Abstract
In Arabidopsis thaliana, cis-regulatory sequences of the floral homeotic gene AGAMOUS (AG) are located in the second intron. This 3-kb intron contains binding sites for two direct activators of AG, LEAFY (LFY) and WUSCHEL (WUS), along with other putative regulatory elements. We have used phylogenetic footprinting and the related technique of phylogenetic shadowing to identify putative cis-regulatory elements in this intron. Among 29 Brassicaceae species, several other motifs, but not the LFY and WUS binding sites identified previously, are largely invariant. Using reporter gene analyses, we tested six of these motifs and found that they are all functionally important for the activity of AG regulatory sequences in A. thaliana. Although there is little obvious sequence similarity outside the Brassicaceae, the intron from cucumber AG has at least partial activity in A. thaliana. Our studies underscore the value of the comparative approach as a tool that complements gene-by-gene promoter dissection but also demonstrate that sequence-based studies alone are insufficient for a complete identification of cis-regulatory sites.
INTRODUCTION
It has been recognized that comparing the regulatory regions of genes that are expressed in similar patterns both within a species and across related taxa can help identify cis elements that confer conserved expression patterns (Gumucio et al., 1992; Wasserman and Fickett, 1998; Jareborg et al., 1999; Dubchak et al., 2000; Wasserman et al., 2000; Bergman and Kreitman, 2001; Kaplinsky et al., 2002). In particular, the analysis of orthologous regulatory regions in multiple species can enhance current attempts to decipher the “cis-regulatory code” (Sumiyama et al., 2001; Berman et al., 2002; Davidson et al., 2002; Dermitzakis and Clark, 2002; Markstein et al., 2002). Conversely, species-specific alterations in expression patterns often are thought to play an important role in generating interspecific variation (Doebley et al., 1997; Wang et al., 1999; Kopp et al., 2000; Sucena and Stern, 2000).
The floral homeotic gene AGAMOUS (AG) of Arabidopsis thaliana has been a paradigm for the study of transcriptional regulation during plant development. Proper expression of AG requires sequences located in a 3-kb intron (Sieburth and Meyerowitz, 1997; Busch et al., 1999; Deyholos and Sieburth, 2000). Two transcription factors, LEAFY (LFY) and WUSCHEL (WUS), that bind to sequences within this intron have been identified (Busch et al., 1999; Lohmann et al., 2001). The plant-specific protein LFY controls floral fate and is expressed throughout floral primordia (Weigel et al., 1992; Parcy et al., 1998). The homeodomain protein WUS is expressed in the center of both shoot and floral meristems (Mayer et al., 1998) and is partly responsible for the region-specific activation of AG by LFY. Binding of both LFY and WUS to AG regulatory sequences is required for the normal activity of the AG enhancer. In addition to LFY and WUS, many other genes that affect the expression pattern of AG or its orthologs in other species have been identified by mutant analysis, but it is not known whether they regulate AG directly (Lohmann and Weigel, 2002). Notably, although almost all of these genes act as repressors of AG, dissection of cis-regulatory sequences by reporter gene analysis has failed to identify specific sites required for the repression of AG (Busch et al., 1999; Deyholos and Sieburth, 2000).
The already extensive characterization of AG orthologs in diverse vascular plants—from gymnosperms to eudicots—encourages comparative evolutionary studies of the regulatory circuits underlying the formation of flowers. Phylogenetic footprinting seeks to identify conserved regulatory sequences by using known species relationships as a rough guide for choosing taxa to be sampled, although individual regulatory elements may evolve at different rates than the genome as a whole. We have analyzed AG noncoding sequences from many species to identify potentially important motifs. The most informative approach has been the comparison of a large number of species that are in the same family as A. thaliana, which circumvents the difficulties associated with aligning long stretches of noncoding sequences from more distantly related species (Clark, 2001). This approach, the identification of largely invariant motifs using sequences from closely related species, has been called “phylogenetic shadowing” (Boffelli et al., 2003), to distinguish it from the use of more distantly related species, which is known as “phylogenetic footprinting” (Gumucio et al., 1992).
Six motifs that were conserved in 29 Brassicaceae species were shown to be important for enhancer activity in A. thaliana, with one of them revealing an unexpected aspect of AG regulation: repression in the shoot apical meristem by MADS domain proteins. On the other hand, the previously identified LFY and WUS binding sites were found to be more variable. Our studies illustrate both the strengths and weaknesses of phylogenetic footprinting and phylogenetic shadowing (Gumucio et al., 1992; Wasserman and Fickett, 1998; Boffelli et al., 2003), which have been promoted as a rapid means of genome-wide identification of regulatory sequences.
RESULTS
Divergence of AG Noncoding Sequences in 29 Brassicaceae Species
To identify candidate sequences for regulatory motifs, we began by sequencing the second AG intron from 28 Brassicaceae species in addition to A. thaliana (Table 1; detailed information on the accessions used as well as GenBank accession numbers are provided in the supplemental data online). The sequence identity for any pair of species ranged from 64 to 94%, with an average of 74%, which was close to the average difference of these species from A. thaliana, 77% (Table 1). Kaplinsky and colleagues (2002) recently reported the use of pair-wise BLAST (Basic Local Alignment Search Tool) analysis (Altschul et al., 1990) to identify short conserved motifs in genes of the Poaceae. This method is not useful in the Brassicaceae, because application of the parameters used by Kaplinsky and colleagues (2002) for comparison of Brassicaceae pairs identifies more than half of the AG intron as conserved.
Table 1.
Species from Which AG Introns Were Compared
| Brassicaceae Species | Percent Identity with A. thaliana |
|---|---|
| Alyssum saxatile | 72 |
| Arabidopsis thaliana cv Columbia | – |
| Arabidopsis (Cardaminopsis) arenosa | 86 |
| Arabidopsis lyrata | 86 |
| Arabis gunnisoniana | 83 |
| Arabis pumila | 83 |
| Barbarea vulgaris | 76 |
| Berteroa incana | 69 |
| Brassica oleracea var oleracea cv A12 | 76 |
| Cakile maritima | 77 |
| Camelina sativa | 78 |
| Capsella bursa-pastoris | 75 |
| Capsella rubella | 75 |
| Cheiranthus cheiri | 81 |
| Conringia orientalis | 80 |
| Coronopus squamatus | 71 |
| Diplotaxis catholica | 75 |
| Draba corrugata var corrugata | 72 |
| Eruca sativa | 75 |
| Erysimum capitatum | 81 |
| Guillenia flavescens | 78 |
| Lepidium africanum | 75 |
| Lepidium phlebopetalum | 72 |
| Lobularia maritima | 72 |
| Nasturtium officinale | 78 |
| Raphanus sativus cv Cherry Bell | 75 |
| Streptanthus insignis | 80 |
| Thlaspi arvense | 78 |
| Thysanocarpus sp | 78 |
| Non-Brassicaceae Species (This Study) | Gene |
| Cucumis sativus (cucumber; Cucurbitaceae) | CUM1 |
| C. sativus | CAG1 |
| C. sativus | CAG2 |
| Lycopersicon esculentum cv Microtom (tomato; Solanaceae) | TAG1 |
| Non-Brassicaceae Species (Known Previously) | Gene |
| Antirrhinum majus (snapdragon; Veronicaceae) | PLE |
| A. majus | FAR |
| Oryza sativa cv japonica (rice; Poaceae) | OsMADS3 |
| Petunia × hybrida (petunia; Solanaceae) | PMADS3 |
| Populus balsamifera subsp trichocarpa (poplar; Salicaceae) | PTAG1 |
| P. balsamifera subsp trichocarpa | PTAG2 |
| Zea mays (maize; Poaceae) | ZMM1 |
| Z. mays | ZAG2 |
See supplemental data online for details.
