Abstract
The EVM1 protein encoded by Ectromelia virus is a member of a highly conserved family of poxvirus chemokine binding proteins that interfere with host immune surveillance processes. EVM1 is abundantly expressed early during mousepox infection and is able to selectively bind CC chemokines and inhibit their interactions with host receptors. Here, we characterize the interaction between EVM1 and the human and murine chemokines CCL3 (MIP-1α), CCL2 (MCP-1), and CCL5 (RANTES). Each of these CC chemokines binds EVM1 with 1:1 stoichiometry and equilibrium dissociation constants ranging from 29 pM to 20 nM. The interactions are characterized by rapid-association kinetics between acidic EVM1 and generally basic chemokines with half-lives enduring up to 30 min. The 2.6-Å crystal structure of EVM1 reveals a globular β sandwich with a large, sequence-conserved surface patch encircled by acidic residues on one face of the protein. To determine whether this conserved cluster of residues is involved in chemokine engagement, a structure-based mutational analysis of EVM1 was employed. Mapping of the mutational results onto the surface of EVM1 reveals that a cluster of five residues (I173, S171, S134, N136, and Y69) emanating from one β sheet is critical for CCL2 and CCL3 sequestration. Additionally, we find that the extended β2-β4 loop flanking this conserved cluster is also essential for high-affinity, lasting interactions with chemokines. This analysis provides insight into the mechanism of CC-chemokine inhibition employed by the poxvirus family of chemokine decoy receptors.
An essential component of host immune surveillance is the migration of immune cells into areas of injury or infection during the inflammatory response. Chemokines are a subset of cytokines that are primarily responsible for orchestrating this leukocyte recruitment. In response to a variety of signals, such as proinflammatory cytokines (interleukin-1, tumor necrosis factor alpha, and gamma interferon), chemokines are secreted from both endothelial and extravascular cells and are thought to establish concentration gradients by interacting with cell surface glycosaminoglycans (GAGs) (34). Subsequent binding of chemokines to their G-protein coupled receptors (GPCRs) on leukocytes stimulates multiple signaling pathways that ultimately result in chemotaxis (25). The induction of particular chemokines, in concert with the differential expression of chemokine receptors, determines which leukocytes migrate during inflammation. The known chemokine network is composed of 45 ligands and 19 receptors that are organized into four structural families, designated CC, CXC, CX3C, and C based on the arrangement of N-terminal cysteine residues in the ligands. Despite the low sequence similarity among chemokines, they adopt a remarkably conserved tertiary structure (25).
The importance of chemokines in the immune response to viral infection is emphasized by the discovery that large DNA viruses have evolved strategies to modulate the chemokine network. For example, poxviruses and herpesviruses encode altered versions of chemokines that can act as agonists or antagonists to host chemokine receptors (26). In addition, virally encoded seven transmembrane chemokine receptors have been shown to signal constitutively or sequester chemokines without signaling in virally infected cells (2). Another immune evasion strategy used by these viruses is based on the secretion of chemokine scavengers that are devoid of endogenous receptor homology. Murine gammaherpesvirus 68 encodes a novel broad-spectrum chemokine decoy, M3, capable of sequestering all four chemokine classes with high affinity (31, 40). The crystal structure of M3 in complex with the human CC chemokine CCL2 (hCCL2) has provided many of the details essential for understanding the mechanism of chemokine sequestration by this virally encoded protein (4). Another example is the M-T7 protein from myxoma virus, which functions as both a soluble gamma interferon receptor and a chemokine binding protein (22). The C-terminal domain of M-T7, which lacks significant sequence similarity to known proteins, is thought to engage the positively charged GAG binding regions of chemokines in a low-affinity interaction.
Members of the Orthopoxvirus and Leporipoxvirus genera encode an additional family of viral chemokine binding proteins (vCKBPs) that bind selectively and with high affinity to CC chemokines and block their interaction with cellular receptors (3, 16, 38). These abundantly secreted 35-kDa glycoproteins are expressed early during viral infection and are conserved in many viral species, including Cowpox virus, Ectromelia virus (EV), Variola virus (VAV), Rabbitpox virus, Myxoma virus (MV), and in some strains of Vaccinia virus (VV). A major biological consequence of vCKBP binding to chemokines appears to be the inhibition of CC chemokine-mediated recruitment of inflammatory cells into infected tissues during the initial phases of poxvirus infection (16, 23, 33). In vitro, MV, Rabbitpox virus, and VV (Lister strain) vCKBPs have been shown to inhibit CC chemokine-stimulated chemotaxis (hCCL3; 50% inhibitory concentration = 6 to 10 nM) and intracellular Ca2+ signaling of monocytes (hCCL3; Ki = 0.1 to 1 nM) (24).
