Abstract
Lutein and zeaxanthin are dihydroxy xanthophylls that are produced from their corresponding carotene precursors by the action of β- and ɛ-ring carotenoid hydroxylases. Two genes that encode β-ring hydroxylases (β-hydroxylases 1 and 2) have been identified in the Arabidopsis genome and are highly active toward β-rings but only weakly active toward ɛ-rings. A third distinct activity required for ɛ-ring hydroxylation has been defined by mutation of the LUTEIN1 (LUT1) locus, but LUT1 has not yet been cloned. To address the individual and overlapping functions of the three Arabidopsis carotenoid hydroxylase activities in vivo, T-DNA knockout mutants corresponding to β-hydroxylases 1 and 2 (b1 and b2, respectively) were isolated and all possible hydroxylase mutant combinations were generated. β-Hydroxylase single mutants do not exhibit obvious growth defects and have limited impact on carotenoid composition relative to the wild type, suggesting that the encoded proteins have a significant degree of functional redundancy in vivo. Surprisingly, the b1 b2 double mutant, which lacks both known β-hydroxylase enzymes, still contains significant levels of β-carotene–derived xanthophylls, suggesting that additional β-ring hydroxylation activity exists in vivo. The phenotype of double and triple hydroxylase mutants indicates that at least a portion of this activity resides in the LUT1 gene product. Despite the severe reduction of β-carotene–derived xanthophylls (up to 90% in the lut1 b1 b2 triple mutant), the double and triple hydroxylase mutants still contain at least 50% of the wild-type amount of hydroxylated β-rings. This finding suggests that it is the presence of minimal amounts of hydroxylated β-rings, rather than minimal amounts of specific β-carotene–derived xanthophylls, that are essential for light-harvesting complex II assembly and function in vivo. The carotenoid profiles in wild-type seeds and the effect of single and multiple hydroxylase mutations are distinct from those in photosynthetic tissues, indicating that the activities of each gene product differ in the two tissues. Overall, the hydroxylase mutants provide insight into the unexpected overlapping activity of carotenoid hydroxylases in vivo.
INTRODUCTION
Xanthophylls are oxygenated carotenoids that perform a variety of critical roles in photosystem structure and assembly, light harvesting, and photoprotection. The xanthophyll content of photosynthetic plant tissues is highly conserved through evolution, with lutein (L) being the most abundant, followed by β-carotene, neoxanthin (N), and violaxanthin (V). Zeaxanthin (Z) and antheraxanthin (A) accumulate to high levels in response to high light stress. L is a critical structural component of the light-harvesting complex II (LHC II) trimers, whereas Z, a structural isomer of L, is best known for its role in nonphotochemical quenching (NPQ). NPQ is a measurement of the dissipation of excess light energy absorbed by the photosystems and is one of the major mechanisms that protect plants from photooxidative damage. Energy-dependent NPQ requires a pH change in the thylakoid lumen, the PsbS protein, and the accumulation of Z (and A) that results from the deepoxidation of V (Li et al., 2000; Müller et al., 2001). L also has been found to be necessary for efficient NPQ, although its role is thought to be more indirect than those of Z and A (Pogson et al., 1998; Lokstein et al., 2002).
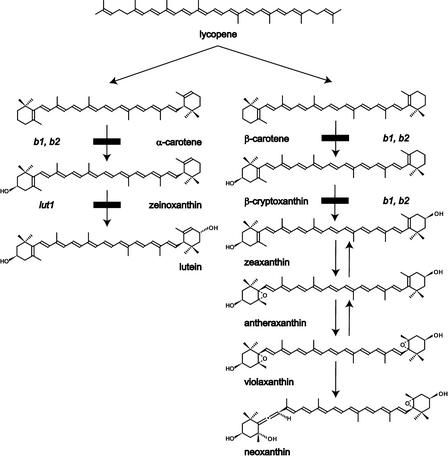
L and Z are dihydroxy xanthophylls that are derived from α-carotene (β,ɛ-carotene) and β-carotene (β,β-carotene), respectively, by the addition of hydroxyl groups to the 3,3′ position of both rings (Figure 1). L and Z have identical chemical formulas and similar structures, but small differences in their ring structures, conjugated double bond systems, and hydroxylation stereochemistry allow for distinct and specialized roles in photosystem structure, light harvesting, and photoprotection (Demmig-Adams et al., 1996; Horton et al., 1996; Niyogi et al., 1998). The enzymes that mediate carotenoid ring hydroxylation reactions are key for the biosynthesis of these two functionally important xanthophylls (Figure 1). Z is widespread in bacteria, fungi, and plants, and β-hydroxylases involved in Z synthesis have been cloned and characterized from all three phyla. All are nonheme di-iron oxidases that contain conserved His motifs required for activity (Bouvier et al., 1998). However, enzymes from the three phyla have otherwise low protein identity and are thought to have evolved independently, but they efficiently catalyze the hydroxylation of both β-rings of β-carotene to form Z (Misawa et al., 1990; Hundle et al., 1993; Bouvier et al., 1998; Masamoto et al., 1998). The formation of L from α-carotene requires the action of a second hydroxylase, the ɛ-hydroxylase, in addition to a β-hydroxylase (Britton, 1998). The ɛ-hydroxylase has not been cloned from any organism, but it has been identified genetically in Arabidopsis (Pogson et al., 1996).
Figure 1.
Xanthophyll Biosynthesis in Arabidopsis.
The positions of the lut1, b1 (CrtR-b1), and b2 (CrtR-b2) mutations in the pathway are indicated.
With regard to the genetics and molecular biology of carotenoid hydroxylation, Arabidopsis is the best-characterized plant system. Two genes that encode β-hydroxylases (β-hydroxylase 1 and 2) are present in the Arabidopsis genome (Sun et al., 1996; Tian and DellaPenna, 2001). The predicted mature proteins share 81% protein identity and are expressed coordinately, although β-hydroxylase 1 mRNA levels are always much higher than those of β-hydroxylase 2 mRNA. When expressed in vitro, both β-hydroxylases are highly active toward β-rings and function poorly with ɛ-ring–containing substrates (Sun et al., 1996; Tian and DellaPenna, 2001). Mutational studies also have identified the LUTEIN1 (LUT1) locus as being essential for the hydroxylation of ɛ-rings in Arabidopsis. Plants homozygous for the lut1 mutation show an 80% reduction in L levels and accumulate the immediate monohydroxy precursor zeinoxanthin. The lut1 mutation does not affect β-ring hydroxylation (Pogson et al., 1996), and lut1 does not map to either the β-hydroxylase 1 or 2 locus. A thorough analysis of the Arabidopsis genome also failed to identify additional paralogs with similarity to β-hydroxylases from bacteria, cyanobacteria, or plants (Tian and DellaPenna, 2001). These combined data suggest that LUT1 defines a novel class of carotenoid hydroxylase enzymes and that β-hydroxylase 1, β-hydroxylase 2, and LUT1 may represent the full complement of carotenoid hydroxylases in Arabidopsis.
The lut1 mutation provided the first demonstration that by manipulating carotenoid hydroxylase activities in vivo, one can modify both the type and amount of xanthophylls that accumulate in plants. More recently, the β-hydroxylase 1 gene was constitutively overexpressed in both the sense and antisense orientation in Arabidopsis leaf tissue. Overexpression resulted in a twofold increase in xanthophyll cycle carotenoids (V + A + Z) without affecting L levels (Davison et al., 2002). Expression of a β-hydroxylase 1 antisense construct reduced the levels of V and N in leaf tissue without affecting L levels; however, it was unclear which of the closely related β-hydroxylase genes were affected and to what levels (Rissler and Pogson, 2001).
To elucidate the in vivo functions of carotenoid hydroxylases, T-DNA knockout mutations in the β-hydroxylase 1 and 2 genes were isolated. Homozygous β-hydroxylase mutants were studied singly and in combination and introduced into the previously isolated lut1 mutant background to determine any specific or overlapping function(s) of the β- and ɛ-hydroxylases in vivo. The effects of various mutant genotypes on carotenoid biosynthesis, xanthophyll composition, and NPQ capacity are presented.
RESULTS
Isolation of Arabidopsis T-DNA Insertion Mutants for β-Hydroxylases and Generation of Double and Triple Mutant Combinations
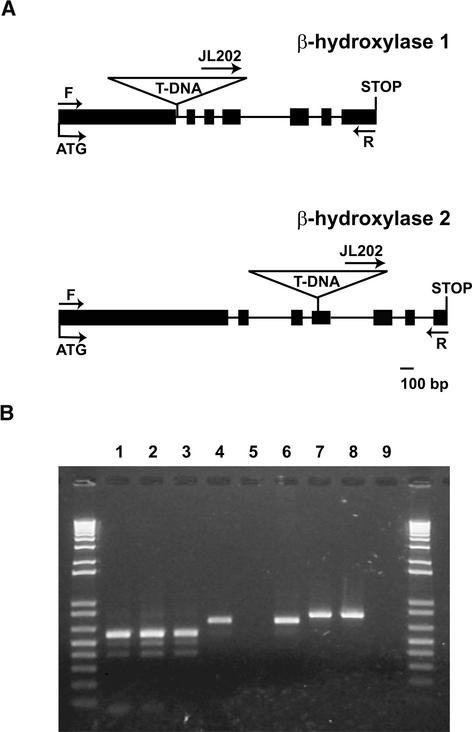
To study the in vivo functions of β-hydroxylase 1 and 2, plants containing T-DNA insertion alleles of each gene were isolated. Segregation of the kanamycin resistance marker indicated that both mutant lines segregated for a single Mendelian locus. Lines homozygous for each insertion were identified and designated b1 and b2 for β-hydroxylase 1 and 2, respectively (CrtR-b1 and CrtR-b2 according to standard nomenclature; Hirschberg, 2001). The genome insertion sites were determined by sequencing products amplified from each mutant. The β-hydroxylase 1 gene (At4g25700) contains an insertion in the first intron 375 bp downstream from the start codon, whereas the β-hydroxylase 2 gene (At5g52570) contains an insertion in the fourth exon 527 bp downstream from the start codon (Figure 2A).
