Abstract
Quantitative PCR of denitrification genes encoding the nitrate, nitrite, and nitrous oxide reductases was used to study denitrifiers across a glacier foreland. Environmental samples collected at different distances from a receding glacier contained amounts of 16S rRNA target molecules ranging from 4.9 × 105 to 8.9 × 105 copies per nanogram of DNA but smaller amounts of narG, nirK, and nosZ target molecules. Thus, numbers of narG, nirK, nirS, and nosZ copies per nanogram of DNA ranged from 2.1 × 103 to 2.6 × 104, 7.4 × 102 to 1.4 × 103, 2.5 × 102 to 6.4 × 103, and 1.2 × 103 to 5.5 × 103, respectively. The densities of 16S rRNA genes per gram of soil increased with progressing soil development. The densities as well as relative abundances of different denitrification genes provide evidence that different denitrifier communities develop under primary succession: higher percentages of narG and nirS versus 16S rRNA genes were observed in the early stage of primary succession, while the percentages of nirK and nosZ genes showed no significant increase or decrease with soil age. Statistical analyses revealed that the amount of organic substances was the most important factor in the abundance of eubacteria as well as of nirK and nosZ communities, and copy numbers of these two genes were the most important drivers changing the denitrifying community along the chronosequence. This study yields an initial insight into the ecology of bacteria carrying genes for the denitrification pathway in a newly developing alpine environment.
Primary successional ecosystems, such as glacier forelands and volcanoes, present an ideal opportunity to study the biological colonization of substrates. Since the ice covers of many glaciers have receded over the past century, glacier forelands have released substrates for soil development. Autotrophic colonizers are expected to be important in the initial stages of primary community assembly. Organic substrates for microbial growth, however, might also come from allochthonous dead organic matter and living invertebrates in these environments. Hodkinson et al. (8) therefore recently proposed a previously unrecognized heterotrophic phase which should allow the initial establishment of functional communities. Accordingly, future studies in microbial ecology must account for both autotrophic and heterotrophic colonization along primary successional gradients such as glacier forelands, land lifts, floodplains, landslides, and volcanoes. In the past few years, studies have focused mainly on the composition and activities of the soil microbiota in primary succession of receding glaciers (19, 21, 24, 25). Only a few studies have employed molecular tools to understand the diversity of archaeal and bacterial community structures along the forefields of receding glaciers (2, 13, 20). Analyses of activity and genetic structures of the nitrate reducer community at the Rotmoosferner glacier have shown that N cycling processes as well as microbial community composition depend on the successional age (2). To this point, no information on the density of the functional community involved in the N cycle in the alpine ecosystem has been available.
Denitrification consists of four reaction steps in which nitrate is reduced to dinitrogen gas. A taxonomically diverse group of bacteria can reduce nitrate into nitrite. Respiratory nitrate reduction, as well as the dissimilatory reduction of nitrate to ammonia, is catalyzed by a membrane-bound or periplasmic nitrate reductase encoded by the narGHJI operon or the napABC operon, respectively (15). The reduction of nitrite (NO2−) to nitric oxide distinguishes denitrifiers from other nitrate-respiring bacteria. This reaction is catalyzed by two different types of nitrite reductases (Nir), either a cytochrome cd1 encoded by nirS or a Cu-containing enzyme encoded by nirK. The reduction of nitrous oxide is the last step in the denitrification pathway and is catalyzed by nitrous oxide reductase encoded by the nosZ gene. Whereas some denitrifiers can reduce nitrate into dinitrogen due to the presence of genes encoding all denitrification reductases, others have a truncated pathway (29).
The objective of this study was to quantify the densities of denitrification genes coding for NO3−, NO2−, and N2O reductase across a receding glacier foreland by using real-time PCR. We therefore used quantitative PCR for the first time to quantify denitrification genes coding for NO3−, NO2−, and N2O reductases along a glacier foreland. The spatial gradient along the receding glacier foreland covered a chronosequence of approximately 130 years of soil development. We focused on the denitrifying community in the rhizosphere of Poa alpina. This plant grows in a loosely organized assemblage early in succession as well as in highly organized assemblages in late successional stages (25). The hypothesis is that the density of early denitrifier communities on young substrates differs from that in the more developed, old substrates. Furthermore, relating our results to 16S rRNA copy numbers should reveal whether the relative abundances of the different denitrification genes involved in the denitrification cascade change during microbial succession.
MATERIALS AND METHODS
Study site.