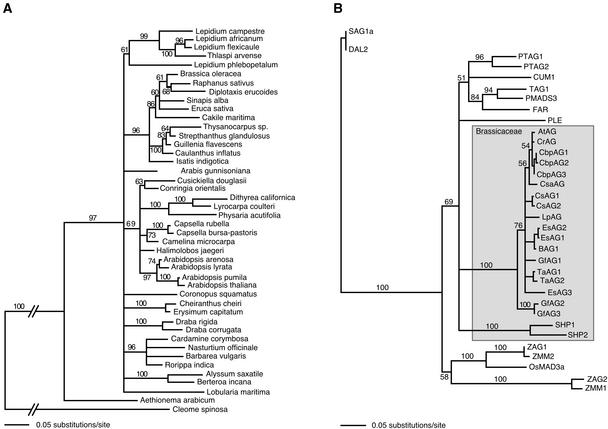
Because many of the species investigated had not been analyzed by molecular phylogeny, we established their relationship using the nuclear ribosomal internal transcribed spacer regions (ITS), including the 5.8S rDNA, a common phylogenetic marker (Figure 1A). The ITS sequences form a monophyletic group with the sequence from Aethionema as an outgroup, although some closely related species, such as Lobularia maritima and Alyssum saxatile, did not cluster together. In addition, we observed that a tree built from the AG intron sequences (data not shown) agrees with the ITS tree within the genera Brassica, Capsella, and Arabidopsis sensu stricto, in accordance with published phylogenetic relationships (Koch et al., 1999, 2000, 2001a; Yang et al., 1999; Mummenhoff et al., 2001). Together, these results indicate that all of the sequences came from the Brassicaceae.
Figure 1.
Brassicaceae ITS DNA Phylogeny and AG Protein Phylogeny.
(A) Rooted neighbor-joining distance tree of 5.8S rDNA ITS sequences using Akaike informational criterion log-likelihood DNA-substitution parameters. Bootstrap support (1000 replicates) is given next to the branches. This tree includes 26 of the 29 species analyzed for AG, plus additional species extracted from GenBank (see supplemental data online for accessions and sequences used). Outgroups were Aethionema, a basal Brassicaceae species, and Cleome, from the Capparaceae family, which some authors have included in the Brassicaceae sensu lato (Judd et al., 1994). The evolutionary distances between A. thaliana and other species used in this study are estimated between 5.8 million years (A. thaliana and A. arenosa) and 40 million years (Aethionema and the rest of the Brassicaceae) (Koch et al., 2001a).
(B) Neighbor-joining distance tree of predicted AG protein sequences. Putative AG orthologs of Capsella rubella (CrAG), Capsella bursa-pastoris (CbpAG), Camelina sativa (CsaAG), Coronopus squamatus (CsAG), Lepidium phlebopetalum (LpAG), Eruca sativa (EsAG), Guillenia flavescens (GfAG), and Thlaspi arvense (TaAG) are from this study. Numbered suffixes designate AG proteins in cases of multiple genomic copies. SHP1 and SHP2 are the closest paralogs of AG in A. thaliana and serve as the outgroup to Brassicaceae AG, whereas the AG-like proteins from monocots and gymnosperms (ZAG1/2, ZMM1/2, OsMAD3a, SAG1a, and DAL2) serve as outgroups to AG homologs from dicotyledons (PTAG1/2, CUM1, TAG1, FAR, PLE, and PMADS3).
The high degree of sequence identity across species not only facilitated unambiguous alignment along the entire AG intron, but it also suggested that these sequences are derived from orthologous genes. For eight species, we confirmed by reverse transcriptase–mediated PCR that the sequenced AG copy is expressed in flowers (data not shown). We then compared the phylogenetic relationships of the predicted protein sequences with those described previously for AG homologs outside of the Brassicaceae (Figure 1B). Because multiple copies of AG introns and cDNAs sometimes were isolated from the same species, it was possible that some of these were paralogs of A. thaliana AG. In A. thaliana, the closest AG paralogs are SHATTERPROOF1 (SHP1) and SHP2, both of which function in fruit development downstream of AG (Liljegren et al., 2000). The SHP1/2 introns cannot be aligned with the AG introns, nor do they contain the CCAATCA and aAGAAT motifs characteristic of AG introns from within and outside the Brassicaceae (see below). In addition, the second introns of SHP1 and SHP2 are only 1.3 and 2 kb in length, respectively, which is much shorter than any of the AG introns from the Brassicaceae. Finally, the partial AG protein sequences from the Brassicaceae form a monophyletic group, with SHP1/2 as a clear outgroup. Together, these results indicate that the sequences that we isolated are from AG orthologs or very recently arisen paralogs.
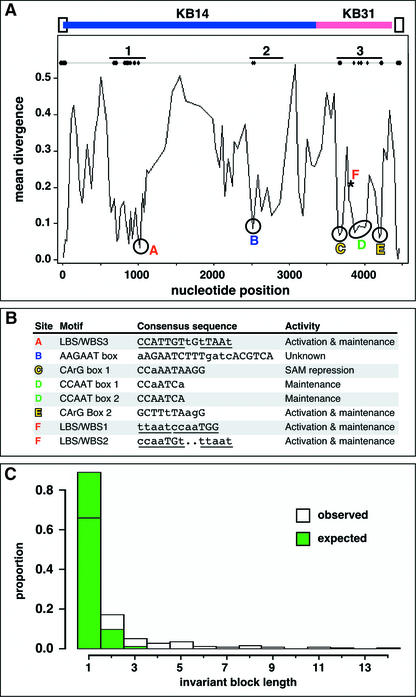
A sliding-window analysis of AG introns from the Brassicaceae revealed three major regions of reduced sequence divergence, which also correspond to clusters of largely invariant sequence blocks that are at least 6 bp long (Figure 2; see supplemental data online for detailed sequence alignments). Because of the generally high sequence identity throughout the Brassicaceae, we tested whether the pattern of identical base-pair blocks (Tang and Lewontin, 1999) was different from a random distribution by permuting the positions of invariant base pairs in the alignment 100 times (see Methods). Invariant blocks that were at least 6 bp long were significantly rarer in the permuted data set than in our observed data (Figure 2C, χ2 p = 0.0055), suggesting that these regions are under substitutional constraint.
Figure 2.
Comparisons of AG Introns from Brassicaceae Species.
(A) Sliding-window analysis. Flanking exon sequences are depicted as open boxes. KB14 and KB31 refer to complementary A. thaliana AG enhancers that confer similar expression patterns in reporter gene assays (Busch et al., 1999). Valleys indicate three regions of reduced sequence divergence and therefore high conservation. These regions also contain the only clusters of highly conserved blocks of at least 6 bp (>90% sequence identity across all positions and species), as indicated by diamonds on the line at top.
(B) Conserved motifs. Invariant positions are shown in uppercase letters. For the consensus sequences of the aAGAAT box and CCAATCA box 1, Draba was excluded, because both motifs are largely deleted in the AG intron sequenced from this species. For the adjacent LFY and WUS binding sites (LBS/WBS), the core motifs to which LFY and WUS bind are underlined. Dots indicate insertions/deletions. Activity refers to effects seen when these sites are mutated in the context of A. thaliana sequences.
(C) Observed distribution of invariant blocks compared with the distribution expected if individual invariant positions were arranged randomly within the sequence alignment. Green bars are shown in the foreground.
Region 1, which is ∼300 bp long, contains adjacent putative LFY and WUS binding sites (LBS/WBS3) that are much less variable among the 29 Brassicaceae species examined than are the two functionally characterized LBS/WBS1 and LBS/WBS2 located in the 3′ intron region of A. thaliana AG (Busch et al., 1999; Lohmann et al., 2001). Only 7 of 29 species have 1- or 2-bp differences in the putative WBS3, whereas LBS3 is invariant (see supplemental data online). Region 2, of ∼300 bp, is near the middle of the aligned sequences and begins with an aAGAAT motif also found outside the Brassicaceae (see below). Region 3, which spans ∼600 bp at the 3′ end of the introns, contains several motifs conserved in the 29 Brassicaceae species examined, including two consensus CArG boxes, CC(A/T)6GG, which are binding sites for MADS domain proteins (Shore and Sharrocks, 1995), and a pair of CCAATCA boxes, which are binding sites for CCAAT-box binding proteins (Mantovani, 1998). CArG box 1 is almost invariant in the Brassicaceae, with only two sequences, from Lepidium africanum and Nasturtium, containing a single A-to-G transition (see supplemental data online). Region 3 also contains the known LBS/WBS1 and LBS/WBS2, which are more variable than several of the other sites, or the putative LBS/WBS3 discussed above. Region 3 is contained within the 744-bp minimal fragment sufficient for early AG expression, KB31 (Busch et al., 1999).
