Abstract
Group II introns are mobile genetic elements that can be redirected to invade specific genes. Here we describe the use of the lactococcal group II intron, Ll.ltrB, to achieve multicopy delivery of heterologous genes into the genome of Lactococcus lactis IL1403-UCD without the need for selectable markers. Ll.ltrB was retargeted to invade three transposase genes, the tra gene found in IS904 (tra904), tra981, and tra983, of which 9, 10, and 14 copies, respectively, were present in IL1403-UCD. Intron invasion of tra904, tra981, and tra983 allele groups occurred at high frequencies, and individual segregants possessed anywhere from one to nine copies of intron in the respective tra alleles. To achieve multicopy delivery of a heterologous gene, a green fluorescent protein (GFP) marker was cloned into the tra904-targeted Ll.ltrB, and the resultant intron (Ll.ltrB::GFP) was induced to invade the L. lactis tra904 alleles. Segregants possessing Ll.ltrB::GFP in three, four, five, six, seven, and eight copies in different tra904 alleles were obtained. In general, increasing the chromosomal copy number of Ll.ltrB::GFP resulted in strains expressing successively higher levels of GFP. However, strains possessing the same number of Ll.ltrB::GFP copies within different sets of tra904 alleles exhibited differential GFP expression, and segregants possessing seven or eight copies of Ll.ltrB::GFP grew poorly upon induction, suggesting that GFP expression from certain combinations of alleles was detrimental. The highest level of GFP expression was observed from a specific six-copy variant that produced GFP at a level analogous to that obtained with a multicopy plasmid. In addition, the high level of GFP expression was stable for over 120 generations. This work demonstrates that stable multicopy integration of heterologous genes can be readily achieved in bacterial genomes with group II intron delivery by targeting repeated elements.
The lactic acid bacteria (LAB) are a large group of gram-positive bacteria which possess similar metabolisms. The LAB are of great importance to the food, medical, and agricultural industries and, as such, are the subject of much research (13). Given their small genomes and relatively simple metabolisms, the LAB are attractive targets for metabolic engineering (12, 18). While genetic techniques to modify LAB chromosomes have improved, methods to generate multiple chromosomal integrations, either for gene disruption or for gene delivery, can be problematic. Traditional methods to generate chromosomal integrations in LAB employ allelic exchange, in which a chromosomal allele is replaced with a cognate modified allele (21). Delivery of heterologous genes thus requires prior construction of flanking regions of homology to allow allelic exchange with a chromosomal target (2, 15). Multiple integrations into the same genome require successive rounds of integration and excision and therefore can be time-consuming.
An alternative mechanism for integrating heterologous genes into bacterial chromosomes is via group II introns. Some group II introns are retroelements that mobilize at high efficiency into specific intronless alleles in a process called homing (14). Homing initiates with intron splicing, resulting in a ligated exon and a ribonucleoparticle comprised of the intron RNA and an intron-encoded protein (IEP). Both intron RNA and IEP components recognize specific DNA sites in the target region and promote reverse splicing of the intron RNA directly into the sense strand of the target DNA (14). The IEP cleaves the antisense strand of the target site and initiates reverse transcription of the intron RNA. The resulting cDNA is then incorporated into genomic DNA via repair mechanisms.
Specificity of the homing reaction is achieved through a combination of both IEP and intron RNA binding to the specific DNA target site. The IEP facilitates unwinding of the double stranded DNA and interacts with specific sites in the target region that flank the insertion site. Sequences within the intron (exon binding sequence 1 [EBS1], EBS2, and δ) anneal with the corresponding target sequences (intron binding sequence 1 [IBS1], IBS2, and δ′) to facilitate reverse splicing of intron RNA into the target site (24, 30).
By rescripting intron EBS1, EBS2, and δ regions to anneal to alternative targets, it is possible to retarget the intron to invade a different site (9, 11), thus providing a novel means for gene invasion. Since integration is independent of host recombination functions (22), intron-mediated inactivation is considered an attractive alternative for bacteria that are recalcitrant to more-standard methods of genomic manipulation.
