genes, vcrA, and tceA per chloride equivalent (Table 2). The 16S rRNA gene yields, reported as the number of 16S rRNA genes per μmol of Cl−, were similar for each of the three subcultures (1.1 × 107 to 1.4 × 107 copies of 16S rRNA gene/μmol of Cl−). The VC-fed subculture results from both Table 1 and Fig. 4 were used to calculate the vcrA yield for the vcrA-containing strain: 1.3 ± 0.3 × 107 copies vcrA/μmol of Cl−. For the cDCE and TCE subcultures, the difference between the observed vcrA copy number and the predicted copy number, based on the vcrA yield defined above, was used to calculate the amount of cDCE or TCE metabolized by the vcrA-containing strain.
TABLE 2.
Comparison of yields and rates for ANAS and published dechlorinating enrichment cultures
| Culture (substrate) | Mean ± SDa
|
Source or reference | |||
|---|---|---|---|---|---|
| No. of copies/μmol of Cl− (107)
|
Rateb (10−10) | ||||
| Deh. 16Sc | vcrA | tceA | |||
| ANAS (VC) | 1.3 ± 0.3 | 1.3 ± 0.3 | 8.6 ± 1.32 | This study | |
| ANAS (cDCE) | 1.1 ± 0.1 | 1.2 ± 0.1* | 1.2 ± 0.11 | 7.6 ± 0.4† | This study |
| ANAS (TCE) | 1.4 ± 0.4 | 1.3 ± 0.4* | 1.6 ± 0.41 | 2.3 ± 0.6† | This study |
| 195 (PCE) | 230 ± 20‡ | 2.2 ± 0.3‡ | 21 | ||
| BAV1 (VC) | 6.3 ± 0.3 | 9 | |||
| VS (VC) | 46 ± 3 | 7.8 | 3 | ||
| VS (TCE) | 58 ± 8 | 7.8 | 3 | ||
| KB1 (VC) | 56 ± 14 | 2.2 ± 0.4 | 9 | ||
| KB1 (TCE) | 3.0 ± 0.3 | 9 | |||
| GT (VC) | 25 ± 2 | 10.1 | 30 | ||
*, yields for vcrA and tceA genes are calculated based on the assumption that vcrA is responsible for VC dechlorination and tceA is responsible for TCE and cDCE dechlorination. †, rates were calculated from the interval of maximum solvent dechlorination observed during culture growth. ‡, yields and rates for D. ethenogenes 195 were calculated by applying the protein-to-cell conversion factor of 4.2 × 10−15 g/cell from Duhamel et al. (9) to published results of Maymo-Gatell et al. (21).
Expressed as μmol of Cl−/(copies of 16S gene · day).
Deh. 16S, Dehalococcoides 16S rRNA gene.
Interestingly, in the cDCE enrichment the cell number of the vcrA-containing strain was approximately what would be predicted from the dechlorination of 132 μmol of VC, suggesting that the vcrA-containing strain did not metabolize a detectable fraction of the cDCE. Assuming that the cDCE to VC dechlorination was used entirely for the increase of the tceA gene, a yield of 1.2 ± 0.1 × 107 copies of tceA/μmol of Cl− can be calculated for the tceA-containing strain.
In the TCE enrichment, the tceA copy number increased by 2.8 × 109 copies whereas the vcrA gene copy number increased by 1.1 × 109. The vcrA copy number predicted from the previously calculated yield for 88 μmol of VC is also 1.1 × 109, suggesting that the vcrA-containing strain did not metabolize a detectable fraction of cDCE or TCE. Assuming that the TCE to VC dechlorination contributed entirely to the increase in tceA gene, a yield of 1.6 ± 0.4 × 107 copies of tceA/μmol of Cl− can be calculated for the tceA-containing strain.
In addition to yields, dechlorination rates per Dehalococcoides 16S rRNA gene were calculated during the period of maximum observed chlorinated solvent consumption using the average number of genes in samples taken at the start and end points of that time period (Table 2). Of the three subcultures, the VC enrichment exhibited the highest molar dechlorination rate of (8.6 ± 1.3) × 10−10 μmol of Cl−/(16S rRNA gene · day), followed by the cDCE subculture with (7.6 ± 0.4) × 10−10 μmol of Cl−/(16S rRNA gene · day), and the TCE subculture with (2.3 ± 0.6) × 10−10 μmol of Cl−/(16S rRNA gene · day).
DISCUSSION
The results of the present study suggest that the ANAS culture contains two functionally different Dehalococcoides strains that share the same 16S rRNA gene sequence. Several studies have noted that 16S rRNA gene sequences vary little or not at all between Dehalococcoides strains with notably different metabolic activities (9, 28), suggesting that the characterization of Dehalococcoides 16S sequences in an unknown community would not be adequate for predicting the activity of the community or for identifying the number of functionally distinct Dehalococcoides strains in that community.