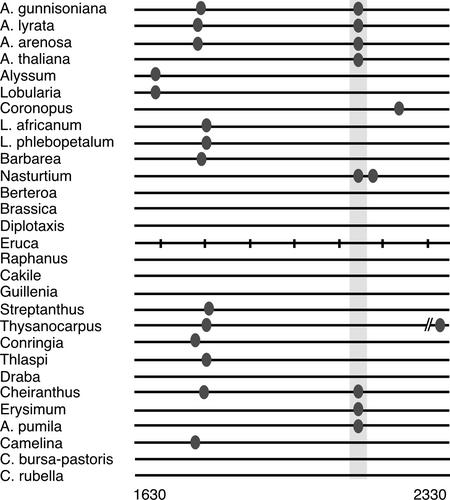
Although conserved motifs within aligned sequences are a good indication of functionally important sites, considerable shuffling of binding sites has been reported for a well-studied developmental enhancer in Drosophila (Ludwig and Kreitman, 1995; Ludwig et al., 1998, 2000). Therefore, we searched individual AG sequences for the presence of putative LFY binding sites using the consensus CCANTG(T/G) (Parcy et al., 1998; Busch et al., 1999). We found a fourth putative LBS in A. thaliana, located in the 5′ enhancer of A. thaliana AG, although this site is more variable than LBS3 (see supplemental data online). Several other species have LBS consensus sequences close by, sometimes in addition to LBS4, whereas other species lack such motifs in this region altogether (Figure 3).
Figure 3.
Putative LFY Binding Site 4 in Brassicaceae AG Introns.
Ovals denote the presence of LFY consensus binding sites (CCANTG[T/G]). The highlighted region indicates the position of LBS4 in A. thaliana (see supplemental data online). Some species have additional consensus motifs, whereas others lack them altogether. Species are ordered according to the phylogenetic relatedness of this region (data not shown). Numbers at bottom refer to the A. thaliana sequence.
Requirement of Candidate Regulatory Motifs for AG Enhancer Activity
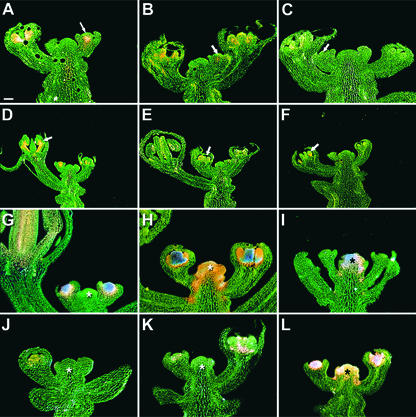
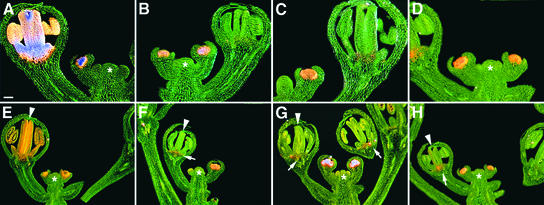
To determine the functions of motifs that are found across the 29 Brassicaceae species examined, we mutated them in the context of two AG reporter constructs, KB14 and KB31, which represent the 5′ and 3′ portions of the A. thaliana AG intron, respectively (Busch et al., 1999).
KB14, which spans the putative LBS3 and LBS4 motifs, is activated in the center of early-stage flowers and is expressed at later stages preferentially in stamens (Busch et al., 1999; Deyholos and Sieburth, 2000). A mutation in putative LBS3 (reporter RH149, CCATTGT to AAATTGT) had a modest effect, reducing reporter gene activity in both early and late stages (Figures 4B and 4E). Even the strongest of the 23 T1 lines tested was weaker than the intermediate KB14 reference line (Busch et al., 1999). By contrast, a mutation in the putative LBS4 (RH141, CCAATGT to AAAATGT) specifically affected early reporter gene activity (0 of 20 T1 plants showed early β-glucuronidase [GUS] expression), whereas the later activity in stamens remained largely unchanged (Figures 4C and 4F).
Figure 4.
Requirement of Putative LFY Binding Sites 3 and 4 and CArG Boxes for the Activity of AG Enhancers.
(A) and (D) KB14, wild-type 5′ AG enhancer reporter (Busch et al., 1999).
(B) and (E) RH149, with LBS/WBS3 mutated.
(C) and (F) RH141, with LBS4 mutated.
(G) KB31, wild-type 3′ AG enhancer reporter (Busch et al., 1999).
(H) MX144, with CArG box 1 mutated, shows ectopic GUS activity in the shoot apical meristem (asterisk).
(I) MX144 in lfy-12.
(J) MX215, with mutations in CArG box 1 and LBS1 and LBS2.
(K) RH155, with CArG box 2 mutated.
(L) RH174, with both CArG box 1 and 2 mutated.
Sections of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide–stained apices are shown. Staining intensities increase from orange to pink to purple. Arrows indicate staining in the center of early floral primordia between stages 3 and 6 and in the stamens, particularly in the developing filaments of stage-6 to -8 flowers. Bar in (A) = 50 μm for (A) to (C) and (G) to (L) and 100 μm for (D) to (F)
Like KB14, KB31 is activated in the center of early-stage flowers, but it is expressed at later stages in both stamens and carpels, with relatively stronger expression in carpels (Busch et al., 1999; Deyholos and Sieburth, 2000). We examined four motifs in the context of KB31, the two CArG boxes and the two CCAATCA boxes. Mutation of CArG box 1 (MX144) resulted in ectopic reporter gene expression in the shoot apical meristem, whereas the spatial and temporal expression patterns in young flowers remained unchanged (Figure 4H). Because the activity of KB31 normally requires the activity of LFY, which is expressed only in floral primordia, we were surprised that the CArG box 1 mutation was sufficient for ectopic activation of the AG enhancer outside of flowers. Consistent with LFY not being expressed in the shoot apical meristem, ectopic expression of MX144 was unaffected in lfy-12 mutants (Figure 4I). However, the CArG box 1 mutation was insufficient for ectopic AG enhancer activity when both LBS1 and LBS2 were mutated (Figure 4J), indicating that activation of the ectopic AG enhancer in the shoot apical meristem requires the LFY binding sites but not LFY protein. This independence from LFY protein but not LFY binding sites reveals an unknown activator of AG that interacts with the LFY binding sites.
In contrast to CArG box 1, a mutation in CArG box 2 caused a reduction in early expression without affecting the spatial pattern of AG enhancer activity (Figure 4K). Mutating both CArG boxes in the context of KB31 had additive effects, suggesting that the two CArG boxes do not interact with each other (Figure 4L).
A pair of directly repeated CCAATCA boxes is present in all Brassicaceae AG, with the exception of Draba, which is missing the region overlapping the 5′ CCAATCA box 1 (see supplemental data online). We deleted CCAATCA box 1, CCAATCA box 2, or the entire 49-bp region containing both CCAATCA boxes in the context of KB31. Reporter gene expression appeared somewhat lower but was otherwise largely normal during early floral stage 3 (Figures 5A to 5D). By contrast, the activity in stamens and carpels of stage-8 and -9 flowers was reduced or abolished in all three mutated reporters (Figures 5E to 5H). In the few lines in which GUS activity was detected in stage-8 and -9 flowers, reporter gene expression was restricted largely to the base of the gynoecium (Figures 5F to 5H).
Figure 5.
Requirement of CCAATCA Boxes for the Maintenance of AG Expression.
(A) and (E) KB31, wild-type 3′ AG enhancer reporter (Busch et al., 1999).
(B) and (F) RH47, with CCAATCA box 1 deleted.
