Abstract
The association of Cryptosporidium oocysts with biofilm communities can influence the propagation of this pathogen through both environmental systems and water treatment systems. We observed the capture and retention of C. parvum oocysts in Pseudomonas aeruginosa biofilms using laboratory flow cells. Biofilms were developed in two different growth media using two different strains of P. aeruginosa, a wild-type strain (PAO1) and a strain that overproduces the exopolysaccharide alginate (PDO300). Confocal laser-scanning microscopy was used in conjunction with image analysis to assess the structure of the biofilms prior to introducing oocysts into the flow cells. More oocysts were captured by the biofilm-coated surfaces than the abiotic glass surface in both media. There was no significant difference in capture across the two strains of P. aeruginosa biofilm, but the fraction of oocysts captured was positively related to biofilm roughness and surface-area-to-volume ratio. Once captured, oocysts were retained in the biofilm for more than 24 h and were not released after a 40-fold increase in the system flow rate. We believe the capture and retention of oocysts by biofilm communities can impact the environmental transmission of C. parvum, and this interaction should be taken into consideration when predicting the migration of pathogens in the environment.
The human pathogen Cryptosporidium parvum is responsible for numerous waterborne disease outbreaks in the United States (15, 20, 34, 41, 43). Outside its host, C. parvum exists as a nonreproductive oocyst, ∼5 μm in diameter, that is resistant to typical environmental stresses (6, 29, 39, 40). C. parvum oocysts originate from the waste of infected hosts and are discharged in large quantities from municipal wastewater treatment facilities, animal agriculture, and wildlife populations (2, 18, 26, 47). Because oocysts are persistent in the environment, the transmission of viable oocysts from sources to public water supplies can result in human infection even over long transport distances. Therefore, protection of public health requires a clear understanding of the factors that control the migration of Cryptosporidium in the environment.
The transport of C. parvum oocysts can be influenced by interactions with surface-attached microbial communities, generally termed biofilms. Biofilms are ubiquitous in aquatic environments, where they form on rocks, plants, and sediments, and are also prevalent in wastewater treatment systems. Biofilm microorganisms are encased in a heterogeneous matrix of extracellular polymeric substances (EPS) composed of polysaccharides, proteins, lipids, and nucleic acids (10, 16). Both the morphology and chemical characteristics of biofilms are expected to promote the deposition and retention of C. parvum oocysts. Previous studies in both laboratory and environmental systems have shown that colloidal particles such as latex beads, bacteria, and virions can be readily transferred to biofilm communities from the surrounding bulk fluid (4, 13, 17, 32, 33, 37, 44, 45). As a result of this accumulation, biofilms can serve as environmental reservoirs of disease, and deposited pathogens can be released back to the water column by detachment or biofilm sloughing. This is particularly a concern with C. parvum because of its persistence under typical environmental conditions.
In this study, we investigated the capture and retention of C. parvum oocysts by Pseudomonas aeruginosa biofilms grown in small flow cell systems. To assess the role of EPS in the capture of oocysts by biofilms, two different biofilms were grown, the P. aeruginosa wild-type strain (PAO1) and a strain that overproduces the extracellular polysaccharide alginate (PDO300). Biofilms were also grown in two separate growth media, as nutrient concentrations and carbon source are known to affect P. aeruginosa biofilm architecture (22, 28, 35). Laser-scanning confocal microscopy coupled with image analysis was used to quantitatively compare the morphology of biofilms grown under different conditions.
MATERIALS AND METHODS
Bacterial strains and media.
Biofilms were developed using both a wild-type strain of Pseudomonas aeruginosa (PAO1) and an alginate-overproducing strain of P. aeruginosa (PDO300), which is a mucA22 derivative of PAO1 (21). Both strains were chromosomally tagged with a green fluorescent protein to facilitate microscopy. To promote the development of different biofilm morphologies, biofilms were grown with each of the two P. aeruginosa strains in both Jensen's medium (27), a defined minimal medium with 7 mM glucose added as a carbon source, and in Luria-Bertani (LB) broth (1/8 strength, 2.5 g/liter), composed primarily of yeast extract, peptone, and sodium chloride.
Flow cell biofilm system.