To investigate the molecular basis of chemokine sequestration, two groups have used panels of CCL2 mutants to determine where this chemokine is contacted by VV Lister vCKBP (6, 36). Several surface residues that mediate binding are conserved among many CC chemokines, despite their varied sequences (36), and residues present in the C termini of chemokines, which may be important for GAG binding, do not appear to be involved in the interaction of CCL2 with vCKBP (3, 6). In addition, deletion of the N-terminal region before the first cysteine of CCL2, which is a key determinant for receptor signaling, had no significant effect on vCKBP binding (36). Most importantly, these studies have demonstrated that vCKBP contacts many of the same residues of CCL2 that are required for its interaction with the cognate host receptor CCR2b (6, 36). Although the structure of cowpox vCKBP has been determined, no detailed information is available regarding its chemokine binding site (11).
We sought to define the structural determinants of chemokine binding for a member of the vCKBP family encoded by Ectromelia virus. EV is a highly virulent rodent pathogen that causes the disease mousepox in mice. Like Variola virus, the causative agent of smallpox in humans, EV has a restricted host range and causes a severe generalized disease with a high mortality rate (10). The genetic similarity between EV and VAV, the common features of the resulting diseases, and the convenience of the mouse as a laboratory animal have led to the use of EV infections in mice as a model of smallpox (15). Furthermore, studies using EV have helped to elucidate critical mechanisms of viral pathogenesis and host defense (1, 21). EV (Moscow strain) encodes EVM1, a homolog of vCKBP that effectively sequesters CC chemokines and is known to inhibit the induction of multiple proinflammatory effects (39).
In order to discover the structural features of vCKBP that allow chemokine binding and inhibition, we have determined the crystal structure of EVM1 and refined it to 2.6-Å resolution. Structural analysis reveals a negatively charged, highly conserved patch that was probed by deletion and single-substitution mutagenesis to identify residues important for interaction with chemokines. The results define a cluster of residues on β sheet II and a large adjacent loop on EVM1 that is used to bind chemokines. Ultimately, this finding enables a better understanding of the mechanism of chemokine sequestration by this novel family of inhibitors.
MATERIALS AND METHODS
Expression and purification of EVM1.
Full-length EVM1 was amplified from Ectromelia virus (Moscow strain) genomic DNA, kindly provided by R. M. L. Buller, and cloned into the pFastBac1 vector (Invitrogen) in frame with a C-terminal thrombin cut site, a BirA consensus sequence, and a six-His tag (GSLVPRGSGLNDIFEAQKIEWHEHHHHHH). Recombinant baculovirus was generated in Sf9 insect cells as described in the Bac-to-Bac System manual (Invitrogen). EVM1 protein was produced by infecting suspension cultures of Hi5 cells in ExCell 405 serum-free medium (JRH Biosciences) with the recombinant baculovirus for 72 h. After centrifugation to remove the insect cells, the medium containing secreted EVM1 was concentrated and exchanged with buffer containing 50 mM sodium citrate, 50 mM sodium phosphate (pH 8.0), and 300 mM NaCl using a Centramate tangential flow system (Pall Life Sciences, Ann Arbor, MI). The protein was purified to homogeneity using Ni-nitrilotriacetic acid Superflow beads (QIAGEN, Valencia, CA) followed by size exclusion chromatography (SEC) on a HiLoad 16/60 Superdex 200 pg column (G.E. Healthcare). Protein purity was confirmed by silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE). The C-terminal tag was removed by incubating EVM1 with thrombin (Sigma) overnight at 4°C, and the protein was purified again by size exclusion chromatography. The molecular mass of the mature, thrombin-cleaved protein was determined by electrospray mass spectrometry at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
Protein crystallization and data collection.
Diffraction quality crystals of EVM1 were obtained by the hanging-drop vapor diffusion method. One-half microliter of protein at 13 mg/ml in 20 mM HEPES (pH 7.5), 0.01% (wt/vol) NaN3 was mixed with an equal volume of solution containing 2 M ammonium sulfate, 100 mM cacodylate (pH 6.2), and 3% (vol/vol) ethylene glycol and equilibrated at 20°C. Crystals, which appeared overnight, were cryoprotected by the addition of 1 μl of precipitant solution containing 30% (vol/vol) ethylene glycol to the 1-μl crystal drop before flash freezing at 100 K. Data were collected by the oscillation method in our laboratory using a Rigaku X-ray source and a Raxis IV detector (Rigaku/MCS, The Woodlands, TX). Diffraction intensities were indexed and scaled to 2.6 Å in the space group P21 using DENZO and SCALEPACK (HKL Suite, HKL Research, Inc., Charlottesville, VA (30).
Structure determination and refinement.
Initial phase estimates were obtained by molecular replacement of the coordinates of cowpox vCKBP (PDB code 1CQ3) using AMORE (CCP4 suite [13]). Map viewing and model building were performed using the program O (20), while refinement and water placement were carried out using CNS (9).
