Abstract
The SCFTIR1 complex is a central regulator of the auxin response pathway in Arabidopsis. This complex functions as a ubiquitin protein ligase that targets members of the auxin/indoleacetic acid (Aux/IAA) family of transcriptional regulators for ubiquitin-mediated degradation in response to auxin. In an attempt to identify additional factors required for SCFTIR1 activity, we conducted a genetic screen to isolate enhancers of the auxin response defect conferred by the tir1-1 mutation. Here, we report the identification and characterization of the eta3 mutant. The eta3 mutation interacts synergistically with tir1-1 to strongly enhance all aspects of the tir1 mutant phenotype, including auxin inhibition of root growth, lateral root development, hypocotyl elongation at high temperature, and apical dominance. We isolated the ETA3 gene using a map-based cloning strategy and determined that ETA3 encodes SGT1b. SGT1b was identified recently as a factor involved in plant disease resistance signaling, and SGT1 from barley and tobacco extracts was shown to interact with SCF ubiquitin ligases. We conclude that ETA3/SGT1b is required for the SCFTIR1-mediated degradation of Aux/IAA proteins.
INTRODUCTION
The hormone auxin regulates many aspects of plant growth and development. Some of the best-characterized examples include stem elongation, lateral branching of roots and shoots, establishment of embryonic polarity, and vascular development (Davies, 1995). Despite the fundamental importance of this hormone, relatively few details of the molecular mechanisms of auxin action are understood.
Previous genetic and molecular studies in Arabidopsis have defined the SCFTIR1 complex as a positive regulator of the auxin response pathway (Gray and Estelle, 2000). Mutations in the TIR1 gene confer reduced auxin response (Ruegger et al., 1998). TIR1 encodes an F-box protein that interacts with the cullin AtCUL1, the RING-H2 protein RBX1, and a SKP1-like protein (ASK1 or ASK2) to form an SCF ubiquitin ligase (Gray et al., 1999, 2002). F-box proteins act as recognition factors that recruit specific substrates to the SCF for ubiquitination. The SCFTIR1 complex is believed to control auxin response by targeting negative regulators of the pathway for ubiquitin-mediated degradation in response to an auxin stimulus. Although SCFTIR1 was the first such complex identified in plants, this mode of regulation is likely to be highly prevalent, because the Arabidopsis genome encodes nearly 700 F-box proteins (Gagne et al., 2002).
The recent demonstration that auxin targets at least certain members of the auxin/indoleacetic acid (Aux/IAA) family of transcriptional regulators to the SCFTIR1 complex, which then facilitates ubiquitin-mediated degradation of the Aux/IAA proteins, provides strong support for this model (Gray et al., 2001; Zenser et al., 2001). Dominant gain-of-function mutations that confer reduced auxin response have been isolated in several Aux/IAA genes (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001). All of these mutations affect a highly conserved motif termed domain II, which acts as a degradation signal that targets the Aux/IAA protein to the SCFTIR1 ubiquitin ligase (Ramos et al., 2001). As a result, the dominant Aux/IAA derivatives fail to interact with the SCFTIR1 complex and thus exhibit increased stability compared with their wild-type counterparts (Gray et al., 2001; Ouellet et al., 2001). The molecular mechanisms involved in the auxin-dependent targeting of Aux/IAA proteins to the SCFTIR1 complex are unknown.
Molecular analysis of the AXR1 gene product further implicates the SCFTIR1 complex as an important regulator of auxin signaling. axr1 mutants exhibit a severe reduction in auxin response (Lincoln et al., 1990). The AXR1 gene encodes an enzyme required for the covalent modification of AtCUL1 with the RUB1 ubiquitin-like protein (Leyser et al., 1993; del Pozo and Estelle, 1999). Although the precise function of this modification is unclear, studies of SCFTIR1 as well as several yeast and mammalian SCF complexes indicate that the RUB1 modification of the cullin SCF subunit is required for proper ubiquitin-ligase activity (Lammer et al., 1998; del Pozo and Estelle, 1999; Podust et al., 2000; Kawakami et al., 2001).
In an effort to identify additional genes required for the SCFTIR1-mediated auxin response, we have isolated several novel mutations that enhance the relatively weak auxin response defect conferred by the tir1-1 mutation. Here, we report our identification and analysis of one of these mutants, eta3 (enhancer of tir1-1 auxin resistance). We have determined that eta3 is an allele of the SGT1b gene. In budding yeast, SGT1 was isolated as a high-copy suppressor of the G2 cell cycle arrest phenotype conferred by the skp1-4 mutation and has been shown to interact with the Skp1p-containing kinetochore and SCF protein complexes (Kitagawa et al., 1999). The yeast SGT1 gene is essential for viability, and temperature-sensitive sgt1 alleles confer defects in kinetochore complex assembly and SCF-mediated ubiquitination. Recently, the Arabidopsis SGT1b gene was found to be a positive regulator of R gene–mediated defense against certain pathogens, raising the possibility that disease resistance signaling may involve an SCF component (Austin et al., 2002; Tör et al., 2002). However, an SCF ubiquitin ligase required for disease resistance has yet to be identified. Our genetic analysis of the eta3 mutant, together with molecular studies examining SCFTIR1 activity, indicate that SGT1b is required for SCF function in Arabidopsis.
