Abstract
A real-time PCR-based method targeting the 18S rRNA gene was developed for the quantitative detection of Hartmannella vermiformis, a free-living amoeba which is a potential host for Legionella pneumophila in warm water systems and cooling towers. The detection specificity was validated using genomic DNA of the closely related amoeba Hartmannella abertawensis as a negative control and sequence analysis of amplified products from environmental samples. Real-time PCR detection of serially diluted DNA extracted from H. vermiformis was linear for microscopic cell counts between 1.14 × 10−1 and 1.14 × 104 cells per PCR. The genome of H. vermiformis harbors multiple copies of the 18S rRNA gene, and an average number (with standard error) of 1,330 ± 127 copies per cell was derived from real-time PCR calibration curves for cell suspensions and plasmid DNA. No significant differences were observed between the 18S rRNA gene copy numbers for trophozoites and cysts of strain ATCC 50237 or between the copy numbers for this strain and strain KWR-1. The developed method was applied to water samples (200 ml) collected from a variety of lakes and rivers serving as sources for drinking water production in The Netherlands. Detectable populations were found in 21 of the 28 samples, with concentrations ranging from 5 to 75 cells/liter. A high degree of similarity (≥98%) was observed between sequences of clones originating from the different surface waters and between these clones and the reference strains. Hence, H. vermiformis, which is highly similar to strains serving as hosts for L. pneumophila, is a common component of the microbial community in fresh surface water.
Hartmannella vermiformis, a free-living amoeba, is widespread in nature and has been isolated from soil, freshwater, air, and a variety of engineered water systems (27, 29, 37). Two distinct life cycle forms are known for H. vermiformis, viz. the trophozoite, an active feeding cell that also multiplies, and cysts, which are inactive dormant cells (28). H. vermiformis has direct and indirect public health significance. The organism has been isolated from the cerebrospinal fluid of a patient with meningoencephalitis and bronchopneumonia (7). Furthermore, it has been suggested that H. vermiformis can cause keratitis, although this is questioned by other investigators (9, 18). The indirect public health significance of the organism is related to its role as a host for Legionella pneumophila, the causative agent of Legionnaires' disease (36).
Traditionally, cultivation methods are applied for the detection of protozoa in aquatic habitats. Cultivated protozoa can subsequently be classified based upon their morphological characteristics and by using biochemical and immunological methods (31). To circumvent the need for cultivation, molecular tools for protozoan detection have been developed during the past decade, mainly using rRNA-targeted fluorescence in situ hybridization with oligonucleotide probes. Probes were designed for the specific detection of the Legionella growth-promoting free-living amoebae Acanthamoeba, Naegleria, and Hartmannella, allowing simultaneous detection and classification of amoebae in situ (15, 30). These techniques are time-consuming, and therefore PCR-based detection methods for the free-living amoebae Acanthamoeba and Naegleria have been developed (19, 35). For the detection of H. vermiformis, however, such a method is still lacking. We developed a real-time PCR-based method to enable investigations of the contribution of H. vermiformis to the occurrence, persistence, and proliferation of L. pneumophila in engineered water systems. The method targets a fragment of the 18S rRNA gene for the specific detection and quantification of H. vermiformis in environmental samples. Subsequently, the developed method was applied to surface water types serving as sources for the production of drinking water in The Netherlands as a first step in elucidating the distribution of H. vermiformis in such freshwater environments.
MATERIALS AND METHODS
Strains of Hartmannella and culture conditions.
Three Hartmannella strains were used in this study. H. vermiformis ATCC 50237 (strain CDC-19) was axenically cultivated in modified PYNFH medium at 30°C (12). Hartmannella abertawensis CCAP 1534/9, obtained from the UK National Culture Collection, was cultivated in MY75S medium without bacteriological agar at 30°C (3) but suspended with Escherichia coli. H. vermiformis KWR-1 (21) was cultivated in Prescott and James's medium including trace elements suspended with heat-killed E. coli at 30°C (3, 34).
Primer design for H. vermiformis.
Using the ARB software package (24), 18S rRNA gene-targeted forward (Hv1227F [5′-TTA CGA GGT CAG GAC ACT GT-3′]) and reverse (Hv1728R [5′-GAC CAT CCG GAG TTC TCG-3′]) primers were designed for the specific detection of H. vermiformis. The primers were checked against all available nucleic acid sequences in the NCBI GenBank database by using the BLAST search program (1). Furthermore, the number and positions of mismatches of the primers with the 18S rRNA gene sequences of the most closely related amoebae were assessed using ClustalW (8). For assessing the number and positions of mismatches of the developed primers with the closely related H. abertawensis, the 18S rRNA gene sequence of this amoeba was determined in this study, using PCR, cloning, and sequence conditions that were described previously (21).
Real-time PCR assay for H. vermiformis.