The most employed group II intron for this purpose is the lactococcal intron Ll.ltrB. Retargeted Ll.ltrB introns have successfully invaded chromosomal loci in a range of hosts including Escherichia coli (11), Shigella (11), Salmonella (11), lactococci (8), and clostridia (5). We recently demonstrated that Ll.ltrB can mediate delivery of heterologous genes, engineered inside the intron, into the lactococcal mleS gene at high frequency, obviating the need for selectable markers (8). Given the absence of selectable markers, the copy number of any delivered gene(s) could then be increased by successive rounds of intron integration. One way to reduce the need for successive rounds of intron invasion is to target repeated genes, such as insertion sequence (IS) elements, within the host genome. IS elements are typically arranged with terminal inverted repeats of 10 to 40 bp flanking a single transposase gene (tra) (20). These elements have been found in multiple copies in numerous LAB (20, 26). L. lactis IL1403-UCD contains six different IS elements, which range in copy number from 1 to 14 copies (3). In this work, Ll.ltrB was retargeted to invade three different IS element transposase genes, tra904, tra981, and tra983, within the IL1403-UCD genome. tra-targeted introns integrated into multiple tra alleles in a single delivery experiment without selection. By cloning a green fluorescent protein (GFP) marker into the tra904-targeted intron, segregants were obtained with different numbers of Ll.ltrB::GFP copies in different sets of tra904 alleles, resulting in a range of GFP expression levels. Thus, this work reveals a strategy for targeting multicopy alleles for intron-mediated delivery of heterologous genes in order to achieve a desired level of integration and expression in bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in the supplemental material (see Table S1 in the supplemental material). Escherichia coli TOP10 was grown at 37°C on brain heart infusion medium (Becton-Dickinson, Franklin Lakes, NJ) and shaken at 250 rpm for broth culture. The lactococcal strains were grown statically at 30°C in GM17 medium (Becton-Dickinson) (33). When appropriate, erythromycin was added to the media at 150 μg/ml and 5 μg/ml for E. coli and L. lactis, respectively. Plating media for either E. coli or L. lactis contained 1.5% agar (Becton-Dickinson).
Molecular biology techniques.
Standard molecular techniques were carried out as described elsewhere (1). For E. coli transformations, One Shot competent cells (Invitrogen, Carlsbad, CA) were used according to the manufacturer's instructions. Transformation of L. lactis was performed as previously described (10). Plasmid DNA was purified from E. coli by use of QIAquick spin columns (QIAGEN, Valencia, CA) following the manufacturer's directions. Chromosomal DNA was prepared from L. lactis as described elsewhere (19). Plasmid DNA was cured from L. lactis by use of ascorbic acid (27). All PCRs were performed using a PTC-200 thermal cycler (MJ Research, San Francisco, CA). Primers and PCR conditions used in this study are listed in the supplemental material (see Tables S2A and S2B, respectively, in the supplemental material). For standard PCRs, including intron rescripting and colony screening, Amplitaq Gold (Applied Biosystems, Foster City, CA) was used. Colony PCR of L. lactis was performed using GeneReleaser (Bioventures Incorporated, Murfreesboro, TN) as previously described (8). PfuTurbo (Stratagene, La Jolla, CA) was used for all inverse PCR and gene amplifications for cloning purposes. Inverse and megaprimer PCRs were purified using Montage PCR filter units (Millipore, Billerica, MA) following the manufacturer's instructions. Restriction enzymes were supplied by New England Biolabs (Beverly, MA). DNA ligations were performed using a Fast-Link DNA ligation kit (Epicentre, Madison, WI) following the manufacturer's instructions. DNA purification from all other PCRs and agarose gels was done using a Geneclean spin kit (Q-Biogene, Irvine, CA) according to the manufacturer's directions. DNA sequencing was performed by the DBS DNA sequencing facility at UC Davis.
Construction of intron donor vector and intron rescripting.
The Ll.ltrB intron was amplified from pLE12 (23) by use of primers IntronUP-F and IntronDN-R, which introduced flanking XbaI and XhoI sites, respectively. The intron fragment was then cloned into the XbaI and XhoI sites of pMSP3535 (4), resulting in pKT0180. Plasmid pKT0180 was then rescripted so that the IBS and EBS1 regions contained MluI and NcoI restriction sites, respectively. This and all subsequent intron rescripting was performed using the two-step PCR method followed by a megaprimer PCR in a fashion analogous to that used previously for Ll.ltrB retargeting (11). The first step in the two-step PCR was performed using primer pairs IBSMluI-EBSR2 and EBS1NcoI-EBS2NruI followed by IBSMluI-EBS2NruI. For the megaprimer PCR, the vector backbone was generated by inverse PCR using pKT0180 as the template and primers pHR057F and pHR057R. The PCR products were purified, and 1 μg of the vector backbone and the two-step PCR product were used in a megaprimer PCR for 10 cycles with an extension of 12 min and an annealing temperature of 55°C. The resulting plasmid DNA was cleaned and transformed into E. coli TOP10 cells (Invitrogen), and clones were screened by sequence analysis to confirm that the correct bases had been incorporated. The resulting plasmid, pKT0185, was used in all subsequent rescripting reactions. Intron rescripting of pKT0185 was performed using two-step PCR primers (Tra904473sIBS, Tra904473sEBS1, Tra904473sEBS2, Tra981555sIBS, Tra981555sEBS1, Tra981555sEBS2, Tra983378sIBS, Tra983378sEBS1, Tra983378sEBS2, EBS2R) to introduce base changes into the Ll.ltrB IBS, EBS1, and EBS2 regions of the intron. Before transformation into E. coli TOP10 cells, the rescripted intron vectors were digested with MluI and NcoI to remove any unrescripted pKT0185 background. The vectors were screened again by sequence analysis to confirm that the correct bases had been incorporated.
