Abstract
The polyketide toxin cercosporin plays a key role in pathogenesis by fungal species of the genus Cercospora. The bacterium Xanthomonas campestris pv. zinniae is able to rapidly degrade this toxin. Growth of X. campestris pv. zinniae strains in cercosporin-containing medium leads to the breakdown of cercosporin and to the formation of xanosporic acid, a nontoxic breakdown product. Five non-cercosporin-degrading mutants of a strain that rapidly degrades cercosporin (XCZ-3) were generated by ethyl methanesulfonate mutagenesis and were then transformed with a genomic library from the wild-type strain. All five mutants were complemented with the same genomic clone, which encoded a putative transcriptional regulator and an oxidoreductase. Simultaneous expression of these two genes was necessary to complement the mutant phenotype. Sequence analysis of the mutants showed that all five mutants had point mutations in the oxidoreductase gene and no mutations in the regulator. Quantitative reverse transcription-PCR (RT-PCR) showed that the expression of both of these genes in the wild-type strain is upregulated after exposure to cercosporin. Both the oxidoreductase and transcriptional regulator genes were transformed into three non-cercosporin-degrading bacteria to determine if they are sufficient for cercosporin degradation. Quantitative RT-PCR analysis confirmed that the oxidoreductase was expressed in all transconjugants. However, none of the transconjugants were able to degrade cercosporin, suggesting that additional factors are required for cercosporin degradation. Further study of cercosporin degradation in X. campestris pv. zinniae may allow for the engineering of Cercospora-resistant plants by using a suite of genes.
Members of the fungal genus Cercospora are pathogenic to plants, causing damaging leaf spot and blight diseases. The genus collectively has a wide host range, with individual species parasitizing specific hosts. Cercospora spp. can infect such hosts as tobacco, corn, soybean, rice, sugar beet, and banana (5, 14, 17, 28, 40). Crop damage and losses can be very high.
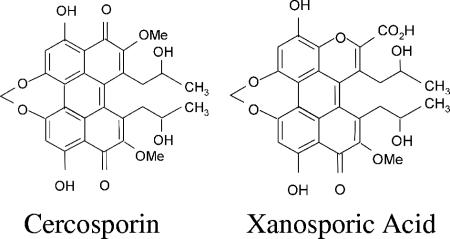
Cercospora spp. are known for the production of the non-host-specific toxin cercosporin. Cercosporin was first isolated from dried mycelium of Cercospora kikuchii, a soybean pathogen, in 1957 by Kuyama and Tamura (23). It is deep red and has the molecular formula C29H26O10 (Fig. 1) (26, 46). Cercosporin is produced by most members of the Cercospora genus and has been isolated from many Cercospora species as well as from infected host plants (2, 4, 17, 18, 23, 28, 31, 43). It is a polyketide secondary metabolite with toxicity to plants, animals, bacteria, and fungi (14). Cercosporin is a photosensitizer and uses light energy to produce the activated oxygen species superoxide and singlet oxygen (11, 47). Although cercosporin has been shown to produce both activated oxygen species, singlet oxygen appears to cause the most damage, leading to lipid peroxidation and the breakdown of cell membranes (11-13, 15, 21, 24).
FIG. 1.
Chemical structure of cercosporin and the breakdown product (xanosporic acid) produced by X. campestris pv. zinniae isolate XCZ-3.
Production of the toxin cercosporin has been associated with parasitism and the development of disease in host plants (14). When cercosporin-deficient mutants of Cercospora kikuchii and Cercospora nicotianae were inoculated onto their host plants, the resulting lesions were fewer and smaller (8, 41). Mutants of Cercospora zeae-maydis disrupted in the CZK3 mitogen-activated protein kinase kinase kinase and deficient in both cercosporin production and conidiation caused only limited small chlorotic lesions on corn rather than the typical large necrotic lesions (37). In addition, in the sugar beet, the disease severity of Cercospora leaf spot disease has been shown to be directly related to light intensity (7). Furthermore, previous studies by Steinkamp et al. showed that ultrastructural changes in the sugar beet caused by infection with Cercospora beticola are almost identical to those caused by treatment with cercosporin (13, 38, 39).
Presently, the control of diseases caused by Cercospora species relies on the use of fungicides and appropriate cultural practices such as tillage and crop rotation (44, 45). The development of resistant cultivars has been attempted, but high levels of resistance to Cercospora diseases are uncommon. Although limited disease resistance has been found in corn, sugar beet, and soybean (9, 33, 36, 44, 45), alternate methods of control would be desirable. One such approach would be to target cercosporin by engineering plants to express genes encoding degradation enzymes.
A patent for a method to identify bacteria that have the ability to degrade cercosporin was issued to Robeson et al. (35). Mixed bacterial populations were harvested from the soil, leaf surfaces, and leaf tissue of C. beticola-infected sugar beet plants and inoculated onto cercosporin-containing medium. After incubation in the dark, bacteria that are able to degrade cercosporin were identified by a clear halo surrounding the colony on the red plates.
In previous work in our laboratory, we followed up on the findings of Robeson et al. and screened bacteria for the ability to degrade cercosporin. Out of 244 isolates that were screened, 43 isolates that were able to degrade cercosporin were identified based on the production of a clear halo when bacteria were cultured on cercosporin-containing medium (30). Of the isolates tested, Xanthomonas species had the largest proportions of cercosporin degraders, and among those, all 32 isolates of Xanthomonas campestris pv. zinniae that were tested were particularly active. We subsequently showed that cercosporin degradation leads to the production of a green breakdown product identified as xanosporic acid (29) (Fig. 1). Xanosporic acid was shown to be nontoxic to a cercosporin-sensitive mutant of C. nicotianae (30). The lactonized derivative of xanosporic acid, xanosporolactone, was used in a plant infiltration assay of tobacco and was also shown to be nontoxic (30).
The goal of the work reported here is to isolate and characterize the gene(s) responsible for cercosporin degradation by a strain of X. campestris pv. zinniae that rapidly degrades cercosporin (XCZ-3). We show that the ability of XCZ-3 to degrade cercosporin requires a region of DNA containing genes encoding an oxidoreductase and a transcriptional regulator.
MATERIALS AND METHODS
Bacterial cultures.
Mutants of strains XCZ-3 (30) and XCZ-3 were maintained on Luria-Bertani (LB) medium (10 g of tryptone, 10 g of NaCl, and 5 g of yeast extract per liter [pH 7.2]) containing rifampin at 100 μg/ml. Xanthomonas axonopodis (pseudonym X. campestris) pv. pruni (34, 42) strains XAP-76, XAP-50, XAP-75, XAP-1, XAP-39, XAP-49, XAP-34, XAP-31, and XAP-79 were obtained from David Ritchie (North Carolina State University) and maintained on LB medium without antibiotics. Isolates of Xanthomonas axonopodis pv. vesicatoria (34, 42) (XAV-79 and XAV-126), also obtained from David Ritchie, were maintained on LB medium containing 200 μg/ml copper sulfate. Pseudomonas syringae pv. tomato DC3000 and Pseudomonas syringae pv. pisi 209-21-3R were obtained from Peter Lindgren (North Carolina State University) and maintained on LB medium containing rifampin (as described above). All of the bacterial isolates were maintained at 28°C in the dark.