Soils of the foreland of the Rotmoosferner glacier (46°50′N, 11°03′E) of the Ötz Valley (Austria) were used for the study. The mean annual temperature is −1.3°C, and the mean annual precipitation is 820 mm (9). The entire foreland lies above the tree line at 2,280 to 2,450 m above sea level. Since 1858, the Rotmoosferner glacier has been retreating and has left a mainly level valley 2 km in length, ascending only in the younger parts of the foreland (<50 years) (9). The well-preserved chronosequence has been described in detail regarding soil formation, vegetational gradient, invertebrate succession, and total soil microbial communities (4, 9, 18, 25). The parent material of the soils is mainly neoglacial moraine till and fluvioglacial sands (25). The texture of the soils is sand to silty sand, without significant differences in texture between the sampling sites (24). The organic carbon (Corg) content increased from 0.37% in 25-year-old rhizospheres of Poa alpina plants to 9.67% in 75-year-old rhizospheres, the pH decreased within the primary succession from 7.53 to 5.56, and less nitrate and ammonium are available in the younger than in the older sites (2).
Soil sampling.
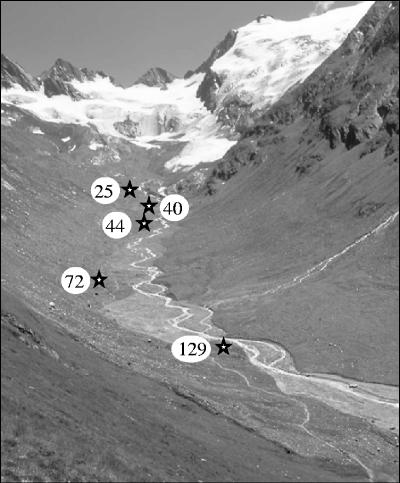
The soil was sampled in mid-August 2004 at the flowering stage of Poa alpina and in late September 2004 after the first snowfalls, to take seasonal variation into account. Five successional sites that had been deglaciated for 25 to 129 years (Rüdiger Kaufmann, personal communication) were selected within the orographic right side of the glacier foreland (Fig. 1). The sampling sites are located at different distances from the glacier: 80 m (25-year-old site), 360 m (40-year-old site), 610 m (44-year-old site), 1,260 m (72-year-old site), and 1,650 m (129-year-old site). Five field replicates were taken from each successional site both in August and in September. For each replicate, the rhizospheres of three to five plants of Poa alpina were bulked by sampling soil adhering to the roots. The rhizospheric samples were sieved directly through a 2-mm sieve on site and stored at −20°C prior to analysis.
FIG. 1.
View of the foreland of the Rotmoosferner glacier, Ötz Valley, Austria. Sampling sites differing in age of succession (in years) are indicated.
DNA extraction.
DNA of each of the 50 samples was extracted from 0.3 g of soil with the FastDNA Spin Kit for soil (BIO 101/Qbiogene), according to the protocol of the manufacturer. The quantity of the DNA extractions was checked with a BioPhotometer (Eppendorf).
Quantitative PCR assay.
Amplification of quantitative PCR products was carried out with an ABI Prism 7900 (Applied Biosystems) by using SYBR green as the detection system in a reaction mixture of 25 μl containing 0.5 μM (each) primer; 12.5 μl of SYBR green PCR master mix, including HotStar Taq DNA polymerase, QuantiTect SYBR green PCR buffer, deoxynucleoside triphosphate mix with dUTP, SYBR green I, ROX, and 5 mM MgCl2 (QuantiTect SYBR green PCR Kit; QIAGEN, France); and 1.25 μl of DNA-diluted template corresponding to 12.5 ng of total DNA. T4gp32 (500 ng/reaction; Q-BIOgene, France) was used to enhance PCR efficiency. Thermal cycling conditions for the 16S rRNA and the narG, nirK, and nosZ genes were as previously described (6, 7, 12). Thermal cycling, fluorescent data collection, and data analysis were carried out with the ABI Prism 7900 sequence detection system according to the manufacturer's instructions. nirS quantitative PCR was performed with the slightly modified nirSCd3aF (5′ AACGYSAAGGARACSGG 3′) and nirSR3cd (5′ GASTTCGGRTGSGTCTTSAYGAA 3′) primers described by Throbäck et al. (22) under the same conditions as those described for the nirK primers (6). In contrast to the nirS, nirK, and nosZ primers, the narG primers used in this study are not universal and were designed to target an unknown group of narG nitrate reducers (12). Two independent quantitative PCRs were performed for each gene and each soil replicate. Standard curves were obtained with serial plasmid dilutions of a known amount of plasmid DNA containing a fragment of the 16S rRNA or the narG, nirK, nirS, or nosZ gene (6, 7, 12).
