Abstract
Nineteen echovirus 11 (E11) and 12 E13 isolates were isolated from three rivers in Toyama Prefecture, Japan, during an environmental surveillance conducted from April 2002 to March 2003. The nucleotide sequences of E13 isolates were closely related to those from patients with aseptic meningitis, with less than 1.3% divergence in the VP1 region of the viral capsid gene, and belonged to the same clade responsible for a worldwide outbreak that started in 2000. In contrast, E11 isolates were clustered into three genomic groups and were not closely related to echovirus strains isolated from patients. These results suggest that the combination of both virus isolation from environmental sources and phylogenetic analysis could be complementary assessment approaches to trace prevalent and minor circulating enteroviruses in the human population.
Environmental surveillance has been conducted in Toyama Prefecture, Japan, four times since 1979 in order to study enteric virus water pollution (18-20). Assessment of enteric viruses found in the environment also plays a role in understanding virus circulation in the community (26, 27). In particular, environmental surveillance for human poliovirus (genus Enterovirus) under an eradication program is reported to be the most sensitive method to detect wild or vaccine-derived polioviruses circulating in the human population (7, 32, 33).
Large outbreaks of aseptic meningitis caused by human echovirus 13 (E13) (genus Enterovirus) have been reported in various areas in the world since 2000 (3, 4, 22, 23, 30). In addition, a nationwide outbreak of aseptic meningitis caused by E13 occurred in Japan during the summer of 2002 (19) after a small outbreak in a limited area in 2001. According to an Infectious Agents Surveillance Report of Japan, E13 was the most common causative agent (67.2%) isolated from aseptic meningitis patients in 2002, followed by human echovirus 11 (E11) (genus Enterovirus) (12.3%) (10). However, E13 had not been isolated clinically during the previous 20 years in Japan (9), and its isolation was rare until 2000. Thus, the outbreaks of aseptic meningitis caused by E13 seem to be a case of reemerging enterovirus infection (21, 29).
Human enterovirus infection is known to be generally asymptomatic, and thus, environmental surveillance has been reported to be a sensitive method to detect silently circulating viruses (26, 27). We show here several enteroviruses isolated during an environmental surveillance conducted in Toyama, and we especially analyzed E13 and E11 phylogenetically and compared their genetic sequences to available clinical isolates.
Virus isolation from rivers.
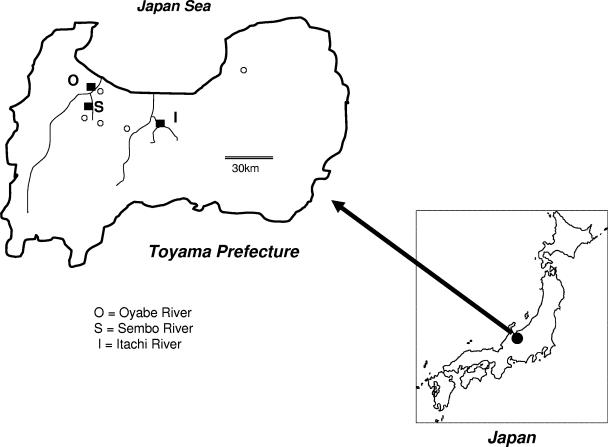
Water samples (800 ml each) were collected from fixed points of three rivers (Oyabe, Itachi, and Sembo) (Fig. 1) twice a month from April 2002 to March 2003, as described previously (18). On the day of collection, samples were concentrated using a negatively charged membrane filter (mixed cellulose ester membrane filter; Advantec Co. Ltd., Tokyo, Japan) as described previously (18). After the initial collection, water samples were centrifuged at 3,000 rpm for 30 min, MgCl2 was added to the supernatant to a final concentration of 0.05 M, and the pH was adjusted to 3.5. The samples were then absorbed to the filter under positive pressure. Absorbents on the filter were next eluted with 3% beef extract solution by sonication for 5 min and then centrifuged at 12,000 rpm for 30 min. The supernatants were collected and stored at 4°C until virus isolation.
FIG. 1.