Figure 2.
Molecular Characterization of β-Hydroxylase 1 and 2 T-DNA Insertional Mutants.
(A) β-Hydroxylase 1 and β-hydroxylase 2 insertional mutations (b1 and b2, respectively). Exons are represented as closed boxes, and introns are represented as lines connecting the boxes. The sizes of the boxes and lines are drawn to scale. Locations of forward (F) and reverse (R) primers for the β-hydroxylase 1 and 2 genes as well as the T-DNA insertion sites are indicated.
(B) RT-PCR with RNA extracted from the wild type (Ws) and homozygous b1 and b2 knockout mutants. Lanes 1, 4, and 7, RT-PCR products from Ws; lanes 2, 5, and 8, RT-PCR products from b1; lanes 3, 6, and 9, RT-PCR products from b2. Arabidopsis ɛ-cyclase–specific primer pairs were used in lanes 1 to 3, β-hydroxylase 1–specific primer pairs were used in lanes 4 to 6, and β-hydroxylase 2–specific primer pairs were used in lanes 7 to 9.
β-Hydroxylases are nonheme di-iron proteins that contain 10 conserved His residues required for iron binding and activity. The mutation of any one of the 10 His residues results in a complete loss of activity (Bouvier et al., 1998). In the b1 mutant, all 10 His residues are downstream of the T-DNA insertion site. Five of the 10 conserved His residues are downstream of the T-DNA insertion site in the b2 mutant. Therefore, both mutants are expected to result in a complete loss-of-function protein. Reverse transcriptase–mediated (RT) PCR using primer pairs designed to span the T-DNA insertion site of each mutant locus was used to determine whether full-length transcripts were produced in each genotype. The Arabidopsis lycopene ɛ-cyclase gene (At5g57030) was used as a control to demonstrate the integrity of all RNA preparations (Figure 2B). Both β-hydroxylase mRNAs were detected in Wassilewskija (Ws), and β-hydroxylase 1 and 2 mRNA was detected in the b2 and b1 mutants, respectively. However, no amplification products were observed for β-hydroxylase 1 and 2 in the b1 and b2 mutants, respectively (Figure 2B). These results confirm that transcripts spanning the T-DNA insertion sites are not present in the respective knockout mutants.
Homozygous b1 and b2 mutants grown under moderate light conditions did not exhibit a whole-plant phenotype different from that of the wild type (Figure 3). This result suggests that significant functional redundancy exists between the two Arabidopsis β-hydroxylases. To further address this question, we attempted to generate a b1 b2 double mutant. Because no additional β-hydroxylase paralogs are present in the Arabidopsis genome, if β-hydroxylase 1 and 2 were the only gene products with β-hydroxylation activity, one would expect a complete absence of β-carotene–derived xanthophylls and likely lethality in a b1 b2 double mutant. b1 and b2 mutants were crossed, and viable, homozygous b1 b2 double mutants were identified by PCR at the expected frequency in F2 progeny. This unexpected result suggests that other gene products also must have β-hydroxylation activity in Arabidopsis.
Figure 3.
Six-Week-Old Wild-Type and Mutant Arabidopsis Plants.
The plants shown were grown under normal light conditions (120 to 150 μmol·m−2·s−1) with a 12-h photoperiod.
Although β-hydroxylase 1 and 2 are most active toward β-rings, each can also add a hydroxyl group to ɛ-rings in vitro, although at much lower efficiency (Sun et al., 1996; Tian and DellaPenna, 2001). Therefore, it is conceivable that the reciprocal effect could occur for the presumed ɛ-hydroxylase encoded by LUT1 (i.e., that it also can function toward β-rings in vivo). To determine whether and to what extent LUT1 is functionally redundant with β-hydroxylase 1 and 2, the b1 and b2 mutations were introduced into the lut1 mutant background, individually and in combination. lut1 is in the Columbia (Col) background, whereas the b1 and b2 mutations are in the Ws background. Therefore, both Col and Ws were used as controls in all experiments that used combinations of lut1 with b1 and b2. Therefore, for a lut1 b1, lut1 b2, or lut1 b1 b2 mutant phenotype to be considered significantly different from the wild-type phenotype, it must be significantly different from both parental wild-type phenotypes.
Homozygous lut1 b1 and lut1 b2 double mutants were selected from the corresponding F2 populations. Neither double mutant showed a visible deleterious whole-plant phenotype compared with the wild type (Figure 3). To generate a line deficient in all three hydroxylase activities, lut1 b1 and lut1 b2 plants were crossed, and viable, homozygous lut1 b1 b2 triple mutants were identified in the F2 progeny. Unlike all other mutant combinations, the lut1 b1 b2 triple mutant exhibited an obvious whole-plant phenotype and was paler and smaller than either wild-type parental ecotype (Figure 3).
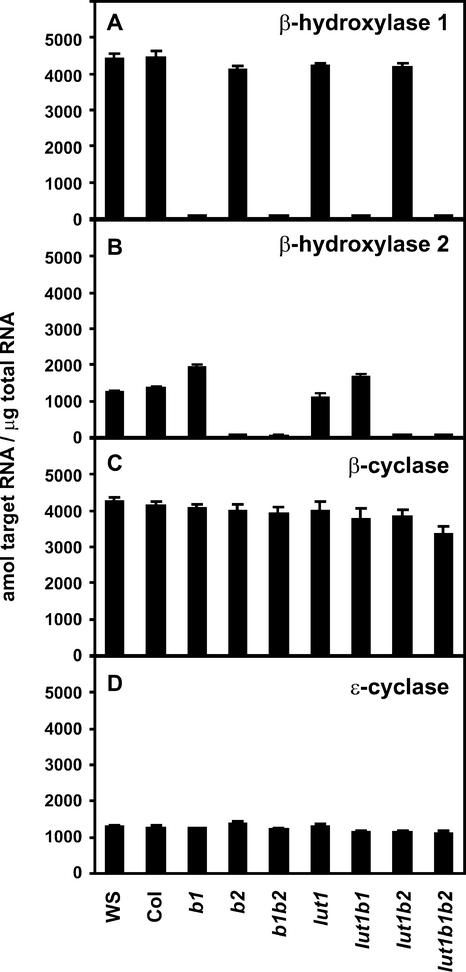
Expression of Carotenoid Hydroxylase and Lycopene Cyclase Genes in Wild-Type and Various Mutant Genotypes
To determine the effect of various mutant combinations on the expression of the remaining unmutated hydroxylases and the two lycopene cyclase enzymes, mRNA levels for β-hydroxylase 1, β-hydroxylase 2, lycopene β-cyclase, and lycopene ɛ-cyclase were determined in wild-type and different mutant genotypes. Because the expression of carotenoid biosynthetic genes is quite low in photosynthetic tissues, we developed TaqMan real-time PCR assays for these studies. β-Hydroxylase 1 and 2 mRNAs were not detected in genotypes that contain disruptions in these genes (Figures 4A and 4B), consistent with the RT-PCR analysis shown in Figure 2B. The steady state β-hydroxylase 1 mRNA level was similar to the wild-type level for all mutant genotypes that contain a functional β-hydroxylase 1 gene. β-Hydroxylase 2 mRNA also was similar to wild-type mRNA in the lut1 mutant but was increased by ∼50 and 30% relative to wild-type mRNA in b1 and lut1 b1, respectively. The expression of lycopene β- and ɛ-cyclase genes also was analyzed to determine whether specific hydroxylase mutant combinations affected cyclase gene expression. Both β- and ɛ-cyclase mRNA levels were nearly identical to wild-type levels in all mutant genotypes except in the lut1 b1 b2 triple mutant, in which the β-cyclase mRNA level was 20% lower than the wild-type level (Figures 4C and 4D).
Figure 4.
TaqMan Real-Time PCR Analysis of β-Hydroxylase 1, β-Hydroxylase 2, Lycopene β-Cyclase, and Lycopene ɛ-Cyclase mRNA Levels in Wild-Type and Mutant Genotypes.
(A) β-Hydroxylase 1.
(B) β-Hydroxylase 2.
(C) Lycopene β-cyclase.
(D) Lycopene ɛ-cyclase.
