Abstract
The potential for humic substances to stimulate the reduction of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) was investigated. This study describes a novel approach for the remediation of RDX-contaminated environments using microbially mediated electron shuttling. Incubations without cells demonstrated that reduced AQDS transfers electrons directly to RDX, which was reduced without significant accumulation of the nitroso intermediates. Three times as much reduced AQDS (molar basis) was needed to completely reduce RDX. The rate and extent of RDX reduction differed greatly among electron shuttle/acceptor amendments for resting cell suspensions of Geobacter metallireducens and G. sulfurreducens with acetate as the sole electron donor. AQDS and purified humic substances stimulated the fastest rate of RDX reduction. The nitroso metabolites did not significantly accumulate in the presence of AQDS or humic substances. RDX reduction in the presence of poorly crystalline Fe(III) was relatively slow and metabolites transiently accumulated. However, adding humic substances or AQDS to Fe(III)-containing incubations increased the reduction rates. Cells of G. metallireducens alone reduced RDX; however, the rate of RDX reduction was slow relative to AQDS-amended incubations. These data suggest that extracellular electron shuttle-mediated RDX transformation is not organism specific but rather is catalyzed by multiple Fe(III)- and humic-reducing species. Electron shuttle-mediated RDX reduction may eventually become a rapid and effective cleanup strategy in both Fe(III)-rich and Fe(III)-poor environments.
Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) is a widely used explosive, and it is recognized as a contaminant of concern at numerous locations, including ammunition depots, production facilities, and live-fire training installations (18, 51). RDX is moderately soluble at low concentrations and can migrate from soil to aquifer material and groundwater. Groundwater contamination, particularly drinking water supply aquifers, with residues of RDX has become a significant problem because RDX is a possible human carcinogen (the lifetime health advisory for exposure to RDX in drinking water is 2 μg/liter) (55). RDX contamination is a significant issue for the U.S. Department of Defense. A recent report from the Massachusetts Military Reservation located in Sandwich, Mass., indicated that RDX was identified in a plume moving offsite at a mean concentration of 290 μg/liter (highest concentration of 370 μg/liter), which is more than 100 times the maximum contaminant level (57).
Few cost-effective technologies exist for the treatment of RDX compounds in groundwater. Pump-and-treat systems are typically inefficient for the remediation of groundwater plumes, largely because they do not address the source of contamination and because of the large volumes of groundwater that must be treated for hydraulic control and regulatory compliance (1). In addition, these strategies merely remove the groundwater and transfer the contaminants to another medium rather than degrading the contaminants. Permeable reactive barriers using zero valent iron show promise in reducing and/or treating nitramine compounds, but the installation of permeable reactive barriers is known to be technically and economically infeasible at sites with deep or wide plumes (53).
RDX has a cyclic, nitrogen-containing molecular structure, and it is moderately resistant to aerobic biodegradation (4, 55). RDX will biodegrade aerobically in the presence of specific microorganisms (14), but specialized enrichment conditions are required for these organisms to proliferate (4), and several reports suggest limited degradation kinetics (19, 60). Aerobic bioremediation is possible in shallow soil or groundwater that has sufficient oxygen but is technically difficult in groundwater that has become anaerobic. Environments with RDX contamination may be cocontaminated with compounds that promote anaerobic conditions.
Most anaerobic strategies for RDX to date have focused on direct microbial reduction of the nitro functional groups on the cyclic structure as the sole terminal electron acceptor (65). This strategy is effective only when specific microorganisms that respire nitramine compounds are widespread in subsurface systems. In the absence of these microorganisms the reactions may be slow, limiting this strategy in many environments.
Extracellular electron shuttling may be one approach for cyclic nitramines. Electron shuttle-mediated contaminant transformation has been demonstrated for the BTEX compounds (11, 39), methyl tert butyl ether (11, 12), carbon tetrachloride (9), and metals such as uranium (12, 15, 25, 26, 30). The role of electron shuttling in environmental reactions was reviewed by Hernandez and Newman (24). To date, the electron shuttle-mediated degradation of cyclic nitramines, including RDX, has not been reported.
Extracellular electron shuttling encompasses all reactions that are catalyzed by microbial reduction of the shuttles, whether it is direct interaction with the reduced shuttle or with Fe(II) resulting from the reaction. Fe(III)- and humic substance (HS)-reducing microorganisms have been identified in shallow and deep aquifer material, freshwater (35) and marine sediment (22), soil (7), and extreme environments such as hot springs and volcanic sediment (27). A few recent reports (17, 59) suggest that RDX was transformed by reactive Fe(II), the product of Fe(III) respiration. The ubiquity of Fe(III)- and HS-reducing microorganisms increases the likelihood that remediation strategies predicated on their physiology will be successful in many subsurface environments (6).
In the present study, indirect or direct electron transfer to RDX via reduced purified HS, reduced anthrquinone-2,6-disulfonate (AQDS), and Fe(II) was investigated. Understanding electron transfer mechanisms via an electron shuttle to RDX (versus direct electron transfer from microbial respiration) is important for establishing strategies for groundwater bioremediation. The objectives of the present study were (i) to verify the specific mechanisms for RDX reduction by Fe(III) reducing microorganisms and HS and (ii) to determine whether stimulating Fe(III) and HS reduction increases the rate and extent of RDX biodegradation. In addition, the present study was designed to determine whether HS-mediated RDX transformation is microorganism-specific or a general phenomenon among different HS-reducing species.
MATERIALS AND METHODS
Chemicals.
RDX (ca. 97% pure) was provided by the U.S. Army Corps of Engineers, Construction Engineering Research Laboratory, Champaign, IL. Hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine (MNX; 99%), hexahydro-1,3-dinitroso-5-nitro-1,3,5-triazine (DNX; 58% pure with 34% MNX and 8% hexahydro-1,3,5-trinitroso-1,3,5-triazine [TNX]), and TNX (>99.9%) were purchased from SRI International (Menlo Park, Calif.). Purified humic acid and AQDS were purchased from Sigma Aldrich (Milwaukee, WI). High-pressure liquid chromatography (HPLC)-grade methanol was purchased from Aldrich Chemicals. All other chemicals used were of reagent-grade quality or higher.
Microorganisms.
Geobacter metallireducens strain GS-15 (ATCC 53774) and Geobacter sulfurreducens strain PCA (ATCC 51573) were originally obtained from the University of Massachusetts at Amherst and maintained using the ferric citrate and AQDS media described below.
Medium and culturing conditions.
