Abstract
Microbial community profiles and species composition associated with two black band-diseased colonies of the coral Siderastrea siderea were studied by 16S rRNA-targeted gene cloning, sequencing, and amplicon-length heterogeneity PCR (LH-PCR). Bacterial communities associated with the surface mucopolysaccharide layer (SML) of apparently healthy tissues of the infected colonies, together with samples of the black band disease (BBD) infections, were analyzed using the same techniques for comparison. Gene sequences, ranging from 424 to 1,537 bp, were retrieved from all positive clones (n = 43 to 48) in each of the four clone libraries generated and used for comparative sequence analysis. In addition to LH-PCR community profiling, all of the clone sequences were aligned with LH-PCR primer sequences, and the theoretical lengths of the amplicons were determined. Results revealed that the community profiles were significantly different between BBD and SML samples. The SML samples were dominated by γ-proteobacteria (53 to 64%), followed by β-proteobacteria (18 to 21%) and α-proteobacteria (5 to 11%). In contrast, both BBD clone libraries were dominated by α-proteobacteria (58 to 87%), followed by verrucomicrobia (2 to 10%) and 0 to 6% each of δ-proteobacteria, bacteroidetes, firmicutes, and cyanobacteria. Alphaproteobacterial sequence types related to the bacteria associated with toxin-producing dinoflagellates were observed in BBD clone libraries but were not found in the SML libraries. Similarly, sequences affiliated with the family Desulfobacteraceae and toxin-producing cyanobacteria, both believed to be involved in BBD pathogenesis, were found only in BBD libraries. These data provide evidence for an association of numerous toxin-producing heterotrophic microorganisms with BBD of corals.
The biodiversity and abundance of reef-building corals are substantially declining in tropical and subtropical waters worldwide (27, 40, 64). Studies indicate that coral diseases are a major cause for this decline (17, 41, 52). There have been increases in the number of coral diseases, the number of coral species affected by disease, and the geographic extent of coral disease (58). Thus far, 18 coral diseases have been reported affecting 150 scleractinians, gorgonians, and hydrozoans in the Caribbean and Indo-Pacific regions (58). Among all coral diseases, black band disease (BBD) is considered one of the most important diseases contributing to the decline of coral reefs on a world-wide basis (52, 58).
Black band disease is characterized as a dark, cyanobacterium-dominated microbial consortium that forms a band which migrates across living coral colonies, actively degrading coral tissue and killing entire colonies over a period of several months (2). It was the first coral disease reported in the literature (3) and now is one of the major diseases affecting both scleractinian and gorgonian corals (25, 45, 58). It has been reported that BBD is most active and prevalent when water temperatures rise to 28°C and above, thus exhibiting a seasonal cycle of disease activity on reefs (10, 18, 32). Although BBD has been a topic of study for more than three decades, a primary pathogen has not yet been identified. Proposed pathogens for BBD thus far include different genera of cyanobacteria (14, 20, 53), sulfate-reducing bacteria including Desulfovibrio spp. (23, 46, 56), sulfide-oxidizing bacteria presumed to be Beggiatoa spp. (16), several other heterotrophs (14, 21, 22), and marine fungi (44).
The BBD pathogens proposed in the 1970s and 1980s were based on optical assessment, including light microscopy, scanning electron microscopy, and transmission electron microscopy (16, 53, 54), whereas more recent studies have used molecular techniques for potential pathogen identification. Together, results reported using these two approaches have led to much controversy over the identification of BBD pathogens. Moreover, only a very few of the pathogens proposed for BBD have been cultured (47, 53, 61), and Koch's postulates have not been addressed or satisfied for any of them (45). BBD was additionally proposed as a pathogenic microbial consortium with no primary pathogen (46).
In recent years, different molecular techniques have been used to characterize the microbes associated with BBD (14, 19-22, 61). Frias-Lopez and colleagues used terminal restriction fragment length polymorphism and molecular cloning based on 16S rRNA genes to characterize the bacteria and cyanobacteria associated with the BBD community. Their studies included comparisons with healthy and dead corals of the Caribbean species Montastraea annularis, Montastraea cavernosa, and Diploria strigosa on the reefs of Curaçao (in the southern Caribbean) as well as Porites lutea from the Indo-Pacific (20-22). These are all massive, reef-forming, thus ecologically important, coral species that are often infected with BBD (58). Another group of investigators, Cooney and colleagues (14), have used amplified 16S ribosomal RNA gene restriction analysis of clone libraries and denaturing gradient gel electrophoresis to characterize the bacterial communities associated with BBD on the corals D. strigosa, M. annularis, and Colpophyllia natans collected on reefs of St. Croix (U.S. Virgin Islands) and Barbados in the eastern Caribbean.
The results reported by these two groups were very different from an existing understanding of the BBD consortium based on traditional microbiological techniques and led to proposals that heterotrophic bacteria may be the potential primary pathogens (14, 21, 22). However, their role in BBD pathogenicity was not described, and no isolates were obtained. Additionally, a previously isolated BBD-associated cyanobacterium, Phormidium corallyticum (47, 53) (now known to be a member of the genus Geitlerinema) (14, 20, 43), and the sulfide-oxidizing bacterium Beggiatoa spp., widely reported as present in BBD (16, 23, 46), were not found in their molecular analyses. Although sulfate-reducing bacteria such as Desulfovibrio spp. were found in both of their studies (14, 21, 22), these were not considered by either group as potential primary pathogens, since they were also present in healthy coral tissues.
The results reported in these molecular studies were not reproducible between or within the research teams. Frias-Lopez et al. (22) first reported that BBD on the corals M. annularis, M. cavernosa, and D. strigosa were dominated by firmicutes, members of the Cytophaga-Flavobacterium-Bacteroides (CFB) group, γ-proteobacteria, and δ-proteobacteria. Marine and terrestrial microorganisms like Clostridium spp. (firmicutes), Campylobacter spp. (ɛ-proteobacteria), Cytophaga fermentans (CFB), Cytophaga columnaris (CFB), and Trichodesmium tenue (cyanobacteria) were also found and newly proposed by this group as BBD pathogens (22). Later studies (20, 21) by the same authors, studying the same coral species sampled on the same reefs (Curaçao), resulted in very different results, with five species of firmicutes, two species of CFB, one species of δ-proteobacteria (21), and three taxa of cyanobacteria (20, 21) reported to be dominant in BBD. The study by Cooney et al. (14) was performed using two of the same coral host species (M. annularis and D. strigosa) studied by Frias-Lopez and colleagues, along with an additional species (C. natans). Cooney et al. (14) confirmed the presence of Desulfovibrio in BBD, reported by Frias-Lopez et al. (22), and also found four additional, previously unreported sulfate-reducing genera (14). Cooney et al. (14) also found two cyanobacterial ribotypes associated with BBD, one of which (GenBank accession no. AF473935) was consistently associated in all of their BBD samples (14) and was also found to be one of three BBD cyanobacteria reported by Frias-Lopez et al. (20).
Bacteria and archaea associated with healthy corals of several BBD-susceptible coral species have also been studied (8, 9, 22, 31, 50, 51, 63). The importance of the coral surface mucopolysaccharide layer (SML) as a niche for bacterial populations is a relatively new area of research, especially in terms of the role of these bacteria in the coral holobiont (50-52). The SML consists of a thin layer of mucus, the thickness of which depends on the coral species, which is secreted by corals and forms a thin, protective covering on the coral surface. When coral surfaces are disturbed, for example, by wounding or by deposition of sand or sediment, copious amounts of mucus are actively released within minutes. This is believed to be a protective strategy on the part of the coral animal (52). The SML is chemically complex and is composed of a variable assortment of carbohydrates, glycoproteins, and lipids, with an abundance of simple sugars (15). It is believed that the SML is an important source of both organic carbon (15) and nutrients (12) that support growth of the bacteria and archaea which live inside the mucus, and it has been proposed that this microbial population is beneficial to the coral (48). Studies to date suggest that there is specificity between a coral species and its population of SML-associated bacteria (48). While some of these bacteria are hypothesized to be potential true symbionts of corals, this question remains to be addressed. In contrast to molecular studies of BBD microorganisms, the studies of bacterial communities associated with healthy corals are much more consistent. In general, these communities are less diverse than those associated with BBD. Bacteria associated with both coral tissue and the SML have been targeted.
The present study was aimed at assessing the microbial diversity associated with BBD and the SML of apparently healthy tissue of a coral host species, Siderastrea siderea, that has previously not been investigated. Samples were collected from reefs near Lee Stocking Island in the Bahamas, Exuma Chain, in an area of the wider Caribbean (northern) that also has not been previously investigated. S. siderea is an important massive reef-building species found throughout the wider Caribbean and is the host species most susceptible to BBD on these Bahamian reefs (62).