Locations of the three rivers (Itachi [I], Oyabe [O], and Sembo [S]) in Toyama Prefecture. Squares (▪) indicate the fixed points for water sampling. Circles (○) indicate the locations of hospitals where aseptic meningitis patients were admitted. Double lines indicate distance.
A total of 0.2 ml of the supernatant was inoculated onto Vero, MA104, RD-18S, and Hep2 cell lines. Eight tubes for each cell line were used for virus isolation (18). Isolates were identified by specific antisera against each enterovirus (Denka Co. Ltd., Tokyo, Japan). Reoviruses (mammalian orthoreovirus) were characterized using neutralization and hemagglutination inhibition tests with type-specific antisera (19).
A total of 171 viruses were isolated at fixed points of three rivers in Toyama twice a month from April 2002 to March 2003 (Table 1). Reoviruses were the most common virus isolates, followed by E11 and E13. Three isolates could not be typed. Cell lines in which viruses were isolated are shown in Table S2 of the supplemental material.
TABLE 1.
Virus isolation from three rivers in the Toyama Prefecture, Japan, between April 2002 and March 2003
| Virusa | No. of virus isolates
|
|||
|---|---|---|---|---|
| Itachi River | Sembo River | Oyabe River | Total | |
| E7 | 3 | 3 | ||
| E11 | 4 | 14 | 1 | 19 |
| E13 | 1 | 8 | 3 | 12 |
| CB2 | 1 | 1 | 2 | |
| CB3 | 4 | 1 | 5 | |
| CB4 | 2 | 2 | ||
| Poliovirus type 2 | 1b | 1 | ||
| Reovirus type 1 | 3 | 1 | 4 | |
| Reovirus type 2 | 33 | 48 | 38 | 119 |
| Reovirus type 3 | 1 | 1 | ||
| Not typed | 3 | |||
| Total | 43 | 84 | 44 | 171 |
E, echovirus; CB, coxsackievirus type B.
A result of intratypic differentiation was Sabin 2.
There were two periods when viruses were frequently isolated: one was from July to September 2002, and the other was from December 2002 to February 2003 (Fig. 2). Twelve E13 strains were isolated from May to December 2002. Nineteen E11 strains were isolated from September 2002 to January 2003. Most reoviruses were isolated in the latter periods, although they were isolated throughout the season.
FIG. 2.
Total numbers of virus isolates from three rivers from April 2002 (Apr) to March 2003 (Mar). Waters from fixed points of three rivers were collected on the indicated dates. CB2, coxsackievirus type B2; Polio2, poliovirus type 2; Reo1, reovirus type 1.
Type 2 poliovirus was isolated in November 2002 after a routine immunization scheduled during the previous month. Differentiation of poliovirus isolates was performed by PCR-restriction fragment length polymorphism and sequencing methods as described previously (34), and isolates were characterized as vaccine type (data not shown).
Virus isolation from patients with aseptic meningitis.
Clinical specimens (stool, cerebral spinal fluid, and throat swab) from seven aseptic meningitis patients diagnosed in Toyama in 2002 were used for virus isolation as described previously (13, 31). Five E13 viruses were also isolated from one stool specimen, two cerebral spinal fluid specimens, and two throat swabs from seven patients with aseptic meningitis in June and July 2002. Eight E11 isolates from aseptic meningitis patients collected between 1993 and 1998 in Hyogo Prefecture, Japan, were also used for analysis.
RT-PCR and nucleotide sequence analysis.
E13 and E11 isolates were used for sequencing analysis. The viral RNA was extracted from virus fluid using a QIAamp Viral RNA Mini kit (QIAGEN, MD) and was then used for reverse transcription-PCR (RT-PCR) (Access RT-PCR system; Promega, WI). For amplification of the partial VP1 region of the viral capsid protein, two sets of panenterovirus degenerate primers described previously by Oberste et al. were used (24). Briefly, for amplification of the region upstream of VP1, sense primer 187 and antisense primer 222 were used, and for amplification of the downstream region, sense primer 012 and antisense primer 011 were used (24). RT-PCR was carried out under the following conditions: reverse transcription at 48°C for 45 min, inactivation at 94°C for 2 min, and 35 cycles of annealing at 50°C for 10 s, polymerization at 65°C for 1 min, and denaturation at 94°C for 10 s. After 35 cycles, an additional elongation step of 65°C for 1 min was done. The PCR product was purified by using a QIAquick PCR purification kit (QIAGEN) and directly sequenced using a PRISM BigDye Terminator cycle sequencing reaction kit on an automated DNA sequencer (Perkin-Elmer Applied Biosystems) as described previously (31).