RESULTS
Identification of the eta3 Mutant
To identify additional genes required for SCFTIR1 function, we mutagenized tir1-1 seeds with ethyl methanesulfonate and screened for mutations that enhance the relatively weak tir1-1 auxin-resistant-root phenotype. We reasoned that the tir1-1 mutation might sensitize the auxin response pathway, such that further perturbations of the pathway would result in severe auxin response defects. Seedlings were screened on nutrient medium containing 0.25 μM 2,4-D, a synthetic auxin. At this concentration of auxin, root elongation was inhibited completely in tir1-1 seedlings. This screen identified 28 eta mutants. Complementation and allelism tests against known auxin resistance mutants revealed that several of the eta mutations affect previously identified loci, including AUX1, AXR1, AXR3, and AXR6. The remaining eta mutants comprise at least seven complementation groups that define novel loci required for normal auxin response. One of these complementation groups consisted of a single allele of the eta3 gene. In addition to enhancing the tir1-1 auxin-resistant root phenotype, the eta3 tir1-1 double mutants displayed a slight reduction of apical dominance, consistent with a further reduction in auxin response (Figure 1). Genetic analysis of the F2 progeny of eta3 tir1-1 backcrossed with tir1-1 plants indicated that eta3 affected a single locus and was recessive, because 58 of 239 F2 seedlings were resistant to 0.25 μM 2,4-D.
Figure 1.
Identification of the eta3 Mutant.
(A) tir1-1 and eta3 tir1-1 seedlings were grown on unsupplemented nutrient medium for 4 days and then transferred to medium containing 0.25 μM 2,4-D and grown for an additional 4 days. Asterisks indicate the position of the root tip at the time of transfer.
(B) tir1-1 and eta3 tir1-1 adult plants. Bars = 5 cm.
Analysis of the eta3 Mutant Phenotype
The eta3 tir1-1 mutant was crossed to wild-type plants to identify eta3 TIR1+ segregants. Thirty of 505 F2 progeny exhibited resistance to 0.25 μM auxin, demonstrating that both the tir1-1 and eta3 mutations are required for resistance at this concentration (15:1; P < 0.01). When assayed on 0.075 μM 2,4-D, 236 of 555 F2 seedlings displayed auxin-resistant root growth, suggesting that the eta3 mutation confers a weak auxin resistance phenotype independent of tir1-1. This possibility was confirmed when several resistant segregants were scored as TIR1+/TIR1+ using a cleaved amplified polymorphic sequence marker specific for the tir1-1 mutation.
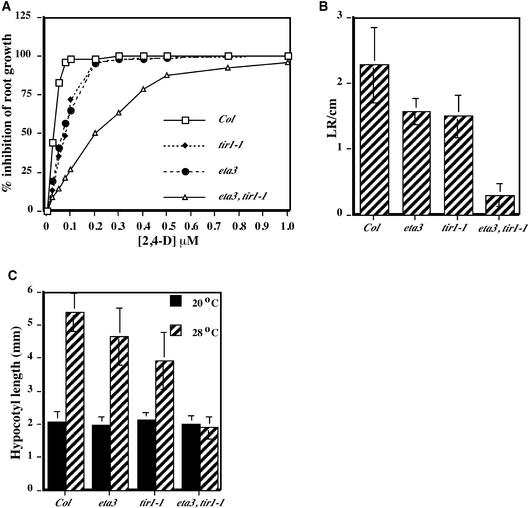
A dose–response assay was conducted to examine more precisely the extent of the eta3 auxin response defect (Figure 2A). Like tir1-1, the eta3 mutation conferred auxin-resistant root growth on low concentrations of auxin, with concentrations of ∼0.07 μM 2,4-D causing 50% inhibition of root growth. By contrast, ∼0.2 μM 2,4-D was needed to produce 50% inhibition in eta3 tir1-1 seedlings. This concentration of auxin conferred nearly complete inhibition of root elongation in both single mutant lines.
Figure 2.
eta3 Enhances the tir1-1 Auxin Response Defect.