MATERIALS AND METHODS
Sampling location and sample collection.
Samples of BBD and the SML of apparently healthy areas of the same BBD-infected corals were collected from two colonies of the coral Siderastrea siderea (identified as colonies 216 and 217) on Horseshoe Reef (23°46.30′N and 76°5.33′W) near Lee Stocking Island in the Exuma Chain of the Bahamas. Both colonies were sampled on 19 July 2004. The two coral colonies (216 and 217) were located at water depths of 27 and 24 m, respectively, and were located 7 m apart on the reef. The water temperature near colony 216 was 29°C and that near colony 217 was 28°C at the time of sample collection. Coral tissue loss in each black band-diseased colony was approximately 30%. The band (BBD) on colony 216 was 18 mm in width, while on colony 217, it was 6 mm in width. On each colony, the SML samples were taken from apparently healthy tissue areas approximately 25 cm from the BBD infection.
Both BBD and SML samples were collected using individual sterile 10-ml syringes while scuba diving. The SML was collected from healthy corals by lightly abrading the surface with the syringe tip, which caused mucus production, and then aspirating the mucus. After collection, samples were allowed to settle to the tips of the syringes (which were held in an upright position on ice and in darkness on the boat). At the Lee Stocking Island field laboratory, BBD and SML samples were carefully extruded from the tips of the syringes, minimizing the amount of seawater (and seawater-associated bacteria), directly into sterile 2-ml cryovials and stored at −20°C until further processing.
Extraction of DNA from SML and BBD samples.
Genomic DNA was extracted from SML and BBD materials by the bead-beating method using the FastDNA SPIN kit for soil (Q-Biogene, Vista, CA) after slight modification of the protocol as described previously (37). It was found during optimization trials in our laboratory that compared various extraction methods that this protocol resulted in high yields of DNA. The frozen SML samples were thawed at room temperature and vacuum filtered onto sterile Versapor 200 filters (25-mm diameter, 0.2-μm pore size; PALL-Gelman Lab, New York), and the filtrate was discarded. The samples on the filters were placed directly into Multimix lysing matrix tubes (BIO 101 Systems, Q-Biogene, California) and processed for DNA extraction. Samples were bead beaten using the FastPrep instrument (Thermo Electron Corporation, Ulm, Germany) for 20 s at the speed of 5.5 m s−1, and the DNA was eluted using 100 μl sterile nuclease-free water. The DNA was verified by electrophoresis in an agarose gel (1% wt/vol) and staining with ethidium bromide. Subsequently, the DNA was quantified using a Bio-Rad fluorometer (Bio-Rad, Hercules, CA) and extracts diluted (if needed) to a uniform concentration of 10 ng/μl. Samples were again stored at −20°C until used for PCR.
16S rRNA gene amplification, cloning, and sequencing.
Bacterial 16S rRNA genes were amplified from the DNA extracts by using the universal bacterial primers 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) (39) (Integrated DNA Technologies, Coralville, IA) in a Peltier thermal cycler (PTC-200; MJ Research, Waltham, MA). The final PCR mixtures contained 1× PCR buffer, 2.5 mM MgCl2, 0.5 U AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 0.1% (wt/vol) bovine serum albumin (fraction V; Fisher, Suwannee, GA), 0.25 mM concentrations of each deoxynucleoside triphosphate (Promega, Madison, WI), 0.5 μM forward and reverse primers, and 10 ng of genomic DNA, and the final volume was made up to 20 μl with nuclease-free water (Fisher, Suwannee, GA). An initial denaturing step at 94°C for 11 min was followed by 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. The PCR products were verified by agarose (1% wt/vol) gel electrophoresis as mentioned above.
Amplified 16S rRNA gene fragments were purified with a QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA). The purified PCR products were cloned using the TOPO TA cloning kit version N (Invitrogen, Carlsbad, CA) per the manufacturer's protocol, and cloned products were transformed into the TOP10 chemically competent Escherichia coli. The clones were screened for inserts using M13F and M13R (34), and the plasmids were extracted from the positive clones (48 to 55 from each library) using the QIAPrep spin miniprep kit (QIAGEN, Inc., Valencia, CA). The plasmid inserts were sequenced with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA) at the DNA Core Facility at Florida International University, Miami, FL, by using the M13F (34) primer. Full-length 16S rRNA genes were sequenced for clones of interest by using the additional primers 518F (38) and M13R (34). The sequences were edited using Sequence Analysis (Applied Biosystems, Foster City, CA) or Sequencher 4.2.2 (GeneCodes, Ann Arbor, MI). The sequences were checked for chimeras (13) and vector contamination (http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html) and were then compared using the BLAST queuing system (http://www.ncbi.nlm.nih.gov/BLAST/) (1) to identify their closest relatives and their tentative phylogenetic positions.
A detailed phylogenetic analysis was done for the sequences obtained in this study that were related to bacteria associated with toxic dinoflagellates and juvenile oyster disease (JOD) by using the ARB software package (http://www.arb-home.de) (33). The sequences were manually aligned in the ARB Edit_Tool by including closest relatives. Maximum parsimony, neighbor-joining, and maximum likelihood phylogenetic analyses were performed, and a consensus tree was produced based on maximum parsimony analysis. The PHYLIP DNA-Parsimony option in ARB software was used to perform the bootstrap analysis (1,000 resamplings) to estimate the confidence of the 16S rRNA gene tree topology.
Amplicon-length heterogeneity (LH) PCR amplification.
Genomic DNA extracted from SML and BBD was used for amplification of the hypervariable V1 plus V2 domains of the 16S rRNA gene. The forward primer was labeled 27F-6-FAM (5′-6-carboxyfluorescein-AGA GTT TGA TCM TGG CTC AG-3′) and used with an unlabeled 355R (5′-GGT GCC TCC CGT AGG AGT-3′) (59) (Integrated DNA Technologies, Coralville, IA). The composition and final concentrations of PCR mixtures were used as mentioned above in the cloning section. The PCR was done in a Peltier thermal cycler (PTC-200; MJ Research, Waltham, MA) with an initial denaturing step at 94°C for 11 min, followed by 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. The PCR product was verified by gel electrophoresis as mentioned above along with a TriDye DNA ladder (New England Biolabs, Ipswich, MA). The PCR products were analyzed in the Forensic DNA Profiling Facility at Florida International University, Miami, FL. The PCR products (0.5 μl each) were denatured using 9.5 μl of 96:1 Hi-dye deionized formamide-GS 500 Rox internal standard (Applied Biosystems [ABI], Foster City, CA) and then separated on a capillary electrophoresis ABI 310 DNA genetic analyzer with the appropriate filter set and run module. For each sample, the PCRs were performed at least three times, and the resulting electropherograms were analyzed using ABI Prism GenemapperHID, version 3.2, software. The marker range was set to analyze amplicons from 300 to 400 bp in length and bins were set to 1 bp in width, with the minimum intensity threshold set to 50 relative fluorescence units.
In silico or virtual alignment of sequences.
The clone sequences obtained from the four clone libraries were imported to the Sequencher program and aligned with the LH-PCR primer (27F and 355R) sequences (59). Based upon alignment of the positions of primers within the clone sequences, the theoretical amplicon lengths (in bp) were calculated. This was correlated with the phylogenetic identification of clones and their presence or absence in the whole-community profiles (36, 37).
Statistical analyses.
Pairwise comparison of variation in the relative abundance of each amplicon within community profiles was tested (Student's t test) between (i) SML profiles from colonies 216 and 217, (ii) BBD profiles from colonies 216 and 217, and (iii) SML and BBD profiles for each colony. Species diversity of clone sequences was calculated using Shannon-Wiener (H′) and Simpson (1 − λ) indices, and richness was calculated using the Margalef (D) richness index.
Nucleotide sequence accession numbers.
The partial and full-length nucleotide sequences determined in this study have been deposited in the GenBank database under the accession numbers DQ445996 to DQ446177 and DQ644006.
RESULTS
Microbial community composition in SML samples.