Soil DNAs were also tested for the inhibitory effects of coextracted substances by determining the 16S rRNA gene copy number in 10-fold dilutions of soil DNA. In addition, standard plasmid DNA was quantified by real-time PCR with and without the addition of environmental DNA, and the obtained values were compared.
Statistical analysis.
The amounts of DNA, 16S rRNA, narG, nirK, nirS, and nosZ in the soil were calculated on an oven-dry weight (105°C) basis. Differences in the mean DNA yields, 16S rRNA copy numbers, and gene copy numbers of narG, nirK, nirS, and nosZ between successional sites were tested by univariate analysis of variance, followed by the multiple range test (Student Newman Keuls test). Measurements of the August and September samples were compared for equality with a standard t test for independent samples. Discriminant function analysis using Wilks' lambda for the stepwise selection of variables (16S rRNA, narG, nirK, nirS, and nosZ) was applied to assess responses of the denitrifying community to the successional stage across the chronosequence. The groups were defined according to the successional age of the site. The discriminant scores of each discriminant function (DF 1, DF 2, DF 3, and DF 4) were tested for significant differences between samples by univariate analysis of variance, followed by the Student Newman Keuls test. The result is a two-dimensional representation that shows every sample as one plot. Relative changes in the density of the denitrifying community are visualized as the distances between the plots. Multiple regression analysis was applied to evaluate the relationship between soil environmental factors (Corg, pH, and NO3−) and the density of the denitrifying community. Significance was accepted at a level of probability (P) of <0.05.
RESULTS
Quantification of 16S rRNA and denitrification genes (narG, nirK, nirS, and nosZ).
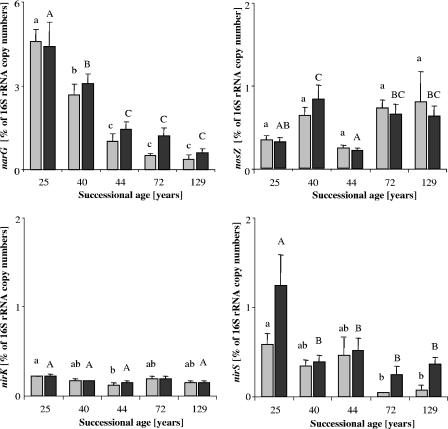
Environmental samples collected at different distances from the terminus of the receding glacier contained amounts of 16S rRNA target molecules ranging from 4.9 × 105 to 8.9 × 105 copies per nanogram of DNA (Table 1). The narG, nirK, nirS, and nosZ target molecules were less abundant than the 16S rRNA: narG ranged from 2.1 × 103 to 2.6 × 104 copies per nanogram of DNA, nirK ranged from 7.4 × 102 to 1.4 × 103, nirS ranged from 2.5 × 102 to 6.4 × 103, and nosZ ranged from 1.2 × 103 to 5.5 × 103 copies per nanogram of DNA (Table 1). The successional age of the environmental sample had a stronger impact on narG and nirS than on nirK and nosZ copy numbers per nanogram of DNA. In August 2004, the 25- and 40-year-old sites showed significantly higher densities of nitrate-reducing microorganisms than the older sites; similar gradients of narG were obtained for soil of the chronosequence sampled in September 2004. The youngest site was also characterized by significantly higher nirS gene copy numbers than in the other successional stages in September 2004 (Table 1). The nirK gene copy numbers based on nanograms of DNA showed a trend for lower values in the 44- and 129-year-old sites, which were significant only for the second sampling date. Similar amounts of the nosZ target molecule were obtained in August. In September, the nosZ gene copy numbers from the 25- and 44-year-old sites were lower than at the other sites.
TABLE 1.