The basal medium consisted of (in g liter−1 unless specified otherwise): NaHCO3 (2.5), NH4Cl (0.25), NaH2PO4 · H2O (0.6), KCl (0.1), modified Wolfe's vitamin and mineral mixtures (each 10 ml liter−1), and 1 ml of 1 mM Na2SeO4. Electron acceptors used with the medium included soluble Fe(III) citrate (45 mM), poorly crystalline Fe(III) (hydr)oxide (50 mmol/liter), or AQDS (5 mM). All cultures were maintained as previously described (10).
Pure phase incubations.
Chemically reduced AQDS (C-AH2QDS) was prepared by sparging the medium bottle with H2-CO2 (80:20 [vol/vol]) in the presence of palladium-covered alumina pellets as previously described (33). The chemically reduced AQDS was filtered through a 0.2-μm-pore-size sterilized polytetrafluoroethylene (PTFE) filter into a presterilized, anaerobic serum bottle. Biologically reduced AQDS (B-AH2QDS) was prepared by incubating Geobacter metallireducens in AQDS medium (5 mM). The B-AH2QDS was filtered through a 0.2-μm-pore-size-sterilized PTFE filter into a presterilized, anaerobic serum bottle to remove cells. Biologically reduced Fe(II) was prepared in a similar manner using 45 mM Fe(III) citrate medium in lieu of AQDS medium.
The concentrations of reduced AQDS (chemical or biological AH2QDS) tested were 500, 300, and 100 μM for the incubations with RDX; 300, 100, and 50 μM for the incubations with MNX; and 200, 100, and 50 μM for the incubations with DNX. Incubations were performed at 25°C. The starting RDX concentration was either 100 or 50 μM, depending on the specific experimental conditions.
Experiments were initiated by injecting RDX into the buffer with AH2QDS. Samples were collected periodically via anaerobic syringe and needle; samples were filtered through sterile, 0.2-μm-pore-size PTFE filters prior to analyses. RDX was quantified at each time point. The AH2QDS concentration was quantified for the AH2QDS or AQDS incubations, and Fe(II) was quantified for the ferrous iron incubations.
Resting cell suspension incubations.
Cell cultures (G. metallireducens or G. sulfurreducens) were grown in freshwater medium with acetate as the sole electron donor and Fe(III) citrate as the sole terminal electron acceptor. One liter of cell culture was harvested during the logarithmic growth phase and centrifuged (at 5,000 rpm for 15 min) to form a dense cell pellet, and cell suspensions were prepared as previously described (42).
Electron acceptors were amended from concentrated, anaerobic, sterile stock solutions. Electron acceptors incubated with the cells included humic acids (0.25g/liter), poorly crystalline Fe(III) oxide (45 mmol/liter), AQDS (5 mM), humic acids plus poorly crystalline Fe(III) oxide, and AQDS plus poorly crystalline Fe(III) oxide. Cells were incubated at 30°C. Acetate was added at a final concentration of 20 mM to each suspension as the sole electron donor. An aliquot (0.3 ml) of the resting cells was added to the sealed pressure tubes to initiate each experiment.
Samples were collected periodically via anaerobic syringe and needle. RDX was quantified at each time point. The AH2QDS concentration was quantified for AQDS incubations and Fe(II) was quantified for ferrous iron incubations. The total cellular protein was determined by using a DC protein kit (Bio-Rad) and a modified Lowry protein assay (40). Cell protein normalized decay rates were calculated based on pseudo first-order degradation coefficients.
Analytical techniques.
Aqueous samples from each experiment were filtered through a 0.2-μm-pore-size sterile PTFE membrane (PALL Life Science) prior to analysis. RDX and its metabolites MNX, DNX, and TNX were analyzed by using HPLC with a variable wavelength photodiode array detector (HPLC/UV; Dionex) at 254 nm as described previously (13, 59). The filtered samples were manually injected into a Supelcosil LC-CN column (25 cm by 4.6 mm, 5-μm inner diameter) at an ambient temperature. A mobile phase consisting of 50% water and 50% methanol was used at a flow rate of 1 ml/min. RDX, MNX, DNX, and TNX were compared to certified analytical standards at known concentrations in acetonitrile. The concentration of reduced AQDS was determined spectrophotometrically at 450 nm versus standards of chemically reduced AH2QDS as previously described (31). Aqueous Fe(II) and total solid-phase iron concentrations were quantified by the Ferrozine assay as described previously (36).
RESULTS
RDX reduction by biologically and chemically reduced AH2QDS.
In order to evaluate whether reduced extracellular electron shuttles transfer electrons to the cyclic nitramine RDX, the HS analog AQDS was biologically and chemically reduced and then incubated in cell-free, anaerobic mixtures with RDX. AQDS was used in lieu of purified HS because it is a defined molecule (molecular mass of 366.32 g/mol) that has been well characterized with respect to its electron-shuttling capacity. In addition, its color changes from opaque pink to vivid orange as it changes from oxidized to reduced state; therefore, it can be quantified spectrophotometrically at 450 nm. Two different concentrations of chemically and biologically reduced AQDS were amended to 50 μM RDX to determine whether the reduction rate of RDX was different with the reduced AQDS produced by different methods. The quantified reduction rate of RDX between chemically and biologically reduced AQDS was not significantly different; therefore, subsequent pure-phase incubations were performed with only biologically reduced AQDS.
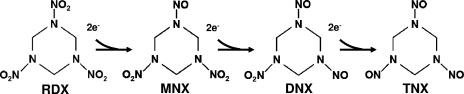
RDX is reduced by a series of stepwise two-electron transfers generating nitroso intermediates (Fig. 1). The stoichiometry of electron transfer between reduced AQDS and RDX was investigated with three different concentrations of microbially reduced AQDS (Fig. 2). RDX at 100 μM was completely transformed within 9 h in the incubations when amended with 500 μM and 300 μM B-AH2QDS. However, RDX was reduced to ca. 61 μM with 100 μM B-AH2QDS (Fig. 2A). This result was within range of the expected stoichiometry for oxidation of AH2QDS coupled to RDX reduction (with TNX as the end product of the reductive pathway). Approximately one-third of the 100 μM RDX was reduced when one-third of the stoichiometric equivalent of AH2QDS was provided, suggesting that RDX was reduced to TNX, with TNX removed by an unidentified reaction pathway. Reduced AQDS transfers two electrons per mole in the coupled oxidation/reduction reaction, and RDX accepts six electrons per mole (to TNX). The half reactions and the full reaction are as follows: C3H6N6O6 (RDX) + 6H+ + 6e− → C3H6N6O3 (TNX) + 3H2O (reduction half reaction); AH2QDS → AQDS + 2H+ + 2e− (oxidation half reaction); and C3H6N6O6 (RDX) + 3AH2QDS → C3H6N6O3 (TNX) + 3AQDS + 3H2O (full reaction).