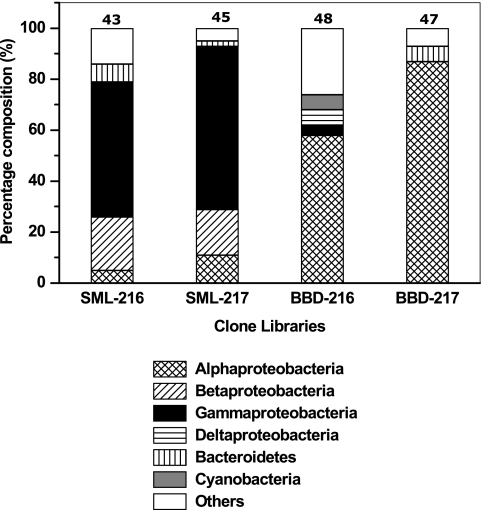
A total of 48 and 55 16S rRNA gene clones from clone libraries SML-216 (SML from colony 216) and SML-217 (SML from colony 217), respectively, were sequenced, and 43 and 45 partial sequences (most >600 bp) were obtained. The overview of the phylogenetic affiliations of the sequences obtained is shown in Fig. 1 and Table 1. In the SML-216 and SML-217 libraries, 5 to 11%, 18 to 21%, and 53 to 64% of the total sequences were closely related to α-, β-, and γ-proteobacteria, respectively. Two percent of the community were representatives of the ɛ-proteobacteria in the SML-216 clone library (Table 1), while no members of this group were found in SML-217. The percent distributions of bacteroidetes (Fig. 1 and Table 1) and firmicutes (Table 1) were in the range of 2 to 7% and 4 to 5%, respectively, in the SML-216 and SML-217 libraries. The γ-proteobacteria dominated both SML clone libraries with members of the orders Alteromonadales and Pseudomonadales representing 52 to 57% and 24 to 26% of the γ-proteobacteria, respectively.
FIG. 1.
16S rRNA gene sequence types affiliated with individual phylogenetic groups present in four different clone libraries produced from SML and BBD on two colonies of Siderastrea siderea (216 and 217). The numbers above the bars give the numbers of sequences in the respective clone libraries.
TABLE 1.
Overview of 16S rRNA gene sequences retrieved from clone libraries produced from the SML of apparently healthy tissues of two black band-diseased Siderastrea siderea colonies (216 and 217)
| SML-216 accession no. (no. of similar clones) | SML-217 accession no. (no. of similar clones) | Sequence length (bp) | Closest relative in GenBank database (accession no.) | Similarity (%) | Phylogenetic affiliation | Virtual V1 + V2 amplicon length (bp) |
|---|---|---|---|---|---|---|
| DQ445996 (1) | 630 | Marine alphaproteobacterium (AF151249) | 99 | α-Proteobacteria | NVMa | |
| DQ445997 (1) | 648 | Alphaproteobacterium SOGA19 (AJ244796) | 94 | α-Proteobacteria | 333 | |
| DQ446039 (1) | 600 | Brevundimonas-like sp. strain LMG 11050 (AJ244650) | 99 | α-Proteobacteria | 313 | |
| DQ446040-41 (2) | 580, 600 | Roseomonas cervicalis (AY150047) | 99 | α-Proteobacteria | 349 | |
| DQ446042 (1) | 610 | Ruegeria sp. (AY712383) | 100 | α-Proteobacteria | NVM | |
| DQ446043 (1) | 610 | Sphingomonas sp. strain AV6C (AF434172) | 99 | α-Proteobacteria | 315 | |
| DQ445998 (1) | 620 | Alcaligenes sp. (X86586) | 90 | β-Proteobacteria | 341 | |
| DQ445999-04 (6) | 580-650 | Comamonas sp. strain D22 (AF188304) | 92-99 | β-Proteobacteria | 341 | |
| DQ446005-06 (2) | DQ446044-50 (7) | 500-620 | Comamonas terrigena (AF078772) | 98 | β-Proteobacteria | 341 |
| DQ446051 (1) | 600 | Dechloromonas sp. (AY515718) | 99 | β-Proteobacteria | NVM | |
| DQ446007-08 (2) | 600, 638 | Gamma proteobacterium (AY580762) | 99 | γ-Proteobacteria | 346 | |
| DQ446052-53 (2) | 630, 620 | Acinetobacter sp. (AY588958) | 98-100 | γ-Proteobacteria | 341 | |
| DQ446009-10 (2) | DQ446054-58 (5) | 470-630 | Aeromonas hydrophila (X87271) | 99 | γ-Proteobacteria | 341 |
| DQ446059 (1) | 600 | Citrobacter sp. (AF025369) | 98 | γ-Proteobacteria | 347 | |
| DQ446011-19 (9) | DQ446060-69 (10) | 595-640 | Shewanella sp. strain H836 (AY369988) | 99 | γ-Proteobacteria | 347 |
| DQ446020 (1) | 640 | Shewanella sp. strain MR-4 (AF005252) | 96 | γ-Proteobacteria | NVM | |
| DQ446021-23 (3) | DQ446070-74 (5) | 554-640 | Shewanella sp. strain ARCTIC-P25 (AY573039) | 98-99 | γ-Proteobacteria | NVM |
| DQ446024-25 (2) | DQ446075-76 (2) | 598-630 | Pseudomonas sp. strain P400Y-1 (AB076857) | 99 | γ-Proteobacteria | 341 |
| DQ446026 (1) | 628 | Pseudomonas sp. strain NN84 (AJ973277) | 98 | γ-Proteobacteria | 347 | |
| DQ446027 (1) | 610 | Pseudomonas sp. strain I-F4 (AY779523) | 99 | γ-Proteobacteria | 347 | |
| DQ446028 (1) | DQ446077 (1) | 628, 619 | Pseudomonas sp. strain NZ103 (AY014821) | 98 | γ-Proteobacteria | 341 |
| DQ446029 (1) | 613 | Pseudomonas putida (AB180734) | 99 | γ-Proteobacteria | 341 | |
| DQ446078 (1) | 610 | Pseudomonas stutzeri (AJ312169) | 99 | γ-Proteobacteria | 341 | |
| DQ446079 (1) | 580 | Pseudomonas syringae (AY574913) | 99 | γ-Proteobacteria | 341 | |
| DQ446080 (1) | 608 | Stenotrophomonas sp. strain LMG 19833 (AJ300772) | 99 | γ-Proteobacteria | 341 | |
| DQ446030 (1) | 650 | Alvinella pompejana symbiont (AF357182) | 99 | ɛ-Proteobacteria | 341 | |
| DQ446081 (1) | 628 | Uncultured bacteroidetes bacterium (AY711048) | 88 | Bacteroidetes | 349 | |
| DQ446031 (1) | 600 | Flavobacteriaceae bacterium (AY987349) | 97 | Bacteroidetes | NVM | |
| DQ446032 (1) | 424 | Flavobacterium sp. strain AS-40 (AJ391201) | 90 | Bacteroidetes | 346 | |
| DQ446033 (1) | 620 | Uncultured Cytophagales bacterium (U70708) | 94 | Bacteroidetes | 348 | |
| DQ446082 (1) | 610 | Carnobacterium sp. strain ARCTIC-P2 (AY573049) | 96 | Firmicutes | 347 | |
| DQ446034-35 (2) | DQ446083 (1) | 600-630 | Staphylococcus sp. strain ARCTIC-P62 (AY573029) | 99 | Firmicutes | NVM |
| DQ446036 (1) | 630 | Symbiodinium sp. strain Fp2 (AJ271761) | 99 | Eukaryota | NVM | |
| DQ446037-38 (2) | 628, 600 | Tubastraea coccinea (AJ133556) | 99 | Eukaryota | NVM |
NVM, no virtual match was found.
Sequence types closely related (98% or greater sequence homology) to Aeromonas hydrophila, Comamonas terrigena, Shewanella sp. strain H836, Shewanella sp. strain ARCTIC-P25, Pseudomonas sp. strain P400Y-1, Pseudomonas sp. strain NZ103, and Staphylococcus sp. strain ARCTIC-P62 were found in both the SML-216 and SML-217 libraries (Table 1). Among the dominant γ-proteobacteria (52 clones), Shewanella sp. strain H836 was identified (99% sequence homology) for 19 of these. Among the β-proteobacteria, Comamonas sp. and Comamonas terrigena represented higher percentages in both the clone libraries (15 of 17 clones). The α-proteobacterial and bacteroidetes sequence types were found in either one or the other of the libraries, with no common types between the libraries. Among firmicutes, the sequence related to Staphylococcus sp. strain ARCTIC-P62 was observed in both libraries. No cyanobacterium-related sequence types were observed in either SML library, and plastid-related sequence types were observed in only one clone library (SML-216).
Microbial community composition in BBD samples.
A total of 49 and 50 16S rRNA gene clones were sequenced from the BBD-216 (BBD from colony 216) and BBD-217 (BBD from colony 217) libraries, respectively, and 48 and 47 partial or full-length sequences (from 690 to 1,537 bp) were obtained. The overview of the phylogenetic affiliations of the sequences obtained from BBD samples from the two colonies is shown in Fig. 1 and Table 2. In contrast to the SML, the α-proteobacteria dominated both of the BBD clone libraries, with contributions of 58% in BBD-216 and 87% in BBD-217. The γ-proteobacteria (dominant in both SML libraries) represented 4% in only one BBD clone library (BBD-216) and were not found in the other. In BBD-216, 4 to 6% of the sequences were related to δ-proteobacteria, spirochetes, firmicutes, and cyanobacteria; these bacterial types were not found in BBD-217 (Table 2). Two to 10% of the verrucomicrobia and 2 to 4% of plastid-related sequences were found in both BBD libraries (Table 2).