Genetic relationships among E13 isolates or E11 isolates were analyzed by MEGA 3.1 software (16) using the partial VP1 region of E13 (703 bp; positions 2579 to 3281 on strain Del Carmen) and of E11 (561 bp; positions 2765 to 3325 on strain Gregory). Phylogenetic trees were constructed by neighbor-joining methods after estimation of genetic distance using the Kimura two-parameter method (14). The transition/transversion rate was set at 2.0, and a bootstrapping test was performed 1,000 times (8).
Sixty-one- and 51-nucleotide sequences of E13 and E11, respectively, were available in GenBank. The strains are represented as accession no./country or city/year/strain code using the reference or Web data in GenBank (1, 2, 5, 12, 15, 25).
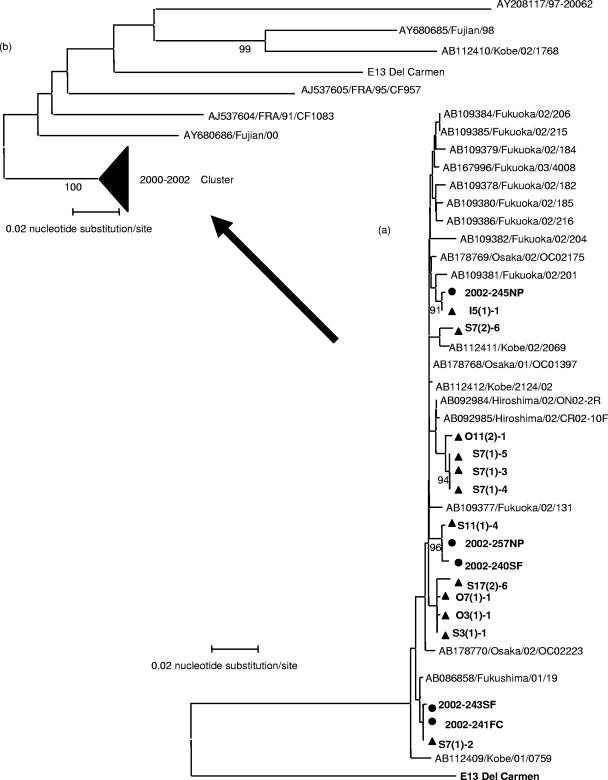
The nucleotide sequences of E11 and E13 isolates were phylogenetically compared. For E13, the nucleotide divergence was less than 1.3% among isolates from the three rivers in the partial VP1 region (703 bp). The nucleotide sequence divergence was 0.9 to 1.3% between 12 environmental isolates and 5 clinical isolates, and at most, one amino acid substitution was found. Therefore, environmental isolates were closely related to clinical isolates in Toyama Prefecture (Fig. 3a). Compared to 19 other isolates from Japan during 2001 to 2002 found in GenBank, the divergence was 1.2 to 1.6%, with, at most, one amino acid substitution. Phylogenetic analysis showed that all E13 isolates in Japan belonged to the same cluster (Fig. 3a).
FIG. 3.
(a) Phylogenetic relationships between the Japanese isolates. Phylogenetic trees for E13 using the partial VP1 region (703 bp) were generated by the neighbor-joining method with 36 strains: 17 Toyama isolates (12 from river samples and 5 from patients) and 19 other Japanese isolates (GenBank accession no. AB086858, AB092984, AB092985, AB109377 to AB109382, AB109384 to AB109386, AB112409, AB112411, AB112412, AB167996, and AB178768 to AB178770). Bootstrap values (in percentages) for 1,000 replicated trees are indicated. Circles and triangles specify patient isolates and environmental isolates, respectively. S, O, and I indicate Sembo, Oyabe, and Itachi, respectively. (b) Phylogenetic trees were reconstructed using 61 strains (accession no. AB112410, AJ537604 to AJ537609, AY227334 to AY227347, AY268561, AY268563 to AY268569, AY268571 to AY268580, AY680685, AY680686, and AJ241427 from GenBank and including the 19 other Japanese isolates). Bootstrap values (in percentages) for 1,000 replicated trees are indicated. The major genomic group isolated during 2000 to 2002 was compressed within the same cluster (2000-2002 cluster). Arrows from tree a to tree b indicate that Japanese isolates in tree a are included in the compressed cluster. The strains are represented as accession no./country or city/year/strain code using the reference or Web data in GenBank.