Data shown are means + sd (n = 4).
Leaf Pigment Compositions in Wild-Type and Various Mutant Genotypes
Wild-type and mutant genotypes were grown under moderate light (120 to 150 μmol·m−2·s−1) for 6 weeks, and their carotenoid compositions were quantified by HPLC. The level of total carotenoids in the mutant genotypes was not significantly different from that of their respective wild-type controls when expressed per mole of chlorophyll or per fresh weight, except that the lut1 b1 b2 triple mutant had 20% less total carotenoids on a fresh weight basis (data not shown). However, although carotenoid levels were unchanged, carotenoid compositions were altered significantly in most mutant lines (Table 1). The b2 mutant was the least affected of all genotypes and did not differ significantly from Ws, with the exception of a small decrease in N. The b1 mutant had a much more severe phenotype, with 30 and 45% decreases in V and N, respectively, and an 18% increase in L relative to Ws. β-Carotene was unchanged (Table 1), and the monohydroxy xanthophyll β-cryptoxanthin (β,β-carotene-3-ol) was not detected in b1 or b2 (data not shown). With the exception of increased L levels, the leaf carotenoid phenotype of the b1 mutant was similar to that reported for a constitutive Arabidopsis β-hydroxylase 1 antisense transgene (Rissler and Pogson, 2001). The lut1 mutant profile is consistent with previously published data (Pogson et al., 1996). lut1 had an 80% decrease in L, accumulated the monohydroxy xanthophyll zeinoxanthin (β,ɛ-carotene-3-ol), and had a twofold larger xanthophyll cycle carotenoid pool (V + A + Z) compared with Col. β-Carotene and N levels were not significantly different in lut1 compared with Col.
Table 1.
Wild Type and Mutant Carotenoid Composition in Leaf Tissue Quantified by HPLC
| Genotype | Lutein | Zeinoxanthin | β-Carotene | Neoxanthin | Violaxanthin | Antheraxanthin | Zeaxanthin | β-Xanthophylls | Total Carotenoids |
|---|---|---|---|---|---|---|---|---|---|
| Ws | 100 ± 9.0 (54)a | — | 31.0 ± 2.5 (17)a | 30.5 ± 5.1 (16)a | 23.1 ± 4.2 (13)a | — | — | 53.6 ± 6.7 (29)a | 184.5 ± 15.0a |
| Col | 104.9 ± 6.9 (58)a | — | 31.2 ± 6.7 (17)a | 25.7 ± 2.5 (14)a,b | 20.8 ± 3.9 (11)a,b | — | — | 46.5 ± 4.6 (25)a | 182.5 ± 14.2a |
| b1 | 118.5 ± 6.2 (64)b | — | 33.7 ± 5.5 (18)a | 16.9 ± 2.0 (9)c | 16.1 ± 0.6 (9)b | — | — | 33.0 ± 3.5 (18)b | 185.1 ± 7.7a |
| b2 | 104.3 ± 3.1 (60)a | — | 26.9 ± 2.9 (15)a | 23.4 ± 2.4 (13)b | 21.2 ± 1.0 (12)a | — | — | 44.6 ± 3.0 (25)a | 175.8 ± 7.8a |
| b1 b2 | 130.9 ± 4.7 (74)c | — | 34.3 ± 4.7 (19)a | 3.2 ± 1.1(2)d | 8.4 ± 1.7 (5)c | — | — | 11.6 ± 2.7 (7)c | 176.8 ± 4.1a |
| lut1 | 20.4 ± 3.6 (11)d | 35.5 ± 3.1 (20)a | 29.8 ± 6.2 (17)a | 21.9 ± 3.2 (12)b,e | 46.5 ± 4.1 (26)d | 19.2 ± 2.9 (11)a | 5.5 ± 1.0 (3)a | 93.1 ± 6.4 (52)d | 178.8 ± 16.7a |
| lut1 b1 | 13.2 ± 1.5 (7)e | 44.1 ± 3.6 (25)b | 40.5 ± 2.6 (23)b | 19.3 ± 1.8 (11)c,e | 35.8 ± 2.1 (20)e | 20.5 ± 3.3 (11)a | 6.3 ± 1.1 (3)a | 81.9 ± 4.6 (46)e | 179.7 ± 6.0a |
| lut1 b2 | 11.5 ± 2.2 (6)e | 32.8 ± 5.0 (18)a | 41.1 ± 3.2 (22)b | 20.2 ± 1.3 (11)c,e | 49.1 ± 1.6 (27)d | 22.3 ± 2.7 (12)a | 6.7 ± 1.0 (4)a | 98.3 ± 5.3 (53)d | 183.8 ± 6.9a |
| lut1 b1 b2 | 14.1 ± 1.5 (8)e | 88.1 ± 8.6 (50)c | 64.0 ± 5.3 (36)c | 1.6 ± 0.1 (1)f | 8.5 ± 1.2 (5)c | — | — | 10.1 ± 1.7 (6)c | 176.3 ± 6.9a |
The amount of carotenoid is expressed as mmol pigment/mol chlorophyll a + b. Each value is the mean result from six experiments ± SD, with the relative molar percentage of each carotenoid given in parentheses. Values marked with the same letters are not significantly different from each other within a column (Student's t test, P > 0.05).
The homozygous b1 b2 double mutant was quite informative with regard to the role of various carotenoid hydroxylases in Arabidopsis. The most surprising result was that, although both β-hydroxylase activities were eliminated in the b1 b2 mutant, β-ring–hydroxylated xanthophylls (L, V, and N) still were synthesized, albeit at lower levels than in Ws or in either single mutant. N and V were reduced by 90 and 65%, respectively, and there was a 30% increase in L relative to Ws. As with the single β-hydroxylase mutants, β-carotene was unchanged relative to the wild type and β-cryptoxanthin did not accumulate. The fact that b1 b2 had a more severe carotenoid phenotype than either single β-hydroxylase mutant indicates that the two mutations are additive and that there is significant functional redundancy between β-hydroxylase 1 and 2. β-Hydroxylase 1 and 2 clearly are the major β-ring hydroxylation activities, because their combined absence decreased the levels of β-carotene–derived xanthophylls by nearly 80%. However, β-carotene–derived xanthophylls still were produced at 20% of wild-type levels, indicating an additional and previously undescribed activity in Arabidopsis that also is capable of producing β-carotene–derived xanthophylls.
The carotenoid compositions in the lut1 b1 and lut1 b2 double mutants were quite different from those of their corresponding single mutant parents. This result indicates a functional compensation or redundancy between LUT1 and the two β-hydroxylases. As with the single β-hydroxylase mutants, b1 had a much greater effect on carotenoid composition than b2 when each was introduced into the lut1 background. The level of zeinoxanthin increased by 25%, V and L levels were reduced by 25 and 35%, respectively, and β-carotene was increased by 35% in lut1 b1 relative to lut1. The only carotenoids changed significantly in lut1 b2 were β-carotene and L.
An even more dramatic alteration in carotenoid composition was observed in the lut1 b1 b2 triple mutant. lut1 b1 b2 had the highest level of β-carotene in any genotype and more than twice the level of either wild-type parent. There were severe reductions in N (95%) and V (60%) relative to either wild-type parent, and unlike in lut1, lut1 b1, or lut1 b2, Z and A were absent in the triple mutant. β-Carotene–derived xanthophylls (V + Z + A + N) accounted for only 5% of the total carotenoid pool in the triple mutant versus 25 to 30% in the wild type. L was unchanged in the triple mutant relative to the lut1 b1 and lut1 b2 double mutants, but zeinoxanthin was more than doubled relative to any other lut1-containing genotype.
Seed Pigment Compositions of the Wild Type and Mutants
To determine the consequence of the various hydroxylase mutant genotypes on seed carotenoid composition, carotenoids from dry seeds of wild-type and the various mutant genotypes were extracted and quantified by HPLC. In general, Arabidopsis seeds contained 5- to 10-fold fewer total carotenoids than leaf tissue on a dry weight basis. The major carotenoid in wild-type seeds was L. A and Z were present at high levels in seeds compared with leaf tissue, but unlike in leaf tissue, there was little β-carotene accumulation in seeds. Ws seeds had twice the level of total carotenoids as Col seeds as a result of significant increases in all carotenoids except A and Z (Table 2). These data indicate that significant quantitative genetic variations for seed carotenoid levels exist between Arabidopsis ecotypes.
Table 2.