(C) and (G) RH48, with CCAATCA box 2 deleted.
(D) and (H) RH49, with both CCAATCA boxes deleted.
Arrowheads in (E) to (H) indicate gynoecia from stage 8 on, and arrows indicate reporter gene activity at the base of the gynoecium. Asterisks indicate shoot apical meristems. Bar in (A) = 50 μm for (A) to (D) and 100 μm for (E) to (H).
Identification of AG cis-Regulatory Motifs Outside the Brassicaceae
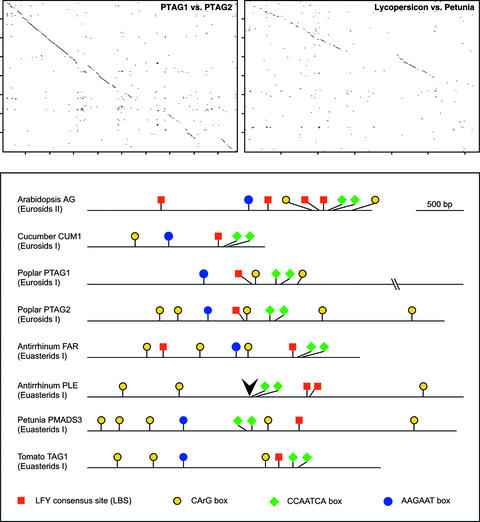
Having compared a large sample of AG enhancers at the family level, we asked whether the motifs identified in the set of 29 Brassicaceae species are found in more distantly related dicotyledons. In addition to previously sequenced AG orthologs and paralogs from poplar, snapdragon, and petunia (Tsuchimoto et al., 1993; Davies et al., 1999; Brunner et al., 2000), we isolated intron sequences from AG orthologs of cucumber and tomato (Table 1; see supplemental data online for GenBank accession numbers). The five species represented by these sequences are in different families, except for petunia and tomato, which are both in the Solanaceae. The length of the large intron immediately downstream of the MADS box ranges from 1993 bp (cucumber CUM1) to 4864 bp (poplar PTAG1). The introns of the two poplar paralogs, which share 71% identity, and the introns of petunia PMADS3 and tomato TAG1, which share 55% identity, can be aligned over much of their lengths (Figure 6).
Figure 6.
Locations of Putative cis-Regulatory Elements in AG Introns from Dicotyledons Outside the Brassicaceae.
Dot plots of poplar paralogs, PTAG1 and PTAG2, and of orthologs from tomato and petunia, which are in different subfamilies of the Solanaceae, are shown at top. The window size was 25 bp, with a minimum of 50% identity and a step size of 1 character. Diagrams of AG introns with putative cis-regulatory motifs are shown at bottom. The position of the Tam3 transposon insertion, which results in ectopic expression of the PLE ovulata mutant (Bradley et al., 1993), is indicated by the arrowhead. Regions of extended similarity that include several motifs are highlighted in gray. The lengths of the introns are as follows: AG, 2999 bp; CUM1, 1993 bp; PMADS3, 4011 bp; PTAG1, 4864 bp; PTAG2, 3882 bp; PLE, 4087 bp; FAR, 2965 bp; and TAG1, 3251 bp.
In the Cucurbitaceae, no motifs with obvious similarity to A. thaliana AG were found in the introns of cucumber AG paralogs CAG1 (=CUM10) and CAG2 (=CUS1), even though both are expressed in reproductive organs (Perl-Treves et al., 1998). Phylogenetic analysis of deduced protein sequences has indicated that CAG1 is related more distantly to other AG homologs than the very similar CUM1/CAG2 pair (Theissen et al., 2000). Based on in situ hybridization results and overexpression phenotypes (Kater et al., 1998, 2001) as well as visual inspection of the second intron, CUM1 is the cucumber gene most closely related to A. thaliana AG.
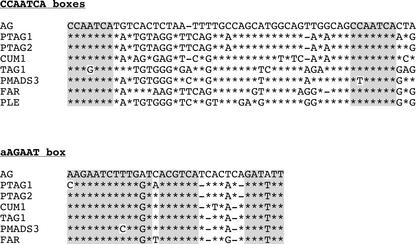
As a group, introns from outside the Brassicaceae are too divergent to be aligned across their whole lengths using software such as CLUSTAL X (Thompson et al., 1997), although CUM1, PTAG1, and PTAG2 introns shared more regions of at least 50% identity than were shared between any of these three and AG. This finding reflects the fact that cucumber (Cucurbitaceae) and poplar (Salicaceae) belong in the Eurosid I superorder, whereas A. thaliana (Brassicaceae) is in Eurosids II. Pair-wise BLAST alignment (Altschul et al., 1990) with the A. thaliana AG intron revealed two short regions that are very similar among all of these sequences, an aAGAAT box and a closely spaced pair of CCAATCA boxes also found in the Brassicaceae (Figure 1). Directly repeated CCAATCA boxes ∼37 bp apart have been noted previously in FAR, PLE, AG, and PTAG1 (Davies et al., 1999), and they are found in all AG introns of dicotyledons (Figures 6 and 7). The A. thaliana genome contains 472 pairs of CCAATCA boxes within 70 bp (http://plantenhancer.org/); if we assume a similar base composition of other dicotyledon genomes, a lower bound estimate for the random occurrence of two closely spaced CCAATCA boxes occurring in the 2- to 5-kb introns of AG orthologs is <2%. A 19-bp motif, AGAATCTNTGNTNACGTCA, corresponding to the aAGAAT motif defined in the comparison of Brassicaceae sequences, is found in all AG orthologs except snapdragon PLE (Figures 6 and 7). This is consistent with the observation that the FAR protein sequence is more similar to that of AG than is the PLE sequence (Davies et al., 1999). The A. thaliana genome contains only one perfect match to this motif.
Figure 7.
Conserved Motifs in Introns of AG Homologs from Outside the Brassicaceae.
Asterisks indicate positions identical to the AG sequence in each alignment. At top, CCAATCA boxes (shaded) are separated by a more variable region. At bottom, the aAGAAT motif is part of a more extended region of similarity. Searches in the AliBaba2.1 (http://www.gene-regulation.com) transcription factor binding site database did not identify obvious candidates that could bind to the core aAGAAT motif.
All introns contain at least one CArG box, (C/G)C(A/T)6GG, and all but that of cucumber CUM1 have at least one pair of CArG boxes within 700 bp (Figure 6). Although CArG boxes occur more often than once every 2 kb in the A. thaliana genome, clusters of at least two CArG boxes within 700 bp are rare; their chance occurrence in a 5-kb sequence is only 10%. We also found putative LFY binding sites, CCANTG(T/G) (Parcy et al., 1998; Busch et al., 1999), in all introns (Figure 6). In A. thaliana, three LBS are adjacent to core homeodomain consensus sites, TTAAT, two of which are known to be bound by WUS (Lohmann et al., 2001). In other dicotyledons, homeodomain consensus sites generally are not found next to putative LBS. Because the putative LFY binding sites are short, finding them in introns of AG orthologs is not significant by itself. More distantly in monocots, intron sequences of the maize AG-like paralogs ZMM1 and ZAG2 (Theissen et al., 1995) and of the rice AG-like gene OsMADS3 (Kang et al., 1998) are available. All three contain very limited similarity to the motif defined by the pair of CCAATCA boxes or to the aAGAAT motif (data not shown).