Cercosporin stocks and chemicals.
Cercosporin was isolated and purified from mycelial cultures of Cercospora nicotianae as previously described (11). Dried cercosporin crystals were dissolved in acetone for use in media. All other chemicals were obtained from Sigma-Aldrich Co. (St. Louis, MO) unless noted otherwise. All restriction enzymes were obtained from Promega Corporation (Madison, WI).
Chemical mutagenesis of Xanthomonas campestris pv. zinniae and screening for mutants unable to degrade cercosporin.
Three of the non-cercosporin-degrading mutants (120-2, MutA, and MutB) were created through ethyl methanesulfonate (EMS) mutagenesis according to the protocol described previously by Thorne et al. (40). The other two non-cercosporin-degrading mutants (MutC and MutD) were obtained by first transforming the wild type with the multicopy vector pLAFR6 (obtained from Brian Staskawicz, University of California, Berkeley, CA) containing both the transcriptional regulator and the oxidoreductase. EMS mutagenesis was then performed to yield the mutants. Screening for non-cercosporin-degrading mutants (Fig. 2) was carried out as previously described (30). For MutC and MutD, vector pLAFR6 was maintained through antibiotic selection until the mutants were discovered and then removed through plasmid curing. Mutant colonies were grown on LB medium containing rifampin at 100 μg/ml to ensure that the mutants retained the RifR marker.
FIG. 2.
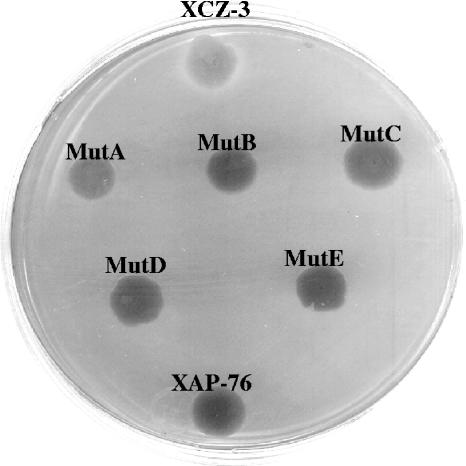
Non-cercosporin-degrading XCZ-3 mutants. Mutants show a lack of a clear halo when grown on cercosporin-containing medium. The positive control (XCZ-3 [wild type]) appears at the top of the plate, and the negative control (XAP-76) appears at the bottom of the plate.
Box-PCR.
The whole-cell method of Box-PCR was performed according to a protocol established previously by Louws et al. (27) in order to confirm that the non-cercosporin-degrading mutants were derived from isolate XCZ-3. The following cycle conditions were used: 95°C for 2 min followed by 35 cycles of 94°C for 3 s, 92°C for 30 s, 50°C for 1 min, and 65°C for 8 min. Ten microliters of each PCR product was separated on a 1.5% agarose gel at 36 V for approximately 15 h at 4°C. The banding patterns on the ethidium bromide-stained gel were viewed under UV light.
Xanthomonas campestris pv. zinniae genomic library construction.
Genomic DNA was purified from XCZ-3 as described previously (3). A genomic library was created based on the protocol described previously by Thorne et al. (40). The genomic DNA was digested with Sau3A, and fragments of approximately 21 kb were selected. The fragments were cloned into the BamHI restriction site of the expression vector pLAFR6, which carries tetracycline resistance as the selectable marker. Plasmids were then transformed into Epicurian Coli JM109 competent cells (Stratagene, La Jolla, CA). Transformants were selected for tetracycline resistance. A total of 2,016 clonal colonies were selected in order to form the library. Colonies were grown in 96-well plates and stored at −80°C.
Complementation assays.
Conjugation of the non-cercosporin-degrading mutants with the library clones was carried out using a triparental mating procedure with a helper cell line containing the pRK2013 plasmid (16). The library clones were cultured in LB liquid medium containing tetracycline at 20 μg/ml and incubated at 37°C. The helper cell line was cultured in LB liquid medium at 37°C. The non-cercosporin-degrading mutants were cultured in LB liquid medium containing 100 μg/ml rifampin at 28°C in the dark. For conjugation, 133 μl of each culture was mixed together and plated onto a 0.45-μm nitrocellulose membrane on LB solid medium using a 96-well replicator. The bacteria were allowed to dry on the membrane for 15 min and were then grown for 48 h at 28°C in the dark. Colonies were then transferred onto LB solid medium containing 100 μg/ml rifampin and 20 μg/ml tetracycline. For screening, a minimum of three colonies per conjugation were collected and transferred onto nutrient agar solid medium containing 50 μM cercosporin. As a control, the colonies were also grown on nutrient agar solid medium containing 0.5% acetone (used to solubilize cercosporin). A density of 12 colonies per plate was used for the screening process. Two controls, the cercosporin-degrading isolate XCZ-3 and the non-cercosporin-degrading isolate XAP-76, were also inoculated onto each plate. The colonies were incubated in the dark at 28°C for 12 days and screened for a clear halo surrounding the colony. Colonies were also screened on M9 minimal medium to identify auxotrophs.
Subcloning of complementing library clone.
Subcloning was carried out through the use of restriction enzyme digests with EcoRI and HindIII. The digested clone was separated on a 1.0% agarose gel, and the individual bands were excised and purified using a QIAGEN (Valencia, CA) gel purification kit. Subclones were ligated into the expression vector pLAFR6 (linearized with EcoRI) and transformed into DH5α chemically competent cells (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Complementation assays were performed with each subclone as described above.
Sequencing of the subclone.
The single complementing subclone (a 7.5-kb fragment obtained from the EcoRI digest) was sheared using a Hydroshear. Sheared fragments were separated on a 1.0% agarose gel, and DNA fragments between 1 and 2 kb were excised and purified using a QIAGEN (Valencia, CA) gel purification kit. The End-Repair kit (Epicenter, Madison, WI) was used to blunt end the fragments. Fragments were ligated to pBluescript II SK(+) (Stratagene, La Jolla, CA), and the ligation was transformed into DH10B electrocompetent cells (Invitrogen, Carlsbad, CA). Colonies were chosen using blue/white screening and ampicillin resistance and then cultured overnight. Plasmids were purified from each colony and were amplified by PCR using the standard primers T3 and T7. The PCR products were sequenced at the North Carolina State University Genome Research Laboratory.