Wild Type and Mutant Carotenoid Composition in Seed Quantified by HPLC
| Genotype | Lutein | β-Carotene | Neoxanthin | Violaxanthin | Antheraxanthin | Zeaxanthin | β-Xanthophylls | Total Carotenoids |
|---|---|---|---|---|---|---|---|---|
| Ws | 108.9 ± 5.2 (76)a | 0.4 ± 0.03 (0.3)a | 7.5 ± 0.2 (5)a | 15.8 ± 0.9 (11)a | 4.8 ± 0.4 (3)a | 5.2 ± 0.1 (4)a | 33.3 ± 1.2 (23)a | 142.6 ± 6.6a |
| Col | 52.6 ± 2.4 (75)b | 0.1 ± 0.03 (0.2)b | 2.9 ± 0.2 (4)b | 3.0 ± 0.2 (4)b | 6.5 ± 0.8 (9)b | 4.6 ± 0.3 (7)a,b | 17.0 ± 0.9 (24)b | 69.7 ± 3.7b |
| b1 | 111.0 ± 2.7 (80)a | 0.4 ± 0.08 (0.3)a | 4.2 ± 0.3 (3)c | 11.4 ± 0.4 (8)c | 7.1 ± 0.2 (5)b,c | 4.0 ± 0.1 (3)b | 26.7 ± 0.8 (19)c | 138.1 ± 3.7a |
| b2 | 96.6 ± 4.4 (72)c | 0.3 ± 0.04 (0.2)a | 6.3 ± 0.6 (5)a | 15.8 ± 1.0 (12)a | 8.5 ± 0.5 (6)c | 6.8 ± 0.3 (5)c | 37.4 ± 1.5 (28)d | 134.3 ± 6.7a |
| b1 b2 | 77.1 ± 2.0 (90)d | 0.4 ± 0.05 (0.4)a | 1.7 ± 0.1 (2)d | 1.3 ± 0.2 (2)d | 2.3 ± 0.2 (3)d | 3 ± 0.2 (3)d | 8.3 ± 0.4 (10)e | 85.7 ± 2.3c |
| lut1 | 35.9 ± 2.9 (57)e | 0.4 ± 0.02 (0.6)a | 4.9 ± 0.4 (8)c | 12.3 ± 1.2 (20)c,e | 5.6 ± 0.5 (9)a,b | 4.0 ± 0.2 (6)b | 26.8 ± 1.5 (42)c | 63.1 ± 5.1b |
| lut1 b1 | 77.2 ± 2.0 (83)d | 0.6 ± 0.05 (0.7)a | 4.2 ± 0.3 (4)c | 6.5 ± 0.5 (7)f | 3.5 ± 0.3 (4)e | 1.5 ± 0.1 (2)e | 15.7 ± 0.8(17)b | 93.6 ± 2.7d |
| lut1 b2 | 67.4 ± 0.3 (68)f | 0.4 ± 0.02 (0.4)a | 8.1 ± 0.7 (8)a | 14.1 ± 0.5 (14)a,e | 6.0 ± 0.4 (6)b | 3.5 ± 0.2 (4)d | 31.7 ± 1.1 (32)a | 99.4 ± 1.6d |
| lut1 b1 b2 | 50.7 ± 3.7 (78)b | 2.3 ± 0.2 (3.5)c | 1.3 ± 0.3 (2)d | 4.1 ± 0.7 (6)b | 3.5 ± 0.6 (5)e | 3.5 ± 0.4 (5)d | 12.4 ± 1.3 (19)f | 65.4 ± 4.2b |
The amount of carotenoid is expressed as nmol pigment/g seed. Each value is the mean result from three experiments ± SD, with the relative molar percentage of each carotenoid given in parentheses. Values marked with the same letters are not significantly different from each other within a column (Student's t test, P > 0.05).
The level of total carotenoids in b1, b2, and lut1 mutant seeds did not differ significantly from that of their corresponding wild-type controls. As with leaf tissue, the b1 mutation had a more significant impact on seed carotenoid composition than b2. L was unchanged, whereas Z, V, and N were decreased significantly and A was increased in b1 seeds relative to wild-type seeds. The carotenoid composition of b2 seeds was quite different from that of both b1 and wild-type seeds, with a 10% decrease in L, a nearly twofold increase in A, and a 30% increase in Z. Unlike in b1, V and N levels in b2 seeds did not differ significantly from those in the wild type. The differential effect of the two single mutations in seeds was most obvious in the total β-carotene–derived xanthophylls accumulated: the b1 mutation decreased and the b2 mutation increased the level of total β-carotene–derived xanthophylls in seeds.
The phenotype of the lut1 mutation in seeds differed dramatically from that in leaves. The most obvious difference was that L levels were reduced by only 30% in seeds versus 80% in leaves, and there was no zeinoxanthin accumulation in lut1 seeds. These surprising results can be understood by considering the total amount of L produced in leaves and seeds as a percentage of dry weight. In Col leaves, the molar amount of L synthesized was 10 times that in seeds (data not shown). Thus, the residual or secondary ɛ-hydroxylase activity that allowed the production of 20% wild-type L levels in lut1 leaf tissue apparently was sufficient to fully convert the much lower level of zeinoxanthin in seeds to L. N and V were increased in lut1 seeds, whereas A and Z were almost unchanged. This finding is in contrast to the large accumulation of A and Z in lut1 leaf tissue relative to the wild type (Table 1).
Unlike in the single mutant parent lines, the carotenoid levels of b1 b2 seeds were reduced by 40% relative to wild-type (Ws) levels. However, as in leaf tissue, β-carotene–derived xanthophylls still were present at 10% of wild-type levels, despite the fact that both β-hydroxylase activities were eliminated in the b1 b2 double mutant. There also was a dramatic shift in carotenoid composition in the b1 b2 mutant, with Z, A, V, and N reduced by 40, 50, 90, and 80%, respectively, relative to the wild type.
Interpretation of seed carotenoid data from lut1 b1, lut1 b2, and lut1 b1 b2 is more difficult because of the large quantitative differences in total and individual carotenoid levels between the two parental ecotypes (Col and Ws). However, assuming the loci responsible for these quantitative differences segregate independently of the mutant hydroxylase loci, F2 progeny should segregate randomly for quantitative differences in seed carotenoids. To minimize the impact of any quantitative loci, 20 F2 individuals homozygous for lut1 b1, lut1 b2, and lut1 b1 b2 were selected and their seeds were pooled for analyses, which should average out quantitative differences in the progeny while maintaining selection for the respective mutant loci. Consistent with this hypothesis, lut1 b1 and lut1 b2 double mutants had similar amounts of total carotenoids relative to each other and were intermediate relative to their Col and Ws parents. lut1 b2 had higher levels of individual and total β-carotene–derived xanthophylls compared with lut1 b1, whereas lut1 b1 had a higher level of L than lut1 b2. The total carotenoid level of the lut1 b1 b2 triple mutant seeds was 35% lower than that of either lut1 b1 or lut1 b2 seeds, but L still accounted for 80% of total carotenoids. Most β-carotene–derived xanthophylls were reduced significantly in the triple mutant relative to lut1 b1 or lut1 b2, and there was a 4- to 20-fold increase in β-carotene relative to other genotypes (Table 2).
Photosynthetic Efficiency and NPQ Capacities in Leaves of Wild-Type and Mutant Genotypes under Normal Light Conditions
Leaf carotenoid compositional analysis showed that the production of α- and β-carotene–derived xanthophylls could be affected severely by disrupting one or more hydroxylase activity (Table 1). To determine the effect of modified xanthophyll profiles on photosystem efficiency and NPQ, chlorophyll fluorescence was measured in leaves of each mutant genotype grown under moderate light conditions (120 to 150 μmol·m−2·s−1). The Fv/Fm ratio (a measurement of maximum photosystem II photochemical efficiency) of the wild type (Col and Ws) and lut1, b1, and b2 mutants did not differ significantly, but b1 b2, lut1 b1, lut1 b2, and lut1 b1 b2 had lower Fv/Fm ratios than did the wild type (Table 3). These results suggest that the altered carotenoid composition resulting from the elimination of more than one hydroxylase activity can affect the efficient transfer of absorbed light energy to the photosystem II reaction center. Chlorophyll a/b ratios in b1 and b2 mutants did not differ significantly from wild-type levels but were increased significantly in the lut1 mutant, as reported previously (Lokstein et al., 2002), and also in all double mutants. The chlorophyll a/b ratio was affected most drastically in the lut1 b1 b2 triple mutant and was significantly higher than in all of the other genotypes (Table 3).
Table 3.
Chlorophyll a/b Ratios and Maximum Photosystem II Efficiency (Fv/Fm) in Wild-Type and Mutant Leaves
| Genotype | Chlorophyll a/b | Fv/Fm |
|---|---|---|
| Ws | 2.47 ± 0.07a | 0.846 ± 0.017a |
| Col | 2.52 ± 0.06a | 0.841 ± 0.006a |
| b1 | 2.54 ± 0.08a | 0.837 ± 0.013a |
| b2 | 2.50 ± 0.05a | 0.847 ± 0.006a |
| b1 b2 | 2.63 ± 0.05b | 0.824 ± 0.011b |
| lut1 | 2.61 ± 0.06b | 0.843 ± 0.010a |
| lut1 b1 | 2.64 ± 0.02b | 0.827 ± 0.010b |
| lut1 b2 | 2.67 ± 0.03b | 0.826 ± 0.015b |
| lut1 b1 b2 | 2.74 ± 0.02c | 0.821 ± 0.007b |
Values marked with the same letters are not significantly different from each other within a column (Student's t test, P > 0.05).