Computational analysis.
Graphical ribbon diagrams were created using RIBBONS (12), while molecular surfaces were generated using GRASP (29). Root mean square deviation (RMSD) values between different proteins were calculated for C-alpha positions using the DALI server (18). Solvent-accessible surface areas were calculated with the program NACCESS (19) using a 1.4-Å probe sphere. Sequence alignment was created with the program ALSCRIPT (5).
Size exclusion chromatography.
Analytical SEC was used to assess the oligomerization state of EVM1 alone and the stoichiometry of its interaction with hCCL2. The experiments were performed by FPLC using a Superdex 200 (10/300 GL) column (G.E. Healthcare) in buffer consisting of 20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.01% (wt/vol) sodium azide. The column was calibrated for molecular-weight determination using standards from LMW and HMW gel filtration calibration kits (G.E. Healthcare) according to the manufacturer's instructions. EVM1 (without tag) was injected onto the column at 1.4 mg/ml. The EVM1 complex was prepared by adding a twofold molar excess of hCCL2 in order to visualize the elution of the complex and free chemokine. The absorbance at 280 nM was monitored, and the fractions from each peak were pooled and concentrated in Centricon YM-3 centrifugal filter devices (Amicon) and analyzed by sodium dodecyl sulfate-PAGE using Coomassie blue stain.
Native PAGE.
Native gel shift experiments were performed using the PhastGel system (G.E. Healthcare) with 20% homogenous gels and native buffer strips, followed by silver staining. EVM1 (without tag) was prepared at a final concentration of 9 μM alone and in complex with human CCL2 or CCL3. Chemokine alone (50 μM) was loaded onto the gel, but it does not enter the gel due to its extremely basic nature. For complex formation, EVM1 was mixed with increasing concentrations of chemokine (2.3 μM, 4.5 μM, 9 μM, 18 μM, and 36 μM), and samples were incubated at room temperature for 20 min prior to gel loading. All proteins were loaded onto the gel in 20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.01% (wt/vol) sodium azide.
Preparation of EVM1 mutants.
Mutations were introduced into the EVM1 pFastBac1 construct by using a QuikChange site-directed mutagenesis kit (Stratagene). DNA sequencing was used to confirm sequences on both strands and exclude other incidental variations. Proteins were produced in the baculovirus expression system as described for wild-type (WT) EVM1. All mutants were expressed and secreted at levels comparable to those for the wild-type protein and eluted from the Superdex 200 column with similar profiles. All variants were assayed by native PAGE alone and with hCCL3 as an initial test for binding.
SPR binding analysis.
Surface plasmon binding (SPR) was used to directly measure the affinity and kinetics of chemokine binding by EVM1 and its variants. EVM1 (wild type or mutant) was immobilized on a CM5 chip (Biacore, Uppsala, Sweden) using standard amine coupling chemistry (BIAcore Amine coupling kit) to a level of 300 to 500 response units (RU) using a Biacore2000 biosensor (BIAcore, Uppsala, Sweden). A control flow cell was prepared by coupling EVM140, a non-chemokine binding protein related to VV A41L (27), to the chip at a similar level. Experiments were performed at 80 μl/min and 25°C using HBS-ET (10 mM HEPES [pH 7.5], 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20 [vol/vol]) as a running buffer. High flow rates and low levels of coupled protein were used to minimize the effects of mass transport (35). For all kinetics experiments, 160 μl of chemokine was injected over the experimental and control flow cells followed by a 4- to 6-min period to monitor dissociation before regeneration was achieved by injecting 80 μl 10 mM glycine (pH 3.4). Because of the long half-life (t1/2) of the EVM1 WT/hCCL3 complex, a control experiment in which dissociation of hCCL3 was monitored for an extended period of time (32 min) was performed. Sensorgrams from the control flow cell, in addition to sensorgrams obtained from injecting buffer alone, were subtracted from the binding curves to remove system noise. Each experiment was performed a minimum of three times using at least four different chemokine concentrations (e.g., 1, 2, 4, and 8 nM for hCCL3). The association (ka) and dissociation (kd) values were determined simultaneously by globally fitting sensorgrams for an entire range of chemokine concentrations to a 1:1 mass transport model with BIAevaluation 4.1 software (BIAcore). This global analysis was performed independently for each series of concentrations, the resulting values were averaged, and the standard deviation was calculated to reflect the experimental error. The apparent equilibrium dissociation constants (KDs) were determined from the mean kinetics values using the equation KD = kd/ka, and the error was propagated using the standard statistical treatment for division of standard deviations. SPR kinetics experiments were performed for WT EVM1 with human and mouse CCL2, CCL3, and CCL5, which were obtained from Peprotech Inc. (Rocky Hill, NJ), and for the EVM1 variants with human CCL3.