FIG. 1.
Structure of RDX and its nitroso metabolites MNX, DNX, and TNX. RDX reduction consists of a series of two-electron transfer steps for each nitro → nitroso reaction.
FIG. 2.
RDX reduction by biologically reduced AQDS (B-AH2QDS) (A) and AH2QDS oxidation (B) in pure-phase incubations (no cells). The results are the means of triplicate analyses, and the bars indicate one standard deviation.
Therefore, three times as much B-AH2QDS (molar basis) is needed to completely reduce RDX. The two likely reaction pathways would be RDX → MNX, in which case the AH2QDS would have been oxidized with stoichiometric MNX accumulation, or RDX → TNX, in which case AH2QDS would be oxidized without MNX or DNX accumulation. MNX, DNX, and TNX did not accumulate; therefore, the second pathway is the likely pathway. The lack of TNX accumulation is discussed below.
The concentration of AH2QDS decreased coupled to RDX reduction. AH2QDS at 500 μM was oxidized to approximately 200 μM, and 300 μM AH2QDS was oxidized to approximately 50 μM concomitantly with RDX reduction (Fig. 2B). When AH2QDS (100 μM) was limiting relative to RDX (at 100 μM), it was completely oxidized within 8 h.
MNX and DNX reduction by biologically reduced AH2QDS.
The metabolites MNX, DNX, and TNX did not significantly accumulate during AH2QDS-mediated RDX reduction; therefore, the individual metabolites were incubated with AH2QDS to determine whether they can be directly reduced. MNX at 50 μM was directly reduced by AH2QDS at all AH2QDS concentrations tested; however, the extent of reduction differed among the treatments (Fig. 3A). Excess AH2QDS (300 μM) completely reduced MNX below detection limits within 8 h. When the stoichiometric equivalent of AH2QDS (100 μM) was provided, reduction was slower and did not proceed to completion. However, previous experiments suggested that the stoichiometric equivalent completely reduced MNX in a much shorter time frame (Fig. 3C). AH2QDS at 50 μM reduced approximately 20 μM MNX (Fig. 3A), which is within the range of the expected stoichiometry.
FIG. 3.
(A and B) Stoichiometric reduction of MNX (A) and DNX (B) with three different concentrations of biologically reduced AQDS (B-AH2QDS). (The DNX stock is not chemically pure; it is ca. 58% DNX, 34% MNX, and 8% TNX.) (C and D) Separate incubations of MNX (C) and DNX (D) with 100 μM biologically reduced AQDS (B-AH2QDS), indicating different reduction rates of DNX than later experiments of MNX or DNX (A and B). The results in panels A and B are the means of triplicate analyses, and the bars indicate one standard deviation; the preliminary experiments in panels C and D are the results of a single analysis.
DNX was also reduced by AH2QDS; however, the DNX stock was not chemically pure, and the reaction did not closely match the expected stoichiometry for DNX (Fig. 3B). The original stock was composed of ca. 58% DNX, 34% MNX, and 8% TNX. AH2QDS in excess of the necessary stoichiometry (200 μM) completely reduced the mixture of DNX plus MNX in approximately 30 h. However, past experiments suggest that MNX and DNX can be reduced in less than 4 h (Fig. 3D). MNX and DNX did not accumulate to a significant extent in any AH2QDS-amended experiments with RDX as the starting material. TNX was transformed as well (data not shown) by an unidentified pathway; the mechanism is currently under investigation using uniformly radiolabeled [14C]RDX. RDX reduction kinetics slowed at 4°C (Table 1). Although MNX and TNX were quantifiable, neither of the nitroso intermediates accumulated in the incubations. DNX was still not quantified at this temperature.
TABLE 1.
Decay rates for pure-phase (no-cell) incubationsa
| Pure-phase (no-cell) incubation | Decay rate [μmol of RDX h−1 at 100 μM AH2QDS or Fe(II)] | Temp (° C) |
|---|---|---|
| AH2QDS alone | 4.050 | 25 |
| 0.400 | 4 | |
| 0.6 mM Fe(II) | 0.042 | 25 |
| 1.2 mM Fe(II) | 0.075 | 25 |
Abiotic degradation rates of RDX by B-AH2QDS at 4°C and by biologically reduced, soluble Fe(II) are shown. Results are the means of triplicate analyses.
RDX reduction by biologically reduced, soluble Fe(II).
RDX at 50 μM was reduced to 19 and 39 μM by 1.2 mM and 600 μM soluble Fe(II), respectively (Table 1).
RDX reduction by Fe(III) and/or extracellular electron shuttles in the presence of Fe(III)-reducing cells (Geobacter metallireducens).
RDX was reduced in all experimental incubations containing resting cells; however, the rate and extent of RDX reduction differed greatly among treatments. AQDS stimulated the fastest rate of RDX reduction (Fig. 4A). RDX was reduced to below detection limits in less than 12 h regardless of the presence of acetate. The initial degradation rate of RDX with the presence of acetate was faster than that of RDX without acetate. Cells of G. metallireducens without AQDS also reduced RDX; RDX was still detectable at 12 h in cell-only incubations. These are the first data demonstrating RDX reduction by cells within the family Geobacteraceae. Later time points demonstrated that cells alone reduced RDX in approximately 50 h, which was comparable to purified humic substance-mediated RDX reduction. The maximum concentration of MNX quantified during the incubations with AQDS was 0.25 μM, and it was no longer detected within 12 h (Fig. 4B). RDX was completely reduced to concentrations below detection within 50 h with purified humic substances (Fig. 4C). MNX transiently accumulated to 0.8 μM in humic substance-amended incubations and quickly decreased at approximately 20 h (Fig. 4D). RDX reduction was the slowest with poorly crystalline Fe(III) oxide as the sole electron acceptor. RDX slowly decreased from 40 to 30 μM with poorly crystalline Fe(III) oxide for 100 h (Fig. 4E). MNX accumulated in these incubations, although the concentration was not a stoichiometric increase, suggesting partial MNX reduction (Fig. 4F). The MNX concentration eventually decreased below the detection limit with longer incubation times. DNX and TNX did not accumulate significantly within the time frame of these experiments. Heat-killed cells did not reduce RDX in the presence or absence of acetate.