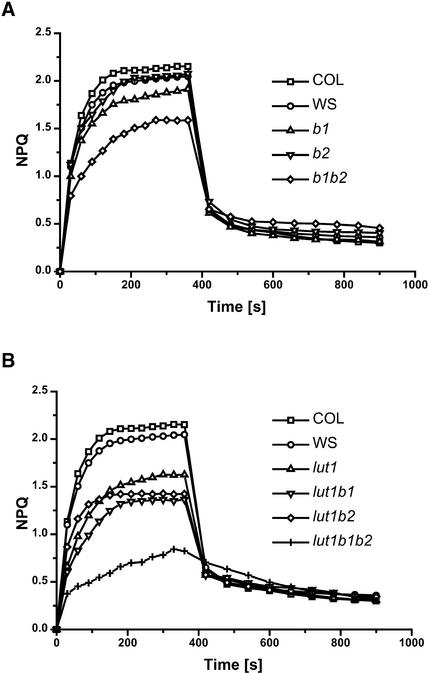
As with wild-type seed carotenoid composition, significant genetic variation was observed for NPQ between Ws and Col. Ws had both a slower induction rate and lower NPQ amplitude compared with Col. The b2 mutant had NPQ induction and amplitude similar to those of Ws, whereas the b1 mutant, in which the relatively highly expressed β-hydroxylase 1 gene was disrupted, had slower induction and a ∼10% reduction in NPQ amplitude. This impact on NPQ was consistent with the greater impact of the b1 mutation on xanthophyll cycle carotenoids (Table 1). Light activation of NPQ also was delayed in lut1 and NPQ amplitude was reduced, consistent with previous reports (Pogson et al., 1998; Lokstein et al., 2002).
In b1 b2, there was a synergistic effect of the two β-hydroxylase mutations on both carotenoid composition and NPQ. NPQ induction and amplitude were reduced to a much greater extent in b1 b2 than in either single mutant parent (Figure 5A). In lut1 b1 and lut1 b2 double mutants, the NPQ phenotypes of the single mutants were additive and likely resulted from the combined deficiency of both L and xanthophyll cycle carotenoids in these double mutants. The fact that the lut1 b1 phenotype was more severe than the lut1 b2 phenotype can be attributed to the respective single mutants. The lut1 b1 b2 triple mutant had a very rapid initial induction of NPQ; however, the NPQ level obtained was reduced greatly compared with levels in the wild type and all other mutant genotypes (Figure 5B).
Figure 5.
NPQ Analysis of Wild-Type and Mutant Genotypes.
(A) Wild type (Ws and Col) and b1, b2, and b1b2 mutants.
(B) Wild type (Ws and Col) and lut1, lut1 b1, lut1 b2, and lut1 b1 b2 mutants.
Chlorophyll fluorescence was measured during 6 min of illumination with a PPFD of 500 μmol·m−2·s−1 followed by 9 min of dark relaxation. Each data point is the mean of six independent experiments, and all standard deviations fell within the symbol sizes.
DISCUSSION
In plants, carotenoid hydroxylases catalyze the formation of α- and β-carotene–derived xanthophylls, which perform a variety of critical roles in the photosynthetic apparatus. Three genes that participate in carotenoid hydroxylation reactions have been identified in the Arabidopsis genome through combined molecular and genetic analyses. Among these, two β-hydroxylases have been cloned and shown to be highly active against β-rings and weakly active against ɛ-rings (Sun et al., 1996; Tian and DellaPenna, 2001). We have isolated T-DNA knockout mutants in the β-hydroxylase 1 and 2 genes and shown that the expression of mRNA for each gene was eliminated fully. A third Arabidopsis gene product required for ɛ-ring hydroxylation has been defined genetically by the lut1 mutation but has not been cloned (Pogson et al., 1996). The effects of these three carotenoid hydroxylase mutations individually and in various combinations have been used to assess the roles, interactions, or functional redundancies of the hydroxylase activities in vivo.
LUT1 Is the Major ɛ-Ring–Hydroxylating Activity in Vivo
The effect of mutating the LUT1 locus, the presumed ɛ-hydroxylase, on leaf carotenoid composition has been described previously (Pogson et al., 1996; Lokstein et al., 2002) and is the most dramatic effect of any single hydroxylase mutation. The fact that ɛ-ring hydroxylation is reduced by 80% in lut1 and zeinoxanthin accumulates indicates that the LUT1 gene product is the primary ɛ-ring hydroxylation activity in photosynthetic tissue. The absence of β,ɛ-carotene-3′-ol (α-carotene with a single hydroxyl group on the ɛ-ring) in lut1 or any other mutant line studied suggests that β-ring hydroxylation precedes ɛ-ring hydroxylation during L synthesis. Both lut1 alleles (lut1-1 and lut1-2) are ethyl methanesulfonate derived and still contain 15 to 20% of wild-type L levels. This finding suggests either that both alleles are leaky and the L made is from residual LUT1 activity or, if the alleles are null, that there is a second ɛ-ring hydroxylation activity in Arabidopsis. The cloning of LUT1 and the identification of a definitive null mutation in the locus is required to distinguish between these possibilities.
β-Hydroxylase 1 and 2 Are Functionally Redundant in Vivo
Neither b1 nor b2 exhibited a whole-plant phenotype, suggesting that significant functional redundancy exists between the two β-hydroxylases. This observation is consistent with our inability to identify β-hydroxylase mutants in various ethyl methanesulfonate mutant screens (data not shown). β-Hydroxylase 1 and 2 are related closely at the protein sequence level, have similar activities in vitro, and are expressed coordinately in all wild-type tissues analyzed, although β-hydroxylase 1 mRNA always is present at much higher levels than β-hydroxylase 2 mRNA (Tian and DellaPenna, 2001). The high relative expression level of β-hydroxylase 1 suggests that it might be responsible for the majority of β-ring hydroxylation activity in leaf tissue. The alterations to carotenoid composition in the β-hydroxylase single mutants support this hypothesis: β-carotene–derived xanthophylls are affected more severely in b1 than in b2 (Table 1).
The function of β-hydroxylase 2 is difficult to discern by single mutant analysis, because it was not significantly different from the wild type in carotenoid composition and NPQ function. However, β-hydroxylase 2 clearly was both functionally additive and complementary to β-hydroxylase 1 in vivo. The b1 b2 double mutant had a more severe effect on β-ring hydroxylations than did the b1 mutation alone (Table 1), indicating that β-hydroxylase 2 can compensate to a significant degree for the absence of β-hydroxylase 1 activity in the b1 mutant. Moreover, β-hydroxylase 2 also was able to compensate for the loss of both LUT1 and β-hydroxylase 1 (in the lut1 b1 double mutant) to almost the same extent as β-hydroxylase 1 in the lut1 b2 background (Table 1). These data indicate that although β-hydroxylase 2 is expressed at a much lower level than β-hydroxylase 1 in the wild type, it plays an important role in functional redundancy in the carotenoid pathway. The functional redundancy of β-hydroxylase 2 in the b1 and lut1 b1 mutants may be enhanced in part by a small increase in β-hydroxylase 2 expression (Figure 4B).
The fact that different b1- and b2-containing carotenoid mutant genotypes are affected preferentially in the accumulation of specific carotenoids (Table 1) indicates that although the two β-hydroxylases can partially compensate for each other, they are not entirely equivalent and likely have specialized functions. This was most apparent when β-hydroxylase 1 or 2 deficiency was present in the wild-type or lut1 background (Table 1). Although the biochemical basis for two closely related but functionally distinct β-hydroxylase enzymes in Arabidopsis is unknown, tomato and pepper also have maintained two highly similar β-hydroxylase genes, suggesting that this may be a common feature of carotenoid synthesis in plants.
β-Carotene–Derived Xanthophylls Still Are Produced in the b1 b2 Double Mutant, Indicating That Additional β-Ring Hydroxylation Activity Exists in Vivo
A major and unexpected outcome of carotenoid hydroxylase double mutant analyses was that when both known Arabidopsis β-hydroxylase activities were eliminated in the b1 b2 double mutant, β-carotene–derived xanthophylls still were produced at significant levels. The 80% reduction in β-carotene–derived xanthophylls in b1 b2 confirmed that β-hydroxylase 1 and 2 were the predominant β-carotene hydroxylation activities in vivo. However, β-carotene–derived xanthophylls still accounted for 7% of the total carotenoids in the b1 b2 mutant versus 29% in the wild type. In addition, β-ring hydroxylation also was necessary for α-carotene to be converted to L, and L was produced at even higher levels in the b1 b2 double mutant than in the wild type. The synthesis of β-carotene–derived xanthophylls and the accumulation of L in b1 b2 indicates that an additional, previously unknown β-ring hydroxylation activity exists in vivo. These data raise the question: what is the nature of this additional β-ring hydroxylation activity?