Sequencing of mutant genes.
The transcriptional regulator and oxidoreductase from each non-cercosporin-degrading mutant were sequenced to identify mutations. Sequences were amplified by high-fidelity PCR using a combination of both Taq DNA polymerase (Promega, Madison, WI) and Pfu Ultra high-fidelity DNA polymerase (Stratagene, La Jolla, CA). PCR products were separated on a 1.0% agarose gel, and the desired bands were excised. The Promega Wizard SV gel and PCR Clean-Up kit were used to purify the DNA from the agarose gel. The bands were then cloned into the pGEM-T Easy vector system (Promega, Madison, WI) according to the manufacturer's protocol. The genes were sequenced using pGEM-T Easy sequencing primers T7 and SP6.
Gene complementation.
The oxidoreductase gene was PCR amplified from XCZ-3 using primers F8kb1 (5′-TCCTGCTCGATGTCAATGC-3′) and R8kb1 (5′-GAAGTTCTAGGGGCGACCAG-3′). The PCR product was cloned into the pGEM-T Easy vector, excised by restriction digestion with EcoRI, and cloned into pLAFR6. A second construct containing both the transcriptional regulator and the oxidoreductase gene was made in the same fashion using primers newsense2 (5′-AGCACCATTGACGCAAGCACC-3′) and R8kb1 (described above). A third construct containing 330 bp at the 3′ end of the transcriptional regulator and all of the oxidoreductase was made by amplification with primers F915 (5′-TGATCGGGCCATTACTGCTGTT-3′) and R8kb1 (described above). All three pLAFR6 constructs were tested for their ability to complement the non-cercosporin-degrading mutants.
Quantitative RT-PCR of oxidoreductase and regulator in wild-type XCZ-3 cells.
XCZ-3 cells were grown in LB liquid medium or LB medium containing 50 μM cercosporin or an equal volume of acetone (0.5%). RNA was isolated at 24 h using the Promega SV Total RNA isolation kit. A total of 1 μg RNA for each sample was used to make cDNA using an Applied Biosystems (Foster City, CA) TaqMan Reverse Transcription Reagents kit. Primers for the oxidoreductase and transcriptional regulator genes were designed using Primer Express (Applied Biosystems, Foster City, CA) software and acquired from Sigma Genosys (The Woodlands, TX). For the oxidoreductase, primers F1 (5′-TGCGCTACGAACGGATCA-3′) and R1 (5′-GGCACGAGCTTGGGAATGT-3′) were used. For the transcriptional regulator, primers F2 (5′-TCTTCAATCGCTTGGTCGAT-3′) and R2 (5′-ATCGCTTCAAGCTTGTCCAG-3′) were used. As an internal control, the internal transcribed sequence that resides between the 16S and 23S rRNAs was amplified using primers F-ITS (5′-GTTCCCGGGCCTTGTACACAC-3′) and R-ITS (5′-GGTTCTTTTCACCTTTCCCTC-3′) (20). Quantitative reverse transcription-PCR (RT-PCR) was carried out using 2× SYBR Green Master Mix (Applied Biosystems, Foster City, CA) with the following conditions: 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and 72°C for 20 s. The sample plate was read after every cycle. Analysis of the quantitative RT-PCR results was carried out according to a method described previously by Livak and Schmittgen (25).
Southern analysis.
A labeled probe was created using digoxigenin (DIG) (Roche, Indianapolis, IN) and primers F8kb1 (5′-TCCTGCTCGATGTCAATGC-3′) and R8kb1 (5′-GAAGTTCTAGGGGCGACCAG-3′) according to the manufacturer's protocol. Genomic DNA was isolated (3) for Southern analysis from XCZ-3, XAP-76, XAV-79, XAV-126, P. syringae pv. tomato DC3000, P. syringae pv. pisi 209-21-3R, XAP-50, XAP-75, XAP-1, XAP-39, XAP-49, XAP-34, XAP-31, and XAP-79. A total of 5 μg of genomic DNA was digested with 10 μl EcoRI from Promega (Madison, WI) in a total volume of 100 μl according to the manufacturer's protocol. The digestion was carried out at 37°C for 18.5 h. The EcoRI enzyme was heat inactivated for 15 min at 65°C. The DNA was cleaned using the Promega Wizard DNA Clean-Up kit, and the entire digest was separated on a 0.7% agarose gel for Southern analysis. Treatment and transfer of the Southern gel were carried out according to a method described previously by Ausubel et al. (3), with the exception that the gel was soaked in 0.25 N HCl for 8 min instead of 30 min. After transfer, the membrane was exposed to hybridization buffer for 4 h at 65°C and then incubated overnight at 65°C with hybridization buffer containing the DIG-labeled probe. The Southern blot was developed according to the manufacturer's protocol for DIG-labeled probes (Roche, Indianapolis, IN).
Transformation of the regulator and oxidoreductase genes into non-cercosporin-degrading bacteria.
The original complementing 23-kb library clone contained in the pLAFR6 vector (p6-23kb) was conjugated into non-cercosporin-degrading bacteria (XAV-79, XAV-126, and P. syringae pv. tomato DC3000) according to the triparental mating protocol described above. Transconjugant bacteria, as well as the corresponding wild-type strains, were grown in LB liquid medium containing 50 μM cercosporin or 0.5% acetone. RNA was isolated at 24 h and used to make cDNA as previously described. Quantitative RT-PCR was carried out using the F1 and R1 primers mentioned above in order to assess the expression of the oxidoreductase gene. Primers F-ITS and R-ITS were used to amplify the internal transcribed spacer region as an internal control.
Time course of cercosporin degradation by transconjugants.
Cercosporin degradation was assessed by growing the transconjugants and the corresponding wild-type strains in 50 ml LB liquid medium containing 60 μM cercosporin. Five-milliliter aliquots were taken at 0, 24, and 48 h and stored at −20°C until ready for use. Each sample was extracted twice with 2 ml chloroform (30), followed by partitioning of the cercosporin into the aqueous phase using 1 N NaOH. Mixtures were vortexed and centrifuged, and the supernatant was collected. The supernatant was treated with 1 N HCl (to partition cercosporin into the organic phase) and extracted with an additional 1 ml of chloroform. The chloroform extractions for the individual samples were combined to yield a total of 5 ml for each culture. Cercosporin degradation was quantified by measuring the absorbance at 472 nm using a Beckman DU 650 spectrophotometer. The experiment was carried out twice.
Nucleotide sequence accession number.
The nucleotide sequence spanning the oxidoreductase and regulator genes has been deposited in GenBank under accession number DQ087176.
RESULTS
Isolation of Xanthomonas campestris pv. zinniae mutants unable to degrade cercosporin.