DNA yields and copy numbers of the 16S rRNA and the narG, nirK, nirS, and nosZ genes from soils of different successional agesa
| Sampling mo | Successional age (yr) | DNA yield (μg g−1 of soil) | No. of gene copies (ng−1 of DNA)
|
||||
|---|---|---|---|---|---|---|---|
| 16S rRNA | narG | nirK | nirS | nosZ | |||
| August | 25 | 7.92 A | 5.1 × 105 A | 2.3 × 104 A | 1.1 × 103 A | 2.9 × 103 A | 1.8 × 103 A |
| 40 | 8.04 A | 8.9 × 105 A | 2.1 × 104 A | 1.4 × 103 A | 2.9 × 103 A | 4.7 × 103 A | |
| 44 | 22.41 A | 7.5 × 105 A | 6.4 × 103 B | 9.1 × 102 A | 4.1 × 103 A | 2.0 × 103 A | |
| 72 | 24.89 A | 6.6 × 105 A | 3.3 × 103 B | 1.3 × 103 A | 2.5 × 102 A | 4.8 × 103 A | |
| 129 | 64.54 B | 6.3 × 105 A | 2.1 × 103 B | 9.5 × 102 A | 5.7 × 102 A | 4.7 × 103 A | |
| September | 25 | 7.81 A | 5.5 × 105 A | 2.6 × 104 A | 1.2 × 103 A | 6.4 × 103 A | 1.7 × 103 AB |
| 40 | 9.15 A | 6.9 × 105 A | 2.1 × 104 B | 1.1 × 103 A | 2.7 × 103 B | 5.5 × 103 D | |
| 44 | 18.15 A | 5.4 × 105 A | 7.3 × 103 C | 7.4 × 102 B | 2.4 × 103 B | 1.2 × 103 A | |
| 72 | 22.24 A | 5.7 × 105 A | 7.2 × 103 C | 1.2 × 103 A | 1.4 × 103 B | 3.7 × 103 C | |
| 129 | 45.27 B | 4.9 × 105 A | 2.9 × 103 C | 7.6 × 102 B | 1.8 × 103 B | 3.2 × 103 BC | |
Suffix letters (A to D) indicate significant differences between successional stages.
The date of soil sampling (August or September) did not significantly influence the 16S rRNA copy number or DNA yield. Calculation of gene copy numbers per gram of soil showed that the copy numbers of the 16S rRNA per gram of soil, as well as of nirK and nosZ, increased significantly with progressing succession, with a maximum in late succession; the narG and nirS copy numbers, however, showed no significant difference with progressing successional ages of the soils (Table 2).
TABLE 2.
Copy numbers of the 16S rRNA and the narG, nirK, nirS, and nosZ genes from different successional stages
| Sampling mo | Successional age (yr) | No. of gene copies (g−1 of soil)a
|
||||
|---|---|---|---|---|---|---|
| 16S rRNA | narG | nirK | nirS | nosZ | ||
| August | 25 | 3.4 × 109 A | 1.6 × 108 A | 7.7 × 106 A | 1.9 × 107 A | 1.3 × 107 A |
| 40 | 7.0 × 109 A | 1.7 × 108 A | 1.1 × 107 A | 2.2 × 107 A | 3.7 × 107 A | |
| 44 | 1.9 × 1010 A | 1.4 × 108 A | 2.2 × 107 AB | 1.1 × 108 A | 5.1 × 107 A | |
| 72 | 1.6 × 1010 A | 7.7 × 107 A | 2.9 × 107 B | 4.9 × 106 A | 1.2 × 108 A | |
| 129 | 4.2 × 1010 B | 1.1 × 108 A | 5.7 × 107 C | 3.1 × 107 A | 2.8 × 108 B | |
| September | 25 | 4.0 × 109 A | 1.9 × 108 A | 8.9 × 106 A | 5.6 × 107 A | 1.4 × 107 A |
| 40 | 6.2 × 109 A | 1.9 × 108 A | 1.0 × 107 A | 2.4 × 107 A | 5.3 × 107 A | |
| 44 | 9.8 × 109 A | 1.2 × 108 A | 1.4 × 107 A | 3.8 × 107 A | 2.3 × 107 A | |
| 72 | 1.1 × 1010 A | 1.2 × 108 A | 2.4 × 107 AB | 1.8 × 107 A | 7.1 × 107 A | |
| 129 | 2.1 × 1010 B | 1.1 × 108 A | 3.2 × 107 B | 7.6 × 107 A | 1.3 × 108 B | |
Suffix letters (A to C) indicate significant differences between successional stages.
Relative abundances of the different denitrification genes (narG, nirK, nirS, and nosZ).
Successional age may affect both the density of denitrifiers and their abundances relative to total bacteria. We therefore calculated ratios of denitrification genes to 16S rRNA from total eubacteria and obtained proportions of around 2%, 0.2%, 0.4%, and 0.5% for narG, nirK, nirS and nosZ, respectively (Fig. 2). The relative contributions of narG and nirS to the total eubacterial community were highest in the youngest site and decreased with ongoing soil development (Fig. 2). The relative abundances of nirK and nosZ showed no significant increase or decrease with soil age. Only a few sites had a lower relative abundance than the others: denitrifying nitrite reductase genes (nirK) in the 44-year-old site were less abundant than in the 25-year-old site in August. In addition, a lower relative abundance of nosZ was detected in the 25- and 44-year-old sites in September.
FIG. 2.