(A) Inhibition of root elongation by increasing concentrations of the synthetic auxin 2,4-D. Data points are averages from 12 seedlings, and standard deviations for all data points were ≤10% of the mean.
(B) Lateral root (LR) initiation was assessed in 10-day-old seedlings grown on unsupplemented ATS nutrient medium (n = 12).
(C) Hypocotyl length of 8-day-old seedlings grown at 20 or 28°C (n = 12).
Error bars indicate standard deviations from the mean. Col, Columbia.
These findings suggest that eta3 and tir1-1 interact synergistically in the auxin response pathway. This possibility was investigated further by examining additional auxin-regulated growth responses, including lateral root development and temperature-induced hypocotyl elongation (Gray et al., 1998). Whereas the eta3 and tir1-1 single mutants exhibited little or no apparent reduction in these auxin-dependent growth processes, the eta3 tir1-1 double mutant was impaired severely in both lateral root development (Figure 2B) and high-temperature-induced hypocotyl elongation (Figure 2C).
eta3 Is an Allele of SGT1b
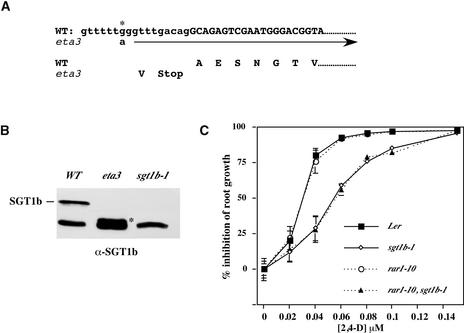
A map-based cloning strategy was used to isolate the ETA3 gene. The eta3 mutation was mapped initially between markers nga8 and g4539 on chromosome 4. Additional mapping narrowed the location of ETA3 to an ∼67-kb interval that spanned BACs T22B4 and F8L21. Inspection of the candidate loci within this interval revealed the presence of the recently identified SGT1b gene. Given the recent findings that SGT1 in both yeast and plants interacts with SCF ubiquitin ligases, we sequenced the SGT1b gene in our eta3 mutant. Sequence analysis detected a 1-bp change in the last intron near the splice acceptor site. RNA gel blot analysis failed to detect a difference in SGT1b mRNA levels between wild-type and eta3 seedlings (data not shown). However, sequence analysis of a reverse transcriptase–mediated PCR product amplified from eta3 seedlings revealed that the mutation in the intron creates a false splice acceptor site that results in a premature stop codon (Figure 3A). Protein gel blot analysis with an SGT1b-specific antibody detected a truncated form of SGT1b in eta3 seedling extracts (Figure 3B). The mobility of this band was consistent with the 36–amino acid truncation predicted by the sequence analysis.
Figure 3.
Positional Cloning of the ETA3 Locus.
(A) Sequencing of a reverse transcriptase–mediated PCR product revealed that the eta3 mutation causes a G→A substitution (asterisk) near the splice acceptor site of the final exon. This substitution creates a false splice acceptor site and the inclusion of the intron sequence AGGTTTGACAG in the eta3 mRNA, resulting in a premature termination codon. Lowercase and uppercase letters indicate intron and exon sequences, respectively.
(B) Protein gel blot analysis of the ETA3/SGT1b protein. The position of the truncated eta3 protein is marked with the asterisk in lane 2. The bottom-most band represents an unknown cross-reacting protein.
(C) Root growth assay with Landsberg erecta (Ler), sgt1b-1, rar1-10, and sgt1b-1 rar1-10 seedlings. Error bars indicate standard deviations from the mean.
To investigate further the possibility that eta3 is allelic to SGT1b, we compared the phenotypes of eta3 and the sgt1b-1 allele, which was isolated previously in a screen for disease resistance components (Austin et al., 2002). Like sgt1b-1 plants, eta3 mutants displayed increased susceptibility to the downy mildew pathogen Peronospora parasitica (data not shown). Likewise, sgt1b-1 mutant seedlings exhibited auxin-resistant root growth (Figure 3C) and reduced lateral root development compared with Landsberg erecta control seedlings. Genetic analysis confirmed that eta3 is allelic to sgt1b-1 (data not shown).