Real-time PCR assays were performed in 96-well plates in an I-Cycler iQ Multi-Color real-time PCR detection system (Bio-Rad, Veenendaal, The Netherlands) with a total reaction volume of 50 μl per well. Each reaction mix contained 25 μl iQ SYBR green supermix (Bio-Rad), 5 μl bovine serum albumin (4 mg/ml; Roche Diagnostics, Almere, The Netherlands), each primer at a concentration of 0.2 μM, and 10 μl template DNA. The thermal cycling conditions included predenaturation at 95°C for 3 min; 40 cycles of denaturation at 95°C for 20 s, annealing at 56°C for 30 s, and extension at 72°C for 40 s; and then a final extension step at 72°C for 10 min. The fluorescence intensity of SYBR green was measured automatically during the annealing steps. At the end of each run, a melting curve analysis was performed. Experiments were performed with undiluted and 10-fold diluted template DNA in duplicate.
For quantification, a cell-based calibration curve for H. vermiformis was included in each PCR assay. This cell-based calibration curve was constructed by preparing 10-fold serial dilutions of DNA extracted from a 1-week-old suspension of H. vermiformis ATCC 50237. The concentration of H. vermiformis in the suspension was determined by direct cell counting as described below. The concentration was 1.14 × 104 ± 2.79 × 103 cells per PCR for the undiluted DNA extract.
Genomic copy number of 18S rRNA gene in H. vermiformis.
To determine the 18S rRNA gene copy number in H. vermiformis, an internal fragment of this gene (from positions 1227 to 1728, according to Weekers et al. [39]) was cloned into a pGem-T Easy vector (Promega, Madison, Wis.). Recombinant plasmid DNA was purified in duplicate, using a Qiaprep spin miniprep kit (QIAGEN, Hilden, Germany). After purification, plasmid DNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, Del.). The number of construct copies in the plasmid solution was calculated, based on plasmid and insert sizes of 3,001 and 502 bp, respectively. A plasmid-based calibration curve was generated with 10-fold serial dilutions of plasmid containing the 18S rRNA gene fragment sequence of the target; to control pipetting steps, three 10-fold serial dilutions were prepared, and concentrations were checked by real-time PCR. This plasmid-based calibration curve, with a concentration of 1.25 × 107 gene copies per PCR for the dilution with the highest copy number, was used for determining the copy number of the 18S rRNA gene in H. vermiformis.
Enumeration of H. vermiformis cells.
The concentration and state (trophozoite or cyst) of H. vermiformis were determined in triplicate by filtering an appropriate volume of H. vermiformis suspension over a 1.2-μm-pore-size RTTP Isopore membrane (Millipore, Bedford, Mass.) in a vacuum not exceeding 0.3 × 105 Pa. The cells were stained with acridine orange as described by Hobbie et al. (17), and fluorescence was detected by use of a Leica DMRXA fluorescence microscope. A total of 100 random fields (with each field having a surface area of 9.604 × 10−3 mm2) per sample were analyzed.
DNA extraction.
Suspensions of H. vermiformis KWR-1, H. vermiformis ATCC 50237, and H. abertawensis and environmental surface water samples were filtered over a 1.2-μm-pore-size RTTP Isopore membrane (Millipore) in a vacuum not exceeding 0.3 × 105 Pa. DNA extraction of the filter-retained cells was done using a FastDNA spin kit for soil (BIO 101, Carlsbad, Calif.) following the instructions supplied by the manufacturer. The following three essential steps in the applied DNA extraction method can be distinguished: (i) concentration of the organism by filtration, (ii) cell disruption by bead beating, and (iii) DNA isolation and purification. The recovery efficiencies of these three different steps were determined for trophozoite and cyst suspensions as described in the supplemental material. The reproducibility of the DNA extraction methods was also tested and, likewise, is described in the supplemental material.
Influence of H. vermiformis growth phase on 18S rRNA gene copy number.
To evaluate the effect of life cycle stage on 18S rRNA gene copy number in H. vermiformis KWR-1 and H. vermiformis ATCC 50237, the two strains were cultivated in a biofilm batch model system (21, 33). Autoclaved Erlenmeyer flasks containing tap water with pieces of plasticized polyvinyl chloride, supplemented with nitrate (KNO3) and phosphate (KH2PO4) from separately autoclaved stock solutions at final concentrations of 59.3 μM and 11.0 μM, respectively, were used. The Erlenmeyer flasks for H. vermiformis KWR-1 were inoculated with a mixed microbial community originating from a hot water system in The Netherlands which was filtered over a 1.2-μm-pore-size cellulose nitrate filter (Sartorius, Goettingen, Germany) and with H. vermiformis KWR-1. The Erlenmeyer flasks for H. vermiformis ATCC 50237 were inoculated with a bacterial strain (Acidovorax sp. originating from a tap water system, cultivated on R2A medium and suspended in sterilized tap water) serving as a food source and with H. vermiformis ATCC 50237. The Erlenmeyer flasks were incubated under static conditions at 37°C for 21 days. The concentration and state of H. vermiformis cells were analyzed at different time points during the experiments from the biomass attached to plasticized polyvinyl chloride. The biofilm microbial community was removed from the material pieces by six 2-min sonication steps in 10 ml of sterilized water (Branson 5510; Bransonic Ultrasonic Cleaner, Danbury, CT) at a frequency of 40 kHz and an average power input of 0.015 W/ml.