Three hypotheses can be proposed for the additional β-ring hydroxylation activity present in the b1 b2 background. (1) It is a third, novel β-hydroxylase enzyme unrelated at the protein sequence level to known β-hydroxylases. (2) It is a secondary, intrinsic activity of the LUT1 gene product (ɛ-hydroxylase) capable of β-ring hydroxylation in vivo. (3) It is a second, LUT1-like enzyme that is active toward both types of rings. Searches of the Arabidopsis genome using representative β-hydroxylase protein sequences from all three phyla as queries failed to identify additional homologs beyond β-hydroxylase 1 and 2 (data not shown). Therefore, if the additional activity resides in a third β-hydroxylase enzyme, it would represent a new structural class of β-hydroxylases in nature. The possibility that LUT1 might hydroxylate β-rings in addition to ɛ-rings in vivo has precedent in the fact that both β-hydroxylase 1 and 2 also can hydroxylate ɛ-rings in vitro, although quite poorly relative to β-ring substrates (Sun et al., 1996; Tian and DellaPenna, 2001). A reciprocal activity also could occur for LUT1. Finally, if LUT1 does not have a secondary β-hydroxylase activity, it is possible that a LUT1 paralog exists that is active toward both β- and ɛ-rings. This would be analogous to the two highly conserved Ara-bidopsis β-hydroxylases; however, this LUT1 paralog likely would be a minor component of total hydroxylase activity because it cannot fully compensate for the ɛ-ring hydroxylation deficiency in lut1. Clearly, the isolation of LUT1 and the biochemical characterization of its product will serve to elucidate these various hypotheses.
Double and Triple Mutants Indicate Overlapping Functions and Interactions between the β- and ɛ-Hydroxylases
To investigate any overlapping functions or interactions between the β- and ɛ-hydroxylases of Arabidopsis, the b1 and b2 mutations were introduced into the lut1 background to generate the corresponding double mutants. As with the single b1 and b2 mutants, b1 had a more pronounced effect in the lut1 background than b2. lut1 b1 had decreased V and increased zeinoxanthin relative to lut1. β-Hydroxylase 1 and 2 also appeared to have a low level of ɛ-ring hydroxylation activity in vivo, because elimination of either activity in the lut1 background further decreased the level of L (Table 1).
The failure to eliminate L in lut1 b1 and lut1 b2 may be attributable to a low level of ɛ-ring hydroxylation activity in vivo from the single functional β-hydroxylase still present in each double mutant. To investigate these possibilities further, we attempted to create a lut1 b1 b2 triple mutant. Assuming that no other carotenoid hydroxylases are present in Arabidopsis and that the lut1-2 allele is null, one would expect the triple mutant to lack all xanthophylls and likely be lethal as a result. Surprisingly, homozygous lut1 b1 b2 mutants were isolated at the expected ratio from crosses, and although they were paler green than the wild type, they were viable and set seed. Because the L level in lut1 b1 b2 (in which both β-hydroxylase 1 and 2 activities were eliminated) was not reduced further relative to lut1 b1 or lut1 b2, we conclude that β-hydroxylase 1 and 2 do not contribute significantly to ɛ-ring hydroxylation in vivo. Therefore, the L synthesized in lut1 b1 b2 must be attributable to either residual ɛ-ring hydroxylation activity from the mutant LUT1 enzyme or another ɛ-hydroxylase. In either case, LUT1 or a second ɛ-hydroxylase also must be active toward β-rings, because β-ring hydroxylation still occurred at significant levels in lut1 b1 b2 (Figure 6).
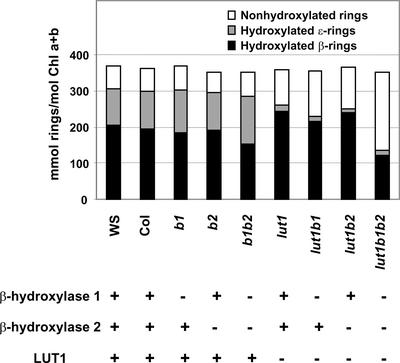
Figure 6.
Hydroxylation Levels of β- and ɛ-Rings in All Genotypes.
The amounts of hydroxylated β- and ɛ-rings and nonhydroxylated rings were calculated from the leaf tissue carotenoid composition for each genotype. The presence or absence of each of the three known carotenoid hydroxylases in each genotype is indicated by a plus or a minus sign, respectively.
Total Hydroxylation Activity and the Synthesis of Specific Xanthophylls Are Affected Differentially in the Hydroxylase Mutants
The impact of hydroxylase deficiencies has been discussed here based on the relative changes to specific carotenoids (e.g., violaxanthin) or groups of α- and β-carotene–derived xanthophylls in each genotype (Table 1). In the case of double and triple mutants, the effects on individual xanthophylls can be quite extreme. However, focusing only on the species or groups of xanthophylls affected leads to the underestimation of total β-ring hydroxylation activity, because hydroxylation of the β-rings of zeinoxanthin and L are overlooked. A more accurate accounting of total hydroxylation activity involves considering the total hydroxylation of each ring type (total moles of hydroxyl groups introduced onto β- and ɛ-rings, respectively; Figure 6). From this perspective, although the b1 mutant decreased N and V significantly (Table 1), the level of hydroxylated β-rings produced by the mutant was hardly affected and total ring hydroxylation (β- plus ɛ-rings) was unchanged relative to that in the wild type (Figure 6). This effect was even more apparent in b1 b2, in which β-carotene–derived xanthophylls were reduced by 80% but β-ring hydroxylations were reduced by only 25% and total hydroxylation was not changed significantly relative to that in the wild type. These data suggests an even greater degree of functional redundancy among the hydroxylases with regard to their ability to perform β- and/or ɛ-ring hydroxylations in the absence of another hydroxylase. The ability of each hydroxylase to compensate functionally for another hydroxylase in the production of specific xanthophylls clearly is much more limited (Table 1).
The differential effects of hydroxylase deficiencies on total hydroxylation levels and the production of specific xanthophylls suggest a level of biochemical regulation in the production of different xanthophylls that has not been considered previously. Whether this regulation is attributable to innate differences in the substrate preference and turnover rates of each hydroxylase or is caused by the differential ability of hydroxylases to participate in biosynthetic complexes for the production of certain xanthophylls is unknown. It also is possible that specific molecular or biochemical regulatory mechanisms could be activated in the mutants to preferentially provide for a minimum level of β-ring hydroxylation.
The surprisingly high level of total β-ring hydroxylation retained in all mutant genotypes suggests that hydroxylated β-rings, rather than specific xanthophylls, might be the structural and/or functional minimal requirements for photosystem assembly in vivo. This finding is consistent with a recent report that 3-hydroxy-β-end groups, rather than β-carotene–derived xanthophylls per se, are the minimal requirements for the binding of xanthophylls to LHC II in vitro (Phillip et al., 2002). Hydroxylated β-rings (monohydroxy or dihydroxy xanthophylls) were able to promote the in vitro assembly of LHC II by facilitating the correct folding of LHC II proteins. In light of this critical role for 3-hydroxy-β-end groups in LHC II assembly, it is easier to understand why the levels of β-ring hydroxylations, rather than specific xanthophylls, were maintained in vivo in the carotenoid hydroxylase mutants. An extreme example of this principle is lut1 b1 b2, which had the lowest level of β-carotene–derived xanthophylls of any genotype (Table 1). In lut1 b1 b2, zeinoxanthin (which contains a single 3-hydroxy-β-end group) was increased more than twofold relative to lut1 and accounted for 75% of the total 3-hydroxy-β-end groups.
Modified Xanthophyll Composition in the Hydroxylase Mutants Correlates with Alterations in NPQ Induction Kinetics
In addition to their essential roles in LHC II assembly and photosystem structure, xanthophylls also have important functions in photoprotection. The severity of xanthophyll compositional changes in the different hydroxylase mutant genotypes coincided with their degree of compromised NPQ induction kinetics (Table 1, Figure 5). Other carotenoid biosynthetic mutants used previously to study xanthophyll function in Arabidopsis in vivo— lut1, lut2, npq1, aba1 (npq2), and their double mutants—block the production of one or more xanthophylls (Niyogi et al., 1998; Pogson et al., 1998; Lokstein et al., 2002). The carotenoid hydroxylase mutants are unique in that the synthesis of multiple xanthophylls is attenuated to different degrees rather than eliminated. This allows for the modification of the types and levels of specific xanthophylls involved in light harvesting and photoprotection under normal growth conditions.
Because chlorophyll b and N are associated primarily with LHC II (Ruban et al., 1999), the increased chlorophyll a/b ratio and decreased N levels in the double and triple hydroxylase mutants (Tables 1 and 3) are consistent with a reduction in LHC II levels. These data also are consistent with the assembly or stability of the peripheral LHC II being affected and are correlated with impaired NPQ induction and amplitude in the double and triple hydroxylase mutants (Figure 5). In addition, the Fv/Fm ratios in all double and triple hydroxylase mutants were decreased significantly, indicating that the quantum efficiency of photosystem II also was affected as a result of their altered carotenoid compositions. Similar observations have been reported for the lut1, lut2, aba1, and lut2 aba1 mutants (Tardy and Havaux, 1996; Pogson et al., 1998; Lokstein et al., 2002). The most severe impact on NPQ and quantum efficiency occurred in the lut1 b1 b2 mutant, in which L and xanthophyll cycle carotenoids were reduced by >80% relative to the wild type. Given the continuum of xanthophyll changes in the full suite of hydroxylase mutants, it will be quite informative for our understanding of xanthophyll functions to assess the responses of the hydroxylase mutants to high light stress.