Molecular analysis in barley cell extracts indicates that SGT1 exists in two distinct pools, one containing RAR1 and a second containing SCF ubiquitin-ligase subunits (Azevedo et al., 2002). We examined whether RAR1 is required for auxin response using root growth assays comparing wild-type, sgt1b-1, rar1-10, and rar1-10 sgt1b-1 seedlings. rar1-10 seedlings displayed wild-type auxin response. Furthermore, the rar1-10 mutation did not confer an increase in the auxin resistance resulting from the sgt1b-1 mutation (Figure 3C).
eta3 Mutants Exhibit Reduced Aux/IAA Protein Degradation
We next examined whether the eta3 mutation affected SCFTIR1 ubiquitin-ligase activity. SCFTIR1 was shown recently to target Aux/IAA proteins for ubiquitin-mediated degradation. We examined Aux/IAA protein stability indirectly using a heat-shock-regulated AXR3/IAA17–β-gluronidase (GUS) reporter construct (HS::AXR3NT-GUS) and directly using AXR2/IAA17 pulse-chase assays with seedlings labeled metabolically with 35S-Met (Gray et al., 2001). Wild-type and eta3 seedlings carrying the HS::AXR3NT-GUS construct were heat shocked for 2 h to induce expression of the transgene. Seedlings were incubated subsequently at room temperature in the presence or absence of auxin and then stained to detect GUS activity. In the absence of applied auxin, the wild-type and eta3 seedlings exhibited similar levels of AXR3NT-GUS staining after a 20-min incubation at room temperature (Figures 4A and 4E). By 40 min, a modest difference was apparent, with the eta3 seedlings displaying slightly more staining (Figures 4B and 4F). This difference was enhanced dramatically when seedlings were treated with 5 μM IAA (Figures 4C, 4D, 4G, and 4H). Auxin has been shown previously to target the AXR3 protein for SCFTIR1-mediated ubiquitination (Gray et al., 2001). Our findings suggest that ETA3/SGT1b is required for the SCFTIR1-targeted ubiquitin-mediated degradation of the AXR3NT-GUS fusion protein.
Figure 4.
eta3 Mutants Exhibit Increased AXR3NT-GUS Stability.
(A) to (H) Wild-type (WT; [A] to [D]) and eta3 ([E] to [H]) seedlings carrying the HS::AXR3NT-GUS reporter construct were heat shocked for 2 h and stained for β-glucuronidase activity after 20- or 40-min incubations at room temperature. IAA (5 μM) was added to the room-temperature incubation where indicated.
(I) AXR2 protein was immunoprecipitated from Columbia (Col), eta3 tir1, eta3, and tir1-1 seedlings labeled with 35S-Met. Precipitations were performed immediately after labeling (t = 0) or after a 15-min chase with medium containing 1 mM Met and 100 μg/mL cycloheximide (t = 15).
AXR2/IAA7 stability was examined by immunoprecipitating the AXR2 protein from metabolically labeled seedlings in a pulse-chase assay. AXR2 stability was increased modestly in the eta3 tir1-1 double mutant (Figure 4I). The average AXR2 half-life was 21.5 ± 1.2 min in eta tir1-1 seedlings compared with 11.0 ± 0.6 min in wild-type seedlings. Neither the tir1-1 nor the eta3 mutation alone conferred a significant change in AXR2 half-life in this assay.
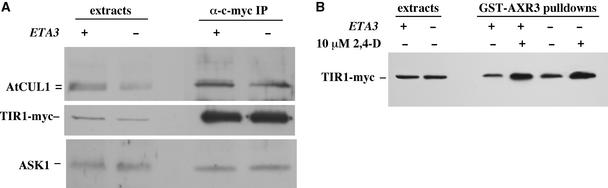
ETA3/SGT1b Is Not Required for SCFTIR1 Assembly or Aux/IAA Binding
SGT1 has been shown to interact with both yeast and barley SCF ubiquitin ligases (Kitagawa et al., 1999; Azevedo et al., 2002). We were unable to detect an interaction between SGT1b and the SCFTIR1 complex specifically by both immunoprecipitation and glutathione S-transferase (GST)–SGT1b pull-down experiments with crude plant extracts. This may simply be a detection problem, because SGT1 in yeast interacts with SCF complexes at substoichiometric levels (Kitagawa et al., 1999). Studies with yeast indicate that SGT1 is required for SCF-mediated ubiquitination. However, the precise role of SGT1 in this process is unclear (Kitagawa et al., 1999). We examined the effect of the eta3 mutation on SCFTIR1 complex assembly and substrate binding. First, the SCFTIR1 complex was immunoprecipitated using the c-myc antibody and extracts prepared from wild-type and eta3 seedlings expressing a TIR1-myc fusion protein. Subsequent protein gel blot analysis detected similar levels of the AtCUL1 and ASK1 subunits in both sets of immunoprecipitates (Figure 5A). This analysis also indicated that RUB modification of AtCUL1 (del Pozo and Estelle, 1999) is not affected by the eta3 mutation. Second, substrate binding was examined using a GST-AXR3 pull-down assay. Recombinant GST-AXR3 fusion protein was incubated with extracts prepared from wild-type and eta3 seedlings expressing c-myc–tagged TIR1. After repurification of the fusion protein, samples were immunoblotted to detect interaction with TIR1-myc. As has been described previously (Gray et al., 2001), auxin was found to stimulate the interaction between GST-AXR3 and TIR1-myc. This interaction appeared to be unaffected by the eta3 mutation, suggesting that SGT1b is not required for SCFTIR1 binding to its Aux/IAA substrates (Figure 5B).