H. vermiformis concentrations were determined by real-time PCR and direct cell counting. For the real-time PCR assay, both the cell-based calibration curve and the plasmid-based calibration curve were included. The state of the amoeba was determined by direct cell counting using an epifluorescence microscope, as described above.
Surface water sampling and DNA extraction.
A total of 28 samples were collected from different surface water types located all across The Netherlands. Most of these water types are used for drinking water production and include rivers (Meuse, Rhine, Waal, and IJssel), open reservoirs for water storage, (artificial) lakes, and a few ditches in national parks. The samples (1 liter) were collected at a depth between 30 and 100 cm, stored on ice, and analyzed within 24 h. Water samples (200 ml) were filtered over one to three 1.2-μm-pore-size RTTP Isopore membranes (Millipore), dependent on silting. DNA extraction of the filter-retained biomass was done using a FastDNA spin kit for soil (BIO 101).
Cloning and sequencing of PCR-amplified products.
PCR amplicons obtained by H. vermiformis-targeted real-time PCR were purified with a MinElute PCR purification kit according to the manufacturer's instructions (QIAGEN). PCR products were cloned into E. coli XL-1 Blue competent cells (Stratagene, Cedar Creek, Tex.) by using the Promega pGEM-T Easy vector system. PCR was performed on cell lysates of ampicillin-resistant transformants by using vector-specific primers T7 and Sp6 (Promega) to confirm the sizes of the inserts. Sequence analysis was done by BaseClear Lab Services (Leiden, The Netherlands).
Phylogenetic analysis.
The determined 18S rRNA gene sequences originating from the surface waters were aligned by using the ARB software package (24). Phylogenetic trees were constructed by different methods and by using different filters as implemented in the ARB software package.
Statistical analysis.
Microsoft Excel 2000 was used for determining average values, standard deviations, standard errors (SE), and regression lines. The standard deviations of the DNA extraction efficiencies were calculated using the equations given in the third edition of Introduction to the Theory of Statistics (26). Wilcoxon's two-sample test was used to determine the difference in the 18S rRNA gene copy numbers between H. vermiformis strains ATCC 50237 and KWR-1.
Nucleotide sequence accession numbers.
The environmental 18S rRNA gene sequences obtained in this study have been deposited in GenBank under accession numbers DQ190242 to DQ190273. The sequence of the H. abertawensis 18S rRNA gene has been deposited in GenBank under accession number DQ190241.
RESULTS
Specificity of PCR assay.
Using the newly designed primers Hv1227F and Hv1728R, specific amplification of a 502-bp fragment of the 18S rRNA gene of the freshwater amoeba H. vermiformis was observed, while no amplification was found for H. abertawensis. A BLAST search showed that the sequences of selected primers matched exactly and only with 18S rRNA gene sequences of H. vermiformis. Furthermore, it was found that the forward primer had eight, four, and seven mismatches and the reverse primer had six, eight, and six mismatches with the most closely related species according to Page (28), i.e., Glaeseria mira (11), Saccamoeba limax (2), and Hartmannella cantabrigiensis (11), respectively. A comparison with the sequence of H. abertawensis, which was obtained in this study, revealed three mismatches for the forward primer, while seven mismatches were found for the reverse primer.
Melting curve analysis was performed after each real-time PCR. This analysis showed that the melting temperature of the obtained amplicon was always 88.3°C (±0.6°C), and no other peaks were present in the melting curve, which implies a high specificity and no primer dimers. Finally, the primer specificity was tested in practice by amplification of DNAs extracted from different environmental surface waters, followed by sequencing and phylogenetic analysis of the obtained amplicons. All sequences showed the highest similarity (≥98%) with the 18S rRNA gene of H. vermiformis, indicating that the primers are indeed specific for H. vermiformis.
Cell-based calibration curve.
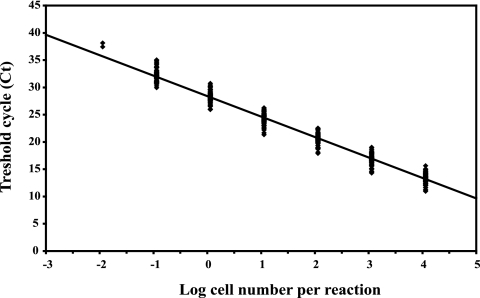
A cell-based calibration curve was constructed with 10-fold serial dilutions of DNA obtained from a 1-week-old culture of H. vermiformis ATCC 50237 (76% ± 19% trophozoites and 24% ± 6% cysts), with an initial concentration of 1.14 × 104 ± 2.79 × 103 cells per PCR for the undiluted DNA extract. Application of the real-time PCR assay with primers Hv1227F and Hv1728R always yielded a linear relationship between the cycle threshold and the log of the starting concentration (Fig. 1). Target DNA was detected at concentrations as low as the equivalent of 1.14 × 10−2 cells per 50 μl of reaction mixture (Fig. 1). The efficiency of each real-time PCR, calculated with the equation E = eln 10/−s − 1 (where E is the efficiency and s is the slope of the calibration curve), which was implemented in the software supplied with the I-Cycler iQ Multi-Color real-time PCR detection system (version 3.1), gave an average value of 84.3% ± 5.3% for the 51 reactions that were performed.