The binding kinetics for EVM1 variants with hCCL2 were not amenable to kinetics analysis, so an equilibrium binding approach was employed. EVM1 was immobilized as described above to a level of 800 to 1,000 RU. A mock-coupled control flow cell in which the sensor surface had been subjected to the coupling reaction in the absence of EVM1 was used as a control. Experiments were performed by injecting 40 μl of CCL2 at 20 μl/min over experimental and control flow cells. For the WT and the tested mutants (Y69R, S134R, N136W, S171W, S171Y, I173Y, I173R, and Δ51-65), each experiment was performed in triplicate using a range of CCL2 concentrations that spans 3 orders of magnitude. For example, 16 concentrations of CCL2 from 1 nM to 2 μM were used for WT, S134R, and N136W EVM1. For the other EVM1 variants, 16 concentrations from 12 nM to 200 μM were used. After subtraction of sensorgrams from the control flow cell and from buffer injections, the equilibrium binding levels were assessed as the average response units over the last 5 seconds of a 2-minute injection at a given CCL2 concentration. These RU values were plotted versus concentration and analyzed by nonlinear curve fitting of a 1:1 Langmuir isotherm model with BIAevaluation 4.1 software (BIAcore) to determine KD.
Protein structure accession number.
The coordinates for EMV1 have been deposited in the RCSB Protein Data Bank (accession code 2GRK).
RESULTS
EVM1 has multiple signal peptide cleavage sites.
The full-length EVM1 gene from the ectromelia virus Moscow strain, with its endogenous signal sequence and a six-histidine tag for purification, was expressed at high levels in a baculovirus expression system. The tag was removed from the purified protein by thrombin cleavage, and this material was used for subsequent studies. Electrospray mass spectrometry was used to identify the signal peptide cleavage site for the mature EVM1 protein. The spectra from two independent samples of EVM1 resulted in an abundant peak of 25,013 Da and an additional peak of 25,668 Da (data not shown), indicating that the insect cell-expressed protein was cleaved at two different sites. The mass of the more abundant peak (25,013 Da) suggests that mature EVM1 begins at residue Q23 with a calculated mass of 25,032 Da; however, the 19-Da discrepancy between the calculated and experimentally determined masses suggests that the N-terminal glutamine residue undergoes an additional posttranslational modification to pyroglutamate. The cyclization of N-terminal glutamine to pyroglutamate has been observed in other proteins. For example, pyroglutamate is essential for the chemotactic activity of some chemokines, and it has been shown to protect these proteins from protease degradation (8). The additional peak of 25,668 Da corresponds to mature EVM1 starting at residue I17, with a calculated mass of 25,671 Da. Interestingly, the SignalP 3.0 program predicted both of these signal peptide cleavage sites (7).
EVM1 is a monomer in solution and binds chemokines with a 1:1 stoichiometry.
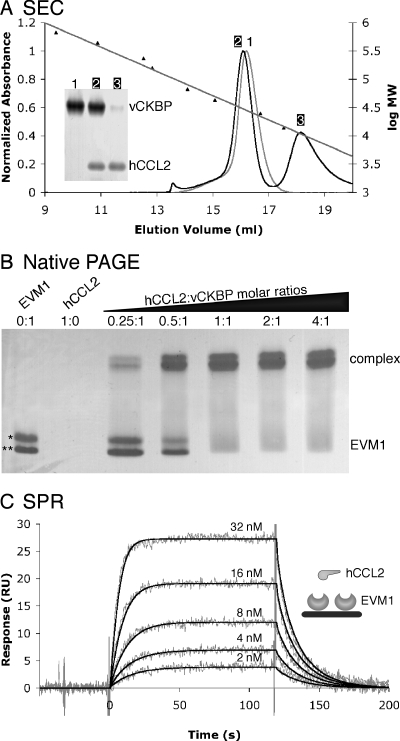
Size exclusion chromatography was used to determine the oligomerization state of EVM1 in solution and the stoichiometry of EMV1 in complex with chemokines. EVM1 eluted from the Superdex 200 column as a single peak with an apparent molecular mass of 28 kDa, suggesting that it exists in solution as a monomer (Fig. 1A). A similar behavior was observed for the related VV vCKBP (36). When EVM1 was mixed with hCCL2 in a 1:2 molar ratio, the initial peak containing EVM1 and hCCL2 had an elution volume slightly higher than that of EVM1 alone (∼30 kDa), while the excess hCCL2 eluted later, indicating the formation of a 1:1 complex. To further evaluate whether EVM1 binds hCCL2 with 1:1 stoichiometry, samples containing various molar ratios of hCCL2 to EVM1 were run on native PAGE (Fig. 1B). Two bands were observed for the unliganded EVM1 as a result of the distinct signal peptide cleavage sites. Both bands shifted upon complex formation, and a complete shift occurred at a 1:1 molar ratio. Similar results from SEC and native PAGE were observed for the EVM1/hCCL3 complex (data not shown).