Biofilms were grown in three-channel flow cells (40 by 4 by 1 mm) with a glass coverslip substratum (7). Either LB broth or Jensen's medium was continuously pumped through sterile flow cell channels at a rate of 0.06 ml/min using a Watson-Marlow 205S peristaltic pump (Watson Marlow Ltd., Falmouth, England). To initiate biofilm growth, the flow of medium through the system was stopped and 0.2 ml of PAO1 or PDO300 liquid culture (optical density at 600 nm [OD600] = 0.50) was injected into each flow cell channel. No-flow conditions were maintained for 1 h after inoculation to allow bacteria to attach to the substratum. After this time, the flow was resumed and the bacteria were cultivated in the flow cell at 30°C until a confluent biofilm developed. This required 3 days in Jensen's medium and 4 days in LB broth. Sterile control channels were also used without injection of bacteria.
Acquisition and analysis of biofilm images.
Confluent biofilms were visualized in situ by means of confocal laser-scanning microscopy. Prior to image acquisition, the biofilms were stained by injecting 0.2 ml of 20 μM Syto 9 (Molecular Probes, Inc., Eugene, OR) into each channel. The biofilm was stained for 40 min in the dark, and then the residual stain was pumped out of the flow cell. Stacks of horizontal-plane images were acquired using a Zeiss LSM 510 (Carl Zeiss, Jena, Germany) equipped with a 488-nm argon laser. The image analysis program COMSTAT was used to quantitatively analyze biofilm architecture, including average thickness, biomass, total surface area, surface-area-to-volume ratio, and roughness (23). Roughness is defined as by the equation:
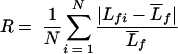 |
where 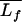 is the mean thickness, Lfi is the ith individual thickness, and N is the number of thickness measurements.
is the mean thickness, Lfi is the ith individual thickness, and N is the number of thickness measurements.
Collection and purification of C. parvum oocysts.
C. parvum oocysts were collected and purified by following procedures described previously (42). Fecal samples were collected in Tulare County, California. from dairy calves naturally infected with C. parvum. Samples were rinsed through a series of mesh sieves and concentrated via sedimentation and centrifugation. Oocysts were purified through a discontinuous sucrose gradient (1) and stored in a 0.01% Tween 20 solution with antibiotics (penicillin G, streptomycin sulfate, and amphotericin B) at 4°C. This method of collection and purification of C. parvum oocysts from naturally infected dairy calves typically produces oocysts that are greater than 90% viable as measured by excystation (24). Oocysts were used in biofilm experiments within 2 months of collection, and the same batch of oocysts was normally used for parallel experiments conducted in the two media.
The concentration of C. parvum oocysts in the purified stock solution was determined through a filtration-direct count method as described by Searcy et al. (42). A sample of the C. parvum oocyst stock solution was vacuum filtered, and the filter was covered with a fluorescein isothiocyanate-conjugated monoclonal antibody solution specific for Cryptosporidium (Waterborne, Inc., New Orleans, LA). The filtered sample was allowed to incubate in the labeling solution for 30 min and then transferred to a microscope slide. The oocysts on the filter were enumerated using a Zeiss Axiophot epifluorescence microscope (Carl Zeiss, Jena, Germany) equipped with a mercury vapor lamp and an excitation/band-pass filter for fluorescein isothiocyanate.
C. parvum oocyst injection and enumeration.
Following the development of a confluent biofilm and the acquisition of confocal microscopy images, C. parvum oocysts were introduced into each flow cell channel. All C. parvum injection experiments were performed at room temperature in a controlled laboratory environment (∼22°C). The oocyst stock solution was diluted with the appropriate medium to a working solution concentration of 5 × 105 oocysts/ml, the flow through the system was stopped, and 0.05 ml of the working solution was injected into the tubing upstream of each flow cell channel. Immediately after the injection, flow was resumed, allowing the oocysts to flow through each flow cell channel. After 1 h, the inflow was stopped and C. parvum oocysts were enumerated in situ. Cy3-conjugated monoclonal antibody solution specific for Cryptosporidium (Waterborne, Inc., New Orleans, LA) was injected into each channel and allowed to incubate for 40 min, after which the residual labeling solution was pumped out of the flow cell. The C. parvum oocysts retained in the biofilm on the glass coverslip were enumerated at a magnification of ×160 with a Zeiss Axiophot epifluorescence microscope equipped with an excitation/band-pass filter for Cy3. A large number of oocysts was counted within the square fields of the microscope's ocular grid across a wide area of the flow cell channel, and the total number of oocysts captured by the biofilm was calculated by normalizing these results to the surface area of the entire flow cell channel. C. parvum oocysts were also resolved within the P. aeruginosa biofilm using a Zeiss LSM 510 equipped with a 488-nm argon laser and a 633-nm helium-neon laser. Horizontal-plane images were obtained at a magnification of ×630.