Intron invasion.
Strains of L. lactis containing the tra-targeted intron donor vectors were grown overnight. A 1/100 dilution was made with media containing 25 ng/ml nisin (Sigma-Aldrich, St. Louis, MO), and the mixture was left overnight. One milliliter of overnight culture was washed in GM17 to remove residual nisin to limit further intron homing. A serial dilution was performed and plated out onto selective media. Colonies were initially screened for the presence of any intron-tra junctions by use of a consensus forward primer designed to the 5′ region of tra904, tra981, or tra983, respectively, combined with an intron-based reverse primer, EBS2R (see Table S2 in the supplemental material). This allowed an estimate of the invasion frequency for tra904, tra981, or tra983, respectively. Between 48 and 192 colonies were screened for each intron in this step. To score which specific tra alleles had been invaded, chromosomal DNA was prepared from colonies which produced amplicons by use of the consensus tra-Ll.ltrB junction PCR and examined by PCR using primers specific to each individual tra allele. To accomplish this, the allele-specific forward primers (see Table S2 in the supplemental material) were paired with reverse primer EBS2R.
Construction of pHR082.
GFP was amplified from plasmid pMSP::GFP3bMut (a gift from G. Dunny) by use of primers pGFPXbaF1 and pGFPXhoRI. The PCR product was digested with XbaI and XhoI and cloned into pMSP3535 digested with the same enzymes. The plasmid, numbered pHR082, contains GFP under control of the nisin promoter.
Construction of Ll.ltrB::GFP.
To enable the cloning of genes into Ll.ltrB, a SacII site was introduced immediately downstream of the stop codon of ltrA in pKT0185. An inverse PCR was performed with pKT0185 as the template by use of primers pKT0185SacIIF and pKT0185SacIIR2. The PCR product was digested with SacII and self-ligated. The resulting plasmid, pHR116, contains an enzyme site for cloning genes into the intron. This plasmid was then rescripted to the tra904 allele as described above, creating pHR120. The nisA promoter and GFP gene were amplified from plasmid pHR082 with primers nisGFPSacIIF5 and nisGFPSacIIR5 and cloned into pHR120, creating pHR131.
Measurement of GFP expression.
Strains were grown overnight and diluted 1/100 into GM17. Growth was monitored until an absorbance at 600 nm of 0.5 was reached. Nisin was added to a final concentration of 25 ng/ml. Samples were then removed hourly to measure GFP levels. One milliliter of cells was removed, washed, and resuspended in phosphate-buffered saline, pH 6.0, to achieve an absorbance at 600 nm of 0.5. One hundred microliters of the washed cells was placed into a 96-well optical plate and covered with an optical cap (Applied Biosystems). The GFP fluorescence was measured using the plate-only read function of an ABI 7700 sequence detection system using an excitation wavelength of 488 nm and an emission wavelength of 520 nm.
RESULTS
tra allele validation.
To validate the presence or absence of all the tra904, tra981, and tra983 alleles in our laboratory strain of IL1403, termed IL1403-UCD, PCRs were performed using allele-specific forward and reverse primers (see Table S2 in the supplemental material). In IL1403-UCD, all alleles were present except for the tra allele found in copy B of IS981 (tra981B) and tra983K.
Construction of tra-targeted introns.
To determine if it was possible to deliver the intron into multiple target sites in a single delivery experiment, introns were retargeted to invade tra904, tra981, and tra983, which are present in the IL1403-UCD genome in 9, 10, and 14 copies, respectively (3). The tra904 allele was targeted using the Targetron software to identify candidate target sites (InGex LLC, St. Louis, MO). Of eight sites analyzed, only the tra904502s intron successfully invaded (data not shown). Target sites for tra981 and tra983 were chosen on the basis of consensus with previously successful intron invasion sites (Fig. 1) (8, 9, 38). The plasmid pKT0185 was rescripted to generate plasmids pHR109, pHR123, and pHR117, containing introns targeted for tra904, tra981, and tra983, respectively.
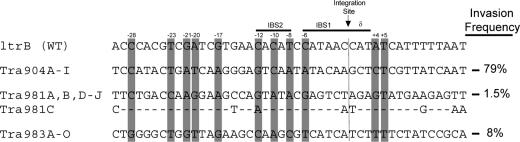
FIG. 1.