Activity of Cucumber AG Sequences in A. thaliana
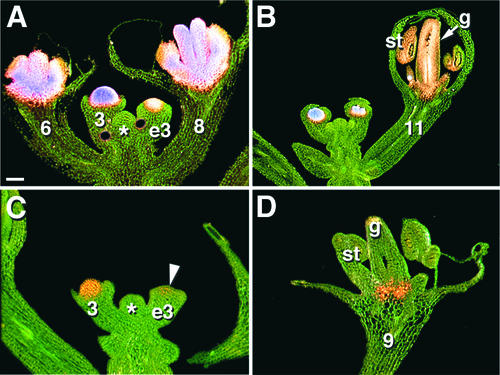
Because there is little overall sequence identity between the AG intron from A. thaliana and orthologs from outside the Brassicaceae, we wanted to determine whether introns of non-Brassicaceae species can perform similar roles in regulating flower-specific transcription. For functional analysis, we selected the CUM1 intron from cucumber, which is 30% shorter than that of AG and has only one CArG box but has a pair of CCAATCA boxes next to motifs with similarity to LBS/WBS from A. thaliana, although these are arranged differently than in AG. Analogous to the A. thaliana reporters, the CUM1 intron was placed upstream of the −46-bp minimal 35S gene promoter of Cauliflower mosaic virus driving a GUS reporter. The CUM1:GUS expression pattern in A. thaliana apices was similar to that of AG:GUS, although the expression levels often were lower (Figures 8A and 8C). Like that of AG, CUM1-driven reporter gene expression began during early stage 3 but reached its highest levels soon after. In contrast to the AG intron, the CUM1 intron did not drive any expression in stamens and carpels of later stage flowers (Figures 8B and 8D).
Figure 8.
Activity of the CUM1 Intron in A. thaliana.
(A) and (B) KB9, full-length A. thaliana AG intron reporter.
(C) and (D) CUM1:GUS apices.
Shoot apical meristems are indicated by asterisks, and numbers indicate floral stages (Smyth et al., 1990). The onset of expression (arrowhead in [C]) during the early stage (e3) is similar in AG:GUS and CUM1:GUS, but CUM1:GUS expression is not maintained as long, except for staining at the base of the gynoecium (D). Staining intensities increase from orange to pink to purple. g, gynoecium; st, stamens. Bar in (A) = 50 μm for (A) and (B) and 100 μm for (C) and (D).
DISCUSSION
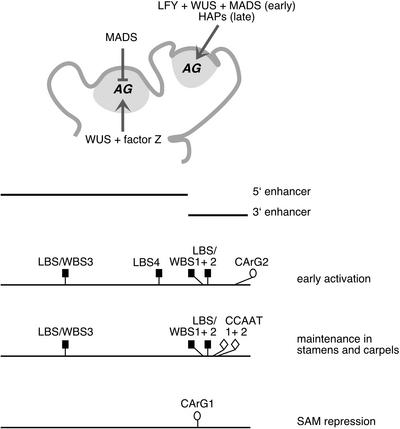
We have examined noncoding sequences of the floral homeotic gene AG in a range of dicotyledons (from Asteridae to Rosidae) and in a large number of Brassicaceae species. We found that comparison of AG sequences between distantly related species reveals only a small number of putative cis-regulatory sequences. On the other hand, within the Brassicaceae, sequence identity is high, and considerably more than two species need to be compared in this variation of the phylogenetic footprinting approach, for which the term phylogenetic shadowing has been coined (Boffelli et al., 2003). At least six of the seven motifs identified by sequence comparison within the Brassicaceae are required for function in A. thaliana, although the degree of variability is not a direct indicator for the importance of a site. For example, two transcription factor binding sites (LBS/WBS1 and LBS/WBS2) shown previously to be essential for enhancer activity in A. thaliana are more variable than several other, newly discovered motifs. Similarly, the invariant LBS3 is less important than the more variable LBS4 for early AG expression, indicating the limitations of this approach. A summary of our findings is shown in Figure 9.
Figure 9.
Summary of AG Regulation.
Initiation and early AG expression in flowers requires all four LFY and LFY/WUS binding sites, as well as CArG box 2. The same LFY/WUS binding sites, along with a pair of CCAATCA boxes, also are required for the maintenance of AG expression in maturing carpels and stamens. In the shoot apical meristem (SAM), CArG box 1 mediates the repression of AG by a MADS domain protein(s). This repression appears to prevent the ectopic activation of AG by WUS, which is expressed in the shoot apical meristem (Mayer et al., 1998), and possibly by another unknown protein, factor Z.
Regulatory Function of AG Intron Sequences Outside the Brassicaceae
We have shown that the ability of the second intron to direct flower-specific expression is shared by at least one species outside the Brassicaceae, cucumber. In snapdragon, a transposon insertion that causes ectopic expression of PLE has been mapped to the second intron (Bradley et al., 1993), also suggesting that this intron regulates flower-specific expression outside the Brassicaceae. Despite the similar function of AG introns, the only extended motifs found throughout the dicotyledons are a pair of CCAATCA boxes, which are required for the maintenance of AG expression and a newly discovered motif, the aAGAAT box.
Shuffling and Divergence of LFY and WUS Binding Sites
Initial studies of the AG intron in A. thaliana suggested that the 5′ and 3′ portions are redundant, because they specify similar early expression patterns (Busch et al., 1999). A more detailed examination has revealed that the 3′ enhancer is more important for the early activation of AG and expression in carpels, whereas the 5′ enhancer appears to be more important for late expression in stamens (Deyholos and Sieburth, 2000). The 3′ enhancer contains two adjacent pairs of LBS and WBS, which are required for its activity (Busch et al., 1999; Lohmann et al., 2001). The 5′ enhancer contains a putative third LBS adjacent to a putative WBS, and both are much less variable than LBS/WBS1 and LBS/WBS2 found in the 3′ enhancer of A. thaliana. LBS/WBS3, which is found throughout the Brassicaceae, appears to be required for both early and late activity of the 5′ enhancer, similar to the requirement of LBS/WBS1 and LBS/WBS2 for the activity of the 3′ enhancer. Although a fourth putative LBS is more variable in both its sequence and its location along the intron than LBS3 and is not adjacent to a putative WUS binding site, mutating LBS4 abolishes all early AG enhancer activity in the context of the 5′ enhancer. Thus, the more variable LBS4 is more important in A. thaliana than the almost invariant LBS/WBS3. Although LBS4 is not found at the same position in all 29 Brassicaceae species, a motif similar to LBS4 is present elsewhere in the 5′ region of the AG intron in most Brassicaceae species. Changes in number and location also have been documented for the BICOID binding sites in the hunchback P2 promoter of higher Diptera species as well as for several other transcription factor binding sites in the even-skipped stripe 2 enhancer from Drosophila (Ludwig et al., 1998; McGregor et al., 2001a, 2001b).
Identification of Additional Regulatory Elements by Phylogenetic Footprinting and Shadowing
By far, the most highly conserved motif in all dicotyledon AG introns includes a pair of CCAATCA boxes, which are required for AG enhancer activity during the later stages of flower development. That these motifs are important for AG regulation is further indicated by the finding that the weak ag-11 allele, in which lateral stamens are transformed into petals, has a point mutation immediately downstream of the second CCAATCA box (S. Liljegren and M. Yanofsky, personal communication). The CCAATCA consensus matches the recognition site of the NF-YA/NF-YB/NF-YC (Nuclear Factor-YA/YB/YC) heterotrimeric complex in vertebrates (also known as HAP2/3/5 in yeast) (Mantovani, 1998), but it is not known which if any of the 23 NF-Y homologs expressed in A. thaliana regulates AG expression (Kwong et al., 2003). The role of the equally conserved aAGAAT motif, which does not match a known consensus binding site of any transcription factor, remains to be determined. Ultimately, it will be important to demonstrate that these motifs have functions in other species similar to those in A. thaliana.
Within the Brassicaceae, two motifs found close to LBS/WBS1 and LBS/WBS2 are CArG boxes, of which one is required for repression in the shoot meristem and the other is required for general activation. CArG box 1 is the only AG intron mutation found to date that causes a dramatic alteration in the spatial pattern of enhancer activity (Busch et al., 1999; Deyholos and Sieburth, 2000; R.L. Hong and D. Weigel, unpublished data). Surprisingly, AG enhancer activity in the shoot apical meristem is independent of LFY protein but dependent on the previously identified LFY binding sites. This unexpected result suggests the presence in the shoot apical meristem of another transcription factor that can bind the AG enhancer but whose effect on AG normally is masked by repressors binding to CArG box 1.