Wild-type XCZ-3, when grown on cercosporin, produces a clear halo surrounding the colony (Fig. 2). By contrast, non-cercosporin-degrading strains do not produce a halo and are purple due to the uptake of cercosporin. XCZ-3 cultures were mutagenized with EMS and screened by transferring individual colonies onto cercosporin-containing medium. The presence or absence of halo formation was determined at 12 days. Two initial screens resulted in the isolation of three non-cercosporin-degrading XCZ-3 mutants. These mutants were named 120-2, MutA, and MutB (Fig. 2). When tested on M9 minimal medium, none of the mutants were auxotrophic. All three of the mutants also retained the rifampin resistance marker. Mutants were screened by Box-PCR to confirm their identities. The banding patters of all three mutants matched that of the wild-type strain (data not shown).
Identification of complementing genes.
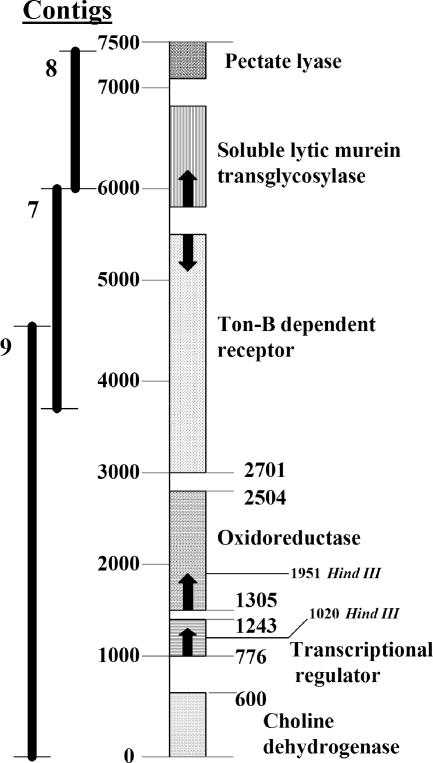
Complementation of the non-cercosporin-degrading mutants with the genomic library was carried out using the triparental mating procedure as outlined in Materials and Methods. The transconjugant colonies were screened for cercosporin degradation on cercosporin-containing medium. Out of 2,016 library clones that were screened, a single clone of 23 kb that restored cercosporin degradation to all three mutants was found. The 23-kb clone was subcloned, and a 7.5-kb subclone that complemented all the mutants was identified. This subclone was sequenced using the shotgun method, the sequences were assembled, and the resulting contig assemblies were compared against the NCBI database using the blastx function. Six genes were identified. These genes had sequence similarities to a pectate lyase (E value = 1e−104), a soluble lytic murein transglycosylase (E value = e−169), a TonB-dependent receptor (E value = e−179), an oxidoreductase (E value = 2e−82), a transcriptional regulator (E value = 2e−37), and a choline dehydrogenase (E value = 2e−96) (Fig. 3).
FIG. 3.
Complementing fragment derived from sequencing the complementing 7.5-kb subclone. Contig 9 alone complemented all five non-cercosporin-degrading mutants.
Initial efforts were focused on the transcriptional regulator and oxidoreductase genes. Subcloning indicated that digestion of the complementing 23-kb library clone with HindIII eliminated complementation ability, and the complementing 7.5-kb subclone has two HindIII digestion sites, one each in the transcriptional regulator and oxidoreductase genes. In addition, the oxidoreductase was a likely candidate based on the proposed reaction for the formation of xanosporic acid from cercosporin, which involves an oxygen insertion followed by the spontaneous rearrangement of the molecule (29). The region spanning the transcriptional regulator and oxidoreductase was amplified by PCR and tested for the ability to complement mutants 120-2, MutA, and MutB; these sequences restored the cercosporin-degrading phenotype to all three mutants.
The nucleotide sequence spanning the oxidoreductase and regulator is shown in Fig. 4. The genes are transcribed in the same direction and are located only 61 bp apart. A blastx search (NCBI) showed that the translated form of the oxidoreductase gene is most similar to an oxidoreductase from Xanthomonas axonopodis pv. citri strain 306 (GenBank accession no. AAM36534; E value = 2e−114) (10), a flavoprotein monooxygenase from Pseudomonas syringae pv. syringae strain B728a (accession no. YP_233205; E value = 9e−105) (19), and a monooxygenase from Pseudomonas syringae pv. tomato strain DC3000 (accession no. NP_790149; E value = e−85) (6). An alignment between the two xanthomonad oxidoreductase sequences, carried out using the programs Vector NTI 8 (Informax, Inc., Frederick, MD) and Clustal W 1.82 (EMBL-EBI, Heidelberg, Germany), showed that the two genes are 63% similar and 51.9% identical at the amino acid level. Despite the similarity, the XCZ-3 oxidoreductase gene does not have any defining domains or motifs known to be associated with monooxygenases according to an ExPASy PROSITE motif scan.
FIG. 4.
Sequence containing the putative transcriptional regulator and oxidoreductase genes of XCZ-3 (GenBank accession no. DQ087176). The dashed line represents the transcriptional regulator, and the solid line represents the oxidoreductase. The dark triangles pointing to specific nucleotides in boldface type show the location of each of the mutations. The shaded area shows the sequence that is homologous to MarR genes and that potentially contains a helix-turn-helix motif. The underlined codons in boldface type are possible alternative start sites that are in frame with the transcriptional regulator. Sites of primers used for amplification are also shown. Nucleotide and amino acid changes resulting from the mutations are as follows: 120-2, T→C and start codon→threonine; MutA, G→A and tryptophan→stop; MutB, MutC, and MutD, all G→A and glycine→aspartic acid. Changes in secondary structure are predicted only for MutB (NNPREDICT analysis).
The translated XCZ-3 transcriptional regulator shows the highest similarity to a transcriptional regulator for cryptic hemolysin from X. axonopodis pv. citri strain 306 (GenBank accession no. AAM36535; E value = e−37) (10). An alignment of these two genes using Vector NTI and Clustal W showed 70.1% similarity and 58.6% identity at the amino acid level. Interestingly, the homologous transcriptional regulator from X. axonopodis pv. citri strain 306 lies directly upstream of the homologous oxidoreductase. As in XCZ-3, these two genes are transcribed in the same direction and are only 30 bp apart. An alignment of the region containing these two genes from XCZ-3 and X. axonopodis pv. citri strain 306 shows 55.2% similarity at the nucleotide level.
The second most similar match to the transcriptional regulator is a MarR transcriptional regulatory protein from Gloeobacter violaceus strain PCC 7421 (GenBank accession no. NP_926807) that shows 53% similarity, with an E value of 5e−12 (32). An ExPASy PROSITE motif scan revealed that the XCZ-3 transcriptional regulator contains a MarR-type helix-turn-helix domain (Fig. 4). The helix-turn-helix domain is often associated with DNA binding in transcriptional regulators (1). The gene marR of Escherichia coli, which is involved in the negative regulation of multiple antibiotic resistance, is the most common representative of the MarR family of helix-turn-helix transcriptional regulators.