FIG. 1.
Calibration curve for real-time PCR results, using primers Hv1227F and Hv1728R and H. vermiformis ATCC 50237 grown in modified PYNFH medium. The indicated line, derived from 42 real-time PCRs, is defined by an average slope of −3.75 ± 0.18 and an intercept of 28.40 ± 0.95. For each concentration, all obtained data points are given. For log cell number −1.94, only two analyses were conducted.
Recovery efficiencies of the analytical procedure.
Application of filtration for retention of cells from a trophozoite suspension resulted in a 14.9% ± 8.6% (n = 2) loss of DNA. No loss (−0.4% ± 16.3%) was observed with a cyst suspension. Cell disruption by bead beating resulted in efficiencies of 99.6% ± 0.1% (n = 3) with the trophozoite cells and 96.0% ± 5.6% (n = 3) for the cyst suspension. These values are not significantly different. The DNA concentrations obtained by real-time PCR before and after a second DNA extraction were compared to determine the efficiency of the kit only, not taking into account the influence of filtration and cell disruption by bead beating. An average efficiency of 54.7% ± 11.7% was calculated from the different determinations, using dilution in DNA-free water to obtain a range of DNA concentrations (Table 1). However, the matrix may influence the DNA purification step, and therefore DNA was spiked, after filtration, into different matrices (surface water, PYNFH medium, and DNA-free water). The DNA recoveries in relation to that for the original spike ranged from 60.8% ± 9.0% to 75.4% ± 14.0% (Table 1). From the three different steps (filtration, bead beating, and the influence of the matrix), overall average DNA extraction efficiencies of 58.7% ± 7.2% and 66.7% ± 7.5% were established by extrapolating to a 100% trophozoite and a 100% cyst suspension, respectively. Since these values are not significantly different, an average efficiency of 62.7% ± 5.7% was used for further calculations.
TABLE 1.
Efficiency of DNA extraction by a FastDNA spin kit for soil, excluding filtration and cell disruption, at different dilutions in DNA-free water (n = 2) and in a few matrices (n = 3)
| Extraction parameter | DNA extraction efficiency (%)a |
|---|---|
| Dilutions in DNA-free water | |
| 100 | 48.8 ± 17.7 |
| 10−1 | 51.8 ± 16.2 |
| 10−2 | 54.5 ± 11.4 |
| 10−3 | 74.5 ± 24.2 |
| 10−4 | 44.0 ± 8.7 |
| Matrices used for dilution (10−1) | |
| Lake IJssel | 72.0 ± 13.9 |
| Lek Canal | 72.9 ± 14.6 |
| River Rhine | 75.4 ± 14.0 |
| Modified PYNFH medium | 60.8 ± 9.0 |
| DNA-free water | 65.0 ± 7.7 |
Values are expressed as average recovery percentages (± standard deviations) for spiked DNA.
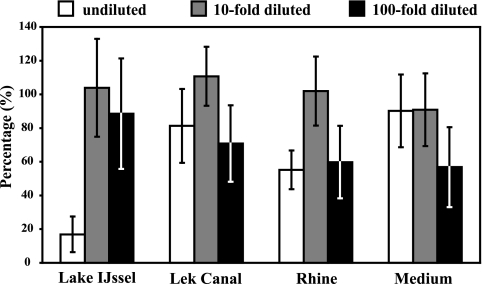
Samples of surface water and sterile PYNFH medium were spiked with H. vermiformis cells, grown as described above, to determine the effect of the matrix during the complete procedure. With this experiment, it was also possible to determine PCR inhibition and the effect of DNA dilution on the inhibitory compounds. The surface waters contained very low indigenous H. vermiformis concentrations (as determined by real-time PCR), as follows: River Rhine, 1.5 × 102 ± 5.8 × 101 cells/liter; Lek Canal, 6.1 × 101 ± 3.8 × 101 cells/liter; and Lake IJssel, 6.6 × 101 ± 2.5 × 101 cells/liter. These concentrations were 0.02% ± 0.008% of the spike concentration and therefore did not influence the calculations. Figure 2 shows the recovery percentages for spiked H. vermiformis cells in different matrices in relation to that for the original spike. Tenfold dilutions of DNA extract in PCR gave recovery rates of about 100%, indicating that a 10-fold dilution is sufficient for removing inhibitory compounds. This finding could be confirmed with the findings for the surface water samples; in 18% of the 28 samples, inhibition of PCR occurred in the undiluted DNA extract. When this inhibition occurred, a 100-fold diluted DNA extract was subsequently analyzed by real-time PCR. Template concentrations calculated from reactions with the 10- and 100-fold dilutions indicated that no inhibition occurred in the 10-fold dilution PCR, and 10-fold dilutions were subsequently used for calculations.