Several studies have used qPCR to compare Dehalococcoides 16S rRNA gene copy numbers to reductive dehalogenase copy numbers (27, 29, 30). These studies have been able to closely track both the total population of Dehalococcoides and the relative populations of different reductive dehalogenase-containing strains (27, 30) despite the 1.5- to 2-fold limit of precision (29).
In the present study, greater precision was required to determine whether the presence of all Dehalococcoides cells in the ANAS enrichment culture could be accounted for by the known reductive dehalogenases. To that end, a plasmid containing fragments of the three identified chlorinated-solvent reductive dehalogenase genes and the Dehalococcoides 16S rRNA gene was developed as a qPCR standard. The increased precision given by using a single standard revealed that two distinct Dehalococcoides strains are present in this culture, one that contains the tceA gene and one that contains the vcrA gene, and that these strains represent the predominant Dehalococcoides cells in the ANAS culture.
To date, only one TCE-RDase and two VC-RDases have been identified to be functional in Dehalococcoides (4, 17-19, 23). It has been suggested, however, that many other functional reductive dehalogenases may be found. The genome of D. ethenogenes 195 has more than a dozen other haloreductase-like genes of unknown function (28), and the presence of multiple reductive dehalogenases is common across Dehalococcoides strains (14, 31).
Further, a recent study found that less than 50% of Dehalococcoides in field samples could be accounted for by the three known reductive dehalogenases (27). Thus, while the two strains in the ANAS culture each utilize one already-known reductive dehalogenase, it is possible that additional uncharacterized reductive dehalogenases are also active in this culture.
The steady-state vcrA/tceA ratio in the TCE-fed ANAS culture has repeatedly been measured to be 2:1. However, when an ANAS subculture was diluted 50-fold and allowed to completely dechlorinate TCE (Table 1), the final ratio was 1:2. The difference in the ratio may relate to the fact that the batch-fed ANAS culture is routinely maintained at a saturated cell density, whereas the subculture was diluted to promote cell growth. Strain 195 has been reported to uncouple dechlorination activity from growth at cell densities of roughly 109 cells/ml (21). The total Dehalococcoides 16S rRNA gene density in the ANAS culture is roughly 7 × 108 cells/ml, whereas the maximum density in the subcultures used in Table 1 was an order of magnitude less: 4 × 107 cells/ml. Thus, the 2:1 ratio in the steady-state ANAS culture may simply reflect the final densities reached by the two strains after uncoupling growth from dechlorination.
An additional interesting observation from the present study was that the vcrA-containing strain in the ANAS culture apparently derived energy only from the VC reduction step, whereas the tceA-containing strain grew from both the TCE and the cDCE reduction steps. Although studies of the tceA-containing D. ethenogenes 195 have consistently shown that this strain derives energy from the reduction of TCE and cDCE but not VC (20, 22), studies of vcrA-containing Dehalococcoides cultures have demonstrated the metabolic degradation of TCE, cDCE, and VC (3, 5, 30).
One possible explanation for this discrepancy is that, while both reductases in the ANAS culture can react with all three chlorinated ethenes, the relative enzyme affinities and reaction rates are such that the tceA enzyme utilizes the majority of available TCE and cDCE, whereas the vcrA enzyme utilizes the available VC. For example, strain VS is reported to have a half-velocity constant for TCE that is four times higher than that for VC, suggesting the culture has a much greater affinity for VC (5). Further, studies with the purified vcrA and tceA proteins reported that the tceA protein exhibited cDCE degradation rates roughly 2 orders of magnitude higher than vcrA (12,100 versus 350 nmol min−1 mg of protein−1) (23). The VC degradation rate for the tceA protein was also roughly 2 orders of magnitude lower than that for the vcrA protein (3.6 versus 350 nmol min−1 mg of protein−1) (18, 23).
The 1:1 vcrA/tceA ratio observed in TCE-fed subcultures likely reflects ability of the vcrA-containing strain to rapidly consume the generated VC before sufficient accumulation triggered flushing of the headspace. This is consistent with the small and transient VC accumulations observed during typical ANAS culture feeding cycles.
The yields for Dehalococcoides from the ANAS culture in the present study are roughly 50-fold lower than the yields reported for strains VS, KB-1/VC, and BAV1 (Table 2) (5). These low yields were based on long-term batch experiments in which Dehalococcoides cells had likely decoupled dechlorination and growth (21). These experiments do illustrate, however, that the yields of the two strains in the ANAS culture are similar despite the involvement of two distinct enzymes and chlorinated substrates.
In contrast, the observed rates of chlorinated solvent degradation in the ANAS culture agree well with those published previously (5). These rates were calculated from the maximum observed dechlorination activity seen in short-term experiments. Interestingly, conversion to units of molecules of Cl−/(16S rRNA genes · s) provides physically plausible rates of several thousand (1,500 to 6,000) solvent molecules dechlorinated per cell per second.