Moreover, isolates in Toyama from both environmental sources and patients were compared with the other E13 sequences available from GenBank, including the above-mentioned 19 strains from Japan. Phylogenetic analysis showed that the Toyama isolates belonged to the major genomic group labeled as the 2000-2002 cluster in Fig. 3b, which was described previously by other studies (1, 2, 5, 12, 15). Divergence of nucleotide sequences between Toyama isolates and others in this group was 1.9 to 2.5% (amino acid divergence, 0.3 to 0.7%), indicating that these Toyama isolates belonged to the major genomic group circulating worldwide.
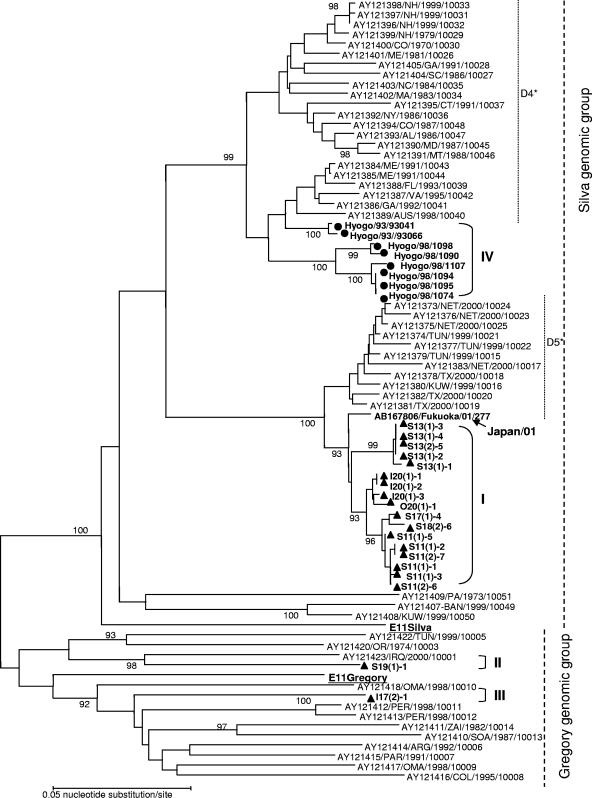
On the other hand, nucleotide sequences of E11 were phylogenetically analyzed and antigenically categorized into two major strains: strain Gregory as the prototype and strain Silva as prime type (Fig. 4). There were several kinds of subgenotypes within these two major strains (6, 25). Although E11 was not isolated from patients in Toyama during the period of this study, 19 E11 samples were isolated from the three rivers. Eight clinical isolates from Hyogo Prefecture between 1993 and 1998 were also used in the analysis together with the 51 E11 sequences available in GenBank.
FIG. 4.
Phylogenetic tree for E11 using the partial VP1 region (561 bp) generated by the neighbor-joining method with 78 strains: 19 Toyama River isolates, 8 patient isolates from Hyogo, and 51 other isolates (accession no. AY121373 to AY121405, AY121407 to AY121418, AY121420, AY121422, AY121423, AB167806, AF295498, and AF081326 from GenBank). Circles and triangles indicate viruses isolated from patients and the environment, respectively. Bootstrap values (in percentages) for 1,000 replicated trees are indicated. Subgenomic groups I to IV in this study are shown, with D4 and D5 clusters being named as suggested previously by Oberste et al. (25). The strains are represented as accession no./country or city/year/strain code using the reference or Web data in GenBank. Isolate identifiers consist of a three-letter country abbreviation (ARG, Argentina; AUS, Australia; BAN, Bangladesh; COL, Colombia; DOR, Dominican Republic; ZAI, Democratic Republic of Congo; IRQ, Iraq; KUW, Kuwait; NET, The Netherlands; OMA, Oman; PAR, Paraguay; PER, Peru; SOA, South Africa; TUN, Tunisia; Tur, Turkey; or USA, United States), with two-letter abbreviations for U.S. states. The viral isolate from Fukuoka City (accession no. AB167806) is indicated by arrows (Japan/01).