TABLE 2.
Overview of 16S rRNA gene sequences retrieved from clone libraries produced from BBD on two colonies of Siderastrea siderea (216 and 217)
| BBD-216 accession no. (no. of similar clones) | BBD-217 accession no. (no. of similar clones) | Sequence length (bp) | Closest relative in GenBank database (accession no.) | Similarity (%) | Phylogenetic affiliation | Virtual V1 + V2 amplicon length (bp) |
|---|---|---|---|---|---|---|
| DQ446131 (1) | 1,500 | Uncultured bacterium (AY354160) | 93 | Unidentified bacteria | 313 | |
| DQ446084 (1) | 1,497 | Alphaproteobacterium (AY701434)b | 96 | α-Proteobacteria | 329 | |
| DQ446132 (1) | 720 | Alphaproteobacterium (AY701455)b | 97 | α-Proteobacteria | 313 | |
| DQ446085-86 (2) | 738, 740 | Alphaproteobacterium (AF473915) | 96 | α-Proteobacteria | 331 | |
| DQ446087-88, DQ644006 (3) | 1,450-1,537 | Alphaproteobacterium PTB1 (AF260726)b | 97 | α-Proteobacteria | 315 | |
| DQ446089 (1) | 690 | Alphaproteobacterium (DQ200416) | 92 | α-Proteobacteria | 316 | |
| DQ446090 (1) | 740 | Antarctobacter sp. strain GWS-BW-H71M (AY515423) | 93 | α-Proteobacteria | 316 | |
| DQ446091 (1) | 720 | Hyphomicrobium sp. (AY499905) | 95 | α-Proteobacteria | 347 | |
| DQ446133 (1) | 1,492 | Oceanicaulis alexandrii (AJ309862) | 98 | α-Proteobacteria | 315 | |
| DQ446134-39 (6) | 1,486-1,516 | Ochrobactrum sp. (AF028733) | 92 | α-Proteobacteria | 315 | |
| DQ446140-41 (2) | 699, 710 | Ochrobactrum sp. strain mp-6 (AY331580) | 91 | α-Proteobacteria | 315 | |
| DQ446092-93 (2) | DQ446142-43 (2) | 699-1,492 | Ochrobactrum sp. strain TD (AY623625) | 91-92 | α-Proteobacteria | 315 |
| DQ446144 (1) | 693 | Rhodobacteraceae bacterium strain A333 (DQ005874) | 98 | α-Proteobacteria | NVMa | |
| DQ446094-101 (8) | DQ446145-51 (7) | 690-740 | Roseobacter sp. strain 8-1 (AJ536670), Roseobacter sp. strain DSS-8 (AF098493) | 96 | α-Proteobacteria | 313 |
| DQ446102 (1) | 1,462 | Roseobacter sp. strain 253-16 (AJ294352)b | 95 | α-Proteobacteria | 307 | |
| DQ446152 (1) | 1,496 | Roseobacter sp. strain 667-19 (AJ294355)b | 95 | α-Proteobacteria | NVM | |
| DQ446153 (1) | 1,501 | Roseobacter sp. strain NAC1-19 (AF245628) | 95 | α-Proteobacteria | 315 | |
| DQ446103-08 (6) | DQ446154-55 (2) | 700-1,493 | Roseovarius crassostreae CV919-312 (AF114484)c | 95-96 | α-Proteobacteria | 313 |
| DQ446156 (1) | 720 | Roseovarius nubinhibens ISM (AF098495) | 97 | α-Proteobacteria | 313 | |
| DQ446157-60 (4) | 1,464-1,482 | Ruegeria sp. strain AS-36 (AJ391197) | 98 | α-Proteobacteria | 315 | |
| DQ446161-62 (2) | 700, 698 | Silicibacter sp. strain E923 (AY369990) | 98 | α-Proteobacteria | 315 | |
| DQ446109 (1) | 1,486 | Silicibacter pomeroyi (AF098491) | 97 | α-Proteobacteria | 315 | |
| DQ446163-65 (3) | 699-750 | Sinorhizobium sp. (AJ012211) | 94-96 | α-Proteobacteria | NVM | |
| DQ446110 (1) | DQ446166-67 (2) | 700-1,482 | Sulfitobacter sp. strain ARCTIC-P49 (AY573043) | 95-96 | α-Proteobacteria | 315 |
| DQ446168-70 (3) | 719-720 | Sulfitobacter pontiacus (Y13155) | 95 | α-Proteobacteria | NVM | |
| DQ446171-72 (2) | 1,462, 1,465 | Thalassobius mediterraneus (AJ878874) | 94 | α-Proteobacteria | 313 | |
| DQ446111 (1) | 720 | Uncultured gamma KTc1119 (AF235120) | 96 | γ-Proteobacteria | 361 | |
| DQ446112 (1) | 1,475 | Ferrimonas marina (AB193751) | 97 | γ-Proteobacteria | 358 | |
| DQ446113-15 (3) | 1,450-1,511 | Desulfobacteraceae bacterium (AJ582696) | 93 | δ-Proteobacteria | 357 | |
| DQ446173 (1) | 1,483 | Adhaeribacter aquaticus (AJ626894) | 88 | Bacteroidetes | 331 | |
| DQ446174 (1) | 1,508 | Flexibacter aggregans (AB078038) | 95 | Bacteroidetes | NVM | |
| DQ446175 (1) | 710 | Flexibacter tractuosus (AB078076) | 91 | Bacteroidetes | NVM | |
| DQ446116-17 (2) | 1,484-1,511 | Leptonema illini (AY714984) | 89 | Spirochaetes | 348 | |
| DQ446118 (1) | 1,480 | Clostridium subatlanticum (AF458779) | 96 | Firmicutes | 360 | |
| DQ446119 (1) | 697 | Desulfitobacter alkalitolerans (AY538171) | 90 | Firmicutes | 357 | |
| DQ446120 (1) | 1,487 | Uncultured Fusibacter sp. (AB189368) | 91 | Firmicutes | 361 | |
| DQ446121 (1) | 1,522 | Fucophilus fucoidanolyticus (AB073978) | 90 | Verrucomicrobia | 347 | |
| DQ446122 (1) | 749 | Verrucomicrobial bacterium (AY499794) | 93 | Verrucomicrobia | 316 | |
| DQ446123 (1) | 1,510 | Verrucomicrobial bacterium (AY028220) | 96 | Verrucomicrobia | NVM | |
| DQ446124 (1) | 720 | Verrucomicrobial bacterium (AY500056) | 92 | Verrucomicrobia | NVM | |
| DQ446125 (1) | 700 | Verrucomicrobiales strain Sva0821 (AJ297461) | 87 | Verrucomicrobia | 360 | |
| DQ446176 (1) | 708 | Verrucomicrobial bacterium (AF424507) | 89 | Verrucomicrobia | NVM | |
| DQ446126-27 (2) | 1,498, 1,502 | Lyngbya hieronymusii var. hieronymusii (AB045906)d | 91 | Cyanobacteria | 316 | |
| DQ446128 (1) | 718 | Uncultured cyanobacterium (AF473911) | 93 | Cyanobacteria | 316 | |
| DQ446129-30 (2) | 739, 700 | Uncultured Coscinodiscophyceae (AY038449) | 97 | Eukaryota | NVM | |
| DQ446177 (1) | 697 | Uncultured prasinophyte (AY038432) | 89 | Eukaryota | NVM |
NVM, no virtual match was found.
Bacterium associated with toxic dinoflagellates.
Pathogen of JOD.
When a partial sequence of this clone was subjected to BLAST search, results showed high sequence homologies to three toxin-producing cyanobacteria (see the text).
Members of the order Rhodobacterales represented 64 to 66% of the α-proteobacteria, followed by Rhizobiales (11 to 32%), in the BBD clone libraries (Table 2). Among genera, the genus Roseobacter represented 22 to 32% of the α-proteobacteria in BBD clone libraries, with Roseobacter sp. strain 8-1 (accession no. AJ536670) and Roseobacter sp. strain DSS-8 (accession no. AF098493) the most dominant among them. The Roseobacter spp. were completely absent in the SML clone libraries. Silicibacter sp. strain E923 (accession no. AY369990), Silicibacter pomeroyi (accession no. AF098491), and the sulfite-reducing bacteria Sulfitobacter sp. strain ARCTIC-P49 (accession no. AY573043) and Sulfitobacter pontiacus (accession no. Y13155) were also found only in the BBD clone libraries. The genus Ochrobactrum (Rhizobiales) represented 7 to 24% of the α-proteobacteria in BBD clone libraries (Table 2). A δ-proteobacterium, Desulfobacteraceae (accession no. AJ582696), was found in one of the BBD clone libraries while it was absent in the SML clone libraries.