Search for additional genes.
Two additional mutants, termed MutC and MutD, were obtained by EMS mutagenesis of XCZ-3 expressing the multicopy vector pLAFR6 containing both the transcriptional regulator and the oxidoreductase in an effort to identify mutations in genes other than the transcriptional regulator and the oxidoreductase. Both MutC and MutD showed the same non-cercosporin-degrading phenotype as the other mutants when grown on cercosporin-containing medium (Fig. 2). Although the mutations in MutC and MutD were generated while carrying multiple copies of the wild-type transcriptional regulator and oxidoreductase genes, they were still complemented by the region containing the wild-type transcriptional regulator and oxidoreductase genes, indicating that the new mutations occurred in the same region already identified.
Sequencing of mutants.
The region spanning the transcriptional regulator and the oxidoreductase genes was sequenced in each of the five non-cercosporin-degrading XCZ-3 mutants. Sequence analysis identified single point mutations in the oxidoreductase gene in all five mutants but no mutations in the transcriptional regulator (Fig. 4). Each mutation in the oxidoreductase gene resulted in an amino acid change. The predicted wild-type oxidoreductase is 400 amino acids long, and homology suggests that the active site resides between amino acids 150 and 340. The mutations in MutA, MutB, and MutD fall within this region. In 120-2, the start codon was changed to a threonine, eliminating the protein start site. In MutA, a tryptophan residue was changed to a stop codon, resulting in a truncated protein. The mutations found in MutB, MutC, and MutD change three different glycines to aspartic acid. When these three mutations were analyzed by NNPREDICT, the secondary structure was predicted to be affected only in MutB. However, this substitution replaces a single hydrogen with a bulky and acidic side group (CH2COO), which is likely to affect the tertiary structure and function of the protein in all three of these mutants.
Importance of regulator sequences in mutant complementation.
As all mutants were defective in the oxidoreductase only, we tested the sequences required to complement the mutants. The entire oxidoreductase gene with the upstream intergenic regions plus 113 bases of the regulator (nucleotides 391 to 1948) (Fig. 4) were amplified and cloned into the pLAFR6 vector. This construct was unable to complement any of the non-cercosporin-degrading mutants. By contrast, a construct containing an additional 217 bases of the regulator (nucleotides 174 to 1948) (Fig. 4) was able to complement all five mutants. Although this construct does not include the full-length gene for the transcriptional regulator, it does include the potential active site (Fig. 4).
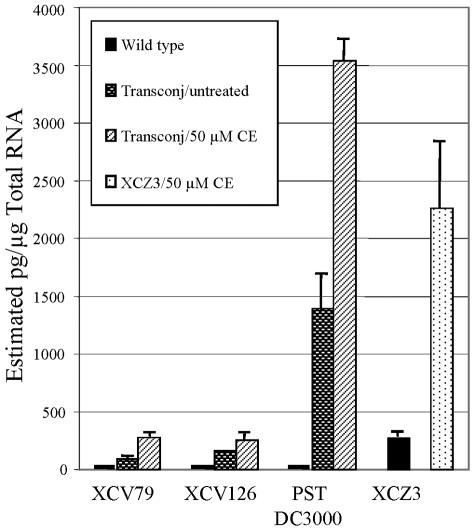
Presence of cercosporin upregulates gene expression.
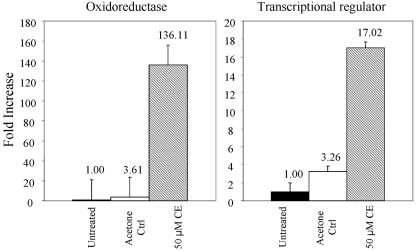
Quantitative RT-PCR was used to determine whether gene expression of either the wild-type oxidoreductase or the wild-type transcriptional regulator was regulated by the presence of cercosporin. Results were compared to data for expression in cells grown in LB medium and in LB medium with 0.5% acetone, used to solubilize cercosporin. Expression of the two genes in the acetone control was increased by approximately three- to fourfold over the untreated control (Fig. 5). When treated with cercosporin, expression of the transcriptional regulator and oxidoreductase were strongly upregulated, 17-fold and 136-fold, respectively, compared to the untreated control.
FIG. 5.
Quantitative RT-PCR analysis of expression of the transcriptional regulator and oxidoreductase genes in XCZ-3 following exposure of the bacterium to cercosporin (CE). Numbers above the bars are increases in expression (n-fold) compared to the untreated control, which was normalized to 1. The acetone control (Acetone Ctrl) contained 0.5% acetone, which was used to solubilize cercosporin.
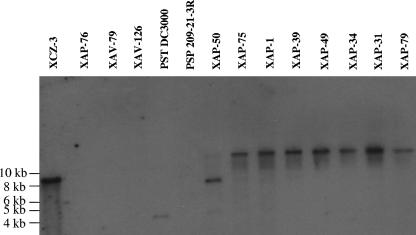
Southern analysis of oxidoreductase homologues in cercosporin-degrading and non-cercosporin-degrading species.
A series of cercosporin-degrading bacteria (XCZ-3, XAP-50, XAP-75, XAP-1, XAP-39, XAP-49, XAP-34, XAP-31, and XAP-79) and non-cercosporin-degrading bacteria (XAP-76, XAV-79, XAV-126, P. syringae pv. tomato DC3000, and P. syringae pv. pisi 209-21-3R) was assayed by Southern analysis for the presence of oxidoreductase homologues using a digoxigenin-labeled probe amplified from wild-type XCZ-3 oxidoreductase (Fig. 6). All of the cercosporin-degrading isolates were found to contain a sequence that hybridized to the oxidoreductase probe. With the exception of P. syringae pv. tomato DC3000, all of the non-cercosporin-degrading isolates lacked a detectable oxidoreductase homologue. Although P. syringae pv. tomato DC300 was unable to degrade cercosporin, Southern analysis detected a band that hybridized to the XCZ-3 oxidoreductase probe. A subsequent NCBI BLAST search identified a similar sequence (GenBank accession no. NP_790149; E value = e−85) (6).
FIG. 6.
Southern hybridization of total DNA isolated from cercosporin-degrading (XCZ-3, XAP-50, XAP-75, XAP-1, XAP-39, XAP-49, XAP-34, XAP-31, and XAP-79) and non-cercosporin-degrading (XAP-76, XAV-79, XAV-126, P. syringae pv. tomato DC3000, and P. syringae pv. pisi 209-21-3R) isolates using a PCR-amplified digoxigenin-labeled oxidoreductase gene as a probe. Wild-type strain XCZ-3 was used in lane 1 as a positive control. Sequences homologous to the oxidoreductase are present in all cercosporin-degrading isolates and in the non-cercosporin-degrading isolate P. syringae pv. tomato DC3000.