Environmental isolates were divided into three genomic groups, groups I, II, and III (Fig. 4). Seventeen isolates fell into major genomic group I, with 1.5 to 2.1% nucleotide divergence within the partial VP1 region (561 bp). Oberste et al. previously described that the genomic group of Silva consisted of five subgroups, subgroups D1 to D5 (25). Toyama isolates had a mean of 18.3% divergence in nucleotides compared to the E11 Silva strain and only 3.9 to 5.1% divergence compared to the 1999-2000 D5 subgroup. Therefore, we classified Toyama isolates as belonging to subgroup D5. Moreover, nucleotide sequences of these isolates were very similar to aseptic meningitis isolates from Fukoaka City in 2001 (nucleotide divergence, 1.8 to 2.3%). Clinical isolates from Hyogo in 1993 and 1998 (group IV) were categorized as belonging to subgenomic group D4 from 1970 to 2001 (Fig. 4). I17(2)-1 (group III) and S19(1)-1 (group II) isolates were classified as belonging the Gregory genomic group and had nucleotide divergences from the genomic group of 21.2 to 24.0% and 18.9 to 21.5%, respectively.
Concluding remarks.
There was a large outbreak of aseptic meningitis caused by E13 in the summer of 2002 in Japan, which coincided with a small outbreak caused by E11 (9). The aim of this study was to assess the performance of an environmental surveillance of river water isolates compared with isolates from clinical samples. These viruses were also isolated from the rivers in Toyama Prefecture by environmental surveillance. E13 was detected not only in Toyama Prefecture but also in other places in Japan simultaneously. The phylogenetic analysis of E13 showed that the isolates from both river waters and patients belonged to the same genomic cluster, one of the major genotypes circulating worldwide since 2000.
E11 was recently detected during the autumn/winter from river water in Toyama and was compared to isolates from elsewhere in Japan. Although there was no outbreak of aseptic meningitis in Toyama, E11 might have been silently circulating in the human population. Phylogenetic analysis showed that the isolates were divided into three genomic groups: isolates in group I belonging to the Silva genomic group, which seemed to be circulating mainly in Toyama, and minor E11 isolates of I17(2)-1 or S19(1)-1 belonging to the Gregory genomic group, which also seemed to be circulating in other areas of Japan. Thus, this environmental survey was capable of detecting enteroviruses of different genomic groups with high sensitivity and was capable of tracing minor circulating viruses.
In addition, many reoviruses were isolated during the surveillance, most frequently between December 2002 and February 2003. Matsuura et al. suggested in a previous study (19) that stool from not only humans but also animals might have contaminated the waters.
Since there were only a few reports of E11 and E13 isolation in Japan between 2003 and 2005 (11) according to the Infectious Agents Surveillance Report, these viruses appeared to temporarily cease circulation or to circulate silently. This report shows that the combination of conventional virus isolation from environmental sources together with phylogenetic analysis of clinical isolates is a useful approach in understanding enterovirus circulation and transmission. Therefore, environmental surveillance should be considered a complementary assessment tool to trace prevalent and minor enteroviruses circulating in the human population.
Nucleotide sequence accession numbers.
The sequence data in this study were deposited in GenBank under accession no. AB239081 to AB239124.
Supplementary Material
Acknowledgments
We are grateful to T. Miyamura (National Institute of Infectious Diseases) for critical review and helpful discussion.