Three cyanobacterium-related sequences were found in one of the BBD clone libraries (BBD-216). Two almost full-length sequences of the clones, DQ446126 (1,498 bp) and DQ446127 (1,502 bp), were related to Lyngbya hieronymusii var. hieronymusii (accession no. AB045906), and one sequence was related to an uncultured cyanobacterium (accession no. AF473911) (Table 2). No cyanobacteria were found in the SML clone libraries.
The results indicate that the microbial composition of the BBD clone libraries was very different than the SML clone libraries from the same colonies. When comparing the SML and BBD clone libraries, there were no sequences common to both. Additionally, the clone sequences from BBD libraries showed higher species diversity and richness than SML clone libraries (Table 3).
TABLE 3.
Species diversity and richness of clone libraries from SML and BBD samples
| Clone library | Shannon-Wiener (H′) index | Simpson (Dsim) index | Margalef (DMar) index |
|---|---|---|---|
| SML-216 | 2.78 | 0.91 | 5.58 |
| SML-217 | 2.54 | 0.89 | 4.73 |
| BBD-216 | 3.02 | 0.97 | 6.75 |
| BBD-217 | 2.90 | 0.97 | 5.71 |
Toxin- and juvenile oyster disease-associated bacterial types.
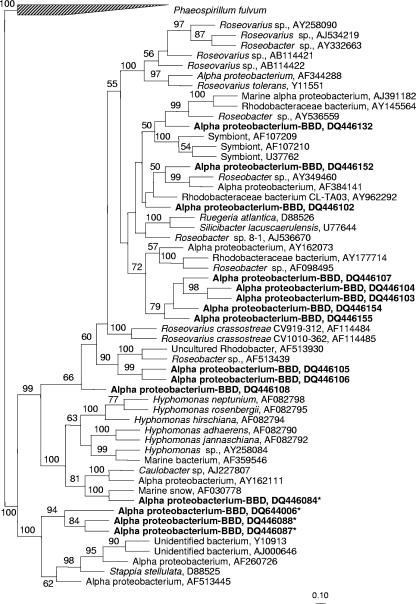
BLAST analysis showed that 6 of 25 of the α-proteobacterial sequence types observed in the BBD libraries were either closely related to bacteria associated with toxin-producing dinoflagellates or the JOD-causing bacterium (Table 2). Of the six, only one, the JOD-associated bacterium, was found in both libraries. At least 5 to 18% (per library) of the α-proteobacterial sequences in BBD clone libraries were closely related (95 to 97% similarity) to bacteria associated with dinoflagellates that produce the neurotoxins commonly known as paralytic shellfish toxin (PST). Three of the sequences in the BBD- 216 clone library (accession no. DQ446087, DQ446088, and DQ644006) showed 97% similarity (by BLAST) to the uncultured α-proteobacterium (accession no. AF260726) associated with the toxin-producing dinoflagellate Alexandrium tamarense. Detailed phylogenetic analysis (Fig. 2) confirmed that these three sequences were closely related to bacteria associated with this toxic dinoflagellate (accession no. AF260726, AJ000646, and Y10913). Bootstrap analysis also supported (100%) a clade consisting of three sequences from the BBD-216 clone library (accession no. DQ446087, DQ446088, and DQ644006), three sequences (from the database) of bacteria associated with a toxic dinoflagellate (accession no. AF260726, AJ000646, and Y10913), and two other sequences from the database (accession no. D88525 and AF513445) (Fig. 2). Two of the sequences (accession no. DQ446084 and DQ446132) in the two BBD clone libraries showed 96 to 97% similarity by BLAST to the uncultured α-proteobacteria (accession no. AY701434 and AY701455) associated with the toxin-producing dinoflagellate Gymnodinium catenatum. The detailed phylogenetic analysis (Fig. 2) showed that one of these sequences (accession no. DQ446084) was strongly supported (bootstrap value of 100%) as a member of a clade including the Hyphomonas group of α-proteobacteria, some members of which are associated with toxin-producing dinoflagellates (accession no. AY258084 and AF359546) (Fig. 2). Although three additional sequences from our BBD clone libraries (accession no. DQ446102, DQ446132, and DQ446152) showed 95 to 97% similarity (by BLAST) to toxic dinoflagellate-associated bacteria, the detailed phylogenetic analysis indicated that they were not associated with such bacterial types (Fig. 2). All sequence types associated with toxin-producing dinoflagellates were observed only in BBD clone libraries and were completely absent in SML libraries.
FIG. 2.
Phylogenetic tree derived from the 16S rRNA gene sequences that were closely related to the bacteria associated with toxin-producing dinoflagellates and JOD-causing bacteria and their neighbors. The tree topology is based on maximum parsimony analysis. Bar, 10% estimated sequence divergence. The numbers at the nodes are bootstrap values (percentages) obtained after 1,000 resamplings. Bootstrap values of less than 50% are not shown. The sequences related to the bacteria associated with toxin-producing dinoflagellates are indicated by asterisks. Boldface type indicates sequences from the present study.
BLAST analysis indicated that eight clones of the α-proteobacterial sequence types were related to the bacterial isolate that causes JOD in cultured Eastern oysters, identified (7) as Roseovarius crassostreae CV919-312 (accession no. AF114484), a member of the Roseobacter clade of α-proteobacteria. The partial or full-length sequences obtained from the present study showed 95 to 96% similarity to the JOD-associated bacterium. The JOD-associated sequence types were found in both BBD clone libraries (Table 2) and were completely absent in the SML clone libraries (Table 1). The detailed phylogenetic analysis (Fig. 2) showed that of these eight clones, none of the sequences was closely related to R. crassostreae (accession no. AF114484 and AF114485). Five sequences (accession no. DQ446103, DQ446104, DQ446107, DQ446154, and DQ446155) were separately clustered within the Roseovarius group. Two other sequences (accession no. DQ446105 and DQ446106) were related to an uncultured Rhodobacter (accession no. AF51930) and Roseobacter sp. (accession no. AF513439), and the sequence with accession no. DQ446108 was separately clustered.
Community profiles of healthy SML and BBD samples.
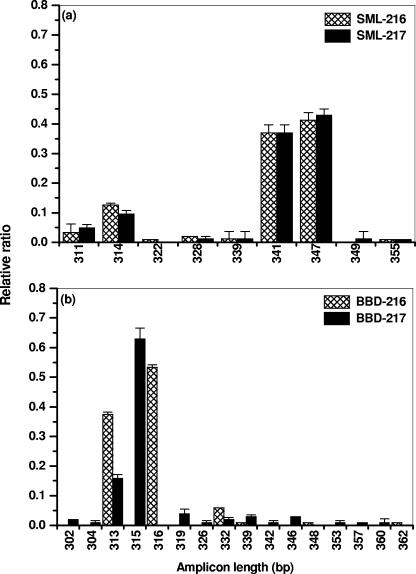
Whole-community LH-PCR profiles were obtained in triplicate for each BBD and SML sample collected from the two colonies (216 and 217) of black band-diseased S. siderea. The relative abundances of amplicon lengths (bp) observed in the BBD and SML samples are shown in Fig. 3. The results showed that the amplicons obtained from both SML samples were significantly different from BBD samples (P < 0.05, t test) (Fig. 3a and b). Both SML samples produced the same dominant amplicons, ranging from 311 to 355 bp in length. There were 7 common amplicons between the two colonies, of which 341 and 347 bp were dominant. The 341 amplicon corresponds to β-, γ-, and ɛ-proteobacteria and that of 347 corresponds to γ-proteobacteria and firmicutes (4, 59). It should be noted that the profiles agree with the clone library sequence data in that the γ-proteobacteria were the most dominant in SML clone libraries, followed by β-proteobacteria (Fig. 1).
FIG. 3.
Microbial community profiles produced using LH-PCR. Two colonies of Siderastrea siderea (216 and 217) were sampled. (a) SML of apparently healthy areas of black band-diseased colonies; (b) BBD samples. The x axis values are amplicon lengths in base pairs; y axis values are relative ratios (± standard deviations) of the peak intensities. Note the different scales on the x axes.
The amplicons obtained from BBD on colony 216 were significantly different from 217 (P < 0.05, t test). Colony 216 produced 6 amplicons ranging from 313 to 362 bp, whereas colony 217 produced 13 amplicons ranging from 302 to 360 bp. Most peaks were different between samples, with only three amplicons found (313, 332, and 339) common to both. Dominant peaks observed at 313, 315, and 316 correspond to the high percentages of α-proteobacteria (4, 59) observed from BBD clone libraries (Fig. 1). This is also the size range for cyanobacteria, which, although visually dominant in the BBD consortium, were not highly represented in the clone libraries.