Alignment of the tra904, tra981, and tra983 target sites with the wild-type ltrB target site region. Shaded bases have been previously shown to predominate (>50% presence) among a range of successful Ll.ltrB targets generated in E. coli (36).
tra intron invasion.
Plasmids pHR109, pHR123, and pHR117 were transformed into IL1403-UCD, and invasion was induced by growing the strains overnight in the presence of nisin. Cells were then washed with GM17 to remove any residual nisin, serially diluted, and plated onto selective media. Primary tra gene integrants were selected by universal tra-Ll.ltrB junction PCR. Secondary PCR analyses of intron-positive integrants determined both copy number and location of the intron invasions (see Materials and Methods).
As seen in Tables 1 2 and 3, the rescripted Ll.ltrB intron was able to invade more than one tra allele in a single invasion event. For all three tra targets, segregants containing different numbers of invaded alleles were obtained, with a maximum of nine alleles invaded for any single tra904, tra981, or tra983 segregant (Tables 1, 2 and 3, respectively). The tra904 invasion showed the most variability, with segregants containing one, two, three, five, and nine copies of the intron in different tra904 alleles. The tra904A, tra904D, and tra904F alleles were invaded at a higher frequency than the other alleles (invaded in 60 to 70% of segregants). All nine of the tra981 alleles present in the IL1403-UCD strain (tra981B is absent) were invaded in two individual segregants, while a third segregant had six alleles harboring an intron. Invasion of tra983 alleles showed some bias. tra983A, tra983B, tra983C, tra983D, tra983E, tra983F, tra983H, and tra983L all were invaded in most of the segregants (>75%). Conversely, tra983G, tra983I, tra983J, and tra983N were invaded less frequently (<25%), and tra983M was not invaded at all. Sequence analysis of the invasion site within tra983M showed that it possessed the same target region sequence as the other tra983 alleles (Fig. 1), indicating that differences in the tra983M target site region of our strain was not the reason for the lack of invasion.
TABLE 1.
Intron invasion of individual tra904 alleles
| Segregant | No. of alleles invaded |
tra904 allele invaded (of 9 alleles)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | ||
| HR268 | 1 | − | − | − | + | − | − | − | − | − |
| HR263 | 2 | + | − | − | + | − | − | − | − | − |
| HR267 | 2 | − | − | − | + | − | + | − | − | − |
| HR266 | 3 | + | + | − | + | − | − | − | − | − |
| HR262 | 3 | + | + | − | − | − | + | − | − | − |
| HR270 | 3 | − | − | + | + | − | + | − | − | − |
| HR264 | 3 | − | − | + | − | + | + | − | − | − |
| HR265 | 5 | + | − | + | + | + | + | − | − | − |
| HR269 | 5 | + | − | − | − | − | + | + | + | + |
| HR261 | 9 | + | + | + | + | + | + | + | + | + |
A to I, tra904A to tra904I, respectively. +, invasion; −, no invasion.
TABLE 2.
Intron invasion of individual tra981 alleles
| Segregant | No. of alleles invaded |
tra981 allele invaded (9 alleles present)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Bb | C | D | E | F | G | H | I | J | ||
| HR296 | 9 | + | NA | + | + | + | + | + | + | + | + |
| HR297 | 6 | + | NA | + | − | + | + | − | + | − | + |
| HR298 | 9 | + | NA | + | + | + | + | + | + | + | + |
A to J, tra981A to tra981J, respectively. +, invasion; −, no invasion.
In our IL1403-UCD strain, tra981B was not present. NA, not applicable.
TABLE 3.
Intron invasion of individual tra983 alleles
| Segregant | No. of alleles invaded |
tra983 allele invaded (of 14 alleles)a
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | Kb | L | M | N | O | ||
| HR299 | 9 | + | + | + | + | + | + | − | + | − | − | NA | + | − | − | + |
| HR300 | 8 | + | + | − | + | − | + | − | + | − | − | NA | + | − | + | + |
| HR301 | 9 | + | + | + | + | + | − | − | + | + | − | NA | + | − | − | + |
| HR302 | 9 | + | + | + | + | + | + | + | − | − | − | NA | + | − | − | + |
| HR303 | 9 | + | + | + | + | + | + | + | + | − | − | NA | + | − | − | − |
| HR304 | 7 | + | − | + | + | + | + | − | − | − | + | NA | + | − | − | + |
| HR305 | 6 | + | + | − | + | + | − | − | + | − | − | NA | + | − | − | − |
| HR306 | 8 | + | + | + | + | + | + | − | + | − | − | NA | + | − | − | − |
A to O, tra983A to tra983O, respectively. +, invasion; −, no invasion.
In our IL1403-UCD strain, tra983K was not present. NA, not applicable.
Multicopy intron-mediated gene delivery into tra904.