FIG. 2.
Recovery of H. vermiformis ATCC 50237 cells spiked into different types of surface water and into PYNFH growth medium. DNA isolations and real-time PCRs were all performed in duplicate. The indicated dilutions are DNA dilutions. Error bars indicate standard deviations.
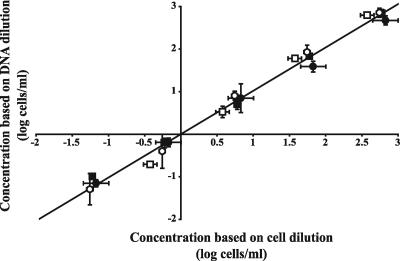
Information about the reproducibility of the applied method (filtering, DNA extraction, and real-time PCR) was obtained by using H. vermiformis KWR-1 cultivated in the biofilm batch model system. At different time points, the amoeba concentrations were determined with direct cell counts and by real-time PCR. The cell suspensions and the DNAs that were obtained at the different time points were 10-fold serially diluted (the cell dilutions were subsequently used for DNA extractions), resulting in cell and DNA dilutions. The obtained concentrations for the DNA dilutions were plotted against the concentrations of the diluted cell suspensions on days 7, 10, 14, and 20 (Fig. 3). Linear regression showed that the concentrations based on the DNA dilutions were equal to expected H. vermiformis concentrations based on the cell dilutions. Hence, the applied PCR method showed a high degree of linearity over a large range of H. vermiformis concentrations (−1 to 3 log cells/ml), and no changes were observed over time, indicating that the method is independent of initial cell concentration. Furthermore, the applied method is highly reproducible, since dilution series from different days showed similar linear regression.
FIG. 3.
Relationship (y = 1.018 [±0.026] x + 0.000 [±0.042]) between the concentrations of H. vermiformis KWR-1 cells in dilutions of cell suspensions, analyzed by direct microscopic cell counting, and the concentrations of cells in DNA dilutions, each of which was analyzed with real-time PCR. Samples were collected at different time points (t = 7 [□], 10 [▪], 14 [○], and 20 [•]) from a biofilm batch model system experiment. Error bars indicate standard deviations.
18S rRNA gene copy number in H. vermiformis.
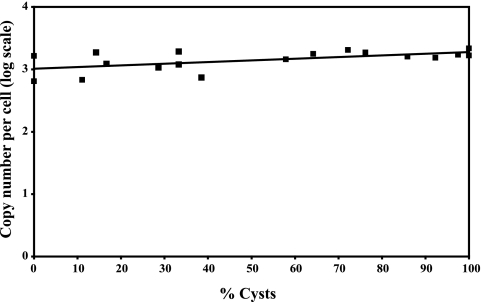
The 18S rRNA gene copy numbers of stage-mixed suspensions of H. vermiformis ATCC 50237 and H. vermiformis KWR-1, grown in the biofilm batch model system and in cultivation medium (only strain ATCC 50237), were determined. For this purpose, a plasmid-based calibration curve was generated, with the plasmid containing a target 18S rRNA gene fragment, and compared with the cell-based calibration curve. In these calculations, the previously determined efficiency of DNA extraction was taken into account. The number of copies of the 18S rRNA gene was not significantly affected by the state of the H. vermiformis ATCC 50237 cells (Fig. 4). An H. vermiformis KWR-1 suspension yielded a regression line with a negative slope; however, the slope of the regression line was also not significant (data not shown). Using the Wilcoxon test, no significant difference was found between the 18S rRNA gene copy numbers for H. vermiformis strains ATCC 50237 and KWR-1 (P ≥ 0.95). The real-time PCR results for a total of 18 cell-based calibration curves for strain H. vermiformis ATCC 50237 and plasmid-based calibration curves generated simultaneously were used to estimate the copy number of the 18S rRNA gene. An average number of 1,330 ± 127 (mean ± SE) copies per cell was derived from these data, taking into account the observed DNA extraction efficiency.
FIG. 4.
18S rRNA gene copy numbers in H. vermiformis ATCC 50237 in relation to maturation state (% cysts). An average DNA extraction efficiency of 62.7% was used in the calculations. The equation of the indicated trend line is y = 0.0026x + 3.0114, and R2 = 0.3314.
Distribution and genetic diversity of H. vermiformis in surface waters in The Netherlands.
H. vermiformis was observed in 75% of the 28 surface water samples collected in autumn (water temperature, 11.7 ± 1.3°C). The concentrations ranged from 5.0 (detection limit) to 75 cells/liter, with a median of 9.6 cells/liter. PCR amplicons originating from eight different surface waters, including drinking water sources, were subjected to cloning and sequence analysis. The sequences (502/503 bp) of 32 clones were compared by phylogenetic analysis with the sequences of different H. vermiformis strains (Fig. 5). A high degree of sequence similarity (≥98%) was obtained for H. vermiformis strains originating from different surface waters in The Netherlands and strains originating from Europe and Northern America. However, despite low bootstrap values, three subgroups could be distinguished (A, B, and C) (Fig. 5).