FIG. 4.
RDX reduction (A) and MNX accumulation (B) with AQDS in cell suspension incubations of G. metallireducens; RDX reduction (C) and MNX accumulation (D) with humic substances in cell suspension incubations of G. metallireducens; RDX reduction (E) and MNX accumulation (F) with poorly crystalline Fe(III) oxide (FeGel) in cell suspension incubations of G. metallireducens. The results are the means of triplicate analyses, and the bars indicate one standard deviation.
RDX reduction was slow with only poorly crystalline Fe(III) oxide (Fig. 4E). Adding humic acids or AQDS- to Fe(III)-containing incubations increased the reduction rates by approximately 5 and 66 times, respectively (Fig. 5A and B). These rates of RDX degradation were calculated from the pseudo first-order RDX decay constants (Table 2). Fe(II) concentrations were higher in AQDS- and HS-amended incubations than poorly crystalline Fe(III) oxide alone (Fig. 5C). RDX was not reduced in the absence of cells; therefore, only the results for the humic-amended “no-cell” control are presented (the results with AQDS were identical).
FIG. 5.
RDX reduction (A), MNX accumulation (B), and Fe(II) accumulation (C) with AQDS plus poorly crystalline Fe(III) oxide (FeGel) and humic substances plus poorly crystalline Fe(III) oxide in cell suspension incubations of G. metallireducens. The results are the means of triplicate analyses, and the bars indicate one standard deviation.
TABLE 2.
Decay rates for cell suspension incubationsa
| Cell suspension incubationb | Decay rate (μmol of RDX h−1 mg of cell protein−1)
|
|
|---|---|---|
| GS15 | PCA | |
| Cells + AQDS + acetate | 0.0312 | 0.0648 |
| Cells + AQDS (no acetate) | 0.0280 | 0.0565 |
| Cells + HS + acetate | 0.0079 | 0.0470 |
| Cells + HS (no acetate) | 0.0065 | 0.0391 |
| Cells + Fe(III) + acetate | 0.0002 | 0.0091 |
| Cells + Fe(III) (no acetate) | 0.0002 | 0.0083 |
| cells + AQDS + Fe(III) + acetate | 0.0131 | 0.0008 |
| Cells + HS + Fe(III) + acetate | 0.0009 | 0.0058 |
| Cells + acetate | 0.0150 | 0.0318 |
| Cells (no acetate) | 0.0174 | NDc |
| Aerated cells + acetate | 0.0142 | ND |
| Aerated cells (no acetate) | 0.0150 | ND |
Biodegradation rates of RDX in the resting cell suspension of G. metallireducens and G. sulfurreducens with or without extracellular electron shuttling compounds (with acetate as the sole electron donor). Results are the means of triplicate analyses.
All cell suspensions were performed at 30°C.
ND, not determined.
RDX reduction by Fe(III) and/or extracellular electron shuttles in the presence of Fe(III)-reducing cells (Geobacter sulfurreducens).
Similar results were obtained with resting cell suspensions of G. sulfurreducens, another member of the Geobacteraceae that has variant physiological properties from G. metallireducens (5, 34). RDX was reduced with cell suspensions of G. sulfurreducens with several electron acceptors including AQDS, HS, and poorly crystalline Fe(III) oxide. Direct electron transfer from AQDS or HS was faster than Fe(III)/Fe(II)-mediated electron transfer to RDX. All G. sulfurreducens results are summarized as individual reaction rates in Table 2.
DISCUSSION
These results suggest that electron shuttle-mediated RDX reduction is favorable and that RDX is reduced faster by extracellular electron shuttles, including HS and the HS analog AQDS, than direct microbial reduction or bound Fe(II). HS reduction has been identified in numerous subsurface environments and is catalyzed by microorganisms that are ubiquitous in their distribution (6, 32, 48). Direct reduction of RDX or its nitroso metabolites has been reported, but the results suggest that activity may differ among contaminated environments (2, 56, 65). Effective remediation strategies for explosive-residue contaminants such as the cyclic nitramines (RDX and HMX) must be effective in many environments before they will be accepted by the regulatory community.
RDX biodegradation has been reported for aerobic and anaerobic systems; anaerobic biodegradation is the preferential pathway because the nitro groups must be reduced prior to ring cleavage and mineralization to CO2 under anaerobic or aerobic conditions (20, 56). Recent data demonstrated that aerobic, methylotrophic bacteria will degrade a ring cleavage product of RDX, 4-nitro-2,4-diazabutanal (14); however, the ring must first be cleaved for this reaction, which is faster under anaerobic conditions. All reported aerobic data are promising, but RDX contaminates anaerobic subsurface and sedimentary environments in which aerobic metabolism is limited and inefficient (54, 58). In addition, given the extent of some RDX plumes (57), no strategy should be overlooked because one plume may encompass several geochemical zones.
Strategies which promote RDX reduction through MNX→ DNX→ TNX have been investigated in more detail than alternative mechanisms (20, 44). RDX biodegradation has been reported for environmental samples (e.g., marine sediment), mixed anaerobic cultures, or pure cultures (44, 62, 63). Mixed enrichment cultures cultivated from marine sediment reduced RDX through its nitroso intermediates and ultimately led to mineralization of nearly 60% of the RDX in one series of incubations (61, 63). The mineralization activity was not consistent, and alternate batches yielded different results (61). However, Shewanella species were the dominant microbial population in the most active enrichment, which suggests that in situ Fe(III)-reducing microorganisms may be responsible for RDX biodegradation. One novel species within the genus Shewanella was isolated from this sediment, which suggests that Fe(III) reduction may have been the dominant terminal electron-accepting process (61). This previous investigation did not explore the link between Fe(III) reduction and RDX transformation, however, and the authors suggested that direct RDX reduction was the primary mechanism. Although the data presented above demonstrate that Geobacteraceae will directly reduce RDX, it is transformed faster by abiotic electron shuttling from reduced quinones. Reports of RDX biodegradation have been sporadic, and no published data have yet to suggest a link between a specific group of microorganisms and RDX reduction. The inconsistent degradation patterns may be due to environmental samples with different dominant terminal electron-accepting processes promoting a wide array of degradation kinetics. However, the most consistent RDX reduction data prior to those reported here have also been in the presence of Fe(III)-reducing bacteria (59).