FIG. 1.
EVM1 binds CC chemokines with 1:1 stoichiometry and high affinity. (A) SEC indicates that EVM1 is a monomer in solution that is able to bind a single chemokine. Elution profiles are shown for EVM1 alone (gray curve) and EVM1 mixed with a twofold molar excess of hCCL2 (black curve). The mean elution volumes for standard proteins used to calibrate the Superdex 200 10/300 GL column are shown with their linear fit versus log molecular weight (log MW). The inset gel shows that peak 1 contains EVM1 alone, peak 2 contains both EVM1 and hCCL2, and peak 3 contains excess hCCL2. (B) Native PAGE provides further evidence for the 1:1 stoichiometry. EVM1 runs as two bands due to multiple signal peptide cleavage sites (*, EVM1 amino acids 1 to 235; **, EVM1 amino acids 7 to 235), and hCCL2 does not enter the gel due to its high pI (∼9). A shift occurs when EVM1 binds to chemokine, and this shift is complete at a 1:1 molar ratio. (C) SPR analysis of hCCL2 binding to EVM1 immobilized on a CM5 chip. Shown are response curves for a typical chemokine titration experiment. The experimental curves (gray lines) were globally fit using a 1:1 mass transport model (black lines) to determine the kinetics parameters presented in Table 1. The inset shows a schematic representation of the experimental setup.
EVM1 binds CC chemokines with high affinity.
Binding studies with several members of the vCKBP family of proteins with CC chemokines demonstrated affinities in the picomolar to low nanomolar range (3, 16, 38). Although chemokine binding proteins from several strains of EV have been shown to bind and inhibit CC chemokines (2), no quantitative affinity measurements have been made for the EV member of this family (EVM1). The apparent binding kinetics and affinities of human and murine CCL2, CCL3, and CCL5 binding to surface-immobilized EVM1 were evaluated using SPR methods (Fig. 1C and Table 1). Strikingly, all six chemokines bind EVM1 with rapid kinetics (5.3 × 105 to 2.3 × 107 M−1 s−1), suggesting the occurrence of electrostatic enhancement between the highly acidic EVM1 and the very basic chemokines (37). The rates of complex dissociation are more varied, with half-lives ranging from 11 seconds to 34 min. EVM1 binds to human CCL3 with the highest affinity (KD = 29 ± 11 pM), exhibiting rapid-association kinetics [ka = (1.2 ± 0.4) × 107 M−1 s−1] and slow-dissociation kinetics [kd = (3.4 ± 0.6) × 10−4 s−1], resulting in the longest half-life of 34 min. To ensure that the slow dissociation of hCCL3 could be fit reasonably over the 6-min period used in the experiments, an experiment in which dissociation was monitored for 32 min was performed. The results were consistent within error, supporting the suitability of the standard measurements (data not shown). Compared to human CCL3, CCL5 and CCL2 bind with an 8-fold to 690-fold reduction in affinity. It is unclear which chemokine is the exact target of EVM1 in vivo, but all tested chemokines bind to EVM1 with affinities comparable to those of their cognate host receptors, 35 pM to 12 nM (17, 28, 32).
TABLE 1.
SPR analysis of human and murine chemokines binding to EVM1a
| Chemokine | ka (106) (M−1s−1) | kd (10−3) (s−1) | KD (nM) | t1/2 (s) |
|---|---|---|---|---|
| hCCL2 | 3.2 ± 0.6 | 66 ± 7 | 20 ± 4 | 11 |
| mCCL2 | 21 ± 5 | 29 ± 7 | 1.4 ± 0.5 | 24 |
| hCCL3 | 12 ± 4 | 0.34 ± 0.06 | 0.029 ± 0.011 | 2,039 |
| mCCL3 | 12 ± 1 | 1.4 ± 0.1 | 0.121 ± 0.015 | 495 |
| hCCL5 | 23 ± 9 | 5 ± 3 | 0.230 ± 0.170 | 139 |
| mCCL5 | 0.53 ± 0.02 | 3.3 ± 0.2 | 6.3 ± 0.5 | 210 |
Values for ka, kd, and KD are means ± standard deviations. KD = kd/ka; t1/2 = 0.693/kd.
Crystal structure of EVM1.