Insights into Carotenoid Pathway Regulation in Photosynthetic Tissues
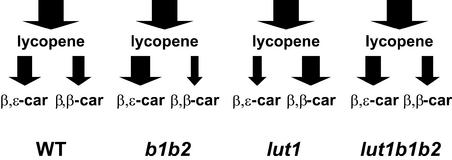
In addition to differentiating the overlapping functions of carotenoid hydroxylases in vivo, the phenotypes of hydroxylase mutants provide further insight into the levels of regulation that exist in the carotenoid pathway. The extreme changes in xanthophyll composition are not the result of a corresponding alteration in total hydroxylation activity (Figure 6) but rather result from a large shift in the accumulation of carotene cyclization products in each mutant line (Figure 7). The increased L level in b1 b2 relative to the wild type indicates that the synthesis of β,ɛ-carotene branch carotenoids was enhanced when β-ring hydroxylation was blocked. Likewise, the synthesis of β,β-carotene branch carotenoids was enhanced by 60% relative to the wild type when ɛ-ring hydroxylation was blocked in lut1. Most significantly, in the lut1 b1 b2 triple mutant, in which all three hydroxylases were absent, the accumulation of β,ɛ- and β,β-carotenoids returned to a wild-type ratio (Figure 7).
Figure 7.
Accumulation of β,β-Carotene and β,ɛ-Carotene Branch Carotenoids in Leaves of the Wild Type and Selected Hydroxylase Mutants.
Total relative amounts of carotenoids accumulated through lycopene and the β,β- and β,ɛ-cyclization branches of the pathway are indicated by arrows. The width of each arrow is proportional to the percentage of each carotenoid accumulated. The arrows leading to lycopene are identical in width because the total amount of carotenoids produced in each line did not differ significantly (see Table 1). WT, wild type.
These results indicate that one response of the carotenoid pathway to the absence of one or more hydroxylases is to shift synthesis between the β,ɛ- and β,β-carotene branches of the pathway, a regulation that can occur only at the level of carotenoid cyclases (Figure 1). This regulation appears to be translational or post-translational, because mRNA levels for both the β- and ɛ-lycopene cyclase were essentially unchanged in all hydroxylase mutant genotypes (Figures 4C and 4D). A model put forth by Cunningham and Gantt (1998) proposed that β,ɛ- or β,β-carotene production is determined by the formation and/or stability of different carotenoid desaturase/cyclase complexes in the thylakoid membrane. Our data are consistent with the carotenoid hydroxylases participating in these proposed complexes to regulate the relative levels of β,ɛ- or β,β-carotene produced in leaf tissue. An alternative explanation is that the hydroxylase deficiencies differentially affect the substrate specificity/affinity of the cyclases by feedback regulation as a result of the altered xanthophylls accumulated in each mutant line.
Impact of Hydroxylase Deficiencies on Seed Carotenoid Composition
The seed carotenoid content of mutants also was analyzed to determine the penetration of each mutant phenotype in a nonphotosynthetic tissue. Lutein is the main carotenoid in both wild-type leaves and seeds (50 and 80%, respectively). β-Carotene accounts for ∼15% of the total carotenoids in wild-type leaf tissue (Table 1) but is present only in trace amounts in seeds (Table 2). This difference may be attributable to more efficient hydroxylation of β-carotene in seeds in the absence of the carotenoid binding proteins present in photosynthetic tissue or to enhanced turnover of β-carotene during seed development relative to leaf tissue. In general, the effect of β-hydroxylase mutations on seed carotenoid composition was similar to that in leaves. The b1 mutation had a more significant impact than b2, and the b1 b2 double mutant was more severe than either single mutant but still synthesized β-carotene–derived xanthophylls. The most surprising result from studies of mutant seeds is that unlike lut1 leaves, lut1 seeds did not accumulate zeinoxanthin, which is a signature phenotype for the lut1 mutation in leaves. It is possible that unlike that in leaf tissue, zeinoxanthin in lut1 seeds is degraded or unbound zeinoxanthin is more readily accessible to the residual ɛ-ring hydroxylation activity of LUT1 or a second ɛ-ring hydroxylation enzyme.
In contrast to leaf tissue, in which individual and total carotenoids do not differ statistically between ecotypes, Ws seeds accumulated more than twice the carotenoid level of Col seeds (Table 2). Interpretation of the impact of combined lut1, b1, and b2 mutations on seed carotenoid profiles was confounded by the segregation of other quantitative loci in the Col (lut1) and Ws (b1 and b2) parental ecotypes. The only genotype combinations that could be compared directly were lut1 b1 and lut1 b2, which were taken from seeds pooled from 20 independent homozygous F2 plants. As observed in leaf tissue, the b1 mutation had a more significant impact in the lut1 background than did b2. As in leaves, β-carotene–derived xanthophylls still were synthesized at reduced levels in b1 b2 and lut1 b1 b2 seeds, despite the absence of both known β-hydroxylases. These results indicate that an additional β-ring hydroxylation activity also is present in nonphotosynthetic tissue.
Conclusions
Our data reveal the multifunctional nature of carotenoid hydroxylases and provide insights into the overlapping functions and interactions of the three known hydroxylases in Arabidopsis and the regulation of carotenoid ring hydroxylations in vivo. The hydroxylase mutants also present additional examples of the remarkable flexibility of plant photosystems with regard to carotenoid composition and will be important tools for furthering our understanding of xanthophyll functions in plants, especially in response to high light stress. Future studies will focus on cloning of the LUT1 gene to determine the biochemical nature of this novel ɛ-hydroxylase and to assess its role in the synthesis of α- and β-carotene–derived xanthophylls.
METHODS
Identification of T-DNA Mutant Lines and Construction of Double and Triple Mutant Combinations
b1 and b2 Arabidopsis thaliana plants were identified from the T-DNA insertion mutant population available through the University of Wisconsin Biotechnology Center (http://www.biotech.wisc.edu/Arabidopsis/) using the screening strategy described previously (Krysan et al., 1999). The T-DNA border primer was used to sequence amplification products to determine the site of insertion into each gene (Figure 2A).
Homozygous b1 and b2 plants were crossed, and individual F1 seeds were grown and self-fertilized to obtain the F2 generation. The b1 and b2 genes are located on chromosome 4 and 5, respectively, and therefore segregate independently in an F2 population. The genotype of the F2 individuals was determined by PCR using primers specific for β-hydroxylase 1 and 2 as well as T-DNA border primer, and plants homozygous for both b1 and b2 were selected. The primer sequences used to screen and identify homozygous β-hydroxylase 1 and 2 T-DNA knockout mutants were as follows: β-hydroxylase 1 (forward, 5′-TTAAACGCTTTTCTGTCTGTT-ACGTCGTC-3′; reverse, 5′-TTGTGATTGTAGGTCACTCCCGATCATAG-3′) and β-hydroxylase 2 (forward, 5′-AATAGAAGTGGAGTGATTCGCTGT-CGATG-3′; reverse, 5′-AAGGACACATCGGTTCCAGAAGAAATAAG-3′).
Homozygous b1 and b2 single mutants were crossed to the ethyl methanesulfonate–mutagenized lut1-2 mutant. Homozygous lut1 b1 and lut1 b2 were selected from the segregating F2 population. Crosses then were performed between homozygous lut1 b1 and lut1 b2 plants, and lut1 b1 b2 triple mutants were isolated from the F2 population.
Reverse Transcriptase–Mediated PCR and TaqMan Real-Time PCR Assay
Total RNA was extracted from the wild type (Wassilewskija), b1, and b2 as described (Tian and DellaPenna, 2001). Ten micrograms of total RNA was used as a template, and first-strand cDNA synthesis was performed using the SuperScript preamplification system (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Subsequent amplification of the first-strand cDNA was performed using Taq DNA polymerase (Promega, Madison, WI) and a MJ Research DNA Engine thermal cycler (Waltham, MA). Primers specific for lycopene ɛ-cyclase (forward, 5′-CGAACAAAAGAATCTCGCC-3′; reverse, 5′-AAACCCTTGCCACAT-CCT-3′) were used as a control to test the integrity of all plant RNA samples. Primer pairs spanning T-DNA insertion sites in β-hydroxylase 1 and 2 open reading frames, β-hydroxylase 1 (forward, 5′-AACCGCCGT-TACATTCAAACC-3′; reverse, 5′-ACCTACAGGGAAACGCTTG-3′) and β-hydroxylase 2 (forward, 5′-TTCTCCGCAAACCACCCTATA-3′; reverse, 5′-CCAACTCTTCTTTTCCTCCCACTTC-3′), were used to determine whether full-length transcripts were synthesized in the corresponding mutants. The PCR program for amplification was as follows: 94°C for 3 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, and a final extension at 72°C for 10 min. The PCR products were analyzed on a 1% agarose gel.
The TaqMan real-time PCR assay (Perkin-Elmer Applied Biosystems, Foster City, CA) was adopted for quantification of steady state mRNA levels in wild-type and hydroxylase mutant plants. First-strand cDNA products from three independent reverse transcription reactions were pooled and used for TaqMan analysis. TaqMan probes and primers (Table 4) were designed from cDNA sequences of Arabidopsis β-hydroxylase 1, β-hydroxylase 2, lycopene β-cyclase, and lycopene ɛ-cyclase using Primer Express software (Perkin-Elmer Applied Biosystems). Reporter (5′ end) and quencher (3′ end) dyes for the TaqMan probes were 6-carboxy-fluorescein and 6-carboxy-tetramethyl-rhodamine, respectively. TaqMan real-time PCR assay conditions and analysis were as described by Tian and DellaPenna (2001).