In silico analysis of clone sequences.
To compare the results of the clone libraries with the LH-PCR community profile data sets, an in silico, or virtual, analysis of cloned sequences was performed (see Materials and Methods). Virtual amplicon lengths obtained are shown in Tables 1 and 2. Virtual analysis produced amplicon lengths in the expected range from 300 to 400 bp and revealed that most of the clone sequences obtained from both SML and BBD samples had matches (bp lengths) with LH-PCR-generated amplicons. Some of the clone sequences showed no virtual matches (NVM) and are indicated in Tables 1 and 2.
In the SML clone libraries, 69 of 88 clone sequences produced virtual amplicon lengths. Of these, the most common were 341 (n = 36) and 347 (n = 23) bp. In the BBD clone libraries, 78 of 94 clone sequences produced virtual amplicon lengths. Of these, the most common were 313 (n = 28), 315 (n = 24), and 316 (n = 6) bp. Thus, the virtual amplicons obtained were in agreement with the LH-PCR profiles.
DISCUSSION
Microbial community composition of SML samples.
In this study, γ-proteobacteria were numerically dominant in the SML clone libraries, followed by α- and β-proteobacteria. Among the γ-proteobacteria, members of the order Pseudomonadales were dominant, followed by Alteromonadales. The dominant γ-proteobacteria we observed in the SML clone libraries were Aeromonas hydrophila, Shewanella sp. strain H836, Shewanella sp. strain ARCTIC-P25, Pseudomonas sp. strain P400Y-1, and Pseudomonas sp. strain NZ103. Earlier studies indicated the presence of some of these genera, including Shewanella sp. (9) and Pseudomonas spp. (9, 51), in apparently healthy coral samples. However, we are not able to directly compare our results with the previous studies, since most of the earlier studies were aimed at investigating microorganisms associated with healthy coral tissues (8, 14, 21, 22, 31, 50, 51) and only rarely the SML (9). In one study (9), clone libraries from both a coral tissue slurry and the SML of the same colony were constructed and compared. In this study, it was found that, while the tissue clone library was dominated by γ-proteobacteria, the SML clone library was dominated by α-proteobacteria.
Microbial community composition in BBD samples.
We found the BBD-associated microbial communities to be highly diverse compared to the SML communities. High bacterial diversity associated with BBD has been reported previously (14, 22) for four different species of corals (M. annularis, M. cavernosa, D. strigosa, and C. natans) in studies that used molecular methods. The present study, which investigated a fifth BBD host coral (S. siderea), also revealed much variability in the species composition in BBD, even between two black band-diseased colonies of the same species on the same reef. A large percentage of members of Rhodobacterales (α-proteobacteria) were observed in our BBD clone libraries, in particular, members of the Roseobacter spp. The Roseobacter clade is one of the major marine groups and comprises more than 20% of the coastal bacterioplankton community (11). It has been reported that these bacteria can be free living, particle associated, or in commensal relationships with marine phytoplankton, vertebrates, and invertebrates (11). Frias-Lopez et al. (22) also reported the presence of some Roseobacter spp. associated with BBD on the coral M. annularis. A reason for the numerical abundance of these bacteria in association with black band-diseased corals has yet to be elucidated.
Members of the α-proteobacteria that are associated with toxin-producing dinoflagellates and the JOD-causing bacterium Roseovarius crassostreae (Rhodobacterales, α-proteobacteria) were observed in our BBD clone libraries (discussed below). Other important members of the α-proteobacteria, Sulfitobacter sp. strain ARCTIC-P49 and Sulfitobacter pontiacus, were also observed in our BBD clone libraries. The presence of S. pontiacus in BBD-associated corals has been reported previously (22). Members of the Sulfitobacter genus have been reported to be involved in oxidation of sulfite (11, 42, 57), which would be important in the active sulfuretum present in BBD. As in the previous studies (14, 21, 22), we detected sulfate-reducing bacteria in (one of) our BBD clone libraries. In our analysis, we found that BLAST searching of partial sequences (used in most previous studies) resulted in high sequence similarity to Desulfovibrio sp., while BLAST searching of full-length sequences resulted in the closest match to a Desulfobacteraceae bacterium (Table 2). In contrast to the earlier studies, we did not find any sulfate-reducing bacteria in the SML of apparently healthy tissue (although the other studies analyzed samples of the tissue and not the SML).
We found sequences related to the division firmicutes and bacteroidetes in both healthy and BBD samples. The members of firmicutes (21, 22) and the bacteroidetes (14, 21, 22) have been previously reported as potential BBD pathogens. Since the present study found these bacterial types in both SML and BBD clone libraries, it is questionable to consider them as BBD pathogens.
As in the previous molecular studies of BBD (14, 21, 22), we did not find Beggiatoa spp.-related sequences in the clone libraries, though their presence was confirmed by microscopy of the BBD samples. It has been suggested that BBD Beggiatoa spp. may be present at cell counts below PCR detection limits. Bissett et al. (5) studied bacterial diversity in sediments and retrieved Beggiatoa-related sequences in some of their clone libraries; however, in others, they did not retrieve Beggiatoa-related sequences, even though Beggiatoa filaments were observed in the samples. The authors concluded that the freezing of sediments prior to DNA extraction lysed the Beggiatoa cells, causing their DNA to be lost (5). While optimizing the DNA extraction procedures for soil and sediment samples, Miller et al. (35) found that refrigerated and frozen samples decreased the DNA yield compared to freeze-dried samples. However, we are not sure of the effect of freezing on DNA in our samples, since the freezing process usually preserves DNA. The lack of all efforts to date to detect BBD Beggiatoa by PCR could also be because (i) the primers do not match the sequences or (ii) the secondary structure of the 16S rRNA gene in Beggiatoa is not allowing the primers to recognize the site. Another possibility postulated by Bissett et al. (5) was that Beggiatoa cells might possess a small amount of chromosomal DNA, and thus are not detected in their PCR, due to the presence of a large central vacuole that occupies much of the cell volume. It has been reported that the vacuole of vacuolated Beggiatoa may occupy up to 80% of the total cell volume (30). It remains unclear why the microscopically dominating Beggiatoa could not be picked up in any of the molecular studies on BBD to date, and it needs further study.
In the present study, three cyanobacterium-related sequence types were observed in one of the two BBD clone libraries, and none in the SML libraries. One sequence (accession no. DQ446128) was related to an uncultured cyanobacterium (accession no. AF473911) that was observed previously in BBD samples by Cooney et al. (14). Two other clones from which full-length sequences (1,498 and 1,502 bp) were retrieved were both most closely related (91%) to Lyngbya hieronymusii var. hieronymusii (accession no. AB045906). BLAST searching of partial (719 and 720) sequences of the same two clones resulted in matches to Lyngbya aestuarii (accession no. AJ000714), aparatoxin-producing Lyngbya sp. (accession no. AY049752), and microcystin-producing Microcystis flos-aquae (accession no. AF139329), with 82, 92, and 93% sequence homology, respectively.
Similar to other molecular studies of BBD, we did not find a sequence related to Geitlerinema sp. (previously reported as Phormidium corallyticum) in the BBD clone libraries in this study, although we have detected this sequence in additional BBD samples (unpublished data). We have confirmed the presence of more than one type of cyanobacteria in one of our BBD samples (216), as reported in earlier studies (14). In this study, we found no cyanobacterial sequences in our clone library from BBD 217, even though BBD is quite obviously dominated by cyanobacteria. This result has also been reported by Frias-Lopez et al. (22), in which one of their three BBD clone libraries had no cyanobacterium-related sequences.
Toxin-producing dinoflagellate or oyster disease-associated bacterial types.
In the present study, sequences related to bacteria associated with toxin-producing dinoflagellates or JOD-causing bacteria were observed in BBD clone libraries. Many dinoflagellates like Alexandrium spp., A. tamarense, and Gymnodinium catenatum (24, 26, 28, 29, 55) are known to produce PST (24). Several bacterial groups, especially α-proteobacteria, are known to be associated with the PST-producing dinoflagellates (28, 29, 55). In the present study, phylogenetic analysis confirmed the presence of sequences related to uncultured α-proteobacteria associated with toxic dinoflagellates in the BBD clone library (Fig. 2). It has been suggested that the bacteria associated with toxic dinoflagellates may play an important role in the accumulation of PST (29) in harmful algal blooms (24) through bacterium-alga interaction. Though the functional roles of these bacteria in black band-diseased corals are unknown, the presence of abundant populations of bacteria associated with toxic dinoflagellates may create a toxic environment that could be lethal to corals. It should be noted that no toxic bacteria were observed in either SML library.