The transcriptional regulator and oxidoreductase are not sufficient for cercosporin degradation in non-cercosporin-degrading species.
To determine if the presence of the transcriptional regulator and oxidoreductase genes alone is sufficient for cercosporin degradation, non-cercosporin-degrading bacteria were conjugated with the 23-kb complementing library clone (in pLAFR6) containing the wild-type transcriptional regulator and oxidoreductase genes. Quantitative RT-PCR was used to assess oxidoreductase gene expression in each of the transconjugants upon exposure to cercosporin. Results showed that the oxidoreductase was expressed in all of the transconjugants and appeared to be regulated similarly to XCZ-3, with increased expression upon treatment with cercosporin (Fig. 7). P. syringae pv. tomato DC3000 transconjugants, in particular, showed high levels of expression, equal to or greater than that in wild-type strain XCZ-3.
FIG. 7.
Quantitative RT-PCR analysis of expression of oxidoreductase in transconjugants of the non-cercosporin-degrading bacteria XAV-79, XAV-126, and P. syringae pv. tomato (PST) DC3000 compared to the cercosporin (CE)-degrading strain XCZ-3. The figure shows estimated total oxidoreductase transcripts in nontransformed cells (wild type) compared to transconjugants (Transconj) grown with and without cercosporin. Untreated indicates the 0.5% acetone control (used to solubilize cercosporin). All transconjugants expressed oxidoreductase and showed increased expression in response to cercosporin.
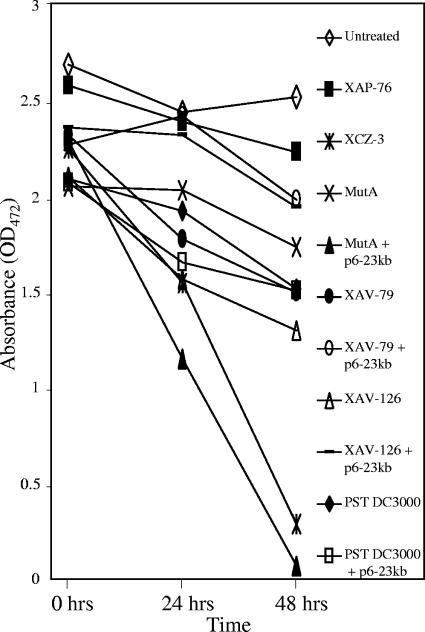
Although the oxidoreductase was expressed in the transconjugants, cercosporin degradation assays in both solid and liquid media failed to detect cercosporin degradation by the oxidoreductase-expressing transconjugants. A time course assay of cercosporin degradation in liquid medium is shown in Fig. 8. Some variability is evident in readings at 0 h due to the inability to completely extract cercosporin from the bacteria. Levels of cercosporin in LB medium alone remained constant over the 48 h of the experiment. Incubation of cercosporin with the wild-type control, XCZ-3, and the complemented mutant, MutA + p6-23kb, resulted in an almost complete loss of cercosporin by 48 h. By contrast, incubation of medium containing cercosporin with MutA and the non-cercosporin-degrading isolates XAV-79, XAV-126, and P. syringae pv. tomato DC3000 resulted in only a small decrease in the apparent cercosporin concentration, likely due to the uptake of cercosporin into the cells and our inability to completely extract the cercosporin from the cells. The transconjugants were unable to degrade cercosporin, showing no more loss of cercosporin than their wild-type counterparts. To ensure that other genes on the 23-kb clone were not interfering with degradation, the pLAFR6 vector containing only the transcriptional regulator and the oxidoreductase genes was conjugated into the non-cercosporin-degrading bacteria. Cercosporin degradation by these transconjugants was still not achieved (data not shown).
FIG. 8.
Degradation of cercosporin in liquid culture by cercosporin-degrading and non-cercosporin-degrading isolates and oxidoreductase-expressing transconjugants. Isolates tested were wild-type strain XCZ-3, XCZ-3 non-cercosporin-degrading mutant MutA, wild-type non-cercosporin-degrading isolates XAP-76, XAV-79, XAV-126, and P. syringae pv. tomato (PST) DC3000, and the oxidoreductase-expressing transconjugants (+p6-23kb) of XAV-79, XAV-126, P. syringae pv. tomato DC3000, and MutA. Cercosporin degradation was assayed by measuring the absorbance at 472 nm after extraction of cercosporin from culture filtrates at 0, 24, and 48 h as described in Materials and Methods. Cercosporin in LB alone (Untreated) was included as a control. Small decreases in apparent cercosporin concentrations with the non-cercosporin-degrading bacteria are due to the uptake of cercosporin by bacteria in the medium. Only XCZ-3 and the complemented MutA mutant were able to degrade cercosporin. Results shown are from one of two experiments performed. OD472, optical density at 472 nm.
DISCUSSION
Many species in the genus Cercospora produce cercosporin and rely on its toxicity to cause disease in their host plants. Research efforts to avoid or limit the exposure of a host plant to the toxin may be of great benefit in disease prevention. We have chosen to focus on toxin degradation as a potential method to achieve toxin and disease resistance. Previous reports of toxin degradation by transgenic plants have shown that this method can be successful in yielding disease resistance. For example, transformation of sugarcane with a gene encoding detoxification of the toxin albicidin provided protection against infection by Xanthomonas albilineans, the causal agent of leaf scald on susceptible sugarcane (48). Also, the Tri101 gene from Fusarium graminearum, which is responsible for self-protection of this fungus from their trichothecene toxins, provided protection against the trichothecene 4,15-diacetoxyscirpenol in transgenic tobacco (22).
In previous work in our laboratory, we identified bacteria that are capable of degrading cercosporin. Of 244 isolates tested, the most efficient degraders were pathovars of X. campestris that are pathogenic on zinnia. X. campestris pv. zinniae isolates were shown to rapidly degrade cercosporin to produce the nontoxic breakdown product xanosporic acid (30). The degradation reaction was hypothesized to be caused by an oxygen insertion into one of the quinoid rings adjacent to the ketone carbonyl (29). The execution of this step would require enzymatic activity, but the subsequent rearrangement that results in xanosporic acid may be spontaneous. We hypothesized that cercosporin degradation is carried out by a single enzyme, perhaps by cytochrome P450.