Table 4.
Sequences of TaqMan Probes and Primers
| Gene | Primer/ Probe |
5′ to 3′ Sequence |
|---|---|---|
| β-Hydroxylase 1 | Forward | CTCGTGCACAAGCGTTTCC |
| Reverse | GGCGACCTTTCGGAGGTAA | |
| Probe | TGTAGGTCCCATCGCCGACGTCC | |
| β-Hydroxylase 2 | Forward | CCCATTGCCAACGTTCCTTA |
| Reverse | TGTCTGTGTGGTGTAGCTGGTG | |
| Probe | CTTCGAAAGGTCGCCGCCGC | |
| β-Cyclase | Forward | TCATTACTGGCACGGATTCTTG |
| Reverse | GAGCGACAACCCGAAGACC | |
| Probe | TCCAGGCTGTTTCTCCCGGAACTG | |
| ɛ-Cyclase | Forward | CCTTTGGTGCTGCCGC |
| Reverse | CTTCAGACAAAGATCTCACAACTGAAT | |
| Probe | AGCATGGTACATCCCGCAACAGGC |
Plant Material and HPLC Analysis of Pigment Content
Two wild-type Arabidopsis ecotypes (Columbia and Wassilewskija) and homozygous b1, b2, lut1, b1 b2, lut1 b1, lut1 b2, and lut1 b1 b2 plants were grown in soil at 120 to 150 μmol·m−2·s−1 under a 12-h-day/12-h-night cycle (21/18°C) in growth chambers. Carotenoids from 6-week-old leaf tissue were extracted in a 96-well format. A well-exposed leaf from each plant was selected, and leaf areas of the same size were excised using a cork borer. A leaf disc was allocated in every tube in a 96-well rack (Dot Scientific, Burton, MI), and 300 μL of acetone:ethyl acetate (60:40, v/v) was added, followed by 200 μL of distilled water. Three 4-mm glass beads (Fisher Scientific, Pittsburgh, PA) were added to each tube, and the plate was sealed, fastened onto a commercial paint shaker (H.E.R.O. Industries, Burnaby, British Columbia, Canada), and shaken for 5 min at the maximum speed. The 96-well plate then was centrifuged in a Sorvall RT centrifuge (Kendro Laboratory Products, Newtown, CT) at 3750g for 5 min to obtain phase separation. The resulting upper ethyl acetate phase was transferred to an Eppendorf tube and dried under vacuum. Once the samples were dried, they were resuspended in 100 μL of acetonitrile:water:triethylamine (900:99:1, v/v/v) for HPLC analysis. Seed carotenoid extractions were performed using similar procedures except that methanol:chloroform (2:1, v/v) was used as the extraction solvent and 500 ng of Tocol [2-methyl-2-(4,8,12-trimethyltridecyl)chroman-6-ol] standard was added to the extraction buffer and used to calculate recovery. HPLC separation of the carotenoids and quantitative analysis of carotenoid composition were performed as described (Tian and DellaPenna, 2001).
Chlorophyll content (chlorophyll a, chlorophyll b, and total) of 11-cm-diameter leaf discs were quantified spectrophotometrically as described (Porra et al., 1989). Briefly, 1 mL of dimethylformamide was added to each leaf disc, and extraction was performed at room temperature for 2 h in darkness. The extraction mixture was centrifuged, the supernatant was transferred to a 1-mL cuvette, absorption at 647 and 664 nm was measured, and chlorophyll a and b contents were calculated as chlorophyll a = 12 A664 − 3.11 A647 and chlorophyll b = 20.78 A647 − 4.88 A664.
Chlorophyll Fluorescence Measurements
Six-week-old wild-type and mutant Arabidopsis plants were dark adapted overnight before chlorophyll fluorescence measurements. In vivo chlorophyll fluorescence was measured using a pulse amplitude modulation fluorometer (FMS2; PP Systems, Haverhill, MA) using attached leaves that had not been shaded during growth. Fv/Fm is the maximum photochemical efficiency of photosystem II in the dark-adapted state. Nonphotochemical quenching was determined as Fm − Fm′/Fm′ (Lokstein et al., 2002).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
Accession numbers for the cDNA sequences mentioned in this article are AY113923 (β-hydroxylase 1), AY117225 (β-hydroxylase 2), U50739 (lycopene β-cyclase), and U50738 (lycopene ɛ-cyclase).
Acknowledgments
We thank the Wisconsin Knockout facility for screening the knockout mutants and the Arabidopsis Stock Center for providing us with mutant seeds. This work was supported by National Science Foundation Grant IBN-0131253.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011403.
References
- Bouvier, F., Keller, Y., D'Harlingue, A., and Camara, B. (1998). Xanthophyll biosynthesis: Molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.). Biochim. Biophys. Acta 1391, 320–328. [DOI] [PubMed] [Google Scholar]
- Britton, G. (1998). Overview of carotenoid biosynthesis. In Carotenoids: Biosynthesis and Metabolism, Vol. 3, G. Britton, S. Liaaen-Jensen, and H. Pfander, eds (Basel, Switzerland: Birkhauser), pp. 13–147.
- Cunningham, F.X., Jr., and Gantt, E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 557–583. [DOI] [PubMed] [Google Scholar]
- Davison, P.A., Hunter, C.N., and Horton, P. (2002). Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418, 203–206. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams, B., Gilmore, A.M., and Adams, W.W., III (1996). Ca-rotenoids. 3. In vivo function of carotenoids in higher plants. FASEB J. 10, 403–412. [DOI] [PubMed] [Google Scholar]
- Hirschberg, J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4, 210–218. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., and Walters, R.G. (1996). Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655–684. [DOI] [PubMed] [Google Scholar]
- Hundle, B.S., O'Brien, D.A., Beyer, P., Kleinig, H., and Hearst, J.E. (1993). In vitro expression and activity of lycopene cyclase and β-carotene hydroxylase from Erwinia herbicola. FEBS Lett. 315, 329–334. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.P., Björkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. [DOI] [PubMed] [Google Scholar]
- Lokstein, H., Tian, L., Polle, J.E., and DellaPenna, D. (2002). Xanthophyll biosynthetic mutants of Arabidopsis thaliana: Altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in photosystem II antenna size and stability. Biochim. Biophys. Acta 1553, 309–319. [DOI] [PubMed] [Google Scholar]
- Masamoto, K., Misawa, N., Kaneko, T., Kikuno, R., and Toh, H. (1998). β-Carotene hydroxylase gene from the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 39, 560–564. [DOI] [PubMed] [Google Scholar]
- Misawa, N., Nakagawa, M., Kobayashi, K., Yamano, S., Izawa, Y., Nakamura, K., and Harshima, K. (1990). Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172, 6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, P., Li, X.P., and Niyogi, K.K. (2001). Non-photochemical quenching: A response to excess light energy. Plant Physiol. 125, 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Grossman, A.R., and Björkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillip, D., Hobe, S., Paulsen, H., Molnar, P., Hashimoto, H., and Young, A.J. (2002). The binding of xanthophylls to the bulk light-harvesting complex of photosystem II of higher plants: A specific requirement for carotenoids with a 3-hydroxy-β-end group. J. Biol. Chem. 277, 25160–25169. [DOI] [PubMed] [Google Scholar]
- Pogson, B., McDonald, K.A., Truong, M., Britton, G., and DellaPenna, D. (1996). Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8, 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson, B.J., Niyogi, K.K., Björkman, O., and DellaPenna, D. (1998). Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 95, 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Rissler, H.M., and Pogson, B.J. (2001). Antisense inhibition of the β-carotene hydroxylase enzyme in Arabidopsis and the implications for carotenoid accumulation, photoprotection and antenna assembly. Photosynth. Res. 67, 127–137. [DOI] [PubMed] [Google Scholar]
- Ruban, A.V., Lee, P.J., Wentworth, M., Young, A.J., and Horton, P. (1999). Determination of the stoichiometry and strength of binding of xanthophylls to the photosystem II light harvesting complexes. J. Biol. Chem. 274, 10458–10465. [DOI] [PubMed] [Google Scholar]
- Sun, Z., Gantt, E., and Cunningham, F.X., Jr. (1996). Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J. Biol. Chem. 271, 24349–24352. [DOI] [PubMed] [Google Scholar]
- Tardy, F., and Havaux, M. (1996). Photosynthesis, chlorophyll fluorescence, light-harvesting system and photoinhibition resistance of a zeaxanthin-accumulating mutant of Arabidopsis thaliana. J. Photochem. Photobiol. B Biol. 34, 87–94. [DOI] [PubMed] [Google Scholar]
- Tian, L., and DellaPenna, D. (2001). Characterization of a second carotenoid β-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol. Biol. 47, 379–388. [DOI] [PubMed] [Google Scholar]