Release of C. parvum oocysts from P. aeruginosa biofilms.
The resuspension of C. parvum oocysts was also investigated. In one set of experiments, the flow was resumed after the initial oocyst enumeration and maintained at this constant rate (0.06 ml/min) for a total of 24 h, after which the oocysts remaining in the biofilm were again enumerated. In a second set of experiments, the flow was increased to 2.5 ml/min after the initial oocyst enumeration and maintained at this higher flow rate for 1 h, after which oocysts retained in the biofilm were enumerated. To monitor possible biofilm sloughing under the higher flow rate, the system effluent was also collected at both flow rates and enumerated for resuspended Pseudomonas aeruginosa cells. To disperse any clusters of P. aeruginosa cells, the effluent samples were sonicated for 1 min (Aquasonic ultrasonic cleaner; VWR Scientific Products, West Chester, PA), placed on ice for 1 min, and vortexed for 15 s. This cycle was repeated three times per sample. The samples were then serially diluted, and viable plate counts were performed.
Zeta potential measurements.
The zeta potentials of C. parvum oocysts in Jensen's medium and in LB broth were measured using a Brookhaven Instruments Corporation (Holtsville, N.Y.) Zeta PALS particle analyzer. Oocyst suspensions were analyzed at a concentration of 5 × 104 oocysts/ml. Electrophoretic mobilities of the oocysts are converted by the instrument to a zeta potential using the Smoluchowski approximation.
Statistical analysis.
Two-way analysis of variance was used to evaluate variation of oocyst capture with surface composition (glass, PAO1 biofilm, and PDO300 biofilm) and growth medium (Jensen's and LB). One-sample t tests were used to assess differences in the number of oocysts retained at 1 h compared to 24 h (both at a flow rate of 0.06 ml/min), at a flow rate of 0.06 ml/min compared to 2.5 ml/min (both maintained for 1 h), and in the release of P. aeruginosa from the biofilm at the two flow rates. Functional relationships between biofilm structural characteristics and the fraction of oocysts captured by the biofilm were determined using linear regression.
RESULTS
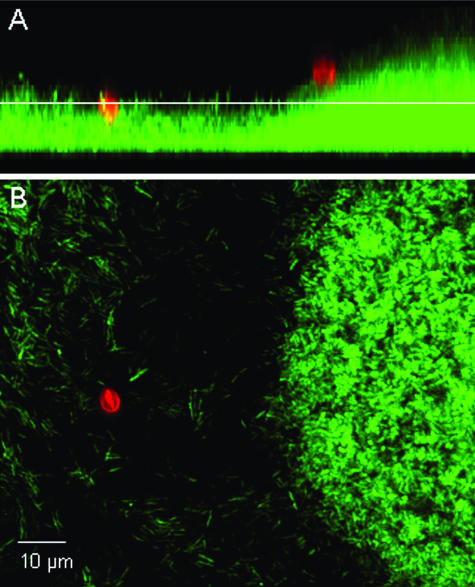
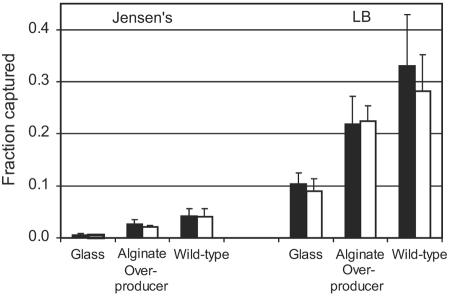
A micrograph of C. parvum oocysts associated with a P. aeruginosa (PDO300) biofilm is provided in Fig. 1. In all cases, oocysts were observed to be retained at the biofilm surface and did not penetrate into the deeper regions of the biofilm. Flow cells hosting a P. aeruginosa biofilm of either the wild-type strain (PAO1) or the alginate-overproducing strain (PDO300) captured a greater fraction of C. parvum oocysts than did the flow cells without biofilms (P < 0.001) (Fig. 2). Only 0.6% of the injected oocysts attached to the glass surface in experiments with Jensen's medium, whereas 2.6% and 4.2% of oocysts were captured by the PDO300 and PAO1 biofilms, respectively, in the same medium. Likewise, 10.4% of the oocysts attached to the glass surface in experiments with LB broth, while 21.9% and 33.1% were captured by the PDO300 and PAO1 biofilms. These results clearly show that the background medium significantly influenced oocyst capture, as there was much greater oocyst deposition in all experiments with LB broth than in experiments with Jensen's medium (P < 0.001). This same trend was observed for the control glass surface, the wild-type biofilm, and the alginate-overproducing biofilm. Oocysts had a more-negative zeta potential in LB medium (−21.5 ± 0.32 mV) than in Jensen's medium (−8.94 ± 2.09 mV). Figure 2 also shows that more oocysts were captured by the wild-type biofilm than the alginate-overproducing biofilm in both media, but this trend was not statistically significant.