Successful strategies for the bioremediation of chlorinated solvents not only effectively transform TCE and PCE but also avoid the accumulation of DCE and VC. The present study demonstrates that quantification of reductive dehalogenase genes can be useful for tracking the relative growth of multiple strains of Dehalococcoides with different metabolic activities. Further, results suggest that the relative ratio of VC- to TCE- degrading strains in microbial communities containing Dehalococcoides may be increased either by enriching to densities at which dechlorination is decoupled from growth or by using cDCE instead of TCE. Preparation of bioaugmentation inocula containing high ratios of VC-degrading Dehalococcoides strains could help to prevent VC accumulation in future field applications.
Acknowledgments
We thank Stephen Zinder of Cornell University for providing Dehalococcoides ethenogenes 195 and Frank Löffler of the Georgia Institute of Technology and the Georgia Tech Research Company for providing Dehalococcoides strain BAV1.
This study was supported by the Lawrence Berkeley National Laboratory, the Laboratory Directed Research and Development Program, the National Science Foundation under grant 0504244, and the Superfund Basic Research Project under NIEHS ES04705.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Gorisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Gorisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 3.Cupples, A. M., A. M. Spormann, and P. McCarty. 2004. Comparative evaluation of chloroethene dechlorination to ethene by Dehalococcoides-like microorganisms. Environ. Sci. Technol. 38:4768-4774. [DOI] [PubMed] [Google Scholar]
- 4.Cupples, A. M., A. M. Spormann, and P. McCarty. 2004. Vinyl chloride and cisdichloroethene dechlorination kinetics and microorganism growth under substrate limiting conditions. Environ. Sci. Technol 38:1102-1107. [DOI] [PubMed] [Google Scholar]
- 5.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl. Environ. Microbiol. 69:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damborsky, J. 1999. Tetrachloroethene-degrading bacteria. Folia Microbiol. 44:247-262. [DOI] [PubMed] [Google Scholar]
- 7.DiStefano, T. D., J. M. Gossett, and S. H. Zinder. 1991. Reductive dechlorination of high concentrations of tetrachloroethene to ethene by an anaerobic enrichment culture in the absence of methanogenesis. Appl. Environ. Microbiol. 57:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duhamel, M., M. Kaiguo, and E. Edwards. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ensley, B. D. 1991. Biochemical diversity of trichloroethylene metabolism. Annu. Rev. Microbiol. 45:283-299. [DOI] [PubMed] [Google Scholar]
- 11.He, J., K. Ritalahti, M. Aiello, and F. Loffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, J., K. Ritalahti, K. Yang, S. Koenigsberg, and F. Loffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 13.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. Loffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 14.Holscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. V. Wintzingerode, H. Gorisch, F. Loffler, and A. Lorenz. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, D. R., P. K. H. Lee, V. F. Holmes, and L. Alvarez-Cohen. 2005. An internal reference technique for quantifying specific mRNAs by real-time PCR with application to the tceA reductive dechlorination gene. Appl. Environ. Microbiol. 71:3866-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krajmalnik-Brown, R., T. Holscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. Loffler. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maymo-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maymo-Gatell, X., Y. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 22.Maymo-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-1,2-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes.” Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 23.Muller, J. A., B. M. Rosner, G. V. Abendroth, G. Meshulam-Simon, P. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman, L. M., and L. P. Wackett. 1997. Trichloroethylene oxidation by purified toluene 2-monooxygenase: products, kinetics, and turnover-dependent inactivation. J. Bacteriol. 179:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pace, N. R. 1973. Structure and synthesis of the ribosomal ribonucleic acid of prokaryotes. Bacteriol. Rev. 37:562-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson, R. E., V. K. Bhupathiraju, D. L. Song, T. A. Goulet, and L. Alvarez-Cohen. 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 36:2652-2662. [DOI] [PubMed] [Google Scholar]
- 27.Ritalahti, K. M., B. K. Amos, Y. Sung, Q. Wu, S. S. Koenigsberg, and F. Löffler. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 29.Smits, T. H. M., C. Devenoges, K. Szynalski, J. Maillard, and C. Holliger. 2003. Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J. Microbiol. Methods 57:369-378. [DOI] [PubMed] [Google Scholar]
- 30.Sung, Y., K. M. Ritalahti, R. P. Apkarian, and F. Löffler. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waller, A. S., R. Krajmalnik-Brown, F. Loffler, and E. Edwards. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, Q., J. E. M. Watts, K. R. Sowers, and H. D. May. 2002. Identification of a bacterium that specifically catalyzes the reductive dechlorination of polychlorinated biphenyls with doubly flanked chlorines. Appl. Environ. Microbiol. 68:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zinder, S. H., and J. M. Gossett. 1995. Reductive dechlorination of tetrachloroethene by a high rate anaerobic microbial consortium. Environ. Health Perspect. 103(Suppl. 5):5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