We have previously shown that Ll.ltrB can deliver genes to the IL1403-UCD chromosome both stably and without the use of selection (8). The ability of the rescripted Ll.ltrB intron to invade multiple tra alleles suggested that these introns could also function as a means for delivering multiple copies of a heterologous gene into the IL-1403-UCD genome. To test this possibility, pKT0185 was modified to introduce a SacII site immediately downstream of the stop codon of ltrA within the intron, producing pHR116. Given that the tra904-targeted intron homed with the highest frequency (79%), pHR116 was rescripted to invade the tra904 target site. The GFP gene (7) under the control of the nisin promoter was amplified from pHR082 and cloned into the SacII site of the tra904-rescripted vector, yielding pHR131. This plasmid was then transformed into L. lactis and induced for tra904 invasion. After the induction, tra904 invasion was screened by tra group PCR using the Tra904consensusF/EBS2R primer set (see Table S2 in the supplemental material), which recognizes all tra904 alleles. Inclusion of the GFP cassette into the tra904-targeted intron reduced the overall invasion frequency to 42%. Segregants identified as positive for tra904 invasion were further examined using allele-specific PCR. This identified segregants containing between three and eight tra904 alleles invaded (Table 4). Interestingly, alleles less frequently invaded by the intron alone (tra904E, tra904G, tra904H, and tra904I) (Table 1) were readily invaded by same intron containing GFP (Table 4), suggesting that allele bias was not a significant factor for tra904 invasion.
TABLE 4.
tra904 alleles invaded by the tra904::GFP intron
| Segregant | No. of alleles invaded |
tra904 allele invadeda
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | ||
| HR341 | 3 | + | − | + | − | + | − | − | − | − |
| HR342 | 3 | + | − | + | − | − | − | − | + | − |
| HR336 | 4 | + | − | + | + | + | − | − | − | − |
| HR312 | 5 | + | − | − | − | + | − | + | + | + |
| HR338 | 5 | + | − | + | − | + | − | − | + | + |
| HR308 | 6 | + | − | + | + | + | + | − | − | + |
| HR311 | 7 | + | + | + | + | + | − | + | + | − |
| HR313 | 7 | + | − | + | + | + | + | + | + | − |
| HR340 | 7 | + | − | + | + | + | + | + | − | + |
| HR337 | 7 | + | − | + | + | + | + | − | + | + |
| HR314 | 8 | + | + | + | + | + | + | + | + | − |
A to I, tra904A to tra904I, respectively. +, invasion; −, no invasion.
Expression of GFP from the chromosome.
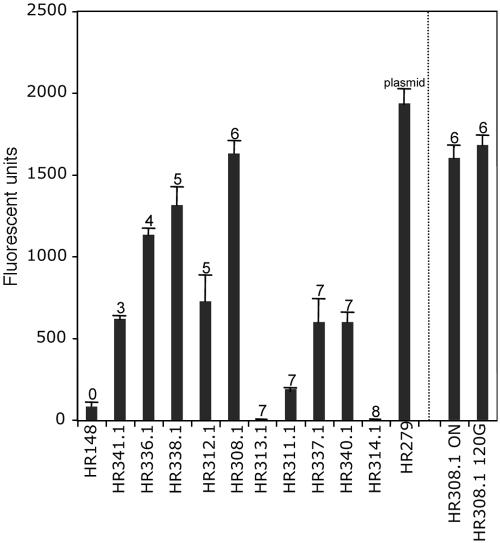
Although multiple copies of GFP were successfully delivered into tra904 alleles, it was important to establish if these copies functionally expressed GFP. To examine this, select segregants containing three, four, five, six, seven, and eight copies of GFP were picked for further analysis. The segregants were cured of pHR131 to remove the plasmid-borne source of GFP. The pMSP3535 vector was then transformed into the segregants to provide the signal transduction genes (nisRK) required for nisin-inducible expression from the nisA promoter upstream of GFP in the delivered introns (4). Analysis of fluorescence from the segregants indicated that the GFP was functional and that higher expression levels were associated with increased copy number (Fig. 2). The highest level of expression was observed for HR308.1, a segregant containing six copies of GFP, and was similar to the level achieved in when GFP was expressed from the multicopy plasmid pHR082 (Fig. 2), which is maintained at approximately six to nine copies per cell (29). Interestingly, significantly lower GFP expression was observed for segregants which harbored seven copies of Ll.ltrB::GFP in tra904 (strains HR311.1, HR337.1, and HR340.1). Moreover, no expression was observed for HR313.1 and HR314.1, strains which contained seven and eight copies of Ll.ltrB::GFP, respectively. However, these last two strains grew poorly upon induction, suggesting that the expression of GFP was detrimental to cell growth. A comparison of the tra904 alleles invaded in the high-expression strain HR308.1 with those invaded in HR313.1 and HR314.1 indicated that GFP expression from within tra904B, tra904G, or tra904H may be responsible for this lack of growth (Table 4).