Figure 5.
The eta3 Mutation Does Not Alter SCFTIR1 Assembly or Substrate Binding.
(A) Anti-c-myc antibody was used to immunoprecipitate the SCFTIR1 complex from extracts prepared from tir1-1 and eta3 seedlings expressing a TIR1-myc fusion gene. Immunoprecipitates (IP) were immunoblotted with anti-ASK1 and anti-AtCUL1 antibodies as described previously (Gray et al., 1999).
(B) Recombinant GST-AXR3 protein was used to pull down the SCFTIR1 complex from tir1-1[TIR1-myc] and eta3[TIR1-myc] seedling extracts. Pull downs were immunoblotted with anti-c-myc antibody to detect the presence of the TIR1-myc fusion protein.
SGT1b Overexpression Partially Suppresses the tir1-1 Auxin Response Defect
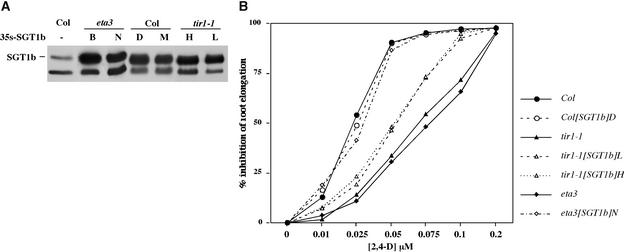
To explore further the role of ETA3/SGT1b in the auxin response, we introduced a 35S-SGT1b construct into wild-type, tir1-1, and eta3 plants. T1 plants were screened by protein gel blot analysis with SGT1b antiserum to identify overexpressing lines (Figure 6A). Plants that expressed high levels of SGT1b exhibited no obvious developmental phenotype, with the exception that overexpression in the eta3 and tir1-1 backgrounds rescued the weak apical dominance defects normally seen in these mutants. Auxin response was assayed in homozygous lines by examining the auxin inhibition of root elongation. Expression of the 35S-SGT1b construct completely restored the wild-type auxin response to the eta3 mutant. Furthermore, overexpression of SGT1b partially suppressed the auxin response defect conferred by the tir1-1 mutation but did not alter the sensitivity of wild-type plants (Figure 6B). Analysis of TIR1-myc immunoprecipitates from 35S-SGT1b seedlings revealed no change in the interactions with the core SCF subunits ASK1 and AtCUL1 (data not shown).
Figure 6.
Analysis of ETA3/SGT1b Overexpression.
(A) Protein gel blot analysis with anti-SGT1b antiserum of crude extracts prepared from single rosette leaves of control and 35S-SGT1b T1 plants. Letters above the lanes indicate specific transgenic lines. Col, Columbia.
(B) Dose–response assay of auxin response in control and homozygous 35S-SGT1b lines. All data points are averages of 12 seedlings, and all standard deviations are ≤10% of the mean.
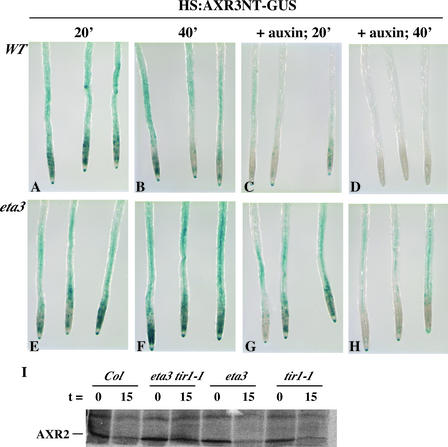
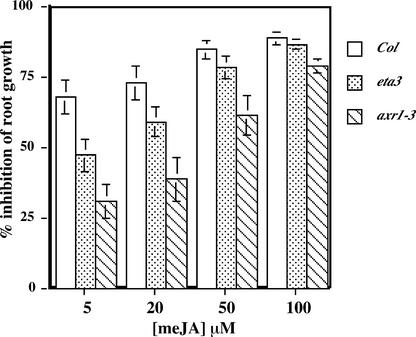
The eta3 Mutation Confers Reduced Jasmonate Sensitivity
To determine whether SGT1b functions in other SCF-regulated pathways, we examined the sensitivity of the eta3 mutant to the phytohormone methyl jasmonate (MeJA). The MeJA response pathway has been shown to require the SCFCOI1 ubiquitin-ligase complex (Xu et al., 2002). eta3 seedlings were slightly more resistant than wild-type seedlings to the inhibition of root growth resulting from MeJA (Figure 7). axr1-3, which was found previously to exhibit a reduced MeJA response (Tiryaki and Staswick, 2002), presumably as a result of a reduction in the RUB modification of the AtCUL1 subunit of SCFCOI1, was included as a positive control.