Gregory et al. demonstrated that reactive biogenic Fe(II) (soluble ferrous iron adsorbed to the surface of magnetite) reduced RDX through its nitroso intermediates (17). The metabolites accumulated for several days prior to their removal, and TNX accumulated in most bottles (17). The data presented above with Fe(III) agree with these past studies, suggesting that Fe(II) will transform RDX; however, soluble Fe(II) reduced RDX much more slowly than Fe(II) generated from poorly crystalline Fe(III) oxide (i.e., magnetite). The past studies did not, however, stimulate Fe(III) reduction by adding soluble electron shuttles. As described above, processes that stimulate the rate of Fe(III) reduction also stimulate the rate of RDX reduction. In all cases electron shuttles reduced RDX and its intermediates faster than biogenic Fe(II) (with reaction rates on the order of hours rather than days) (Fig. 3 to 5 and Tables 1 and 2).
HS have been reported to facilitate the degradation of several organic contaminants (31, 52) as an electron acceptor or shuttle to Fe(III) which promoted oxidation of organic molecules such as benzene, toluene, or methyl tert butyl ether (11, 12, 38). HS and quinone analogs such as AQDS, juglone, and lawsone also reduce inorganic and organic molecules (32, 46, 47). AQDS was reported to reduce U(VI) in solution or adsorbed to the surface of oxide solids (16, 26). Juglone and lawsone reduced trinitrotoluene in aqueous suspension (47). Although high-molecular-weight HS are insoluble and can merely adsorb organic contaminants, low-molecular-weight HS are soluble and will likely promote these reactions in situ. Remediation strategies predicated on low-molecular-weight HS are relatively benign in that the HS are catalytic and only a low concentration is needed to promote these reactions (32).
Microorganisms within the family Geobacteraceae are generally reported as model Fe(III)-reducing bacteria (8, 37). They have been identified in pristine and contaminated subsurface and sedimentary environments and are implicated in iron- and HS-mediated processes—including contaminant transformation—both in pure culture and in situ (6, 8). A large body of data exists for their physiology, which is why two species within the Geobacter genus were selected to first test the electron shuttle-RDX biodegradation pathway. The data presented here are the first demonstrating cyclic nitramine reduction by a member of the Geobacteraceae; however, the electron shuttle-mediated mechanism was significantly faster than direct electron transfer from high-density resting cells. In addition, the two species tested did not conserve energy for growth coupled to RDX reduction (data not shown). Numerous genera are reported to reduce Fe(III), AQDS, or natural HS; alternative genera that reduce HS include Desulfitobacterium, Anaeromyxobacter, Shewanella, and Rhodoferax (10, 23, 32). Initial experiments with the genera mentioned here suggest that all of these organisms promote the same reactions (data not shown). It is the ubiquity of Fe(III) and HS reduction (and the diversity of the microorganisms involved) that makes the proposed bioremediation strategy an attractive alternative to RDX reduction by direct cellular electron transfer processes.
The data presented here are the first demonstrating RDX reduction by reduced HS or the reduced HS analog AH2QDS. Reduction rates were similar for chemically reduced or biologically reduced AQDS; however, the eventual strategy is based on microbial HS reduction, and therefore the biological mechanism is more relevant to the ongoing investigation. Past studies suggest RDX reduction on the orders of days, weeks, or months. AH2QDS and HS mediated RDX reduction was significantly faster, on the order of hours for complete degradation. Although the number of reactive pathways for reduced quinones will increase in environmental media, these data suggest that HS and Fe(III) reduction stimulated by microorganisms will attenuate RDX in situ.
RDX is reduced through three nitroso metabolites; remediation strategies must attenuate these compounds, as well as the parent compound, because the mono- and di-nitroso forms (MNX and DNX, respectively) are also relatively stable and pose a groundwater hazard. TNX, the trinitroso metabolite, is less stable and may degrade via several putative pathways in situ (29). However, several reports have demonstrated that TNX may accumulate during RDX biodegradation (41, 44, 50), which suggests that the ultimate fate of the nitroso metabolites depends on the site conditions. Initial experiments with MNX and DNX suggested reduction in less than 2 h (Fig. 3C and D). However, experimental conditions were different (higher temperature, lower pH), which may have altered the reduction kinetics. Later experiments (Fig. 3A and B) run at more biologically relevant conditions demonstrated MNX and DNX reduction over 8 to 12 h, which is still a reasonable time frame for bioremediation.
TNX was not recovered at stoichiometric concentration in any of the incubations within this research, suggesting that TNX is either (i) unstable and degrades autocatalytically or (ii) degraded by an as-yet-unidentified reductive mechanism. These HS- and AQDS-mediated reactions were pH and temperature specific. As expected, a lower temperature decreased the rate of reaction with RDX and all intermediates (Table 1). Small fluctuations in the pH (±0.2 pH units) did not alter the reaction stoichiometry or rate. Larger pH fluctuations may change the reaction rate and will be tested.
RDX reduction kinetics did not change significantly in the presence of resting cell mass with either Geobacter metallireducens or G. sulfurreducens. Cell-free AH2QDS completely reduced RDX in approximately 8 h; both AQDS-amended cell suspensions reduced RDX completely within 12 h. However, this does suggest that the rate-limiting step is likely microbial AQDS reduction rather than abiotic electron transfer from AH2QDS to RDX. Cells reduced RDX in the presence of AQDS whether acetate was added as an electron donor or not. This has been previously described for several metal-reducing microorganisms, including Thermus strain SA and Deinococcus radiodurans R1 (15, 28). It has been referred to as “endogenous respiration” and refers to the cellular capacity to utilize electron donors from cell mass (either storage molecules or cellular lysate debris) to continue metabolism. Given that Geobacteraceae are not reported to generate storage molecules under these conditions, it is likely that the acetate-free respiration was promoted by cellular debris. It was not nonspecific electron transfer from reduced cytochromes (or equivalent electron transfer molecules) to AQDS or HS because air-oxidized cell suspensions behaved in a similar manner, with RDX being reduced in the absence of acetate (Table 2).