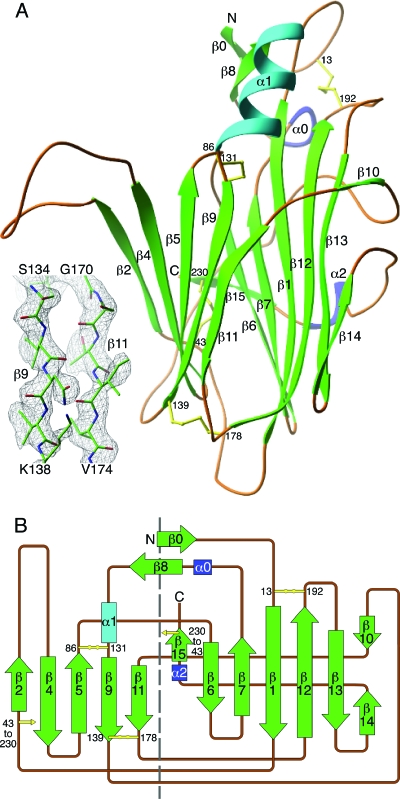
To understand the structural basis of chemokine binding, EVM1 was crystallized and the structure was determined. EVM1 crystals belong to a monoclinic space group, P21, with unit cell dimensions as follows: a, 49.2 Å; b, 55.8 Å; c, 85.8 Å; and β, 106.3°; there were two molecules per asymmetric unit. The crystal structure of the decoy was determined at 2.6 Å by molecular replacement using atomic coordinates from the closely related structure of cowpox vCKBP (1CQ3) as a starting model (11). The structure was built and refined to generate a final model (Fig. 2) that includes residues 8 to 231 with an Rcryst of 21.7% and an Rfree of 28.6% (Table 2). Four of the last six residues (GSLV), which are part of the C-terminal tag remaining after thrombin cleavage, were also visible in the electron density maps. In addition, the electron density for residues 51 to 62 in a large extended loop of EVM1 is relatively weak, especially in the second molecule of the asymmetric unit.
FIG. 2.
Crystal structure of EVM1. (A) The ribbon diagram is colored according to secondary structure (green β strands and cyan and blue helices), and the structural elements are numbered according to the cowpox vCKBP structure (11). The disulfide bonds are shown in yellow, with the cysteine positions labeled. The inset shows an example of the experimental electron density from the 2.6-Å composite omit map contoured at 1.2 σ, with the refined atomic model superimposed. (B) Topology diagram of the EVM1 structure. A dashed line separates the two β sheets of the sandwich.
TABLE 2.
Summary of data collection and refinement statistics for crystal space group P21
| Parameter | Value(s)a |
|---|---|
| Unit cell dimensions | |
| a (Å) | 49.2 |
| b (Å) | 55.8 |
| c (Å) | 85.8 |
| β (°) | 106.3 |
| Data processingb | |
| Resolution | 20.0-2.6 (2.72-2.60) |
| No. of observations to 2.6 Å/no. unique | 41,056/13,573 |
| Completeness (%) | 97.6 (96.4) |
| Rsym | 0.112 (0.426) |
| I/σ | 9.5 (2.4) |
| Refinementc | |
| Data range (Å) | 20.0-2.6 (2.76-2.60) |
| Reflections (F > 0) | 12,565 (1,773) |
| Completeness (%) | 90.3 (80.7) |
| Reflections in Rfree set | 640 (95) |
| Nonhydrogen atoms | 3,438 |
| Solvent molecules | 122 |
| RMSD for: | |
| Bond lengths (Å) | 0.007 |
| Bond angles (degrees) | 1.3 |
| Dihedral angles (degrees) | 26.0 |
| Improper angles (degrees) | 0.75 |
| Avg B factor for protein (Å2) | 41 |
| Avg B factor for water molecules (Å2) | 60 |
| Estimated coordinate error from Luzzati plot (Å) | 0.32 |
| Rcryst (%) | 21.7 (30.1) |
| Rfree (%) | 28.6 (39.6) |
| Ramachandran plot | |
| Most favored/additional (%) | 82.4/17.4 |
| Generously allowed/disallowed (%) | 0.2/0.0 |
Values in parentheses are for data in the highest resolution shell.
Statistics as defined in SCALEPACK.
Statistics as defined in CNS.
The structure of EVM1, a globular β sandwich composed of two β sheets, is very similar to that of its cowpox relative vCKBP (11), exhibiting a RMSD of 2.0 Å for 220 amino acids. The first β sheet (β sheet I) contains seven antiparallel β strands that are mostly solvent inaccessible due to two large loops (β9-β10 and β14-β15), which wrap around this half of the molecule. The second β sheet (β sheet II) has five solvent-exposed β strands and includes a prominently extended loop (β2-β4). The largest α helix (α1) is located above β sheet II and is adjacent to two small β strands (β0 and β8) and the N terminus of the molecule, while two 310 helices, α0 and α2, precede the β8 and β15 strands, respectively. Eight conserved cysteine residues form four disulfide bridges; one connects strands in β sheet I (C13 to C192); two connect strands in β sheet II (C86 to C131 and C139 to C178), and the final disulfide bridge connects the two β sheets (C43 to C230).