Relative densities of narG, nirK, nirS, and nosZ gene fragments in soils across a receding glacier foreland. Gray bars, August 2004; black bars, September 2004. Data of sampling sites not sharing a common capital letter are statistically different for September (P < 0.05); data of sampling sites not sharing a common lowercase letter are statistically different for August (n = 5; results are means and standard errors).
Soil environmental interrelations between denitrifier densities and environmental properties.
Multiple regression analysis, including Corg, pH, and NO3− as independent factors and copy numbers per gram of soil as dependent factors, was used to relate changes in denitrifier community densities to environmental properties. The increasing organic matter during soil development was positively related to DNA yields and to 16S rRNA gene copies, as well as to the three denitrification genes (narG, nirK, and nosZ) (Table 3). Multiple regression also revealed a significant influence of pH on narG. The nitrate content of soils could not explain the abundance of 16S rRNA and narG, nirK, nirS, and nosZ genes. None of the environmental properties analyzed could explain the abundance of nirS genes.
TABLE 3.
Multiple regression analysis including Corg, pH, and NO3 as independent factors
| Parameter | Standardized coefficient (β)a
|
Model P value | ||
|---|---|---|---|---|
| Corg | pH | NO3− | ||
| DNA yield | 0.881*** | 0.017 | −0.050 | 0.000 |
| 16S rRNA level | 0.827*** | −0.065 | 0.113 | 0.000 |
| narG level | 0.400* | 0.694*** | 0.023 | 0.005 |
| nirK level | 0.689*** | −0.272* | 0.000 | 0.000 |
| nirS level | 0.065 | 0.077 | 0.275 | 0.381 |
| nosZ level | 0.697*** | −0.244 | −0.149 | 0.000 |
Significance levels (P) of the multiple regression model are as follows: *, P < 0.05; ***, P < 0.001.
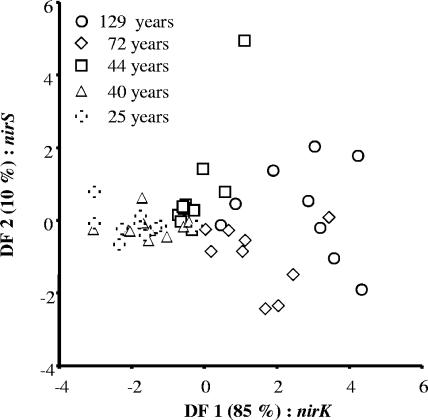
Discriminant analysis including all data (16S rRNA and the narG, nirK, nirS, and nosZ genes per gram of soil) was performed to test whether the density of the denitrifying community had changed along the chronosequence. Figure 3 shows that the successional age of the soil strongly affected the density of the denitrifying community. Along axis 1, the successional sites were significantly separated from each other according to age, except for the 25- and 40-year-old sites, which were grouped together. Along axis 2, the 72-year-old site differed significantly from the 44-year-old and 129-year-old sites. DF 3 and DF 4 did not significantly contribute to site differentiation. DF 1 already explained 85.2% of the variation within the data set; DF 2 contributed an additional 10.1% (Fig. 3; Table 4). The high standardized function coefficients of nirK and nosZ revealed that the densities of these two functional genes were mainly responsible for the separation along axis 1. nirS density was the most important property for the separation of the data along axis 2. In addition, the variability of replicates within each site differed depending on site age. Whereas the young sites grouped closely together, older sites were characterized by increased intrasite heterogeneity of their replicates.
FIG. 3.
Discriminant analysis of quantitative PCR data of gene copy numbers for 16S rRNA and denitrification genes (narG, nirK, nirS, and nosZ) from selected sites within the glacier foreland differing in successional age. The labels of the two axes give the percentage of explained variance as well as the most important factor responsible for the separation of the data along the corresponding axis.
TABLE 4.
Results of discriminant analyses of the 16S rRNA and the narG, nirK, nirS, and nosZ genes
| Parameter (unit) | Discriminant functiona
|
|||
|---|---|---|---|---|
| DF 1 | DF 2 | DF 3 | DF 4 | |
| Wilks' lambda | 0.147 | 0.617 | 0.849 | 0.994 |
| Eigenvalue | 3.184 | 0.377 | 0.171 | 0.006 |
| Degrees of freedom | 20 | 12 | 6 | 2 |
| Cumulative variance (%) | 85.2 | 95.3 | 99.8 | 100 |
| Canonical correlation coefficient | 0.87 | 0.52 | 0.38 | 0.08 |
| Correlation coefficientb | ||||
| 16S rRNA | 0.74** | 0.38* | 0.30* | 0.05 |
| narG | −0.54** | 0.11 | 0.62** | 0.56** |
| nirK | 0.81** | 0.04 | 0.22 | 0.36* |
| nirS | 0.07 | 0.70** | −0.13 | 0.68** |
| nosZ | 0.75** | −0.07 | 0.62** | 0.15 |
August and September measurements were pooled; 10 measurements were taken for each successional age. Significance in the model is accepted at a P value of ≤0.05. *, P < 0.05; **, P < 0.01.