Natural HS also stimulated RDX reduction in the presence of both cell suspensions. HS-mediated RDX reduction was slower than AQDS-mediated RDX reduction (Fig. 3 to 5 and Tables 1 and 2). This is not surprising given that AQDS is a small molecule with a single, easily accessible (i.e., not sterically hindered) quinone moiety that transfers two electrons per mole (31). Natural HS are large, undefined molecules with varying quinone content, and each individual quinone functional group may be slightly less accessible (for either cell-quinone interaction or hydroquinone-RDX interaction) than the comparable AQDS or AH2QDS reaction mechanisms. Previous investigations with electron shuttling demonstrated that AQDS, natural (Aldrich) humics, and purified International Humic Substance Society humics shuttle electrons at different rates to Fe(III) and other electron acceptors (32, 43). Follow-up experiments with International Humic Substance Society humics will be performed. However, these data suggest that different HS (or analogs and/or quinones) will reduce RDX at different rates with slightly different metabolite accumulation dynamics and that for in situ applications different HS sources must be tested to identify an option that is both mechanistically feasible and cost efficient. Based on past data these options include river sediment humic and/or fulvic extracts, leaf litter extracts, and possibly military smoke dyes that contain quinones (43, 45).
In addition, this was a species-specific phenomenon. HS significantly increased RDX reduction for G. sulfurreducens (Table 2) relative to cells alone, but RDX reduction by G. metallireducens was not stimulated by HS at the concentrations tested. In all cases the HS- and AQDS-facilitated reactions were faster in the absence of bioavailable, poorly crystalline Fe(III), and RDX was completely removed from the system in HS-amended incubations; AQDS-amended incubations were RDX depleted at a faster rate.
Bioavailable Fe(III) has a dual role with respect to electron shuttles and RDX. In both cases the eventual outcome is RDX reduction and therefore degradation; however, the mechanisms to reach that point differ, and thus the rates vary in the presence or absence of Fe(III). Fe(III) readily accepts electrons from reduced HS and AH2QDS; Fe(III) reduction in this manner is very fast (31, 32). As demonstrated here, Fe(III) at times competed for electrons from the reduced HS and AH2QDS, and RDX was only reduced once Fe(II) had been generated. In the absence of Fe(III) electrons are transferred directly from the reduced shuttle to RDX, with very little degradation lag time. In the presence of Fe(III) the lag time increased and RDX degraded only once Fe(III) had been significantly reduced. Therefore, the “dual role” is a slight inhibitory period, followed by a stimulatory period, with respect to RDX reduction. Fe(III)-reducing biomass generated in these environments will reduce available HS; therefore, any increase in Fe(III) reduction eventually benefits site restoration. However, direct electron transfer from electron shuttles will be faster than from cells or Fe(II).
MNX, DNX, and TNX accumulated only transiently in either of the resting cell suspensions without Fe(III) present. In several experiments metabolites were reduced so quickly that they were not even quantified above the error range of the method detection limit. However, MNX accumulated for longer periods of time when Fe(III) was present. MNX eventually degraded in all Fe(III) amended incubations, but MNX degradation was within several days rather than a single day, which was the case in AQDS or HS-amended incubations that lacked Fe(III). DNX and TNX did not accumulate.
Bioremediation options for RDX have been limited to electron donor amendment to stimulate native microorganisms to potentially reduce RDX, most likely by direct electron transfer from cells to the nitro groups of the molecule (3, 21, 49, 64, 65). However, this approach can be described as moderately successful at best, with rates and extent of RDX transformation varying widely among the treated material. The data presented above describe a novel approach for RDX reduction: targeting HS and/or Fe(III) reduction by adding soluble electron shuttles to promote extracellular electron transfer from Fe(III)/HS-reducing biomass to RDX (and its nitroso intermediates). Targeting a group of microorganisms known to be ubiquitous in their distribution circumvents the need for specialized cells capable of direct RDX reduction. This approach may become an effective and efficient bioremediation strategy for subsurface environments contaminated by RDX or other cyclic nitramine compounds.
Acknowledgments
We thank Scott R. Drew of GeoSyntec Consultants for technical assistance and logistical management for field sampling and acquisition, for manuscript review, and for thoughtful discussions. We thank Pam Sheehan of the Picatinny Arsenal and Paul Hatzinger of Shaw Group for aquifer material sampling and preservation. We also acknowledge the assistance of Oi Fei Choi (UIUC) and Anna Knussmann (UIUC) for laboratory technical support.
This study was supported by the Department of Defense Strategic Environmental Research and Development Program under project number ER-1377.
REFERENCES
- 1.Abdelouas, A., Y. Li, W. Lutze, and H. E. Nuttall. 1998. Reduction of U(VI) to U(IV) by indigenous bacteria in contaminated ground water. J. Contam. Hydrol. 35:217-233. [Google Scholar]
- 2.Bhushan, B., A. Halasz, J. Spain, S. Thiboutot, G. Ampleman, and J. Hawari. 2002. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine catalyzed by a NAD(P)H: nitrate oxidoreductase from Aspergillus niger. Environ. Sci. Technol. 36:3104-3108. [DOI] [PubMed] [Google Scholar]
- 3.Bhushan, B., A. Halasz, S. Thiboutot, G. Ampleman, and J. Hawari. 2004. Chemotaxis-mediated biodegradation of cyclic nitramine explosives RDX, HMX, and CL-20 by Clostridium sp. EDB2. Biochem. Biophys. Res. Commun. 316:816-821. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, P. M., and F. H. Chapelle. 1995. Factors affecting microbial 2,4,6-trinitrotoluene mineralization in contaminated soil. Environ. Sci. Technol. 29:802-806. [DOI] [PubMed] [Google Scholar]
- 5.Caccavo, F., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate- oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coates, J. D., D. J. Ellis, E. L. Blunt-Harris, C. V. Gaw, E. E. Roden, and D. R. Lovley. 1998. Recovery of humic reducing bacteria from a diversity of environments. Appl. Environ. Microbiol. 64:1504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coates, J. D., D. J. Lonergan, E. J. P. Philips, H. Jenter, and D. R. Lovley. 1995. Desulfuromonas palmitatis sp. nov, a marine dissimilatory Fe III reducer that can oxidize long-chain fatty acids. Arch. Microbiol. 164:406-413. [PubMed] [Google Scholar]
- 8.Coates, J. D., E. J. P. Philips, D. J. Lonergan, H. Jenter, and D. R. Lovley. 1996. Isolation of Geobacter species from diverse sedimentary environments. Appl. Environ. Microbiol. 62:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doong, R. A., and H. C. Chiang. 2005. Transformation of carbon tetrachloride by thiol reductants in the presence of quinone compounds. Environ. Sci. Technol. 39:7460-7468. [DOI] [PubMed] [Google Scholar]