FIG. 1.
Micrograph of C. parvum oocysts (red) associated with a P. aeruginosa (PDO300) biofilm (green). C. parvum oocysts are labeled with a Cy3-conjugated monoclonal antibody solution specific for Cryptosporidium, and P. aeruginosa cells are chromosomally tagged with a green fluorescent protein. (A) Sagittal view of the biofilm. (B) Planar view of the biofilm at the depth indicated by the white line in the sagittal view.
FIG. 2.
The fraction of C. parvum oocysts captured by the glass surface, alginate-overproducing P. aeruginosa biofilm, and wild-type P. aeruginosa biofilm 1 h (filled bars) and 24 h (open bars) after injection in Jensen's medium and LB broth.
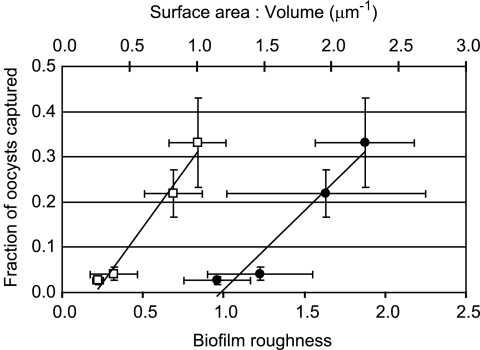
Structural characteristics of biofilms formed by both P. aeruginosa strains grown in both media are provided in Table 1. Note that the average biofilm thickness was negligible compared to the thickness of the flow cell channel in all cases, so that biofilm growth did not appreciably change the volume of the pore space in the flow cells. Oocyst capture was found to be positively related to biofilm roughness (P < 0.01) and surface-area-to-volume ratio (P < 0.05) (Fig. 3). No relationships were found between oocyst capture and other biofilm characteristics, including biomass, total surface area, and average thickness.
TABLE 1.
Structural characteristics of PAO1 and PDO300 biofilms grown in Jensen's and LB media
| Strain | Medium | Avg thickness (μm) | Biomass (μm3) | Total surface area (μm2) | Surface area/vol ratio (μm−1) | Roughness |
|---|---|---|---|---|---|---|
| PAO1 | Jensen's | 7.68 ± 1.99 | 7.31 ± 1.85 | 2.27E+05 ± 4.56E+04 | 1.47 ± 0.39 | 0.32 ± 0.15 |
| PDO300 | Jensen's | 4.75 ± 0.86 | 5.27 ± 0.75 | 1.33E+05 ± 2.95E+04 | 1.15 ± 0.25 | 0.22 ± 0.03 |
| PAO1 | LB | 2.81 ± 0.80 | 2.26 ± 0.59 | 1.10E+05 ± 2.62E+04 | 2.25 ± 0.37 | 0.84 ± 0.18 |
| PDO300 | LB | 5.72 ± 1.21 | 4.67 ± 1.57 | 1.82E+05 ± 2.52E+04 | 1.96 ± 0.74 | 0.69 ± 0.18 |
FIG. 3.
The fraction of C. parvum oocysts captured by biofilms versus biofilm roughness (open squares) and surface-area-to-volume ratio (filled circles).