Denotes the correlation between the discriminating variables and the scores of each discriminant function.
DISCUSSION
Quantification of 16S rRNA and denitrification genes by real-time PCR.
Quantitative PCR of denitrification genes encoding NO3− reductase, NO2−, and N2O reductase was used to study the ecology of denitrifiers across a glacier foreland. DNA extraction yielded slightly more DNA than previously reported for primary, developing soils (20). 16S rRNA copy numbers were in the range of 3.4 × 109 to 4.2 × 1010 copies g−1 of soil and were higher than reported for total counts of soil bacteria of 1.13 × 108 to 5.93 × 109 cells g−1 of dry soil determined by DAPI (4′,6′-diamidino-2-phenylindole) staining in the Rotfirn forefield (Göscherneralp, Switzerland) (20). Absolute values of 16S rRNA copy numbers, however, cannot yet be accurately compared with cell numbers because the numbers of 16S rRNA copies per genome (ranging between 1 and 13) are variable (5). The date of soil sampling did not significantly influence the 16S rRNA, narG, nirK, nirS, and nosZ copy numbers, suggesting that differences in microclimatic conditions or in the ecophysiology of plant communities (e.g., flowering stage in August versus senescence in September) did not affect microbial abundance.
For the denitrifying genes, only single copies per genome have been found so far. An exception is the narG gene, which can be present in up to three copies (14). Calculation of results for the target molecules of the three denitrification genes per nanogram of DNA revealed narG, nirK, and nosZ densities similar to previously reported values (6, 7, 12). Calculation of nirK gene copies per gram of soil also revealed densities of around 106 to 107 in our glacier samples, which are in the range of those reported by Henry et al. (7). Using an alternative approach based on competitive PCR, Qiu et al. (17) detected around 108 to 109 nirK gene copies per gram of soil in an A horizon from eastern Tennessee. This difference of 1 to 2 orders of magnitude between the two investigations may reflect differences in the nutritional statuses of the soils.
Expressing the copy numbers of denitrification genes as a percentage of 16S rRNA copy numbers, our study yields 0.5% to 5% narG copies (Fig. 2). Similar percentages of the unknown group of nitrate reducers targeted with our narG primers were found in a high-altitude soil from the Himalayas (7) as well as in some agricultural French soils (12). The relative abundances of nirK and nosZ were 0.2% and 0.5%, respectively, and were roughly in the same order previously found in turf grass rhizosphere and in agricultural soils (6, 7, 27, 28). The average level of nirS was 0.4%, suggesting that denitrifiers having the cd1 nitrite reductase are more abundant than copper nitrite reductase denitrifiers in the studied glacier foreland. These results are also in accordance with previous culture-based studies showing that the proportions of denitrifiers to total bacteria range from 0.1 to 5% (1, 23).
Successional pattern of denitrification genes across the glacier foreland.
The densities and relative abundances of different denitrification genes provide evidence that different denitrifier communities developed under primary succession (Fig. 2). The abundance of eubacteria and denitrifiers increased with the development of the newly formed soils (Table 1). Significant differences were recorded only between sites up to 72 years and the oldest site (129 years). Similar trends were found by Sigler and Zeyer (20), who described a DNA concentration ranging from 1.61 μg g−1 of dry soil in samples from debris on the glacial ice to 13.5 μm g−1 of dry soil 150 m away from the terminus of the Damma glacier. The significantly higher 16S rRNA gene copy numbers in the oldest sites were also supported by measurements of microbial biomass using the fumigation extraction method, total phospholipid fatty acids, and enzyme activities of soil samples at the same site (24, 25, 26). The present study showed that densities of nirK and nosZ followed this general trend by increasing during progressing soil development. In contrast, the densities of narG and nirS remained similar in all sites. A previous study reported that the activity of the nitrate reducer community increased significantly with successional age (2). The lack of agreement between the density and activity of the nitrate reducer community is not surprising, since (i) the density of a functional gene may not be linked to enzyme expression, (ii) nutrient availability may have contributed more to nitrate-reducing activities than to the density of the nitrate reducer community, and (iii) the unknown nitrate-reducing group targeted here represents only 10 to 20% of the total nitrate reducer community in this alpine ecosystem (2) and therefore contributes little to the overall activity. A previous study of the foreland of the Rotmoosferner glacier reported that soil development opened new environmental niches and especially carbon resources (25). Accordingly, the higher intrasite heterogeneity of denitrifying genes in the older sites than in the younger ones (Fig. 3) is probably due to an increase in microhabitat diversity with progressing soil development.