Figure 7.
The eta3 Mutation Confers a Reduced MeJA Response.
Wild-type Columbia (Col), eta3, and axr1-3 seedlings were grown for 10 days on nutrient medium supplemented with MeJA. Error bars indicate standard deviations from the mean.
DISCUSSION
ETA3/SGT1b Is Required for Auxin Response
The SCFTIR1 ubiquitin ligase plays a central role in the auxin response pathway by targeting members of the Aux/IAA family of transcriptional regulators for ubiquitin-mediated degradation in response to auxin. To identify additional factors required for auxin response, we screened for mutations that enhance the weak auxin response defect conferred by the tir1-1 mutation. This analysis identified several novel auxin response genes, including ETA3, which is described in this report.
We used map-based cloning and genetic strategies to determine that eta3 is an allele of the SGT1b gene. SGT1 interacts with SCF ubiquitin-ligase complexes in yeast and in barley and tobacco extracts (Kitagawa et al., 1999; Azevedo et al., 2002; Liu et al., 2002b). Furthermore, sgt1 mutants in yeast exhibit reduced SCF-mediated ubiquitination (Kitagawa et al., 1999). The strong genetic interactions we observed between the eta3 and tir1-1 mutations suggest that SGT1b also is required for SCF function in Arabidopsis. Consistent with this possibility, we found that the eta3 mutant exhibited a reduction in the auxin-induced degradation of the AXR3NT-GUS fusion protein. We also found that AXR2 stability was increased in eta3 tir1-1 double mutants, although we detected no significant effect on AXR2 stability in eta3 single mutant seedlings. The reason for the different findings between these two assays for measuring Aux/IAA stability is unclear. One possibility is that the HS::AXR3NT-GUS construct overexpresses its SCFTIR1 substrate, making these plants more sensitive to perturbation of the SCFTIR1 complex. As in the eta3 mutant, we detect increased Aux/IAA stability in tir1-1 seedlings with the AXR3NT-GUS reporter (Gray et al., 2001) but not with the AXR2 pulse-chase assay.
Recently, mutational screens identified Arabidopsis SGT1b as a regulator of R gene–mediated defenses against the downy mildew pathogen P. parasitica (Austin et al., 2002; Tör et al., 2002). Gene-silencing experiments in barley and tobacco also revealed a role for SGT1 in R gene–mediated and nonhost pathogen resistance (Azevedo et al., 2002; Peart et al., 2002; Liu et al., 2002b). The RAR1 gene product is required by some of these R gene–regulated pathways (Austin et al., 2002; Liu et al., 2002a; Muskett et al., 2002). Molecular analysis in barley cell extracts indicates that SGT1 exists in two distinct pools, one containing RAR1 and a second containing SCF subunits (Azevedo et al., 2002). We examined the possibility that RAR1 is required for the auxin response but observed no effect of rar1 mutations on the auxin response pathway in either the SGT1b or the sgt1b-1 background. These findings are consistent with the notion that SGT1b regulates the auxin response through the SCFTIR1 complex.
Function of SGT1b in SCF-Mediated Ubiquitination
Studies in yeast, and now Arabidopsis, indicate that SGT1 is required for SCF-mediated ubiquitination. However, the findings that SGT1 associates with SCF complexes at substoichiometric levels and apparently is dispensable for SCF ubiquitin-ligase activity in vitro (Kitagawa et al., 1999) make the role of SGT1 in this process unclear. A number of recent studies noted a resemblance between the SGT1 N-terminal tetratricopeptide repeat and central CHORD-containing proteins and SGT1 domains with the three-dimensional folds of the Hop and p23 families of Hsp70 and Hsp90 cochaperones, respectively (Dubacq et al., 2002; Garcia-Ranea et al., 2002; Stemmann et al., 2002). Thus, one hypothesis is that SGT1 facilitates SCF assembly. However, SCFTIR1 assembly appeared unaffected in our studies with the eta3 mutant (Figure 5) and 35S-SGT1b plants (data not shown). Likewise, the ability of the SCFTIR1 complex to bind its Aux/IAA substrates in an in vitro assay was not affected by the eta3 mutation or SGT1b overexpression. These data suggest that SGT1b may operate at another level of SCF function, possibly by assisting the ubiquitination process once the substrate is bound. Analysis of the SCFTIR1–Aux/IAA interaction in vivo, as well as the development of an in vitro Aux/IAA ubiquitination assay, will be helpful in elucidating the role of SGT1b in SCF function. This analysis is complicated by the fact that the Arabidopsis genome contains a second SGT1 homolog, SGT1a, that displays ∼87% amino acid identity with SGT1b (Austin et al., 2002). This high degree of similarity, along with the fact that both SGT1a and SGT1b can complement yeast sgt1 mutations, suggests that the two Arabidopsis proteins likely exhibit some degree of functional redundancy (Azevedo et al., 2002). The identification and analysis of sgt1a mutants should be helpful in clarifying this issue.