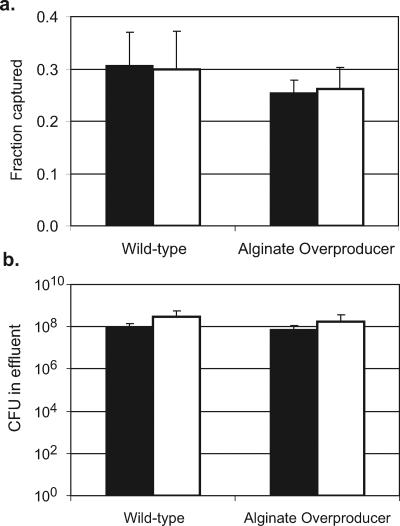
In experiments designed to observe the release of C. parvum oocysts, there was no significant difference in the number oocysts occurring in biofilms or on the abiotic glass surface under any strain or medium conditions 24 h after oocyst injection compared to 1 h after injection (Fig. 2). The fraction of oocysts captured by the biofilm also did not change following the increase in flow, indicating that there was not a significant release of oocysts from the biofilms after the initial deposition (Fig. 4). Detachment or sloughing of P. aeruginosa biofilm cells was monitored through viable plate counts of the flow cell system effluent. There was no significant change in the total number of CFU in the system effluent across the different flow rates (Fig. 4), indicating no increase in bacterial cell detachment or biofilm sloughing as a result of the increased flow rate.
FIG. 4.
(A) The fraction of oocysts captured by wild-type and alginate-overproducing P. aeruginosa biofilms at a flow rate of 0.06 ml/min (solid bars) and remaining in the biofilm after the flow rate is increased to 2.5 ml/min (open bars). (B) The total P. aeruginosa CFU enumerated from the effluent of the flow cell system during 1 h at flow rates of 0.06 ml/min (solid bars) and 2.5 ml/min (open bars).
DISCUSSION
The experimental results clearly demonstrate that biofilms significantly increase the deposition of C. parvum oocysts relative to abiotic mineral surfaces. Our results are congruent with those of other investigations that have observed the deposition of various abiotic and biotic colloidal particles in biofilm communities in a variety of experimental systems (13, 17, 32, 37, 45). The heterogeneous structure of biofilms, which can contain pores and voids, is believed to facilitate the capture and retention of colloid particles (3, 14, 36). Although the P. aeruginosa biofilms developed in our study were relatively thin and the C. parvum oocysts deposited only on the biofilm surface, the roughness coefficient and surface-area-to-volume ratio, both indicators of biofilm heterogeneity, were shown to be positively correlated with oocyst capture. Similar behavior was observed by Battin et al. (3), who found a relation between particle deposition in biofilms and biofilm sinuosity, and by Drury et al. (14), who found that microbead capture by biofilms was proportional to the standard deviation of biofilm thickness.
The growth medium had a large effect on the fraction of oocysts captured by the biofilms, with greater oocyst capture consistently found in LB broth than in Jensen's medium. This difference was not due solely to the change in biofilm architecture as a result of medium type because more oocysts also attached to the abiotic glass surface in LB broth than in Jensen's medium. This difference in oocyst capture is most likely a result of the different chemical properties of the media, which mediate oocyst-surface interactions. C. parvum oocysts have a negative surface charge under typical aqueous conditions (8, 12, 25, 30, 38), and the oocyst surface is believed to contain proteins that extend out into solution, causing both electrostatic and steric forces to be involved in oocyst-surface associations (8, 30, 31). The zeta potential of C. parvum oocysts was −21.5 ± 0.32 mV in LB medium and −8.94 ± 2.09 mV in the Jensen's medium. Glass surfaces are normally negatively charged (11, 30), as are the surfaces of biofilms because of the presence of anionic carboxyl, sulfate, and phosphoryl groups in EPS (16). Electrostatic repulsion cannot explain the observed oocyst association patterns because oocyst deposition was greater in the LB medium than in the Jensen's medium, while the oocyst zeta potential was more negative in the LB medium than in the Jensen's medium. An alternative explanation could be a reduction in steric repulsion, which would promote oocyst-surface association. Dissolved cations, such as Ca2+, have been reported to bind to oocyst surface proteins, thereby neutralizing and collapsing surface proteins (30). It is plausible that solution conditions in the LB medium reduced steric repulsion between the oocysts and surfaces, facilitating attachment. However, the lack of knowledge of the exact composition of the LB medium, derived from peptone and yeast extract, prevents firm definition of conclusions regarding the chemical effects of this medium on oocyst-surface interactions.