The goal of this work was to isolate the gene(s) required for cercosporin degradation by X. campestris pv. zinniae. We identified two genes, encoding a putative transcriptional regulator and an oxidoreductase, required for cercosporin resistance. Homology searches showed that the transcriptional regulator homologue has similarity to a transcriptional regulator for cryptic hemolysin and a MarR transcriptional regulator. The oxidoreductase homologue has similarity to an oxidoreductase and two monooxygenases. Although the regulator and oxidoreductase are most similar to a region present in X. axonopodis pv. citri, the function of these homologous genes has not yet been experimentally confirmed in X. axonopodis pv. citri.
Sequence analysis of the region spanning the transcriptional regulator and the oxidoreductase in the mutants identified a single point mutation present in the oxidoreductase of each mutant and no mutations in the transcriptional regulator. Our studies suggest, however, that the transcriptional regulator may play an important role in the regulation of oxidoreductase. A construct containing the wild-type oxidoreductase gene plus the intergenic region and ca. 100 bases of the regulator did not complement the non-cercosporin-degrading mutants, whereas a construct containing both the transcriptional regulator and the oxidoreductase genes did complement the mutants. It is not clear if the requirement is for the regulator or for upstream regulatory sequences. Transformation of the mutants with a construct containing the oxidoreductase gene and upstream sequences spanning 330 bases into the transcriptional regulator gene proved to be as efficient in complementing the mutants as did the construct containing the intact regulator. This construct did contain the active site of the transcriptional regulator as well as sequences encoding five possible start sites that may allow the terminal portion of the regulator to be translated in frame with the original sequence, resulting in the expression of the potentially necessary active site. It is interesting that we were unable to clone the oxidoreductase gene alone into high-expression vectors; recovered clones were slow to grow and died after only 5 days (data not shown). When the gene for the transcriptional regulator was included upstream of the oxidoreductase in constructs in the high-expression vectors, however, clones that grew normally were readily recovered. These results suggest that the transcriptional regulator plays a critical role in regulating the expression of oxidoreductase. Finally, our quantitative RT-PCR studies showed that both the regulator and the oxidoreductase genes were upregulated after exposure to cercosporin, supporting our hypothesis that expression of both genes is necessary to yield cercosporin degradation.
The region containing both the transcriptional regulator and the oxidoreductase was transformed into other non-cercosporin-degrading bacteria in an attempt to determine if these genes were sufficient for degradation. Quantitative RT-PCR analysis confirmed that the oxidoreductase was expressed and was upregulated in the presence of cercosporin. However, the transconjugants were unable to degrade cercosporin, indicating that the genes are not sufficient for cercosporin degradation in bacteria. Although we attempted to identify additional genes involved in cercosporin degradation, the two newer mutants, MutC and MutD, also contained a point mutation in the oxidoreductase gene. These results suggest that if additional genes are involved in the degradation process, mutations in these genes may not be recoverable, perhaps due to lethality.
Interestingly, Southern analysis showed that all bacterial isolates tested that are able to degrade cercosporin contain a sequence that hybridizes to the XCZ-3 oxidoreductase gene that was used as a probe. With one exception, the bacterial isolates tested that are unable to degrade cercosporin do not contain a detectable oxidoreductase homologue. The exception is P. syringae pv. tomato DC3000, which does not have the ability to degrade cercosporin but does contain a sequence homologous to the oxidoreductase. These results are consistent with our hypothesis that additional genes may be necessary for cercosporin degradation. Further study is needed to identify additional genes. Once found, expression of these genes in plants may provide a novel strategy for the control of Cercospora spp. and the diseases that they cause.
Acknowledgments
We thank David Ritchie and Peter Lindgren (North Carolina State University, Raleigh, NC) for providing the bacterial strains and Brian Staskawicz (University of California, Berkeley, CA) for use of the pLAFR6 vector. We also thank Frank Louws and Heriberto Velez (North Carolina State University, Raleigh, NC) for assistance with the Box-PCR and oxidoreductase expression experiments, respectively.
This work was supported in part by Pioneer Hi-Bred International, Inc., and an NIH Biotechnology Training Grant fellowship.
REFERENCES
- 1.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 36:1394-1404. [DOI] [PubMed] [Google Scholar]
- 2.Assante, G., R. Locci, L. Camarda, L. Merlini, and G. Nasini. 1977. Screening of the genus Cercospora for secondary metabolites. Phytochemistry 16:243-247. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2002. Current protocols in molecular biology, 5th ed. Wiley Interscience, New York, N.Y.
- 4.Balis, C., and M. G. Payne. 1971. Triglycerides and cercosporin from Cercospora beticola: fungal growth and cercosporin production. Phytopathology 61:1477-1484. [Google Scholar]
- 5.Batchvarova, R. B., V. S. Reddy, and J. Bennett. 1992. Cellular resistance in rice to cercosporin, a toxin of Cercospora. Phytopathology 82:642-646. [Google Scholar]
- 6.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calpouzos, L., and G. F. Stalknecht. 1967. Symptoms of Cercospora leaf spot of sugar beets influenced by light intensity. Phytopathology 57:799-800. [Google Scholar]
- 8.Choquer, M., K. L. Dekkers, H. Chen, L. Cao, P. P. Ueng, M. E. Daub, and K. R. Chung. 2005. The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae. MPMI 18:468-476. [DOI] [PubMed] [Google Scholar]
- 9.Coates, S. T., and D. G. White. 1994. Sources of resistance to gray leaf spot of corn. Plant Dis. 78:1153-1155. [Google Scholar]
- 10.da Silva, A. C. R., J. A. Ferro, F. C. Relnach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 11.Daub, M. E. 1982. Cercosporin, a photosensitizing toxin from Cercospora species. Phytopathology 72:370-374. [Google Scholar]