- 10.Finneran, K. T., C. V. Johnsen, and D. R. Lovley. 2003. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 53:669-673. [DOI] [PubMed] [Google Scholar]
- 11.Finneran, K. T., and D. R. Lovley. 2001. Anaerobic degradation of methyl tert-butyl ether (MTBE) and tert-butyl alcohol (TBA). Environ. Sci. Technol. 35:1785-1790. [DOI] [PubMed] [Google Scholar]
- 12.Finneran, K. T., D. R. Lovley, and E. Moyer. 2001. Anaerobic strategies for enhanced MTBE and TBA bioremediation. J. Contam. Soil Sediment Water Spring 2001:91-94. [Google Scholar]
- 13.Fournier, D., A. Halasz, J. Spain, P. Firuasek, and J. Hawari. 2002. Determination of key metabolites during biodegradation of hexahydro-1,3,5-trinitro-triazine with Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 68:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier, D., S. Trott, J. Hawari, and J. Spain. 2005. Metabolism of the aliphatic nitramine 4-nitro-2,4-diazabutanal by Methylobacterium sp. strain JS178. Appl. Environ. Microbiol. 71:4199-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredrickson, J. K., H. M. Kostandarithes, S. W. Li, A. E. Plymale, and M. J. Daly. 2000. Reduction of Fe(III), Cr(VI), U(VI), and Tc(VII) by Deinococcus radiodurans R1. Appl. Environ. Microbiol. 66:2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fredrickson, J. K., J. M. Zachara, D. W. Kennedy, M. C. Duff, Y. A. Gorby, S. W. Li, and K. M. Krupka. 2000. Reduction of U(VI) in goethite suspensions by a dissimilatory metal-reducing bacterium. Geochim. Cosmochim. Acta 64:3085-3098. [Google Scholar]
- 17.Gregory, K. B., P. Larese-Casanova, G. F. Parkin, and M. M. Scherer. 2004. Abiotic transformation of hexahydro-1,3,5-trinitro-1,3,5-triazine by Fe(II) bound to magnetite. Environ. Sci. Technol. 38:1408-1414. [DOI] [PubMed] [Google Scholar]
- 18.Haas, R., I. Schreiber, E. U. Low, and G. Stork. 1990. Conception for the investigation of contaminated munition plants. Fresenius J. Anal. Chem. 338:41-45. [Google Scholar]
- 19.Harvey, S. D., R. J. Fellows, D. A. Cataldo, and R. J. Bean. 1991. Fate of the explosives hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) in soil and bioaccumulation in bush bean hydroponics plants. Environ. Toxicol. Chem. 10:845-855. [Google Scholar]
- 20.Hawari, J., S. Beaudet, A. Halasz, S. Thiboutot, and G. Ampleman. 2000. Microbial degradation of explosives: biotransformation versus mineralization. Appl. Microbiol. Biotechnol. 54:605-618. [DOI] [PubMed] [Google Scholar]
- 21.Hawari, J., A. Halasz, T. Sheremata, S. Beaudet, C. Groom, L. Paquet, C. Rhofir, G. Ampleman, and S. Thiboutot. 2000. Characterization of metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with municipal anaerobic sludge. Appl. Environ. Microbiol. 66:2652-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes, L. A., K. P. Nevin, and D. R. Lovley. 1999. Role of prior exposure on anaerobic degradation of naphthalene and phenanthrene in marine harbor sediments. Organic Geochem. 30:937-945. [Google Scholar]
- 23.He, Q., and R. A. Sanford. 2003. Characterization of Fe(III) reduction by chlororespiring Anaeromxyobacter dehalogenans. Appl. Environ. Microbiol. 69:2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez, M. E., and D. K. Newman. 2001. Extracellular electron transfer. Cell. Mol. Life Sci. 58:1562-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes, D., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of Geobacteraceae associated with stimulation of dissimilatory Fe(III) reduction during stimulated bioremediation of uranium-contaminated subsurface sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon, B. H., S. D. Kelly, K. M. Kemner, M. O. Barnett, W. D. Burgos, B. A. Dempsey, and E. E. Roden. 2004. Microbial reduction of U(VI) at the solid-water interface. Environ. Sci. Technol. 38:5649-5655. [DOI] [PubMed] [Google Scholar]
- 27.Kashefi, K., and D. R. Lovley. 2003. Extending the upper temperature limit for life. Science 301:934. [DOI] [PubMed] [Google Scholar]
- 28.Kieft, T. L., J. K. Frederickson, T. C. Onstott, Y. A. Gorby, H. M. Kostandarithes, T. J. Bailey, D. W. Kennedy, S. W. Li, A. E. Plymale, C. M. Spadoni, and M. S. Gray. 1999. Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl. Environ. Microbiol. 65:1214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larson, S. L., J. L. Davis, G. Fabian, K. Watts, G. O'Connor, and W. A. Martin. 2005. Grenade range management using lime for dual role of metals immobilization and explosives transformation, abstr. 40, p. F-21. Presented at the SERDP/ESTCP Partners in Environmental Technology, Washington, D.C.
- 30.Lloyd, J. R., and D. R. Lovley. 2001. Microbial detoxification of metals and radionuclides. Curr. Opin. Biotechnol. 12:248-253. [DOI] [PubMed] [Google Scholar]
- 31.Lovley, D. R., J. D. Coates, E. L. Blunt-Harris, E. J. P. Phillips, and J. C. Woodward. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445-448. [Google Scholar]
- 32.Lovley, D. R., J. L. Fraga, E. L. Blunt-Harris, L. A. Hayes, E. J. P. Phillips, and J. D. Coates. 1998. Humic substances as a mediator for microbially catalyzed metal reduction. Acta Hydrochim. Hydrobiol. 26:152-157. [Google Scholar]
- 33.Lovley, D. R., J. L. Fraga, J. D. Coates, and E. L. Blunt-Harris. 1999. Humics as an electron donor for anaerobic respiration. Environ. Microbiol. 1:89-98. [DOI] [PubMed] [Google Scholar]
- 34.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. P. Philips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 35.Lovley, D. R., and E. J. P. Phillips. 1986. Availability of ferric iron for microbial reduction in bottom sediments of the freshwater tidal Potomac river. Appl. Environ. Microbiol. 52:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovley, D. R., and E. J. P. Phillips. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 38.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1996. Rapid anaerobic benzene oxidation with a variety of chelated Fe(III) forms. Appl. Environ. Microbiol. 62:288-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1994. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature 370:128-131. [DOI] [PubMed] [Google Scholar]
- 40.Lowry, O. H., N. J. Rosbrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 41.McCormick, N. G., J. H. Cornell, and H. S. Kaplan. 1981. Biodegradation of hexadro-1,3,5-trinitro-1,3,5-triazine. Appl. Environ. Microbiol. 42:817-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl. Environ. Microbiol. 68:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol. J. 19:141-159. [Google Scholar]
- 44.Oh, B. T., C. L. Just, and P. J. J. Alvarez. 2001. Hexahydro-1,3,5-trinitro-1,3,5-triazine mineralization by zerovalent iron and mixed anaerobic cultures. Environ. Sci. Technol. 35:4341-4346. [DOI] [PubMed] [Google Scholar]
- 45.Rubin, I. B., and M. V. Buchanan. 1985. Carbon-13 NMR spectra of anthraquinone-derived dyes. Magn. Reson. Chem. 23:161-165. [Google Scholar]
- 46.Schwarzenbach, R. P., P. M. Gschwend, and D. M. Imboden. 2003. Environmental organic chemistry, 2nd ed. John Wiley & Sons, Ltd., London, United Kingdom.