This report was supported by grants-in-aid for research on re-emerging infectious diseases from the Ministry of Health and Welfare, Japan.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Archimbaud, C., J. L. Bailly, M. Chambon, O. Tournilhac, P. Travade, and H. Peigue-Lafeuille. 2003. Molecular evidence of persistent echovirus 13 meningoencephalitis in a patient with relapsed lymphoma after an outbreak of meningitis in 2000. J. Clin. Microbiol. 41:4605-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avellon, A., I. Casas, G. Trallero, C. Perez, A. Tenorio, and G. Palacios. 2003. Molecular analysis of echovirus 13 isolates and aseptic meningitis, Spain. Emerg. Infect. Dis. 9:934-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. 2001. Echovirus type 13—United States, 2001. Morb. Mortal. Wkly. Rep. 50:777-780. [PubMed] [Google Scholar]
- 4.CDR. 2000. Viral meningitis associated with increase in echovirus type 13. Commun. Dis. Rep. CDR Wkly. 10:277, 280. [PubMed] [Google Scholar]
- 5.Cheon, D. S., J. Lee, K. Lee, S. Lee, K. Park, J. Ahn, Y. Jee, J. Yoon, and H. Cho. 2004. Isolation and molecular identification of echovirus 13 isolated from patients of aseptic meningitis in Korea, 2002. J. Med. Virol. 73:439-442. [DOI] [PubMed] [Google Scholar]
- 6.Chevaliez, S., A. Szendroi, V. Caro, J. Balanant, S. Guillot, G. Berencsi, and F. Delpeyroux. 2004. Molecular comparison of echovirus 11 strains circulating in Europe during an epidemic of multisystem hemorrhagic disease of infants indicates that evolution generally occurs by recombination. Virology 325:56-70. [DOI] [PubMed] [Google Scholar]
- 7.Deshpande, J. M., S. J. Shetty, and Z. A. Siddiqui. 2003. Environmental surveillance system to track wild poliovirus transmission. Appl. Environ. Microbiol. 69:2919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-789. [DOI] [PubMed] [Google Scholar]
- 9.Infectious Disease Surveillance Center. 2002. The trend of enterovirus isolation in association with aseptic meningitis, 1999-2002. Infect. Agents Surveill. Rep. 23:193-194. (In Japanese.) [Google Scholar]
- 10.Infectious Disease Surveillance Center. 2005.Virus isolation from aseptic meningitis cases, 1997-2002. [Online.] http://idsc.nih.go.jp/iasr/virus/graph/asmen9702.gif.
- 11.Infectious Disease Surveillance Center. 2005.Virus isolation from aseptic meningitis cases, 2003-2005. [Online.] http://idsc.nih.go.jp/iasr/virus/graph/asmen0305.gif.
- 12.Kaida, A., H. Kubo, N. Iritani, T. Murakami, and K. Haruki. 2004. Isolation of echovirus type 13 in Osaka City during 2001-2002. Jpn. J. Infect. Dis. 57:127-128. [PubMed] [Google Scholar]
- 13.Keino, M., M. Kanno, K. Hirasawa, T. Watari, M. Mikawa, K. Saito, K. Kato, M. Katayose, and H. Yoshida. 2001. Isolation of echovirus type 13 from patients of aseptic meningitis. Jpn. J. Infect. Dis. 54:249-250. [PubMed] [Google Scholar]
- 14.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, K., T. Haruta, M. Kubota, K. Akiyoshi, T. Suga, and M. Ito. 2005. Clinical spectrum in hospitalized children with echovirus type 13 infection. Pediatr. Int. 47:185-189. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Matsuura, K., S. Hasegawa, T. Nakayama, O. Morita, and H. Uetake. 1984. Viral pollution of the rivers in Toyama City. Microbiol. Immunol. 28:575-588. [DOI] [PubMed] [Google Scholar]
- 19.Matsuura, K., M. Ishikura, T. Nakayama, S. Hasegawa, O. Morita, and H. Uetake. 1988. Ecological studies on reovirus pollution of rivers in Toyama Prefecture. Microbiol. Immunol. 32:1221-1234. [DOI] [PubMed] [Google Scholar]