Cooney et al. (14) reported the consistent presence of Roseovarius crassostreae, the JOD bacterium (6, 7), in diseased coral samples, including BBD, and have proposed that this species should be investigated as a potentially important coral pathogen. In the present study, the BLAST analyses showed the presence of JOD-associated bacteria in the BBD clone library (Table 2). However, the detailed phylogenetic analysis did not confirm the presence of such a bacterial type in the BBD clone libraries (Fig. 2). While eight of our sequences (700 to 1,493 bp) from BBD clone libraries showed 95 to 96% BLAST similarity to the JOD-associated bacterium (Table 2), none of the sequences was found to be closely related to the R. crassostreae sequence as a result of phylogenetic analysis. Analysis of some of our clones using partial sequences (data not shown) showed 98 to 99% BLAST homology to the JOD bacterium, similar to reports by Cooney et al. (14) for their partial sequences (516 to 586 bp) in which they reported 98% similarity to this bacterium. The use of partial versus full sequences may be contributing to some of the discrepancies in the results of molecular analysis of BBD.
Community profiles of healthy and BBD-associated microbes.
Microbial community profiling was carried out using amplicon LH-PCR. This is a simple, robust, and highly reproducible profiling technique which has been successfully applied to study the microbial communities in different environmental samples such as bacterioplankton of freshwaters, estuaries, and the ocean (4, 59), biofilms (60), and soil (37, 49). In the present study, LH-PCR revealed much variation between the SML and BBD samples. LH-PCR analysis of the SML samples showed 7 common amplicons, which indicates that perhaps only a few groups of organisms were dominant in the samples. Amplicons with lengths of 341 and 347 bp were present at high relative abundance and correlated (via the in silico analysis) with the higher percentages of β- and γ-proteobacteria in the SML clone libraries. This is in agreement with the results of Suzuki et al. (59) and Bernhard et al. (4), who showed that amplicon sizes ranging from 340 to 350 bp correspond to β- and γ-proteobacteria. LH-PCR analysis of our BBD samples showed 16 amplicon lengths, with dominant peaks in the range of 313 to 316. This region correlates to the α-proteobacteria and cyanobacteria (4, 59) in general and also with the JOD-associated bacterium R. crassostreae. All of these are implicated as being important members of the BBD community. There was much variation between the community profiles of the two BBD samples for amplicons that were not in the 313- to 316-bp ranges. This revealed that the BBD-associated microbial communities were quite different, even between two coral colonies of the same species on the same reef sampled within minutes of each other.
LH-PCR community profiling can be conducted in combination with cloning and sequencing to promote identification of amplicon lengths, as we have done in this study. The presence of a specific amplicon length in both the clone library and the community profile is supportive evidence that the clone may be a member of the profiled community. It must be noted that since different bacterial species (and groups) can have amplicons with the same lengths, the detection of one specific amplicon length in both a profile and a clone library does not confirm that that amplicon is derived from only that particular species.
Our results clearly showed high variability in the microbial community composition and profiles between different BBD samples and the SML of apparently healthy areas of the same black band-diseased coral colonies; thus, they are in agreement with the results of other studies. While we did not find the same microbial communities as found in other reports, this finding is also similar to results of different studies (14, 20-22, 61), and even within the same research group (20-22), as discussed previously. The similarity of the bacterial communities in the SML of two colonies of S. siderea is consistent with the idea of bacterium-coral species specificity (51). We have provided a new data set for this area of investigation, specifically by study of a previously unstudied coral species.
The variability between BBD bacterial communities appears to be a more complex problem. As discussed above, all studies to date have provided inconsistent results in terms of characterization of the entire BBD community, although evidence for dominant members continues to accumulate. The reason for this variability may be multifold. While we found indications that there are similar BBD community members in our community analysis of BBD on S. siderea, we also found some variability between the two BBD samples. Since, to date, only 5 of the 19 Caribbean scleractinian coral species reported to be susceptible to BBD have been studied in terms of molecular characterization of BBD microorganisms, the question of the degree of diversity and variability of BBD microorganisms is only beginning to be addressed. There may also be a regional difference in the BBD microbial community. Our study is the first to use molecular techniques to characterize the BBD community on reefs of the northern Caribbean; the other studies have been conducted in the eastern (14) and southern (19-22) regions. Finally, our novel finding of bacteria associated with toxin-producing dinoflagellates is an important area of BBD etiology that has thus far not been explored.
Acknowledgments
This research was funded by the NIH (NIH/NIGMS SO6GM8205 to L.L.R. and D.K.M.) and NSF (NSF ADVANCE 0340695 to D.K.M.). Field collection was supported by NOAA's Caribbean Marine Research Center (CMRC-04-PRJV-01-04C to J.D.V. and L.L.R.) and the Perry Institute of Marine Science on Lee Stocking Island.
We thank Longin Kaczmarsky and two anonymous reviewers for helpful comments on the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Antonius, A. 1981. The ‘band’ diseases in coral reefs, p. 7-14. In E. D. Gomez et al. (ed.), The reef and man. Proceedings of the Fourth International Coral Reef Symposium, vol. 2. Marine Sciences Center, University of Philippines, Quezon City, Philippines. [Google Scholar]
- 3.Antonius, A. 1973. New observations on coral destruction in reefs, p. 3. Abstr. 10th Meet. Asso. Isl. Mar. Lab. Caribbean University of Puerto Rico, Mayaguez, Puerto Rico.
- 4.Bernhard, A. E., D. Colbert, J. McManus, and K. G. Field. 2005. Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiol. Ecol. 52:115-128. [DOI] [PubMed] [Google Scholar]
- 5.Bissett, A., J. Bowman, and C. Burke. 2006. Bacterial diversity in organically-enriched fish farm sediments. FEMS Microbiol. Ecol. 55:48-56. [DOI] [PubMed] [Google Scholar]
- 6.Boettcher, K. J., B. J. Barber, and J. T. Singer. 2000. Additional evidence that juvenile oyster disease is caused by a member of the Roseobacter group and colonization of nonaffected animals by Stappia stellulata-like strains. Appl. Environ. Microbiol. 66:3924-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boettcher, K. J., K. K. Geaghan, A. P. Maloy, and B. J. Barber. 2005. Roseovarius crassostreae sp nov., a member of the Roseobacter clade and the apparent cause of juvenile oyster disease (JOD) in cultured Eastern oysters. Int. J. Syst. Evol. Microbiol. 55:1531-1537. [DOI] [PubMed] [Google Scholar]
- 8.Bourne, D. G. 2005. Microbiological assessment of a disease outbreak on corals from Magnetic Island (Great Barrier Reef, Australia). Coral Reefs 24:304-312. [Google Scholar]
- 9.Bourne, D. G., and C. B. Munn. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7:1162-1174. [DOI] [PubMed] [Google Scholar]
- 10.Bruckner, A. W., R. J. Bruckner, and E. H. Williams. 1997. Spread of a black-band disease epizootic through the coral reef system in St Ann's Bay, Jamaica. Bull. Mar. Sci. 61:919-928. [Google Scholar]
- 11.Buchan, A., J. M. Gonzalez, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffroth, M. A. 1990. Mucus sheet formation on poritid corals: an evaluation of coral mucus as a nutrient source on coral reefs. Mar. Biol. 105:39-49. [Google Scholar]
- 13.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooney, R. P., O. Pantos, M. D. A. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 15.Ducklow, H. W., and R. Mitchell. 1979. Composition of mucus released by coral reef coelenterates. Limnol. Oceanogr. 24:706-714. [Google Scholar]
- 16.Ducklow, H. W., and R. Mitchell. 1979. Observations on naturally and artificially diseased tropical corals: a scanning electron microscopy study. Microb. Ecol. 5:215-223. [DOI] [PubMed] [Google Scholar]