It is believed that the sticky matrix of EPS secreted by biofilm microorganisms serves as a protective layer for the encased cells by capturing and binding harmful solutes such as metals and antibiotics (21, 46). We investigated the role of EPS in the capture of C. parvum oocysts using a P. aeruginosa strain that overproduces the exopolysaccharide alginate. Alginate is a polysaccharide composed of mannuronic acid and guluronic acid monomers, and the EPS from the alginate-overproducing biofilm represents 21% of the total soluble biofilm biomass. The EPS from the wild-type P. aeruginosa biofilm consists primarily of glucose, rhamnose, and nucleic acids and represents only 4 to 5% of the total soluble biomass (5, 48). Fewer C. parvum oocysts were captured by the mucoid strain than by the wild-type P. aeruginosa biofilm in both growth media used, though these trends were not statistically significant. These results clearly show that biofilm architecture was more important than EPS composition in controlling C. parvum deposition with the P. aeruginosa strains used here. It is not clear why there should be less deposition in the alginate-overproducing biofilm than in the wild-type biofilm. One possibility could be a difference in electrostatic interactions between oocysts and each of the two different biofilms. Uronic acids are known to be relatively negatively charged and can be expected to make the alginate biofilm more negative than the wild-type, and this, in combination with the negative charge of C. parvum oocysts, would be expected to hinder oocyst deposition to the alginate biofilm.
Following capture of the C. parvum oocysts by the biofilms, no release of the oocysts from the biofilm was observed after a 24-h period or after a 40-fold increase in the water flow rate through the system. This result does not mean that oocysts are permanently retained in biofilms but rather indicates that release should be expected only over longer time scales or greater increases in the overlying flow rate. Previous studies of oocyst deposition on sand grains have shown that while only small concentrations of oocysts are typically observed immediately after the initial injection is terminated, substantial numbers of oocysts can by released over a much longer time period (9, 19). We also observed that there was no statistically significant detachment of Pseudomonas cells or sloughing of biofilm clusters at the higher flow rate. These results are limited, however, by the fact that the flow field within the flow cell remained laminar even at the highest experimental flow rate. It is apparent that the maximum flow rate that could be achieved in this apparatus was insufficient to generate the hydrodynamic shear necessary to detach oocysts from the biofilm surface or to cause sloughing of the bulk biofilm. The more extreme changes in flow conditions typically found in dynamic surface water systems such as rivers are still expected to release biofilm-resident pathogens under high flow conditions.
This study demonstrated that hydrodynamic transport processes cause C. parvum oocysts to migrate from the bulk fluid to surface-attached microbial communities, where they are retained and concentrated. The results presented here indicate that biofilm architecture and surface-chemical interactions, as mediated by the background water chemistry, play a significant role in mediating the deposition of pathogens from suspension into biofilms. This association is expected to decrease the concentration of oocysts in surface waters and cause biofilms to become reservoirs of C. parvum. Deposited oocysts can then be resuspended during events that promote oocyst detachment or biofilm sloughing. Furthermore, in the event of biofilm sloughing, C. parvum oocysts will most likely be released in association with suspended cell clusters, which can cause them to exhibit different transport behavior than free oocysts. We believe that these oocyst-biofilm interactions play an important role in regulating the migration of C. parvum in aquatic systems and should be considered when predicting the fate and transport of pathogens in the environment.
Acknowledgments
This project was supported by USDA National Research Initiative grant CSREES-2004-00854. This work was conducted in part under the auspices of the Bernice Barbour Communicable Disease Laboratory, with financial support from the Bernice Barbour Foundation, Hackensack, N.J., as a grant to the Center of Equine Health, University of California—Davis.
We thank Lissa Dunbar for assistance in the collection and purification of C. parvum oocysts and Matthew Parsek and Grant Balzer for providing the P. aeruginosa strains and for assistance with the flow cell apparatus.
REFERENCES
- 1.Arrowood, M. J., and C. R. Sterling. 1987. Isolation of Cryptosporidium parvum and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J. Parasitol. 73:314-319. [PubMed] [Google Scholar]
- 2.Atwill, E. R., R. Phillips, M. Das Gracas, C. Pereira, X. Li, and B. McCowan. 2004. Seasonal shedding of multiple Cryptosporidium genotypes in California ground squirrels (Spermophilus beecheyi). Appl. Environ. Microbiol. 70:6748-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battin, T. J., L. A. Kaplan, J. D. Newbold, and C. M. E. Hansen. 2003. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426:439-441. [DOI] [PubMed] [Google Scholar]
- 4.Buswell, C., Y. Herligy, L. Lawerence, J. McGuiggan, P. Marsh, C. Keevil, and S. Leach. 1998. Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 64:733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, S. I., M. R. Parsek, and K. A. Gray. Unpublished data.