FIG. 5.
Phylogenetic tree for different H. vermiformis sequences (502/503 bp) amplified with the specific primers described in this study. Trees were calculated using different methods, as implemented in the ARB software package (24), and yielded the same topology as that of the neighbor-joining tree shown in the figure. However, the bootstrap values of the tree were low. The overall tree is shown at the left, with Echinamoeba exundans (2) as the outgroup. The bar within the box indicates 10% sequence divergence. The rest of the figure provides an expanded view of the three subgroups, with the bar at the top of the figure indicating 10% sequence divergence. The numbers in parentheses indicate the numbers of clones. Strains Koblenz (X75514), Nijmegen (X75515), and Atlanta (X75513) were previously sequenced by Weekers et al. (39); strain Balamuth (ATCC 30966; M95168) was sequenced by Gunderson et al. (16); strain C3/8 (AF426157) was sequenced by Walochnik et al. (38); strains JK-1 and ATCC 50256 were sequenced by Kuchta et al. (20); and strains KWR-1, -2, and -3 were sequenced by Kuiper et al. (21).
DISCUSSION
Primer design.
Until now, laborious and time-consuming methods have been used for the detection and quantification of H. vermiformis, which is a potential host for L. pneumophila. In this study, a method for the quantitative detection of H. vermiformis by 18S rRNA gene-targeted real-time PCR was developed. The 18S rRNA gene is repeated in tandem in high copy numbers and is highly conserved (23), thus potentially providing a high detection sensitivity. Theoretically, a single-copy gene, for example, the actin-encoding gene, may provide a better target for quantification, but sequences of this gene were not available for H. vermiformis. On the other hand, extensive application of rRNA as the prime marker for microbial detection and identification in ecological and diagnostic studies has resulted in large sequence databases, including at least 18 entries of 18S rRNA gene sequences for H. vermiformis strains, providing a sound basis for the design of specific primers. Hence, estimation of the potential copy number variation with life cycle stage was needed to obtain an accurate method. Furthermore, the presence of highly variable segments within the 18S rRNA gene fragment amplified by the primers used in this study allows further identification of H. vermiformis microdiversity by cloning and sequence analysis of amplicons retrieved from environmental samples.
PCR efficiency, reproducibility, and inhibition.
The developed real-time PCR assay showed a high PCR efficiency (84.3%) and a high reproducibility (100%). Furthermore, the method is highly sensitive, as evidenced by the detection of concentrations of 18S rRNA gene fragments corresponding to less than a single cell (Fig. 1). Complex matrices such as surface waters may contain organic and inorganic compounds which interfere with several steps in the isolation and amplification protocols, including cell lysis and polymerase activity during the amplification of target DNA. Such compounds may even degrade the DNA (41). The FastDNA spin kit for soil used for DNA extraction in this study was able to eliminate most of the inhibitory compounds and is therefore recommended for further applications.
DNA extraction efficiency.
The real-time PCR assay developed in this study was used for the detection and quantification of H. vermiformis in environmental samples. However, to be quantitative, the method requires reproducible and efficient recovery at all stages of the analytical procedure. The efficiency of the entire procedure (62.7% ± 5.7%) is lower than the 92 to 96% extraction efficiency reported for bacterioplankton in seawater, using another DNA extraction method (4). Nevertheless, the efficiency obtained in this study is high compared with the 20% to 61% efficiencies obtained in other studies (13, 40).
Copy number of 18S rRNA gene.
In most eukaryotes, rRNA genes are organized in tandemly repeated units (23). Furthermore, it is known that the DNA content can vary substantially from species to species but also between growth phases. For example, the dinoflagellate Alexandrium minutum is generally haploid (n), with only the resting cysts (planozygotes) being diploid (2n) (14). Therefore, it has been suggested that the copy number of the 18S rRNA gene might vary during the growth cycle of protozoa (5). Our results with H. vermiformis strains ATCC 50237 and KWR-1 indicate that the copy number of the 18S rRNA gene in cysts did not differ significantly from that in trophozoites. The estimated copy number for the 18S rRNA gene on the genome of H. vermiformis (1,330 ± 127 [mean ± SE] copies/cell) exceeds the values reported for other unicellular eukaryotes (14, 22) but is well within the range of about 50 to 10,000 found for other eukaryotic cells (23).