Relative abundances of denitrification genes in the glacier forefield.
Calculation of the relative abundances of different denitrification genes based on ratios of denitrification genes to 16S rRNA revealed higher percentages of narG copy numbers in the early stage of primary succession than in later stages of soil development (Fig. 2). The highly standardized function coefficient of the narG gene also revealed that copy numbers of the narG gene distinguished mainly the early versus late succession forefield denitrifying population (Fig. 3; Table 4). Plant growth is very limited in the early primary successional stage: plants cover only up to 5% of the soil surface and do not significantly change the activity and biomass of rhizosphere microorganisms (26). Therefore, the initial energy and nutrients for these microorganisms might have been provided not only by rhizodeposition but also by aerial and fluvioglacial depositions of organic detritus, as well as by residues from soil animals (8, 9, 10, 11). Data on the total carbon input of aerial deposition and transport from particulate organic matter from the melting ice into the ecosystem are currently lacking, but fluvial deposition of organic matter onto the soil surface of the young sites clearly took place. Differences in carbon acquisition could explain the higher competitiveness of the unknown nitrate-reducing group targeted in our study at the young site. This hypothesis is supported by the phospholipid fatty acid pattern of the rhizosphere flora of Poa alpina, where a low ratio of gram-positive to gram-negative bacteria in the young soils indicated preferential colonization by r strategists (25). Stress tolerance could also explain this higher competitiveness, as previously demonstrated by Sigler et al. (19) and Sigler and Zeyer (20) from studies of temperature and antibiotic resistance from successional soils at the forefield of the Dammaglacier (Switzerland). The relative abundances of narG and nirS decreased with soil development, while those of nirK and nosZ did not (Fig. 2). Multiple regression provided evidence that the amount of organic substances is the most important factor in the abundance of eubacteria, as well as for nirK and nosZ denitrifiers (Table 3). Surprisingly, no pH effect on nirS and nosZ was observed, while several studies have reported that pH strongly affected the denitrifying community in forested upland and wetland soils (16), as well as in agricultural soils (3).
In conclusion, quantification of the densities of denitrification genes encoding the NO3−, NO2−, and N2O reductases across a glacier foreland using quantitative PCR allowed us to characterize the primary succession of the denitrifying communities. Soil development influenced both the densities and relative abundances of 16S rRNA and denitrification genes in the glacier foreland. Thus, recorded densities of 16S rRNA and nirK and nosZ gene copies were higher in old than in young soils, while higher percentages of narG and nirS copies were observed in the early stage of primary succession. At this point, the consequences of such modifications of the densities and relative abundances of the denitrification genes for N2O and N2 fluxes in these ecosystems are still unclear. In the future, this question should be addressed by linking quantification of denitrification genes, analysis of the composition of the denitrifying community, and in situ measurement of denitrification rates.
Acknowledgments
We thank Meinhard Strobel for support at the alpine research center (Obergurgl, Ötz Valley) as well as Stéphanie Hallet for technical assistance with the laboratory analysis in Dijon.