We also found that overexpression of ETA3/SGT1b partially suppressed the auxin response defect conferred by the tir1-1 mutation. Because SGT1b overexpression did not result in heightened auxin response in a wild-type background, it seems unlikely that SGT1b is a limiting factor for SCFTIR1 activity. Rather, suppression of the tir1-1 phenotype may be the result of an SGT1b-mediated increase in the activity or assembly of the crippled SCF complex containing the mutant tir1-1 protein. Alternatively, it should be noted that Arabidopsis encodes three TIR1-like proteins that exhibit 60 to 69% identity to TIR1 (Ruegger et al., 1998; M. Estelle, unpublished data). Overexpression of SGT1b may compensate for the reduction in SCFTIR1 activity resulting from the tir1-1 mutation by increasing the activity of these related SCF complexes.
The SGT1-Specific Domain Is Required for SGT1b Function
The eta3 mutation causes errant splicing of the SGT1b mRNA, resulting in a 36–amino acid truncation. All of the previously identified mutations in sgt1b apparently are null alleles (Austin et al., 2002; Tör et al., 2002). Our analysis of the eta3 allele indicates that the highly conserved C terminus of SGT1b, previously designated the SGT1-specific domain (Azevedo et al., 2002), is required for SGT1b function. Genetic and molecular analysis of sgt1/git7 mutant strains of Saccharomyces cerevisiae and Schizosaccharomyces pombe reveal that SGT1 is an activator of cAMP signaling through association between the SGT1-specific domain of SGT1 and the C-terminal Leu-rich repeats of adenylyl cyclase CYR1/CDC35 (Dubacq et al., 2002; Schadick et al., 2002). It is possible that SGT1b interacts transiently with the Leu-rich repeats of TIR1 as a conformational activator in the ubiquitination process. However, because SGT1 is required for the degradation of multiple SCF substrates, it seems more likely that SGT1 functions through one of the core SCF subunits.
The demonstration that SGT1b is required for the auxin response is an important step forward in our understanding of SGT1 function in plants. Interpretation of the role of SGT1b in previous studies of plant disease resistance has been clouded by the fact that an SCF complex required for R gene–mediated disease resistance has not been identified. SGT1b may regulate R gene–mediated signaling via an SCF ubiquitin ligase; this hypothesis is supported by our preliminary findings indicating that mutations that affect core SCF subunits confer increased susceptibility to P. parasitica (data not shown). Confirmation of this possibility awaits the identification of a specific SCF complex. Our finding that SGT1b functions in the SCFTIR1-mediated ubiquitination of Aux/IAA proteins provides convincing support for the hypothesis that SGT1b is required for SCF activity in plants. Furthermore, we observed that the eta3 mutant exhibited a reduced response to jasmonic acid. Because MeJA response is regulated by the SCFCOI1 ubiquitin ligase (Xu et al., 2002), SGT1b is likely a key component of multiple SCF-regulated pathways.
METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana lines used in this study are in the Columbia (Col) ecotype with the exception of the sgt1b-1 and rar1-10 lines, which are in the Landsberg erecta (Ler) background. The tir1-1 line used for the eta enhancer screen contains a Gly→Glu substitution at amino acid 147 of the TIR1 protein (Ruegger et al., 1998). Seedlings were grown under sterile conditions on vertically oriented ATS nutrient medium (Lincoln et al., 1990). For hypocotyl elongation studies, seedlings were germinated and grown for 8 days under constant light (60 μmol·m−2·s−1) at 20 or 28°C. Adult plants were grown in soil under long-day conditions. 35S-SGT1b plants were generated by Agrobacterium tumefaciens–mediated transformation with the SGT1b cDNA cloned into the plant transformation vector pROKII.