FIG. 2.
GFP expression from segregants containing Ll.ltrB::GFP in tra904 alleles. The “0.1” designation after the strain name indicates that the intron delivery plasmid was removed and the vector pMSP3535 was introduced as a source of nisRK required for nisin-inducible expression of GFP. The number above each column indicates the number of tra904 copies invaded. The “ON” and “120G” designations represent cultures grown overnight and for 120 generations, respectively.
Stability of tra904::Ll.ltrB::GFP segregants.
To determine if the tra904::Ll.ltrB::GFP segregants were stable, HR308.1 was grown for 120 generations in the absence of nisin, and then GFP induction was performed. As seen in Fig. 2, the GFP expression levels observed after serial passage were similar to that obtained after overnight growth of the same culture. This suggests that the multiple copies of the intron-GFP cassette in the lactococcal chromosome are stable, a critical feature when engineering cells for overproduction of heterologous proteins.
DISCUSSION
Recently, lactococci have received much attention as a pliable host for oral delivery of therapeutic or prophylactic proteins (25, 32). Using L. lactis as an oral delivery vehicle is an attractive approach for a number of reasons. L. lactis is consumed frequently in numerous fermented products and possesses a “generally recognized as safe” (GRAS) designation. Moreover, due to its prevalent use in commercial food and beverage fermentations, there is a wealth of knowledge pertaining to L. lactis fermentation characteristics, strain preservation systems, and shelf-life behavior(s) (34).
When L. lactis is engineered for this purpose, a common strategy is to incorporate the expression cassette into the genome of the target strain (28). This approach is taken for a variety of reasons, including the need to eliminate all vector-associated antibiotic resistance genes as well as the potential stability and containment issues inherent with plasmid-borne expression. Chromosomal incorporation typically involves selection of crossover events between cloned genomic DNAs that flank the heterologous gene of choice and cognate chromosomal loci.
We previously demonstrated an alternative means for delivery of genes into the genome of L. lactis IL1403 by use of the Ll.ltrB intron (8). Intron-mediated delivery occurred at high frequencies, obviating the need for antibiotic selection to identify the appropriate integrant (8). In the present work, we examined intron delivery targeted toward repeated elements in L. lactis as a means to increase the copy number of the delivered gene. Introns were retargeted to invade tra904, tra981, and tra983 alleles present in the IL1403 genome. Rescripted introns invaded multiple alleles in a single induction event, although invasion frequencies varied between 1.5 and 79% for various tra genes. This variation is likely due to the differences in the specific target sites and their respective capacities to serve as a viable partner for the intron ribonucleoparticle-based invasion (Fig. 1). Target nucleotide requirements for Ll.ltrB homing have been extensively examined for E. coli, and 11 specific bases were shown to be favored (>50% predominance) in Ll.ltrB target sites (36). The rescripted Ll.ltrB introns utilized in this study possessed a maximum of five bases in common with those selected for in E. coli (Fig. 1). Of the three sites targeted, the similarity of the tra gene target sequence to the wild-type ltrB target sequence appeared related to invasion efficiency, with targets of greater similarity to the wild type being invaded more efficiently. tra904, which has 17 nucleotides in common with ltrB, homed at 79%, while tra983 and tra981, with 12 and 6 nucleotides shared with ltrB, homed at 8 and 1.5%, respectively. A more comprehensive survey of intron target site preferences in lactococci will enable better prediction of introns that invade at high efficiency.
After screening a wide range of segregants from tra904, tra981, and tra983 intron invasions, we noticed that a maximum of nine tra allele integrations could be accomplished in a single step. It is unclear why no more than nine introns could be integrated in one induction event. Perhaps a screening of more segregants would reveal additional insertions. Alternatively, polar expression from the intron-encoded ltrA promoter (39) into genes downstream of a specific tra allele might have made integration into a larger complement of target alleles problematic.
By cloning into the tra904-targeted intron, as many as eight copies of Ll.ltrB::GFP were delivered into the chromosome in a single step without the use of selection. The general invasion frequency of Ll.ltrB::GFP into the tra904 alleles (42%) was reduced in comparison to that of Ll.ltrB invasion alone (79%), suggesting that the addition of heterologous DNA inside the intron impedes invasion. As expected, the copy number of tra904::GFP directly correlated with the level of GFP fluorescence. The level of fluorescence observed for a strain containing six copies of tra904::GFP (strain HR308.1) was similar to that observed for a GFP-containing plasmid maintained at approximately the same copy number (six to nine copies per cell). Growth was inhibited upon nisin induction in strains HR313.1 and HR314.1, which carried seven and eight copies of GFP, respectively. The cause for this growth inhibition remains unknown. Perhaps the expression of GFP from the chromosome at a copy number of seven or more produces a significant metabolic burden on the cells. This is unlikely, given the lack of previous reports of diminished growth of L. lactis due to expression of heterologous proteins from other multicopy vectors (18, 25). An alternative explanation is that the expression of GFP has a polar effect on genes downstream of the intron insertion.