REFERENCES
- 1.Chèneby, D., L. Philippot, A. Hartmann, C. F. Hènault, and J. C. Germon. 2000. 16S rRNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microbiol. Ecol. 34:121-128. [DOI] [PubMed] [Google Scholar]
- 2.Deiglmayr, K., L. Philippot, D. Tscherko, and E. Kandeler. Microbial succession of nitrate-reducing bacteria in the rhizosphere of Poa alpina across a glacier foreland in the Central Alps. Environ. Microbiol., in press. [DOI] [PubMed]
- 3.Enwall, K., L. Philippot, and S. Hallin. 2005. Activity and composition of the bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 71:8335-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erschbamer, B., W. Bitterlich, and C. Raffl. 1999. Die Vegetation als Indikator für die Bodenbildung im Gletschervorfeld des Rotmoosferners (Obergurgl, Ötztal, Nordtirol). Ber. Naturwiss.-Med. Verein Innsbruck 86:107-122. [Google Scholar]
- 5.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokariotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 6.Henry, S., E. Baudoin, J. C. López-Gutiérrez, F. Martin-Laurent, A. Brauman, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. (Erratum, 61:289-290.) [DOI] [PubMed] [Google Scholar]
- 7.Henry, S., D. Bru, B. Stres, S. Hallet, and L. Philippot. Quantitative detection of the nosZ gene encoding the nitrous oxide reductase and comparison of the abundance of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 8.Hodkinson, I. D., N. R. Web, and S. J. Coulson. 2002. Primary community assembly on land—the missing stages: why are the heterotrophic organisms always there first? J. Ecol. 90:569-577. [Google Scholar]
- 9.Kaufmann, R. 2001. Invertebrate succession on an alpine glacier foreland. Ecology 82:2261-2278. [Google Scholar]
- 10.Kaufmann, R. 2002. Glacier foreland colonisation: distinguishing between short-term and long-term effects of climate. Oecologia 130:470-475. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann, R., M. Fuchs, and N. Gosterxeier. 2002. The soil fauna of an alpine glacier foreland: colonization and succession. Arct. Alp. Res. 34:242-250. [Google Scholar]
- 12.López-Gutiérrez, J. C., S. Henry, S. Hallet, F. Martin-Laurent, G. Catroux, and L. Philippot. 2004. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57:399-407. [DOI] [PubMed] [Google Scholar]
- 13.Nicol, G. W., D. Tscherko, T. M. Embley, and J. I. Prosser. 2005. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ. Microbiol. 7:337-347. [DOI] [PubMed] [Google Scholar]
- 14.Philippot, L. 2002. Denitrifying genes in bacterial and Archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 15.Philippot, L., and O. Hojberg. 1999. Dissimilatory nitrate reductases in bacteria. Biochim. Biophys. Acta 1577:1-23. [DOI] [PubMed] [Google Scholar]
- 16.Priemé, A., G. Braker, and J. M. Tiedje. 2002. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl. Environ. Microbiol. 68:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu, X. Y., R. A. Hurt, L. Y. Wu, C. H. Chen, J. M. Tiedje, and J. Z. Zhou. 2004. Detection and quantification of copper-denitrifying bacteria by quantitative competitive PCR. J. Microbiol. Methods 59:199-210. [DOI] [PubMed] [Google Scholar]
- 18.Raffl, C., and B. Erschbamer. 2004. Comparative vegetation analysis of two transects crossing a characteristic glacier valley in the Central Alps. Phytocoenologia 34:225-240. [Google Scholar]
- 19.Sigler, W. V., S. Crivii, and J. Zeyer. 2002. Bacterial succession in glacial forefield soils characterized by community structure, activity and opportunistic growth dynamics. Microb. Ecol. 44:306-316. [DOI] [PubMed] [Google Scholar]
- 20.Sigler, W. V., and J. Zeyer. 2002. Microbial diversity and activity along the forefields of two receding glaciers. Microb. Ecol. 43:397-407. [DOI] [PubMed] [Google Scholar]
- 21.Sigler, W. V., and J. Zeyer. 2004. Colony-forming analysis of bacterial community succession in deglaciated soils indicates pioneer stress-tolerant opportunists. Microb. Ecol. 48:316-323. [DOI] [PubMed] [Google Scholar]
- 22.Throbäck, I. N., K. Enwall, A. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK and nos Z genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 23.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. Zehnder (ed.), Biology of anaerobic microorganisms. Wiley, New York, N.Y.
- 24.Tscherko, D., J. Rustemeier, A. Richter, W. Wanek, and E. Kandeler. 2003. Functional diversity of the soil microflora in primary succession across two glacier forelands in the Central Alps. Eur. J. Soil Sci. 54:685-696. [Google Scholar]
- 25.Tscherko, D., U. Hammesfahr, M.-C. Marx, and E. Kandeler. 2004. Shifts in rhizosphere microbial communities and enzyme activity of Poa alpina across an alpine chronosequence. Soil Biol. Biochem. 36:1685-1698. [Google Scholar]
- 26.Tscherko, D., U. Hammesfahr, G. Zeltner, E. Kandeler, and R. Böcker. 2005. Plant succession and rhizosphere microbial communities in a recently deglaciated alpine terrain. Basic Appl. Ecol. 6:367-383. [Google Scholar]
- 27.Wang, G., and H. D. Skipper. 2004. Identification of denitrifying rhizobacteria from bentgrass and bermudagrass golf greens. J. Appl. Microbiol. 97:827-837. [DOI] [PubMed] [Google Scholar]
- 28.Zumft, W. G. 1992. The denitrifying prokaryotes, p. 554-582. In A. Balowa, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 29.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]