CArG boxes are bound by MADS domain proteins (Norman et al., 1988; Huang et al., 1993, 1996; Acton et al., 1997), and several genes that encode MADS domain proteins are expressed in the shoot apical meristem, especially after the transition to flowering (Mandel and Yanofsky, 1995; Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; M. Yanofsky, personal communication). Using a gain-of-function approach, we have identified AGL6 as a candidate factor that mediates AG repression in the shoot apical meristem (R.L. Hong, F. Godard, and D. Weigel, unpublished results).
Prospects for Phylogenetic Footprinting and Shadowing in Plants
There has been much interest recently in the use of genome-wide sequence comparisons to identify conserved regulatory elements as a scalable alternative to tedious gene-by-gene promoter dissection (Jareborg et al., 1999; Dubchak et al., 2000; Wasserman et al., 2000; Bergman and Kreitman, 2001; Levy et al., 2001; Kaplinsky et al., 2002). We have found for AG that the divergence of A. thaliana and Brassica oleracea, another Brassicaceae species with extensive genome sequence information (Colinas et al., 2002), is itself not very informative (Table 1). Even the most divergent pair we found, Lepidium phlebopetalum and Berteroa incana, still share 64% sequence identity. Similarly, the degree of divergence that we found between tomato and petunia, which are in different dicotyledon families, is only slightly less than that seen with members of the Brassicaceae. On the other hand, comparisons between dicotyledon families identify only a small number of conserved elements of obvious significance. It is possible that a judicious choice of a species outside, but still close to, the Brassicaceae would have partially overcome this limitation. Examples of such species include members of the Caricaceae, which could be papaya, and the Malvaceae.
It was reported recently that conserved noncoding motifs can be identified by comparing sequences from the monocots maize and rice, both of which are members of the Poaceae (Kaplinsky et al., 2002). The families Brassicaceae and Poaceae are of similar age, likely having arisen ∼40 and 50 million years ago, respectively (Kellogg, 2001; Koch et al., 2001a). Within the Poaceae, the number of motifs identified by maize and rice comparison is small, only one or two for most genes (Kaplinsky et al., 2002). These findings suggest that the divergence of noncoding sequences among members of the Poaceae is similar to that found for members of different dicotyledon families, rather than within a single dicotyledon family, perhaps as a result of a faster mutational rate in the grasses (Song et al., 2002).
An important question is whether our findings can be extrapolated to other genes, because we analyzed regulatory sequences located in an intron. In Drosophila, patterns of sequence divergence are similar for intergenic and intron sequences (Bergman and Kreitman, 2001). In A. thaliana, the regulatory sequences of another floral homeotic gene, APETALA3 (AP3), are located in the promoter, and its sequence has been compared between A. thaliana and B. oleracea (Hill et al., 1998) and, more recently, among 14 other Brassicaceae species (Koch et al., 2001b). The general pattern observed was similar to the pattern reported here. The AP3 promoter of A. thaliana contains three CArG boxes with different roles in AP3 regulation, of which only CArG box 2 is identical between B. oleracea and A. thaliana (Hill et al., 1998; Tilly et al., 1998). Yet, when additional species are considered, CArG box 3 is not more variable than CArG box 2, thus underscoring the importance of examining several species (Koch et al., 2001b).
The value of using several closely related species for the identification of functionally important motifs has been noted (Dubchak et al., 2000; Cliften et al., 2001; Dermitzakis and Clark, 2002). After this study was completed, a similar study appeared in which sequence divergence within primates was exploited (Boffelli et al., 2003); the authors of that study came to similar conclusions as we did here. Although our approach can identify important elements, it will miss at least some, such as the previously characterized LFY/WUS binding sites 1 and 2. Thus, neither phylogenetic footprinting nor phylogenetic shadowing is a panacea for rapidly understanding transcriptional regulation on a genomic scale, but they can be effective when combined with traditional reporter mutational analysis. In the future, our comparisons would benefit from a more extensive molecular framework for understanding how regulatory sequences evolve, for which both metazoan and plant models should be considered.
METHODS
Growth Conditions
Seeds of different species were kindly provided by the individuals and institutions listed in the supplemental data online. Seeds were stratified at 4°C in the dark for 3 days and sown directly onto soil in the greenhouse or in growth rooms under a 16-h-light/8-h-dark cycle at 23°C. Seeds that did not readily germinate on soil, most notably from Capsella and Lepidium species, were sown on half-strength Murashige and Skoog (1962) agar containing 30 μM gibberellic acid, and the seedlings were transplanted onto soil. For DNA extraction, we used one individual per accession. When possible, seeds derived from selfing were collected from the individual used as the DNA source.
Oligonucleotide Primers
Sequences of oligonucleotide primers used for PCR amplification, site-directed mutagenesis, and genotyping are listed in the supplemental data online.
PCR Amplification of AG Introns
Genomic DNA from leaves of a single plant was isolated using a modified cetyl-trimethyl-ammonium bromide protocol (Lukowitz et al., 2000). Nested oligonucleotide primers flanking the second intron were designed based on the Brassica napus BAG1 cDNA sequence (primary primers oRH1050 and oRH1051, secondary primers oRH1034 and oRH1035) (Mandel et al., 1992), tomato TAG1 cDNA (primary primers oRH1046 and oRH1047, secondary primers oRH1040 and oRH1041) (Pnueli et al., 1994), and cucumber CUM1 (CAG3) (primary primers oRH1044 and oRH1045, secondary primers oRH1038 and oRH1039), CAG1 (primary primers oRH1044 and oRH1045, secondary primers oRH1038 and oRH1039), and CAG2 cDNA (primary primers oRH1058 and oRH1059, secondary primers oRH1056 and oRH1057) (Kater et al., 1998; Perl-Treves et al., 1998). Primary PCR for 26 cycles was performed using ExTaq polymerase (Takara, Shiga, Japan). After 1:200 dilution of the primary reaction product as a template, secondary PCR was performed for 35 cycles. Conditions were as follows: 92°C for 1 min, 55 to 60°C for 1 min, and 72°C for 3 min per cycle.
PCR Amplification of Internal Transcribed Spacers
Primers N-471 and N-472 (Koch et al., 1999) were used, with the same conditions as for the AG intron, except for a shorter extension time of 1 min.
Cloning and Sequencing
In general, PCR products were gel isolated, cloned into pGEM-T Easy (Promega, Madison, WI), and sequenced with Applied Biosystems BigDye Terminators version 3 on ABI Sequencers 3700 or 3100 (Foster City, CA). Sequences were assembled with Autoassembler version 2.0 (Applied Biosystems). For AG introns, two clones from each PCR initially were sequenced with flanking SP6 and T7 primers. Additional clones were sequenced with these primers if the two initial clones showed substantial differences. One clone representing each species was sequenced completely by primer walking. For internal transcribed spacer regions (ITS), a single clone was sequenced on both strands. For reverse transcriptase–mediated PCR of AG cDNA, RNA was extracted from two to three floral buds from a single plant using the Qiagen Plant RNeasy kit (Valencia, CA). After reverse transcription using oligo(dT) primers with AMV-RT Polymerase (Promega) to generate single-stranded cDNA, PCR was performed with ExTaq (Takara). Primers RH1028 and RH1029 amplified AG mRNA transcripts beginning 28 bases downstream of the AG translational start codon (ACG) in the MADS box until 2 bases before the last codon (GTG) to yield ∼730-bp fragments. PCR products from one to two reactions were cloned into the pGEM-T Easy vector (Promega), and at least four clones were sequenced in both directions with T7 and SP6 primers. Only one type of insert (RH184) was isolated from Lepidium phlebopetalum, which shared an AA insertion 42 bases from the last base, likely because of AMV-RT or ExTaq polymerase error. This insertion was removed manually to predict the LpAG coding sequence.