- 17.Dustan, P. 1999. Coral reefs under stress: sources of mortality in the Florida Keys. Nat. Resour. Forum 23:155. [Google Scholar]
- 18.Edmunds, P. J. 1991. Extent and effect of black band disease on a Caribbean reef. Coral Reefs 10:161-165. [Google Scholar]
- 19.Frias-Lopez, J., G. T. Bonheyo, and B. W. Fouke. 2004. Identification of differential gene expression in bacteria associated with coral black band disease by using RNA-arbitrarily primed PCR. Appl. Environ. Microbiol. 70:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frias-Lopez, J., G. T. Bonheyo, Q. S. Jin, and B. W. Fouke. 2003. Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific Reefs. Appl. Environ. Microbiol. 69:2409-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frias-Lopez, J., J. S. Klaus, G. T. Bonheyo, and B. W. Fouke. 2004. Bacterial community associated with black band disease in corals. Appl. Environ. Microbiol. 70:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frias-Lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrett, P., and H. Ducklow. 1975. Coral diseases in Bermuda. Nature 253:349-350. [Google Scholar]
- 24.Green, D. H., L. E. Llewellyn, A. P. Negri, S. I. Blackburn, and C. J. S. Bolch. 2004. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 47:345-357. [DOI] [PubMed] [Google Scholar]
- 25.Green, E. P., and A. W. Bruckner. 2000. The significance of coral disease epizootiology for coral reef conservation. Biol. Conserv. 96:347-361. [Google Scholar]
- 26.Groben, R., G. J. Doucette, M. Kopp, M. Kodama, R. Amann, and L. K. Medlin. 2000. 16S rRNA targeted probes for the identification of bacterial strains isolated from cultures of the toxic dinoflagellate Alexandrium tamarense. Microb. Ecol. 39:186-196. [DOI] [PubMed] [Google Scholar]
- 27.Hayes, R. L., and N. I. Goreau. 1998. The significance of emerging diseases in the tropical coral reef ecosystem. Rev. Biol. Trop. 46:173-185. [Google Scholar]
- 28.Hold, G. L., E. A. Smith, T. H. Birkbeck, and S. Gallacher. 2001. Comparison of paralytic shellfish toxin (PST) production by the dinoflagellates Alexandrium lusitanicum NEPCC 253 and Alexandrium tamarense NEPCC 407 in the presence and absence of bacteria. FEMS Microbiol. Ecol. 36:223-234. [DOI] [PubMed] [Google Scholar]
- 29.Hold, G. L., E. A. Smith, M. S. Rappe, E. W. Maas, E. R. B. Moore, C. Stroempl, J. R. Stephen, J. I. Prosser, T. H. Birkbeck, and S. Gallacher. 2001. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol. Ecol. 37:161-173. [Google Scholar]
- 30.Kalanetra, K. M., S. B. Joye, N. R. Sunseri, and D. C. Nelson. 2005. Novel vacuolate sulfur bacteria from the Gulf of Mexico reproduce by reductive division in three dimensions. Environ. Microbiol. 7:1451-1460. [DOI] [PubMed] [Google Scholar]
- 31.Klaus, J. S., J. Frias-Lopez, G. T. Bonheyo, J. M. Heikoop, and B. W. Fouke. 2005. Bacterial communities inhabiting the healthy tissues of two Caribbean reef corals: interspecific and spatial variation. Coral Reefs 24:129-137. [Google Scholar]
- 32.Kuta, K. G., and L. L. Richardson. 1996. Abundance and distribution of black band disease on coral reefs in the northern Florida Keys. Coral Reefs 15:219-223. [Google Scholar]
- 33.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 35.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mills, D. K. 2000. Molecular monitoring of microbial populations during bioremediation of contaminated soils. Ph.D. dissertation. George Mason University, Fairfax, Va.
- 37.Mills, D. K., K. Fitzgerald, C. D. Litchfield, and P. M. Gillevet. 2003. A comparison of DNA profiling techniques for monitoring nutrient impact on microbial community composition during bioremediation of petroleum-contaminated soils. J. Microbiol. Methods 54:57-74. [DOI] [PubMed] [Google Scholar]
- 38.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 40.Porter, J. W., and J. I. Tougas. 2001. Reef ecosystems: threats to their biodiversity, p. 73-95. In S. A. Levin (ed.), Encyclopedia of biodiversity, vol. 5. Academic Press, San Diego, Calif. [Google Scholar]
- 41.Porter, J. W., P. Dustan, W. C. Jaap, K. L. Patterson, V. Kosmynin, O. W. Meier, M. E. Patterson, and M. Parsons. 2001. Patterns of spread of coral disease in the Florida Keys. Hydrobiologia 460:1-24. [Google Scholar]
- 42.Pukall, R., D. Buntefuss, A. Fruhling, M. Rohde, R. M. Kroppenstedt, J. Burghardt, P. Lebaron, L. Bernard, and E. Stackebrandt. 1999. Sulfitobacter mediterraneus sp. nov., a new sulfite-oxidizing member of the alpha-proteobacteria. Int. J. Syst. Bacteriol. 49:513-519. [DOI] [PubMed] [Google Scholar]
- 43.Ragoonath, D. N. 2005. Heterotrophic capabilities and the molecular identification of a cyanobacterium found in black band disease of coral reefs. M.S. thesis. Florida International University, Miami.
- 44.Ramos-Flores, T. 1983. Lower marine fungus associated with black line disease in star corals (Montastrea annularis). Biol. Bull. 165:429-435. [DOI] [PubMed] [Google Scholar]
- 45.Richardson, L. L. 2004. Black band disease, p. 325-349. In E. Rosenberg and Y. Loya (ed.), Coral health and disease. Springer-Verlag, Berlin, Germany.
- 46.Richardson, L. L., K. G. Kuta, S. Schnell, and R. G. Carlton. 1997. Ecology of the black band disease microbial consortium, p. 597-600. In H. A. Lessios and I. G. Macintyre (ed.), Proceedings of the Eighth International Coral Reef Symposium, vol. 1. Smithsonian Tropical Research Institute, Balboa, Panama. [Google Scholar]
- 47.Richardson, L. L., and K. G. Kuta. 2003. Ecological physiology of the black band disease cyanobacterium Phormidium corallyticum. FEMS Microbiol. Ecol. 43:287-298. [DOI] [PubMed] [Google Scholar]
- 48.Ritchie, K. B., and G. W. Smith. 2004. Microbial communities of coral surface mucopolysaccharide layers, p. 259-264. In E. Rosenberg and Y. Loya (ed.), Coral health and disease. Springer-Verlag, Berlin, Germany.
- 49.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohwer, F., M. Breitbart, J. Jara, F. Azam, and N. Knowlton. 2001. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20:85-91. [Google Scholar]
- 51.Rohwer, F., V. Seguritan, F. Azam, and N. Knowlton. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1-10. [Google Scholar]
- 52.Rosenberg, E., and Y. Loya. 2004. Coral health and disease. Springer-Verlag, Berlin, Germany.
- 53.Rützler, K., and D. Santavy. 1983. The black band disease of Atlantic reef corals. I. Description of a cyanophyte pathogen. PSZNI Mar. Ecol. 4:301-319. [Google Scholar]
- 54.Rützler, K., D. L. Santavy, and A. Antonius. 1983. The black band disease of Atlantic reef corals. III. Distribution, ecology and development. PSZNI Mar. Ecol. 4:329-358. [Google Scholar]
- 55.Sala, M. M., V. Balague, C. Pedros-Alio, R. Massana, J. Felipe, L. Arin, H. Illoul, and M. Estrada. 2005. Phylogenetic and functional diversity of bacterioplankton during Alexandrium spp. blooms. FEMS Microbiol. Ecol. 54:257-267. [DOI] [PubMed] [Google Scholar]
- 56.Schnell, S., S. Assmus, and L. L. Richardson. 1996. Role of sulfate reducing bacteria in the black band disease of corals, p. 116. Abstr. Annu. Meet. VAAM (Ver. Allg. Angew. Mikrobiol.) GBCH (Ges. Biol. Chem.). Elsevier, Cologne, Germany.
- 57.Sorokin, D. Y. 1995. Sulfitobacter pontiacus gen. nov., sp. nov., a new heterotrophic bacterium from the Black Sea, specialized on sulfite oxidation. Microbiology 64:295-305. [Google Scholar]
- 58.Sutherland, K. P., J. W. Porter, and C. Torres. 2004. Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266:273-302. [Google Scholar]
- 59.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiirola, M. A., J. E. Suvilampi, M. S. Kulomaa, and J. A. Rintala. 2003. Microbial diversity in a thermophilic aerobic biofilm process: analysis by length heterogeneity PCR (LH-PCR). Water Res. 37:2259-2268. [DOI] [PubMed] [Google Scholar]
- 61.Viehman, S., D. K. Mills, G. W. Meichel, and L. L. Richardson. 2006. Culture and identification of Desulfovibrio spp. from black band disease of corals on reefs of Florida Keys and Dominica. Dis. Aquat. Organ. 69:119-127. [DOI] [PubMed] [Google Scholar]
- 62.Voss, J. D., and L. L. Richardson. 2006. Coral diseases near Lee Stocking Island, Bahamas: patterns and potential drivers. Dis. Aquat. Organ. 69:33-40. [DOI] [PubMed] [Google Scholar]
- 63.Wegley, L., Y. N. Yu, M. Breitbart, V. Casas, D. I. Kline, and F. Rohwer. 2004. Coral-associated archaea. Mar. Ecol. Prog. Ser. 273:89-96. [Google Scholar]
- 64.Wilkinson, C. (ed.). 2002. Status of coral reefs of the world: 2002. Australian Institute of Marine Science, Townsville, Australia.