Ethyl Methanesulfonate Mutagenesis and Mutant Screen
Approximately 10,000 tir1-1 gl seeds were mutagenized with ethyl methanesulfonate as described previously (Timpte et al., 1994). M2 seeds were germinated in ATS nutrient medium, and 4- to 6-day-old seedlings were transferred to ATS plates containing 0.25 μM 2,4-D. Resistant seedlings were potted as putative enhancers of tir1-1 auxin resistance (eta) mutants, and their progeny were subjected to additional physiological and genetic analysis. All eta3 lines used in this study were backcrossed to tir1-1 or Col a minimum of three times.
eta3 Mapping Analysis
A total of 436 auxin-resistant F2 seedlings from a cross between eta3 (ecotype Col) and Ler were identified and used to prepare DNA for PCR-based mapping with codominant cleaved amplified polymorphic sequence and simple sequence length polymorphism markers. We initially mapped the eta3 mutation to an interval between markers nga8 and g4539 described in TAIR (The Arabidopsis Information Resource; http://www.arabidopsis.org). For fine mapping, we generated several additional markers in this interval using the Cereon Arabidopsis Polymorphism Collection (Jander et al., 2002). Simple sequence length polymorphism markers defining our final mapping interval were CER466317 (5′-GAGTTACTTCGAGAAACTTAC-3′ and 5′-TTGTGTGGCTCACCCATC- 3′), which amplifies products of 179 bp (Col) and 164 bp (Ler), and CER466322 (5′-GGATAAATCTTATCACTCCTC-3′ and 5′-GATACATCA-ACATGCTGTAG-3′), which yields products of 291 bp (Col) and 265 bp (Ler). These and additional markers are available upon request.
β-Glucuronidase Histochemical Staining
The HS::AXR3NT–β-glucuronidase transgene (Gray et al., 2001) was crossed into the eta3 mutant. Six-day-old Col and eta3 seedlings homozygous for the reporter construct were heat shocked for 2 h at 37°C to induce expression of the transgene. Seedlings then were transferred to 20°C medium and incubated for 20 or 40 min before staining for β-glucuronidase activity (Stomp, 1991). Indoleacetic acid (5 μM) was added to the 20°C medium where indicated (Figure 4).
Antibodies, Coimmunoprecipitation, and Protein Gel Blot Analysis
All antibodies used in this study have been described previously (Gray et al., 1999; Austin et al., 2002). Protein gel blot, glutathione S-transferase pull-down, AXR2 pulse-chase, and coimmunoprecipitation analyses were performed as described by Gray et al. (2002).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Mark Estelle for his support during the early stages of this work, Ken Shirasu for communicating unpublished results, Bill Crosby for the gift of α-ASK1 antisera, and Cereon Genomics for access to its Arabidopsis Polymorphism Collection. We also are grateful to Mark Estelle and Neil Olszewski for helpful comments on the manuscript. This work was funded by The Max Planck Society, the Alexander von Humboldt Foundation, Biological and Biotechnology Research Council Grant P11874 to J.E.P., and National Institutes of Health Grant GM067203-01 to W.M.G.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010884.
References
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Davies, P.J., ed. (1995). Plant Hormones: Physiology, Biochemistry, and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq, C., Guerois, R., Courbeyrette, R., Kitagawa, K., and Mann, C. (2002). Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1, 568–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ranea, J.A., Mirey, G., Camonis, J., and Valencia, A. (2002). p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529, 162–167. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Hellmann, H., Dharmasiri, S., and Estelle, M. (2002). Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14, 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Ostin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4, 21–33. [DOI] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M., Lincoln, C.A., Timpte, C., Lammer, D., Turner, J., and Estelle, M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364, 161–164. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002. a). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.-W., and Dinesh-Kumar, S.P. (2002. b). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene–mediated resistance response to Tobacco mosaic virus. Plant Cell 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D.J., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene–mediated defences against multiple pathogens. Plant Cell 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99, 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust, V.N., Brownell, J.E., Gladysheva, T.B., Luo, R.S., Wang, C., Coggins, M.B., Pierce, J.W., Lightcap, E.S., and Chau, V. (2000). A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. USA 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, O., and Callis, J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadick, K., Fourcade, H.M., Boumenot, P., Seitz, J.J., Morrell, J.L., Chang, L., Gould, K.L., Partridge, J.F., Allshire, R.C., Kitagawa, K., Hieter, P., and Hoffman, C.S. (2002). Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgtlp family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation. Eukaryot. Cell 1, 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann, O., Neidig, A., Kocher, T., Wilm, M., and Lechner, J. (2002). Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc. Natl. Acad. Sci. USA 99, 8585–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomp, A.-M. (1991). Histochemical localization of β-glucuronidase. In GUS Protocols, S.R. Gallagher, ed (London: Academic Press), pp. 103–113.
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Timpte, C., Wilson, A.K., and Estelle, M. (1994). The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryaki, I., and Staswick, P.E. (2002). An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tör, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., Can, C., Dangl, J.L., and Holub, E.B. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98, 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]