EVM1 crystallizes with two molecules in the asymmetric unit (RMSD, 0.5 Å) of the monoclinic unit cell. Carfi et al. also observed two cowpox vCKBP monomers within the asymmetric unit (11); however, the interactions appear to be distinct for the two poxvirus proteins. The cowpox vCKBPs interact between the β11 strand of one molecule and a strand (β3) that forms in the large β2-β4 loop of the second molecule. This head-to-tail configuration results in N termini with opposite orientations and a buried surface area of 1,797.54 Å2 (9). In contrast, a tail-to-tail interaction occurs between the two EVM1 monomers in which the β11 and β14 strands along the edge of one β sandwich contact the corresponding strands of the second molecule. The total buried surface area at the interface of the two EVM1 monomers is 1,578.48 Å2. For EVM1, the N termini splay at an angle of about 100°, while the two β2-β4 loops come together and form a smaller contact area. Notably, the EVM1 extended β2-β4 loop does not form the β3 strand seen in the cowpox decoy receptor, and it is significantly different in position by an average of 10 Å for the α carbon atoms of residues S52 to V62. All available solution studies of poxviral vCKBPs indicate that the protein is monomeric both free and bound to chemokines, suggesting that these crystallization results are unlikely to be of any functional significance (11, 36).
Interestingly, EVM1 is a distant structural relative of the N-terminal domain of M3, the chemokine binding protein encoded by gammaherpesvirus 68. The crystal structure of M3 reveals a two-domain protein that forms a head-to-tail dimer (4). Chemokines are able to bind in the cleft at the dimer interface, making contacts with loops on the N-terminal domain of one monomer and on the C-terminal domain of the other. The M3 N-terminal domain is a 210-residue β sandwich that is completely distinct from EVM1 in amino acid sequence and β-strand connectivity. However, EVM1 and the M3 N-terminal domain can be structurally aligned, resulting in 81/199 matched residues with an RMSD of 3.3 Å and 4% sequence identity. Based on this alignment, the face of the M3 domain that interacts with chemokines is analogous to β sheet II of EVM1. M3 primarily uses the s2b-s3 loop of the N-terminal domain to contact CCL2; this roughly corresponds to the loops on the bottom of the EVM1 structure (e.g., the β11-β12 loop [Fig. 2A]). The large, acidic β2-β4 loop of EVM1 is equivalent to the smaller s5-s6a loop in M3, which is buried at the dimer interface.
Structure-based prediction of the EVM1 chemokine binding site.
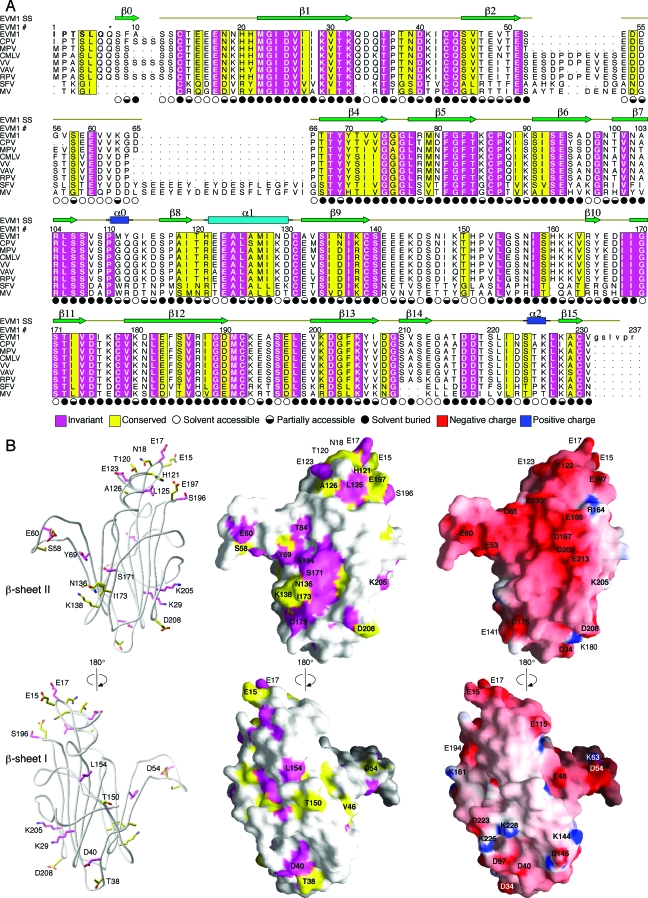
Further structural analysis examining sequence conservation was performed to provide clues about the chemokine binding interface on EVM1. The sequence alignment of EVM1 to six orthopox and two more-distantly related leporipox chemokine binding proteins (Fig. 3A) was used to identify regions of conservation that were mapped onto the surface of the EVM1 structure (Fig. 3B). While this family of proteins has a high degree of sequence similarity (35% to 84% identity), most conserved residues are buried in the core of the β sandwich and therefore would appear to be important for the overall fold of this family of proteins. One region of highly conserved, surface-accessible side chains is located near the N terminus of the molecule and includes residues of the β0-β1 loop, the α1 helix, and the β12-β13 loop, many of which are acidic (E15, E17, E123, and E197). The more prominent cluster of conserved, surface-exposed side chains maps to β sheet II and contains mostly polar residues and one notable hydrophobic residue (I173). This region of conservation is located between a hydrophobic groove that runs along the edge of the β sandwich and the extended β2-β4 loop. Interestingly, although it contains three conserved amino acids (D54, S58, and E60), the β2-β4 loop is the most variable region of the vCKBPs (Fig. 3A). EVM1 has one of the smaller loops in the family (14 residues), while many of the other orthopox members (e.g., VV and VAV) have an insertion of up to 10 amino acids, and the more-distantly related leporipox viruses (e.g., Shope fibroma virus and MV) have approximately 30 additional residues.