Multicopy intron-mediated delivery is unlike other methods used to obtain multiple chromosomal copies of heterologous genes in other bacteria. Others have achieved multicopy integrants through forcing multiple, tandem integrations of suicide or conditionally replicating plasmids by increasing selection for a plasmid-encoded antibiotic resistance gene (37). This method utilizes Campbell-type integration to amplify the integrated plasmid carrying a gene of interest and has been previously performed with L. lactis (6, 16, 17). Leenhouts et al. (15) modified this method to enable food-grade delivery of multiple gene copies by using sucrose as the selection system. Using this technique, these authors were able to insert up to 20 copies of the plasmid into the chromosome. However, the integrated plasmids were lost at a rate of 7.5 × 10−2 to 15 × 10−2 copies per generation (15). Intron-mediated delivery does not rely on homologous recombination to achieve multicopy integration, and unlike tandem integration of plasmids, the delivered genes were stable over 120 generations, as was previously observed for singly integrated introns (8).
There are relatively few reports that show the impact of the copy number of chromosomal-borne heterologous genes on the overall expression achieved from those strains. Srinivasan et al. (31) delivered up to three copies of organophosphohydrolyase into the chromosome of Ralstonia eutropha and observed a linear increase in protein expression as the copy number increased. Wang et al. (35) optimized expression of a heterologous keratinase gene (kerA) through multiple, random integrations into the Bacillus licheniformis genome by use of a suicide plasmid. Interestingly, those authors found that the highest level of secreted keratinase was achieved with strains containing between three and five integrated kerA gene copies, while strains possessing higher numbers exhibited lower levels of keratinase production. We observed an analogous result in this work, in which reduced GFP expression was demonstrated for strains containing more than six tra904 alleles possessing the intron-GFP cassette.
Given the increased use of lactococcal strains as oral delivery vehicles for prophylactic or therapeutic proteins, versatile tools for both chromosomal stabilization and optimal protein expression are needed. The lactococcal group II intron has already been shown to facilitate stable chromosomal gene delivery without selection. By targeting repeated elements in lactococci, this work demonstrates how these introns can be used to achieve a desired copy number, thereby facilitating protein overexpression aims. Given that group II intron invasion is operable in various bacterial and even mammalian environments (9), the general strategy of targeting repeated elements should be a viable method to construct production strains that harbor a specified number of chromosomally borne expression cassettes in other organisms as well.
Supplementary Material
Acknowledgments
This work was supported by the UC Discovery Program (Biostar01-10204) and the California Dairy Research Foundation.
D.A.M. thanks Sharon Clark for technical assistance. David Sela is acknowledged for his assistance with the manuscript preparation.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. Wiley, New York, N.Y.
- 2.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., B. A. McClane, D. J. Fisher, J. I. Rood, and P. Gupta. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71:7542-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopin, M. C., A. Chopin, A. Rouault, and N. Galleron. 1989. Insertion and amplification of foreign genes in the Lactococcus lactis subsp. lactis chromosome. Appl. Environ. Microbiol. 55:1769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 8.Frazier, C. L., J. San Filippo, A. M. Lambowitz, and D. A. Mills. 2003. Genetic manipulation of Lactococcus lactis by using targeted group II introns: generation of stable insertions without selection. Appl. Environ. Microbiol. 69:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, H., M. Karberg, M. Long, J. P. Jones, B. Sullenger, and A. M. Lambowitz. 2000. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science 289:452-457. [DOI] [PubMed] [Google Scholar]
- 10.Holo, N., and I. F. Ness. 1989. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karberg, M., H. Guo, J. Zhong, R. Coon, J. Perutka, and A. M. Lambowitz. 2001. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 19:1162-1167. [DOI] [PubMed] [Google Scholar]
- 12.Kleerebezem, M., P. Hols, and J. Hugenholtz. 2000. Lactic acid bacteria as a cell factory: rerouting of carbon metabolism in Lactococcus lactis by metabolic engineering. Enzyme Microb. Technol. 26:840-848. [DOI] [PubMed] [Google Scholar]