H. vermiformis in surface water.
H. vermiformis was observed in 75% of samples (200 ml) from freshwater environments used for water supply and cooling purposes in The Netherlands. In contrast, only 2% of 330 samples (50 ml) collected from the James River in Virginia were Hartmannella positive, and the organism was mainly found in samples collected in late summer and autumn (10). For the present study, all samples were taken in late autumn (water temperature, 11.7°C ± 1.3°C). The difference in detection rate can be attributed partly to the high sensitivity of the method used in this study, with a detection limit of 5 cells/liter. The amoebae of the genus Hartmannella observed in the James River were more associated with the sediment than with the water column (10). Hence, analysis of water samples only, as done in our survey, may give an underestimation of positive sites. H. vermiformis has also frequently been found in samples collected from natural waters (rivers, 36.4%; lakes, 16.7%) and from man-made environments (artificial lake, 20%; swimming pools, 6.3%) in Bulgaria (32) and in water at different stages of water treatment in Germany (25). Furthermore, H. vermiformis has been observed as the predominant amoeba in warm tap water (6, 29). These reports and our observations indicate that this amoeba is a common component of natural freshwater environments and water installations. The majority of the surface water types included in this study serve as sources for drinking water production in The Netherlands. Cysts may survive and proliferate during different steps of water treatment, may enter the distributing system, and subsequently may multiply in biofilms attached to the pipe walls.
The high degree (≥98%) of sequence similarity between H. vermiformis strains originating from different surface waters in The Netherlands and the reference strains indicates that the observed organisms may be potential hosts for Legionella. Our phylogenetic analysis did not support the previously suggested separation of European strains from strains originating from North America (20, 39). This supports other findings obtained when the whole 18S rRNA gene was taken into account (21, 38). Despite the high level of sequence similarity, three subgroups of H. vermiformis could be distinguished. Elucidation of the ecophysiological characteristics of these different subgroups requires further research.
Supplementary Material
Acknowledgments
The Water Supply Companies in The Netherlands financed this study in the framework of the Joint Research Program.
We thank Johannes Hackstein, Marcel Zwietering, and Rijkelt Beumer for valuable discussions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral Zettler, L. A., T. A. Nerad, C. J. O'Kelly, M. T. Peglar, P. M. Gillevet, J. D. Silberman, and M. L. Sogin. 2000. A molecular reassessment of the leptomyxid amoebae. Protist 151:275-282. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 2001. Catalogue of the UK National Culture Collection (UKNCC). List of algae and protozoa, 1st ed. UK National Culture Collection, Ambleside, United Kingdom.
- 4.Boström, K. H., K. Simu, Å. Hagström, and L. Riemann. 2004. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol. Oceanogr. Methods 2:365-373. [Google Scholar]
- 5.Bowers, H. A., T. Tengs, H. B. Glasgow, Jr., J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breiman, R. F., B. S. Fields, G. N. Sanden, L. Volmer, A. Meier, and J. S. Spika. 1990. Association of shower use with Legionnaires' disease. Possible role of amoebae. JAMA 263:2924-2926. [PubMed] [Google Scholar]
- 7.Centeno, M., F. Rivera, L. Cerva, V. Tsutsumi, E. Gallegos, A. Calderon, R. Ortiz, P. Bonilla, E. Ramirez, and G. Suarez. 1996. Hartmannella vermiformis isolated from the cerebrospinal fluid of a young male patient with meningoencephalitis and bronchopneumonia. Arch. Med. Res. 27:579-586. [PubMed] [Google Scholar]
- 8.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Jonckheere, J. F., and S. Brown. 1998. There is no evidence that the free-living ameba Hartmannella is a human parasite. Clin. Infect. Dis. 26:773. [DOI] [PubMed] [Google Scholar]
- 10.Ettinger, M. R., S. R. Webb, S. A. Harris, S. P. McIninch, G. C. Garman, and B. L. Brown. 2003. Distribution of free-living amoebae in James River, Virginia, USA. Parasitol. Res. 89:6-15. [DOI] [PubMed] [Google Scholar]
- 11.Fahrni, J. F., I. Bolivar, C. Berney, E. Nassonova, A. Smirnov, and J. Pawlowski. 2003. Phylogeny of lobose amoebae based on actin and small-subunit ribosomal RNA genes. Mol. Biol. Evol. 20:1881-1886. [DOI] [PubMed] [Google Scholar]
- 12.Fields, B. S., T. A. Nerad, T. K. Sawyer, H. King, J. M. Barbaree, W. T. Martin, W. E. Morrill, and G. N. Sanden. 1990. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J. Protozool. 37:581-583. [DOI] [PubMed] [Google Scholar]
- 13.Fuhrman, J. A., D. E. Comeau, Å. Hagström, and A. M. Chan. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garces, and M. Magnani. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm, D., W. Ludwig, B. C. Brandt, R. Michel, K.-H. Schleifer, J. Hacker, and M. Steinert. 2001. Development of 18S rRNA-targeted oligonucleotide probes for specific detection of Hartmannella and Naegleria in Legionella-positive environmental samples. Syst. Appl. Microbiol. 24:76-82. [DOI] [PubMed] [Google Scholar]