Phylogenetic Analysis
Sequences were aligned with CLUSTAL X (Thompson et al., 1997), resulting in lengths of 4447 and 643 characters for AG introns and ITS, respectively. Sequences were further aligned manually using Se-Al version 2.0 (Andrew Rambaut, Oxford University, UK). Likelihood settings for AG introns (TrN+I+G; Tamura-Nei model with proportion of invariable sites and gamma distribution [Tamura and Nei, 1993]) and ITS (SYM+I+G; symmetrical model with proportion of invariable sites and gamma distribution [Zharkikh, 1994]) were selected using the Akaike informational criterion for best-fit DNA substitution models based on log-likelihood scores produced by Modeltest version 3.06 (Posada and Crandall, 1998). These substitution models then were used to generate neighbor-joining distance trees bootstrapped with 1000 replicates in Paup4.0b8 (Swofford, 1993). Phylogenetic trees using predicted AG amino acid sequences also were constructed in PAUP using the neighbor-joining distance method. We obtained a similar tree using sequences without the highly conserved MADS domain. Sliding-window analysis was performed with DNA SP version 3.51 (Rozas and Rozas, 1999) with a window size of 20 bp at steps of 20 characters. To determine whether invariant positions were arranged randomly within the aligned Brassicaceae sequences, we converted the alignment into a string of 0s and 1s, indicating variable and invariant positions (Tang and Lewontin, 1999). Permutations of the positions of 0s and 1s were generated using a script implemented in R (with help from B. Schönfisch, University of Tübingen, and N. Warthmann, Max Planck Institute). Pair-wise distances from dot matrices were generated with MacVector 6.0.1 (Oxford Molecular Biology Group PLC) with a window size of 25 bp and a minimum of 50% identity. Long sequence alignments between Arabidopsis thaliana and non-Brassicaceae species were visualized with the World Wide Web version of VISTA, using as a parameter 50% identity over 25 bp (Dubchak et al., 2000).
Site-Directed Mutagenesis
PCR-based mutagenesis was performed using mutagenic primers or primers flanking the deletion sites. Mutated bases are underlined in the supplemental data online. After PCR with Turbo-Pfu (Stratagene), the vector templates were digested with DpnI and transformed into Escherichia coli. Mutations were verified by sequencing.
For LBS3, PCR mutagenesis using primers oN-583 and oN-584 was performed on the SpeI fragment of the 5′ enhancer (pRH134) to yield pRH135, which then replaced the same region in the full-length AG intron to yield pRH147. For LBS4, PCR mutagenesis was performed on pKB8 (full AG intron) using primers oN-654 and oN-655 to yield pRH140. Mutations in the 3′ AG enhancer were first introduced in pKB42, containing the 3′ XbaI-HindIII fragment (Busch et al., 1999). For CArG box 1 mutation (pMX144), oMX1133 and oMX1134 were used; for CArG box 2 mutation (pRH155), oN-601 and oN-602 were used; for CArG box 1+2 mutations (pRH174), oN-601 and oN-602 were used on pMX141 containing mutated CArG box 1; for CCAATCA box 1 deletion (pRH47), oRH1082 and oRH1083 were used; for CCAATCA box 2 deletion (pRH48), oRH1084 and oRH1085 were used; for deletion of both CCAATCA boxes (pRH49), oRH1095 and oRH1096 were used. For simultaneous mutations in LBS1, LBS2, and CArG box 1 (pMX215), oMX1133 and oMX1134 were used on pMX68 carrying LBS1 and LBS2 mutations (Busch et al., 1999).
Plant Vectors and Transformation
β-Glucuronidase (GUS) reporters were in the pDW294 background, with the −46-bp 35S promoter of Cauliflower mosaic virus driving GUS (Busch et al., 1999). Mutations in LBS3 and LBS4 were introduced into plants in the context of the 5′ enhancer (present in a 2.2-kb HindIII-BamHI fragment) in the forward direction, pKB14 (Busch et al., 1999). For mutated variants of the A. thaliana 3′ enhancer, 0.7-kb BamHI-HindIII fragments were cloned into pDW294, yielding mutated versions of the pKB31 reporter (Busch et al., 1999). The CUM1 intron was cloned into pDW294 in the reverse orientation, because this orientation gave more consistent and stronger GUS expression with the A. thaliana AG intron (Busch et al., 1999). Transgenic lines in the Columbia ecotype were generated by floral dipping and selected on kanamycin medium (Weigel and Glazebrook, 2002).
Histology
Inflorescences were stained for GUS activity as described (Blázquez et al., 1997), embedded in Paraplast, and sectioned at 10 μm thickness. Between 10 and 25 individual T1 lines were examined for each reporter construct.
Availability of Material
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers (AY253235–AY253268, AY254527–AY254546, and AY254702–AY254705) of AG introns, partial cDNA sequences of newly isolated AG copies from Brassicaceae, and ITS sequences are listed in the supplemental data online. cDNA accession numbers are as follows: Brassica napus BAG1 (M99415 [Mandel et al., 1992]), tomato TAG1 (AW035543.2 [Pnueli et al., 1994]), cucumber CUM1 (CAG3), CAG1, and CAG2 (AF035438, AF022378, and AF022377 [Kater et al., 1998; Perl-Treves et al., 1998]). Other accession numbers are X81199 and X80206 for ZMM1 and ZAG2, respectively (Theissen et al., 1995), and P0489A05.5 for the rice AG-like gene OsMADS3.
Supplementary Material
Acknowledgments
We are grateful for the help of Kristina Gremski in isolating and sequencing several Brassicaceae AG introns and for the help of Birgitt Schönfisch and Norman Warthmann with statistical analyses. We thank David Baum, Justin Borevitz, Des Bradley, José Dinneny, Marcus Koch, Sarah Liljegren, Jan Lohmann, Julin Maloof, Tom Mitchell-Olds, Bob Schmidt, Marty Yanofsky, Phillip Wigge, and Xuelin Wu for discussions, sharing unpublished results, and manuscript review. The manuscript was improved by comments from anonymous molecular evolutionists who acted as reviewers. We also thank the many individuals and institutions who provided seeds of various Brassicaceae species. This work was supported by a National Institutes of Health Training Grant to R.L.H., a fellowship from the Human Frontier Science Program Organization to M.A.B., grants from the National Institutes of Health (GM62932) and the U.S. Department of Energy (DE-FG03-98ER20317) to D.W., and the Max Planck Society. D.W. is a Director of the Max Planck Institute.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009548.
Footnotes
Online version contains Web-only data.