- 6.Chauret, C., K. Nolan, P. Chen, S. Springthorpe, and S. Sattar. 1998. Aging of Cryptosporidium parvum oocysts in river water and their susceptibility to disinfection by chlorine and monochloramine. Can. J. Microbiol. 44:1154-1160. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilms physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 8.Considine, R. F., D. R. Dixon, and C. J. Drummond. 2002. Oocysts of Cryptosporidium parvum and model sand surfaces in aqueous solutions: an atomic force microscope (AFM) study. Water Res. 36:3421-3428. [DOI] [PubMed] [Google Scholar]
- 9.Cortis, A., T. Harter, L. Hou, E. R. Atwill, A. I. Packman, and P. G. Green. A CTRW filtration model for Cryptosporidium parvum oocyts in porous media. Water Resour. Res., in press.
- 10.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 11.Dai, X., J. Boll, M. E. Hayes, and D. E. Aston. 2004. Adhesion of Cryptosporidium parvum and Giardia lamblia to solid surfaces: the role of surface charge and hydrophobicity. Colloids Surf. B 34:259-263. [DOI] [PubMed] [Google Scholar]
- 12.Drozd, C., and J. Schwartzbord. 1996. Hydrophobic and electrostatic cell surface properties of Cryptosporidium parvum. Appl. Environ. Microbiol. 62:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drury, W., P. Stewart, and W. Characklis. 1993. Transport of 1-μm latex particles in Pseudomonas aeruginosa biofilms. Biotechnol. Bioeng. 42:111-117. [DOI] [PubMed] [Google Scholar]
- 14.Drury, W. J., W. G. Characklis, and P. S. Stewart. 1993. Interactions of 1 μm latex particles with Pseudomonas aeruginosa biofilms. Water Res. 27:1119-1126. [DOI] [PubMed] [Google Scholar]
- 15.Dworkin, M. S., D. P. Goldman, T. G. Wells, J. M. Kobayashi, and B. L. Herwaldt. 1996. Cryptosporidiosis in Washington state: an outbreak associated with well water. J. Infect. Dis. 174:1372-1376. [DOI] [PubMed] [Google Scholar]
- 16.Flemming, H.-C. 1995. Sorption sites in biofilms. Water Sci. Technol. 32:27-33. [Google Scholar]
- 17.Flood, J., and N. Ashbolt. 2000. Virus-sized particles can be entrapped and concentrated one hundred fold within wetland biofilms. Adv. Environ. Res. 3:403-411. [Google Scholar]
- 18.Hansen, J. S., and J. E. Ongerth. 1991. Effects of time and watershed characteristics on the concentration of Cryptosporidium oocysts in river water. Appl. Environ. Microbiol. 57:2790-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harter, T., S. Wagner, and E. R. Atwill. 2000. Colloid transport and filtration of Cryptosporidium parvum on sandy soils and aquifer sediments. Environ. Sci. Technol. 34:62-70. [Google Scholar]
- 20.Hayes, E. B., T. D. Matte, T. R. O'Brien, T. W. McKinley, G. S. Logsdon, J. B. Rose, B. L. P. Ungar, D. M. Word, P. F. Pinsky, M. L. Cummings, M. A. Wilson, E. G. Long, E. S. Hurwitz, and D. D. Juranek. 1989. Large community outbreak of Cryptosporidiosis due to contamination of a filtered public water supply. N. Engl. J. Med. 320:1372-1376. [DOI] [PubMed] [Google Scholar]
- 21.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heydorn, A., B. K. Ersboll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 23.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 24.Hou, L., X. Li, L. Dunbar, R. Moeller, B. Palermo, E. R. Atwill. 2004. Neonatal mice infectivity of intact Cryptosporidium parvum oocysts isolated after optimized in vitro excystation. Appl. Environ. Microbiol. 70:642-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu, B.-M., and C. Huang. 2002. Influence of ionic strength and pH on hydrophobicity and zeta potential of Giardia and Cryptosporidium. Colloids Surf. A 201:201-206. [Google Scholar]