- 16.Gunderson, J. H., S. J. Goss, and M. L. Sogin. 1994. The sequence of the Hartmannella vermiformis small subunit rRNA coding region. J. Eukaryot. Microbiol. 41:481-482. [DOI] [PubMed] [Google Scholar]
- 17.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy, S. M., P. Devine, C. Hurley, Y.-S. Ooi, and L. M. T. Collum. 1995. Corneal infection with Hartmannella vermiformis in contact-lens wearer. Lancet 346:637-638. [PubMed] [Google Scholar]
- 19.Kilvington, S., and J. Beeching. 1995. Development of a PCR for identification of Naegleria fowleri from the environment. Appl. Environ. Microbiol. 61:3764-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuchta, J. M., S. J. States, R. M. Wadowsky, and T. J. Byers. 1998. Interactions of Legionella pneumophila with Hartmannella vermiformis including the efficacy of chlorine or copper and silver ions to disrupt the intra-amoebic multiplication of L. pneumophila. Recent Results Dev. Microbiol. 2:405-425. [Google Scholar]
- 21.Kuiper, M. W., B. A. Wullings, A. D. L. Akkermans, R. R. Beumer, and D. van der Kooij. 2004. Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl. Environ. Microbiol. 70:6826-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Blancq, S. M., N. V. Khramtsov, F. Zamani, S. J. Upton, and T. W. Wu. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 23.Long, E. O., and I. B. Dawid. 1980. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49:727-764. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel, R., R. Hoffman, A. Giese, and K.-D. Muller. 1995. Untersuchung von drei Grundwasserwerken auf Vorkommen von Acanthamoeben, Naeglerien und anderen freilebenden Amoben. Acta Hydrochim. Hydrobiol. 23:202-211. [Google Scholar]
- 26.Mood, A. M., F. A. Graybill, and D. C. Boes. 1974. Introduction to the theory of statistics, 3rd ed. McGraw-Hill, New York, N.Y.
- 27.Page, F. C. 1974. A further study of taxonomic criteria for limax amoebae with descriptions of new species and a key to genera. Arch. Protistenkd. 116:149-184. [Google Scholar]
- 28.Page, F. C. 1988. A new key to freshwater and soil gymnamoebae. Freshwater Biological Association, Ambleside, United Kingdom.
- 29.Rohr, U., S. Weber, R. Michel, F. Selenka, and M. Wilhelm. 1998. Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl. Environ. Microbiol. 64:1822-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stothard, D. R., J. Hay, J. M. Schroeder-Diedrich, D. V. Seal, and T. J. Byers. 1999. Fluorescent oligonucleotide probes for clinical and environmental detection of Acanthamoeba and the T4 18S rRNA gene sequence type. J. Clin. Microbiol. 37:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szénasi, Z., T. Endo, K. Yagita, and E. Nagy. 1998. Isolation, identification and increasing importance of ‘free-living’ amoebae causing human disease. J. Med. Microbiol. 47:5-16. [DOI] [PubMed] [Google Scholar]
- 32.Tsvetkova, N., M. Schild, S. Panaiotov, R. Kurdova-Mintcheva, B. Gottstein, J. Walochnik, H. Aspöck, M. Lucas, and N. Müller. 2004. The identification of free-living environmental isolates of amoebae from Bulgaria. Parasitol. Res. 92:405-413. [DOI] [PubMed] [Google Scholar]
- 33.Van der Kooij, D., H. R. Veenendaal, N. P. G. Slaats, and D. Vonk. 2002. Biofilm formation and multiplication of Legionella on synthetic pipe materials in contact with treated water under static and dynamic conditions, p. 460. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. ASM Press, Washington, D.C.
- 34.Vishniac, W., and M. Santer. 1957. The thiobacilli. Bacteriol. Rev. 21:195-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vodkin, M. H., D. K. Howe, G. Visvesvara, and G. McLaughlin. 1992. Identification of Acanthamoeba at the generic and specific levels using the polymerase chain reaction. J. Protozool. 39:378-385. [DOI] [PubMed] [Google Scholar]
- 36.Wadowsky, R. M., L. J. Butler, M. K. Cook, S. M. Verma, M. A. Paul, B. S. Fields, G. Keleti, J. L. Sykora, and R. B. Yee. 1988. Growth-supporting activity for Legionella pneumophila in tap water cultures and implication of hartmannellid amoebae as growth factors. Appl. Environ. Microbiol. 54:2677-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, P. L., P. Provic, W. G. Gardiner, and D. E. Moorhouse. 1986. Isolation of free-living amoebae from air samples and an air-conditioner filter in Brisbane. Med. J. Aust. 145:175. [DOI] [PubMed] [Google Scholar]
- 38.Walochnik, J., R. Michel, and H. Aspock. 2002. Discrepancy between morphological and molecular biological characters in a strain of Hartmannella vermiformis Page 1967 (Lobosea, Gymnamoebia). Protistology 2:185-188. [Google Scholar]
- 39.Weekers, P. H. H., R. J. Gast, P. A. Fuerst, and T. J. Byers. 1994. Sequence variations in small-subunit ribosomal RNAs of Hartmannella vermiformis and their phylogenetic implications. Mol. Biol. Evol. 11:684-690. [DOI] [PubMed] [Google Scholar]
- 40.Weinbauer, M. G., I. Fritz, D. F. Wenderoth, and M. G. Hofle. 2002. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.