- 13.Konings, W. N., J. Kok, O. P. Kuipers, and B. Poolman. 2000. Lactic acid bacteria: the bugs of the new millennium. Curr. Opin. Microbiol. 3:276-282. [DOI] [PubMed] [Google Scholar]
- 14.Lambowitz, A. M., and S. Zimmerly. 2004. Mobile group II introns. Annu. Rev. Genet. 38:1-35. [DOI] [PubMed] [Google Scholar]
- 15.Leenhouts, K., A. Bolhuis, G. Venema, and J. Kok. 1998. Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Appl. Microbiol. Biotechnol. 49:417-423. [DOI] [PubMed] [Google Scholar]
- 16.Leenhouts, K. J., J. Gietema, J. Kok, and G. Venema. 1991. Chromosomal stabilization of the proteinase genes in Lactococcus lactis. Appl. Environ. Microbiol. 57:2568-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leenhouts, K. J., J. Kok, and G. Venema. 1989. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55:394-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Loir, Y., V. Azevedo, S. C. Oliveira, D. A. Freitas, A. Miyoshi, L. G. Bermudez-Humaran, S. Nouaille, L. A. Ribeiro, S. Leclercq, J. E. Gabriel, V. D. Guimaraes, M. N. Oliveira, C. Charlier, M. Gautier, and P. Langella. 2005. Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb. Cell Fact. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewington, J., S. D. Greenaway, and B. J. Spillane. 1987. Rapid small-scale preparation of bacteria genomic DNA; suitable for cloning and hybridization analysis. Lett. Appl. Microbiol. 5:51-53. [Google Scholar]
- 20.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills, D. A. 2001. Mutagenesis in the post genomics era: tools for generating insertional mutations in the lactic acid bacteria. Curr. Opin. Biotechnol. 12:503-509. [DOI] [PubMed] [Google Scholar]
- 22.Mills, D. A., D. A. Manias, L. L. McKay, and G. M. Dunny. 1997. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol. 179:6107-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohr, G., D. Smith, M. Belfort, and A. M. Lambowitz. 2000. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev. 14:559-573. [PMC free article] [PubMed] [Google Scholar]
- 25.Nouaille, S., L. A. Ribeiro, A. Miyoshi, D. Pontes, Y. Le Loir, S. C. Oliveira, P. Langella, and V. Azevedo. 2003. Heterologous protein production and delivery systems for Lactococcus lactis. Genet. Mol. Res. 2:102-111. [PubMed] [Google Scholar]
- 26.Polzin, K. M., D. Romero, M. Shimizu-Kadota, T. R. Klaenhammer, and L. L. McKay. 1993. Copy number and location of insertion sequences ISS1 and IS981 in lactococci and several other lactic acid bacteria. J. Dairy Sci. 76:1243-1252. [DOI] [PubMed] [Google Scholar]
- 27.Ramesh, A., P. M. Halami, and A. Chandrashekar. 2001. Ascorbic acid-induced loss of a pediocin-encoding plasmid in Pediococcus acidilactici CFR K7. World J. Microbiol. Biotechnol. 16:695-697. [Google Scholar]
- 28.Sanders, M. E., F. Guarner, D. Mills, B. Pot, J. Rafter, B. Rastall, G. Reid, Y. Ringel, I. Rowland, M. Saarela, and K. Tuohy. 2005. Selected topics in probiotics and prebiotics: meeting report for the 2004 international scientific association for probiotics and prebiotics. Curr. Issues Intest. Microbiol. 6:55-68. [PubMed] [Google Scholar]
- 29.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 30.Singh, N. N., and A. M. Lambowitz. 2001. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol. 309:361-386. [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan, S., G. C. Barnard, and T. U. Gerngross. 2003. Production of recombinant proteins using multiple-copy gene integration in high-cell-density fermentations of Ralstonia eutropha. Biotechnol. Bioeng. 84:114-120. [DOI] [PubMed] [Google Scholar]
- 32.Steidler, L. 2003. Genetically engineered probiotics. Best Pract. Res. Clin. Gastroenterol. 17:861-876. [DOI] [PubMed] [Google Scholar]
- 33.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teuber, M. 1995. The genus Lactococcus, p. 173-234. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria. Blackie Academic & Professional, London, United Kingdom.
- 35.Wang, J. J., K. Rojanatavorn, and J. C. Shih. 2004. Increased production of Bacillus keratinase by chromosomal integration of multiple copies of the kerA gene. Biotechnol. Bioeng. 87:459-464. [DOI] [PubMed] [Google Scholar]
- 36.Yao, J., J. Zhong, and A. M. Lambowitz. 2005. Gene targeting using randomly inserted group II introns (targetrons) recovered from an Escherichia coli gene disruption library. Nucleic Acids Res. 33:3351-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, M. 1984. Gene amplification in Bacillus subtilis. J. Gen. Microbiol. 130:1613-1621. [DOI] [PubMed] [Google Scholar]
- 38.Zhong, J., M. Karberg, and A. M. Lambowitz. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 31:1656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, L., D. A. Manias, and G. M. Dunny. 2000. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol. Microbiol. 37:639-651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.