- 26.Jellison, K. L., H. F. Hemond, and D. B. Schauer. 2002. Sources and species of Cryptosporidium oocysts in the Wachusett Reservoir watershed. Appl. Environ. Microbiol. 68:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, S. E., I. T. Fecycz, and J. N. Campbell. 1980. Nutritional factors controlling exocelluar protease production by Pseudomonas aeruginosa. J. Bacteriol. 144:844-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella, and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 29.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:4123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuznar, Z. A., and M. Elimelech. 2004. Adhesion kinetics of viable Cryptosporidium parvum oocysts to quartz surfaces. Environ. Sci. Technol. 38:6839-6845. [DOI] [PubMed] [Google Scholar]
- 31.Kuznar, Z. A., and M. Elimelech. 2005. Role of surface proteins in the deposition kinetics of Cryptosporidium parvum oocysts. Langmuir 21:710-716. [DOI] [PubMed] [Google Scholar]
- 32.Langmark, J., M. V. Storey, N. J. Ashbolt, and T.-A. Stenstrom. 2005. Accumulation and fate of microorganisms and microspheres in biofilms formed in a pilot-scale water distribution system. Appl. Environ. Microbiol. 71:706-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackay, W., L. Gribbon, M. Barer, and D. Reid. 1998. Biofilms in drinking water systems—a possible reservoir for Helicobacter pylori. Water Sci. Technol. 38:181-185. [DOI] [PubMed] [Google Scholar]
- 34.MacKenzie, W. R., N. J. Hoxie, M. E. Proctor, M. S. Gradus, K. A. Blair, D. E. Peterson, J. J. Kazmierczak, D. G. Addiss, K. R. Fox, J. B. Rose, and J. P. Davis. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161-667. [DOI] [PubMed] [Google Scholar]
- 35.Moller, S., D. R. Korber, G. M. Wolfaardt, S. Molin, and D. E. Caldwell. 1997. Impact of nutrient composition on a degradative biofilm community. Appl. Environ. Microbiol. 63:2432-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okabe, S., H. Kuroda, and Y. Watanabe. 1998. Significance of biofilm structure on transport of inert particulates into biofilms. Water Sci. Technol. 38:136-170. [Google Scholar]
- 37.Okabe, S., T. Yasuda, and Y. Watanabe. 1997. Uptake and release of inert fluorescence particles by mixed population biofilms. Biotechnol. Bioeng. 53:459-469. [DOI] [PubMed] [Google Scholar]
- 38.Ongerth, J. E., and J. P. Pecoraro. 1996. Electrophoretic mobility of Cryptosporidium oocysts and Giardia cysts. J. Environ. Eng. 122:228-231. [Google Scholar]
- 39.Parker, J. F. W., and H. V. Smith. 1993. Destruction of oocysts of Cryptosporidium parvum by sand and chlorine. Water Res. 27:729-731. [Google Scholar]
- 40.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1992. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 58:3494-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roefer, P. A., J. T. Monscvitz, and D. J. Rexing. 1996. The Las Vegas cryptosporidiosis outbreak. J. Am. Water Works Assoc. 88:95-106. [Google Scholar]
- 42.Searcy, K. E., A. I. Packman, E. R. Atwill, and T. Harter. 2005. Association of Cryptosporidium parvum with suspended particles: impact on oocyst sedimentation. Appl. Environ. Microbiol. 71:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SoloGabriele, H., and S. Neumeister. 1996. US outbreaks of cryptosporidiosis. J. Am. Water Works Assoc. 88:76-86. [Google Scholar]
- 44.Storey, M., and N. Ashbolt. 2001. Persistence of two model enteric viruses (B4-8 and MS-2 bacteriophages) in water distribution pipe biofilms. Water Sci. Technol. 43:133-138. [PubMed] [Google Scholar]
- 45.Stott, R., and C. C. Tanner. 2005. Influence of biofilm on removal of surrogate faecal microbes in a constructed wetland and maturation pond. Water Sci. Technol. 51:315-322. [PubMed] [Google Scholar]
- 46.Teitzel, G. M., and M. R. Parsek. 2003. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker, M. J., C. D. Montemagno, and M. B. Jenkins. 1998. Source water assessment and nonpoint sources of acutely toxic contaminants: a review of research related to survival and transport of Cryptosporidium parvum. Water Resour. Res. 34:3383-3392. [Google Scholar]
- 48.Wozniak, D. J., T. J. O. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]