- 12.Daub, M. E. 1982. Peroxidation of tobacco membrane lipids by the photosensitizing toxin, cercosporin. Plant Physiol. 69:1361-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daub, M. E., and S. P. Briggs. 1983. Changes in tobacco cell membrane composition and structure caused by the fungal toxin, cercosporin. Plant Physiol. 71:763-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daub, M. E., and M. Ehrenshaft. 2000. The photoactivated Cercospora toxin cercosporin: contributions to plant disease and fundamental biology. Annu. Rev. Phytopathol. 38:437-466. [DOI] [PubMed] [Google Scholar]
- 15.Daub, M. E., and R. P. Hangarter. 1983. Light-induced production of singlet oxygen and superoxide by the fungal toxin, cercosporin. Plant Physiol. 73:855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duvick, J. 1987. Detection of cercosporin in grey leaf spot-infected maize leaf tissue. Phytopathology 77:1754. [Google Scholar]
- 18.Fajola, A. O. 1978. Cercosporin, a phytotoxin from Cercospora spp. Physiol. Plant Pathol. 13:157-164. [Google Scholar]
- 19.Feil, H., W. S. Feil, P. Chain, F. Larimer, G. Dibartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 102:11064-11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonçalves, E. R., and Y. B. Rosato. 2002. Phylogenetic analysis of Xanthomonas species based upon 16S-23S rDNA intergenic spacer sequences. Int. J. Syst. Evol. Microbiol. 52:355-361. [DOI] [PubMed] [Google Scholar]
- 21.Hartman, P. E., W. J. Dixon, T. A. Dahl, and M. E. Daub. 1988. Multiple modes of photodynamic action by cercosporin. Photochem. Photobiol. 47:699-703. [DOI] [PubMed] [Google Scholar]
- 22.Higa, A., M. Kimura, K. Mimori, T. Ochiai-Fukuda, T. Tokai, N. Takahashi-Ando, T. Nishiuchi, T. Igawa, M. Fujimura, H. Hamamoto, R. Usami, and I. Yamaguchi. 2003. Expression in cereal plants of genes that inactivate Fusarium mycotoxins. Biosci. Biotechnol. Biochem. 67:914-918. [DOI] [PubMed] [Google Scholar]
- 23.Kuyama, S., and T. Tamura. 1957. Cercosporin. A pigment of Cercospora kikuchii Matsumoto et Tomoyasu. I. Cultivation of fungus, isolation and purification of pigment. J. Am. Chem. Soc. 79:5725-5726. [Google Scholar]
- 24.Leisman, G. B., and M. E. Daub. 1992. Singlet oxygen yields, optical properties, and phototoxicity of reduced derivatives of the photosensitizer cercosporin. Photochem. Photobiol. 55:373-379. [Google Scholar]
- 25.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 26.Lousberg, R. J. J. C., U. Weiss, C. A. Salemink, A. Arnone, L. Merlini, and G. Nasini. 1971. The structure of cercosporin, a naturally occurring quinone. Chem. Commun. 22:1463-1464. [Google Scholar]
- 27.Louws, F. J., D. W. Fulbright, C. T. Stephens, and F. J. de Bruijn. 1994. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 60:2286-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch, F. J. 1977. Production of cercosporin by Cercospora species. Trans. Br. Mycol. Soc. 69:496-498. [Google Scholar]
- 29.Mitchell, T. D., F. Alejos-Gonzalez, H. S. Gracz, D. A. Danehower, M. E. Daub, and W. S. Chilton. 2003. Xanosporic acid, an intermediate in bacterial degradation of the fungal phototoxin cercosporin. Phytochemistry 62:723-732. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, T. D., W. S. Chilton, and M. E. Daub. 2002. Biodegradation of the polyketide toxin cercosporin. Appl. Environ. Microbiol. 68:4173-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mumma, R. O., F. L. Lukezic, and M. G. Kelly. 1973. Cercosporin from Cercospora hayii. Phytochemistry 12:917-922. [Google Scholar]
- 32.Nakamura, Y., T. Kaneko, S. Sato, M. Mimuro, H. Miyashita, T. Tsuchiya, S. Sasamoto, A. Watanabe, K. Kawashima, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, C. Takeuchi, M. Yamada, and S. Tabata. 2003. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 10:181-201. [DOI] [PubMed] [Google Scholar]
- 33.Orth, C. E., and W. Schuh. 1994. Resistance of 17 soybean cultivars to foliar, latent, and seed infection by Cercospora kikuchii. Plant Dis. 78:661-664. [Google Scholar]
- 34.Rademaker, J. L. W., F. J. Louws, M. H. Schultz, U. Rossbach, L. Vauterin, J. Swings, and F. J. de Bruijn. 2005. A comprehensive species to strain taxonomic framework for Xanthomonas. Phytopathology 95:1098-1111. [DOI] [PubMed] [Google Scholar]
- 35.Robeson, J. R., M. A. F. Jalal, and R. B. Simpson. November 1993. U.S. patent 5,262,306.
- 36.Rossi, V. 1995. Effect of host resistance in decreasing infection rate of Cercospora leaf spot epidemics on sugarbeet. Phytopathol. Mediterr. 34:149-156. [Google Scholar]
- 37.Shim, W. B., and L. D. Dunkle. 2003. CZK3, a MAP kinase kinase kinase homolog in Cercospora zeae-maydis, regulates cercosporin biosynthesis, fungal development, and pathogenesis. Mol. Plant-Microbe Interact. 16:760-768. [DOI] [PubMed] [Google Scholar]
- 38.Steinkamp, M. P., S. S. Martin, L. L. Hoefert, and E. G. Ruppel. 1979. Ultrastructure of lesions produced by Cercospora beticola in leaves of Beta vulgaris. Physiol. Plant Pathol. 15:13-26. [Google Scholar]
- 39.Steinkamp, M. P., S. S. Martin, L. L. Hoefert, and E. G. Ruppel. 1981. Ultrastructure of lesions produced in leaves of Beta vulgaris by cercosporin, a toxin from Cercospora beticola. Phytopathology 71:1272-1281. [Google Scholar]
- 40.Thorne, L., L. Tansey, and T. Pollock. 1987. Clustering of mutations blocking synthesis of xanthan gum by Xanthomonas campestris. J. Bacteriol. 169:3593-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Upchurch, R. G., D. C. Walker, J. A. Rollins, M. Ehrenshaft, and M. E. Daub. 1991. Mutants of Cercospora kikuchii altered in cercosporin synthesis and pathogenicity. Appl. Environ. Microbiol. 57:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:472-489. [Google Scholar]
- 43.Venkataramani, K. 1967. Isolation of cercosporin from Cercospora personata. Phytopathol. Z 58:379-382. [Google Scholar]
- 44.Ward, J. M. J., E. L. Stromberg, D. C. Nowell, and J. F. W. Nutter. 1999. Gray leaf spot: a disease of global importance in maize production. Plant Dis. 83:884-895. [DOI] [PubMed] [Google Scholar]
- 45.Windels, C. E., H. A. Lamey, D. Hilde, J. Widner, and T. Knudsen. 1998. A Cercospora leaf spot model for sugar beet. In practice by an industry. Plant Dis. 82:716-726. [DOI] [PubMed] [Google Scholar]
- 46.Yamazaki, S., and T. Ogawa. 1972. The chemistry and stereochemistry of cercosporin. Agric. Biol. Chem. 1707-1718.
- 47.Yamazaki, S., A. Okubo, Y. Akiyama, and K. Fuwa. 1975. Cercosporin, a novel photodynamic pigment isolated from Cercospora kikuchii. Agric. Biol. Chem. 39:287-288. [Google Scholar]
- 48.Zhang, L., J. Xu, and R. G. Birch. 1999. Engineered detoxification confers resistance against a pathogenic bacterium. Nat. Biotechnol. 17:1021-1024. [DOI] [PubMed] [Google Scholar]