- 20.Matsuura, K., M. Ishikura, H. Yoshida, T. Nakayama, S. Hasegawa, S. Ando, H. Horie, T. Miyamura, and T. Kitamura. 2000. Assessment of poliovirus eradication in Japan: genomic analysis of polioviruses isolated from river water and sewage in Toyama Prefecture. Appl. Environ. Microbiol. 66:5087-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuta, K., C. Abiko, T. Murata, T. Itagaki, N. Katsushima, T. Akiba, M. Sakamoto, K. Ootani, and S. Murayama. 2003. Re-emergence of echovirus type 13 infections in 2002 in Yamagata, Japan. J. Infect. 47:243-247. [DOI] [PubMed] [Google Scholar]
- 22.Mullins, J. A., N. Khetsuriani, W. A. Nix, M. S. Oberste, A. LaMonte, D. R. Kilpatrick, J. Dunn, J. Langer, P. McMinn, Q. S. Huang, K. Grimwood, C. Huang, and M. A. Pallansch. 2004. Emergence of echovirus type 13 as a prominent enterovirus. Clin. Infect. Dis. 38:70-77. [DOI] [PubMed] [Google Scholar]
- 23.Narkeviciute, I., and D. Vaiciuniene. 2004. Outbreak of echovirus 13 infection among Lithuanian children. Clin. Microbiol. Infect. 10:1023-1025. [DOI] [PubMed] [Google Scholar]
- 24.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberste, M. S., W. A. Nix, D. R. Kilpatrick, M. R. Flemister, and M. A. Pallansch. 2003. Molecular epidemiology and type-specific detection of echovirus 11 isolates from the Americas, Europe, Africa, Australia, southern Asia and the Middle East. Virus Res. 91:241-248. [DOI] [PubMed] [Google Scholar]
- 26.Sedmak, G., D. Bina, and J. MacDonald. 2003. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected august 1994 to December 2002. Appl. Environ. Microbiol. 69:7181-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sellwood, J., J. V. Dadswell, and J. S. Slade. 1981. Viruses in sewage as an indicator of their presence in the community. J. Hyg. (London) 86:217-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somekh, E., K. Cesar, R. Handsher, A. Hanukoglu, I. Dalal, A. Ballin, and T. Shohat. 2003. An outbreak of echovirus 13 meningitis in central Israel. Epidemiol. Infect. 130:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toho, M., M. Nakamura, T. Asada, K. Matsumoto, and T. Horikawa. 2002. Outbreak of aseptic meningitis by echovirus 13 and sero-epidemiological study in Fukui Prefecture. Infect. Agents Surveill. Rep. 23:172-173. (In Japanese.) [Google Scholar]
- 30.Trallero, G., I. Casas, A. Avellon, C. Perez, A. Tenorio, and A. De La Loma. 2003. First epidemic of aseptic meningitis due to echovirus type 13 among Spanish children. Epidemiol. Infect. 130:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, H., Z. Hong, T. Yoneyama, K. Yoshii, H. Shimizu, K. Ota, T. Murakami, N. Iritani, M. Tsuchiya, S. Takao, K. Uchida, S. Yamanishi, M. Hamazaki, S. Yoshino, M. Oseto, K. Abe, M. Hamano, K. Sakae, H. Tsuzuki, S. Chiya, H. Onishi, T. Fujimoto, T. Munemura, A. Kawamoto, T. Miyamura, et al. 1999. Phylogenic analysis of echovirus type 30 isolated from a large epidemic of aseptic meningitis in Japan during 1997-1998. Jpn. J. Infect. Dis. 52:160-163. [PubMed] [Google Scholar]
- 32.Yoshida, H., H. Horie, K. Matsuura, T. Kitamura, S. Hashizume, and T. Miyamura. 2002. Prevalence of vaccine-derived polioviruses in the environment. J. Gen. Virol. 83:1107-1111. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, H., H. Horie, K. Matsuura, and T. Miyamura. 2000. Characterisation of vaccine-derived polioviruses isolated from sewage and river water in Japan. Lancet 356:1461-1463. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida, H., J. Li, T. Yoneyama, K. Yoshii, H. Shimizu, T. H. Nguyen, K. Toda, T. L. Nguyen, V. T. Phan, T. Miyamura, and A. Hagiwara. 1997. Two major strains of type 1 wild poliovirus circulating in Indochina. J. Infect. Dis. 175:1233-1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.