References
- Acton, T.B., Zhong, H., and Vershon, A.K. (1997). DNA-binding specificity of Mcm1: Operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol. 17, 1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bergman, C.M., and Kreitman, M. (2001). Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11, 1335–1345. [DOI] [PubMed] [Google Scholar]
- Berman, B.P., Nibu, Y., Pfeiffer, B.D., Tomancak, P., Celniker, S.E., Levine, M., Rubin, G.M., and Eisen, M.B. (2002). Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez, M.A., Soowal, L., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844. [DOI] [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. (2003). Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299, 1391–1394. [DOI] [PubMed] [Google Scholar]
- Borner, R., Kampmann, G., Chandler, J., Gleissner, R., Wisman, E., Apel, K., and Melzer, S. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Carpenter, R., Sommer, H., Hartley, N., and Coen, E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72, 85–95. [DOI] [PubMed] [Google Scholar]
- Brunner, A.M., Rottmann, W.H., Sheppard, L.A., Krutovskii, K., DiFazio, S.P., Leonardi, S., and Strauss, S.H. (2000). Structure and expression of duplicate AGAMOUS orthologues in poplar. Plant Mol. Biol. 44, 619–634. [DOI] [PubMed] [Google Scholar]
- Busch, M.A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587. [DOI] [PubMed] [Google Scholar]
- Clark, A.G. (2001). The search for meaning in noncoding DNA. Genome Res. 11, 1319–1320. [DOI] [PubMed] [Google Scholar]
- Cliften, P.F., Hillier, L.W., Fulton, L., Graves, T., Miner, T., Gish, W.R., Waterston, R.H., and Johnston, M. (2001). Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11, 1175–1186. [DOI] [PubMed] [Google Scholar]
- Colinas, J., Birnbaum, K., and Benfey, P.N. (2002). Using cauliflower to find conserved non-coding regions in Arabidopsis. Plant Physiol. 129, 451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E.H., et al. (2002). A genomic regulatory network for development. Science 295, 1669–1678. [DOI] [PubMed] [Google Scholar]
- Davies, B., Motte, P., Keck, E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18, 4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E.T., and Clark, A.G. (2002). Evolution of transcription factor binding sites in mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 19, 1114–1121. [DOI] [PubMed] [Google Scholar]
- Deyholos, M.K., and Sieburth, L.E. (2000). Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12, 1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Loots, G.G., Pachter, L., Mayor, C., Rubin, E.M., and Frazer, K.A. (2000). Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10, 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumucio, D.L., Heilstedt-Williamson, H., Gray, T.A., Tarle, S.A., Shelton, D.A., Tagle, D.A., Slightom, J.L., Goodman, M., and Collins, F.S. (1992). Phylogenetic footprinting reveals a nuclear protein which binds to silencer sequences in the human gamma and epsilon globin genes. Mol. Cell. Biol. 12, 4919–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, T.A., Day, C.D., Zondlo, S.C., Thackeray, A.G., and Irish, V.F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125, 1711–1721. [DOI] [PubMed] [Google Scholar]
- Huang, H., Mizukami, Y., Hu, Y., and Ma, H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Tudor, M., Su, T., Zhang, Y., Hu, Y., and Ma, H. (1996). DNA binding properties of two Arabidopsis MADS domain proteins: Binding consensus and dimer formation. Plant Cell 8, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jareborg, N., Birney, E., and Durbin, R. (1999). Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 9, 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd, W.S., Sanders, R.W., and Donoghue, M.J. (1994). Angiosperm family pairs: Preliminary phylogenetic analyses. Harv. Pap. Bot. 5, 1–51. [Google Scholar]
- Kang, H.G., Jeon, J.S., Lee, S., and An, G. (1998). Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. 38, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kaplinsky, N.J., Braun, D.M., Penterman, J., Goff, S.A., and Freeling, M. (2002). Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99, 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater, M.M., Colombo, L., Franken, J., Busscher, M., Masiero, S., Van Lookeren Campagne, M.M., and Angenent, G.C. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater, M.M., Franken, J., Carney, K.J., Colombo, L., and Angenent, G.C. (2001). Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 13, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, M., Bishop, J., and Mitchell-Olds, T. (1999). Molecular systematics and evolution of Arabidopsis and Arabis. Plant Biol. 1, 529–537. [Google Scholar]
- Koch, M., Haubold, B., and Mitchell-Olds, T. (2001. a). Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am. J. Bot. 88, 534–544. [PubMed] [Google Scholar]
- Koch, M.A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498. [DOI] [PubMed] [Google Scholar]
- Koch, M.A., Weisshaar, B., Kroymann, J., Haubold, B., and Mitchell-Olds, T. (2001. b). Comparative genomics and regulatory evolution: Conservation and function of the Chs and Apetala3 promoters. Mol. Biol. Evol. 18, 1882–1891. [DOI] [PubMed] [Google Scholar]
- Kopp, A., Duncan, I., and Carroll, S.B. (2000). Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553–559. [DOI] [PubMed] [Google Scholar]
- Kwong, R.W., Bui, A.Q., Lee, H., Kwong, L.W., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S., Hannenhalli, S., and Workman, C. (2001). Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics 17, 871–877. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., and Weigel, D. (2002). Building beauty: The genetic control of floral patterning. Dev. Cell 2, 135–142. [DOI] [PubMed] [Google Scholar]
- Ludwig, M.Z., Bergman, C., Patel, N.H., and Kreitman, M. (2000). Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564–567. [DOI] [PubMed] [Google Scholar]
- Ludwig, M.Z., and Kreitman, M. (1995). Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Mol. Biol. Evol. 12, 1002–1011. [DOI] [PubMed] [Google Scholar]
- Ludwig, M.Z., Patel, N.H., and Kreitman, M. (1998). Functional analysis of eve stripe 2 enhancer evolution in Drosophila: Rules governing conservation and change. Development 125, 949–958. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis: Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A., Bowman, J.L., Kempin, S.A., Ma, H., Meyerowitz, E.M., and Yanofsky, M.F. (1992). Manipulation of flower structure in transgenic tobacco. Cell 71, 133–143. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani, R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26, 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein, M., Markstein, P., Markstein, V., and Levine, M.S. (2002). Genome-wide analysis of clustered dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99, 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F.X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McGregor, A.P., Shaw, P.J., and Dover, G.A. (2001. a). Sequence and expression of the hunchback gene in Lucilia sericata: A comparison with other Dipterans. Dev. Genes Evol. 211, 315–318. [DOI] [PubMed] [Google Scholar]
- McGregor, A.P., Shaw, P.J., Hancock, J.M., Bopp, D., Hediger, M., Wratten, N.S., and Dover, G.A. (2001. b). Rapid restructuring of bicoid-dependent hunchback promoters within and between Dipteran species: Implications for molecular coevolution. Evol. Dev. 3, 397–407. [DOI] [PubMed] [Google Scholar]
- Mummenhoff, K., Brüggemann, H., and Bowman, J.L. (2001). Chloroplast DNA phylogeny and biogeography of Lepidium (Brassicaceae). Am. J. Bot. 88, 2051–2063. [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Norman, C., Runswick, M., Pollock, R., and Treisman, R. (1988). Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55, 989–1003. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M.A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. [DOI] [PubMed] [Google Scholar]
- Perl-Treves, R., Kahana, A., Rosenman, N., Xiang, Y., and Silberstein, L. (1998). Expression of multiple AGAMOUS-like genes in male and female flowers of cucumber (Cucumis sativus L.). Plant Cell Physiol. 39, 701–710. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Hareven, D., Rounsley, S.D., Yanofsky, M.F., and Lifschitz, E. (1994). Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D., and Crandall, K.A. (1998). MODELTEST: Testing the model of DNA substitution. Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and Rozas, R. (1999). DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Shore, P., and Sharrocks, A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca, V., and Messing, J. (2002). Mosaic organization of orthologous sequences in grass genomes. Genome Res. 12, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena, E., and Stern, D.L. (2000). Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. USA 97, 4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama, K., Kim, C.B., and Ruddle, F.H. (2001). An efficient cis-element discovery method using multiple sequence comparisons based on evolutionary relationships. Genomics 71, 260–262. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (1993). PAUP: A Computer Program for Phylogenetic Inference Using Maximum Parsimony. (Washington, DC: Smithsonian Institute).
- Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. [DOI] [PubMed] [Google Scholar]
- Tang, H., and Lewontin, R.C. (1999). Locating regions of differential variability in DNA and protein sequences. Genetics 153, 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J.T., Münster, T., Winter, K.U., and Saedler, H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. [PubMed] [Google Scholar]
- Theissen, G., Strater, T., Fischer, A., and Saedler, H. (1995). Structural characterization, chromosomal localization and phylogenetic evaluation of two pairs of AGAMOUS-like MADS-box genes from maize. Gene 156, 155–166. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL-X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly, J.J., Allen, D.W., and Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125, 1647–1657. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto, S., van der Krol, A.R., and Chua, N.-H. (1993). Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.L., Stec, A., Hey, J., Lukens, L., and Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. [DOI] [PubMed] [Google Scholar]
- Wasserman, W.W., and Fickett, J.W. (1998). Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 278, 167–181. [DOI] [PubMed] [Google Scholar]
- Wasserman, W.W., Palumbo, M., Thompson, W., Fickett, J.W., and Lawrence, C.E. (2000). Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 26, 225–228. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Yang, Y.-W., Lai, K.-N., Tai, P.-Y., Ma, D.-P., and Li, W.-H. (1999). Molecular phylogenetic studies of Brassica, Rorippa, Arabidopsis and allied genera based on the internal transcribed spacer region of 18S-25S rDNA. Mol. Phylogenet. Evol. 13, 455–462. [DOI] [PubMed] [Google Scholar]
- Zharkikh, A. (1994). Estimation of evolutionary distance between nucleotide sequences. J. Mol. Evol. 39, 315–329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.