- 47.Schwarzenbach, R. P., R. Stierli, K. Lanz, and J. Zeyer. 1990. Quinone and iron porphyrin mediated reduction of nitroaromatic compounds in homogeneous aqueous solution. Environ. Sci. Technol. 24:1566-1574. [Google Scholar]
- 48.Scott, D. T., D. M. McKnight, E. L. Blunt-Harris, S. E. Kolesar, and D. R. Lovley. 1998. Quinone moieties act as electron acceptors in the reduction of humic substances by humics-reducing microorganisms. Environ. Sci. Technol. 32:2984-2989. [Google Scholar]
- 49.Sheremata, T., and J. Hawari. 2000. Mineralization of RDX by the white rot fungus Phanerochaete chrysosporium to carbon dioxide and nitrous oxide. Environ. Sci. Technol. 34:3384-3388. [Google Scholar]
- 50.Sheremata, T. W., A. Halasz, L. Paquet, S. Thiboutot, G. Ampleman, and J. Hawari. 2001. The fate of the cyclic nitramine explosive RDX in natural soil. Environ. Sci. Technol. 35:1037-1040. [DOI] [PubMed] [Google Scholar]
- 51.Spalding, R. F., and J. W. Fulton. 1988. Groundwater munition residues and nitrate near Grand Island, Nebraska, USA. J. Contaminant Hydrol. 2:139-153. [Google Scholar]
- 52.Stevenson, F. J. 1982. Humus chemistry genesis, composition, reactions. Wiley Interscience, New York, N.Y.
- 53.U.S. Department of Energy. 2002. Passive reactive barrier: subsurface contaminants focus area. Department of Energy innovative technology summary report DOE/EM-0623. U.S. Department of Energy, Washington, D.C.
- 54.U.S. Department of Energy. 1998. Treatment of HMX and RDX Contamination, Amarillo National Resource Center for Plutonium, March 1998, ANRCP-1998-2. U.S. Department of Energy, Washington, D.C.
- 55.U.S. Environmental Protection Agency. 2004. 2004 edition of the drinking water standards and health advisories. U.S. Environmental Protection Agency, Washington, D.C.
- 56.Wani, A. H., and J. L. Davis. 2003. RDX biodegradation column study: influence of ubiquitous electron acceptors on anaerobic biotransformation of RDX. J. Chem. Technol. Biotechnol. 78:1082-1092. [Google Scholar]
- 57.Wani, A. H., B. R. O'Neal, J. L. Davis, and L. D. Hansen. 2002. Treatability study for biologically active zone enhancement (BAZE) for in situ RDX degradation in groundwater. Environmental Security Technology Certification Program, U.S. Army Engineer Research and Development Center, Vicksburg, Miss. [Online.] http://el.erdc.usace.army.mil/elpubs/pdf/trel102-35.pdf
- 58.Weissmahr, K. W., M. Hildenbrand, R. P. Schwarzenbach, and S. B. Haderlein. 1999. Laboratory and field scale evaluation of geochemical controls on groundwater transport of nitroaromatic ammunition residues. Environ. Sci. Technol. 33:2593-2600. [Google Scholar]
- 59.Williams, A. G. B., K. B. Gregory, G. F. Parkin, and M. M. Scherer. 2005. Hexahydro-1,3,5-trinitro-1,3,5-triazine transformation by biologically reduced ferrihydrite: evolution of Fe mineralogy, surface area, and reaction rates. Environ. Sci. Technol. 39:5183-5189. [DOI] [PubMed] [Google Scholar]
- 60.Yinon, J. 1990. Toxicity and metabolism of explosives. CRC Press, Inc., Boco Raton, Fla.
- 61.Zhao, J.-S., C. W. Greer, S. Thiboutot, G. Ampleman, and J. Hawari. 2004. Biodegradation of the nitramine explosives hexahydro-1,3,5-trinitro-1,3,5-triazine and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine in cold marine sediment under anaerobic and oligotrophic conditions. Can. J. Microbiol./Rev. Can. Microbiol. 50:91-96. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, J.-S., A. Halasz, L. Paquet, C. Beaulieu, and J. Hawari. 2002. Biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine and its mononitroso derivative hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine by Klebsiella pneumoniae strain SCZ-1 isolated from an anaerobic sludge. Appl. Environ. Microbiol. 68:5336-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, J.-S., D. Manno, C. Beaulieu, L. Paquet, and J. Hawari. 2005. Shewanella sediminis sp. nov., a novel Na+-requiring and hexahydro-1,3,5-trinitro-1,3,5-triazine-degrading bacterium from marine sediment. Int. J. Syst. Evol. Microbiol. 55:1511-1520. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, J. S., L. Paquet, A. Halasz, and J. Hawari. 2003. Metabolism of hexahydro-1,3,5-trinitro-1,3,5-triazine through initial reduction to hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine followed by denitration in Clostridium bifermentans HAW-1. Appl. Microbiol. Biotechnol. 63:187-193. [DOI] [PubMed] [Google Scholar]
- 65.Zhao, J. S., J. Spain, S. Thiboutot, G. Ampleman, C. Greer, and J. Hawari. 2004. Phylogeny of cyclic nitramine-degrading psychrophilic bacteria in marine sediment and their potential role in the natural attenuation of explosives. FEMS Microbiol. Ecol. 49:349-357. [DOI] [PubMed] [Google Scholar]