FIG. 3.
Sequence conservation can be used to predict the chemokine binding site on EVM1. (A) The sequence of EVM1 is aligned with eight other members of this chemokine binding protein family from other poxviruses. CPV, cowpox; MPV, monkeypox; CMLV, camelpox; VAV, variola virus; RPV, rabbitpox virus; SFV, Shope fibroma virus. The secondary structure of EVM1 is shown above the alignment. Regions of sequence conservation are highlighted. Magenta, invariant residues; yellow, residues with a conservation index of 7 or greater as determined by ALSCRIPT (5). Symbols indicating the solvent accessibility of each side chain are shown under the alignment. Filled circles, <30% accessible; half-filled circles, 30 to 60% accessible; open circles, >60% accessible. The asterisk above residue 7 indicates the position of the first residue of the molecule, a pyroglutamate, when the second signal peptide cleavage site is used. (B) Ribbon diagram (left) highlights the conserved, surface-exposed side chains suggesting a potential binding site. Sequence conservation is mapped to the EVM1 surface (middle). Electrostatic surface potential of EVM1 as calculated by GRASP (29) is mapped to the surface (right). Surface colors are contoured from red (−15 kT) to blue (15 kT).
Chemokines in general are very basic, and certain clusters of basic residues are known to be important for interactions with GAGs, GPCRs, and viral decoy receptors (25). We reasoned that an examination of the electrostatic properties of EVM1 could be insightful for locating the chemokine interaction site. The surfaces and electrostatic potentials calculated with GRASP (29) reveal that the entire face of β sheet II has a strong electronegative character, whereas the opposite side of the molecule has no remarkable electrostatic properties (Fig. 3B). Based on this finding, we hypothesized that β sheet II is involved in chemokine binding. While many of the acidic residues on β sheet II are not conserved spatially, the large concentration of acidic residues on β sheet II is maintained in vCKBP family members. This suggests that the exact position of charged residues may not be particularly important for general chemokine binding. Alternatively, the spatial positioning of acidic residues on vCKBP family members may alter chemokine binding specificity.
Identification of the EVM1 chemokine binding site.
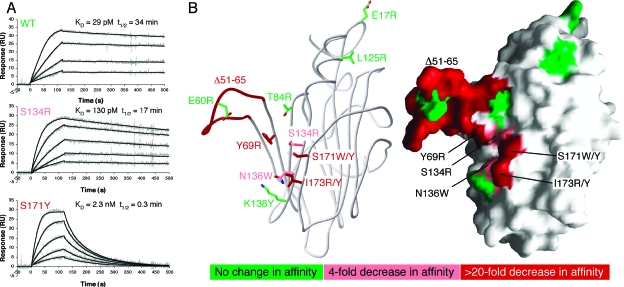
The cluster of sequence conservation surrounded by a concentration of acidic residues makes β sheet II a likely candidate for the chemokine binding site. To test this hypothesis, kinetics and affinity parameters were determined for a series of EVM1 single-point mutants and for one deletion mutant. The strategy for mutant design was to substitute the surface-exposed conserved residues with large, bulky side chains, often positively charged, with the intention of sterically and electrostatically hindering chemokine binding. Single mutations were introduced around the N-terminal conserved patch and on β sheet II. In addition, the entire β2-β4 loop (Δ51-65) was replaced with a short GSG linker. All mutant EVM1 proteins were secreted from insect cells using the baculovirus expression system. Purified variants migrated in SEC experiments similarly to wild-type EVM1 and behaved like wild-type EVM1 on a native gel, suggesting that they were properly folded. SPR binding experiments were performed to assess binding of EVM1 variants to hCCL2 and hCCL3, the highest affinity chemokine tested (Table 1). Table 3 and Fig. 4 summarize the results of kinetics and affinity measurements.
TABLE 3.
SPR results for binding of chemokines to mutant EVM1
| EMV1 variant | Binding parameters for hCCL3a
|
Affinity for hCCL2 (KDb [nM]) | ||||
|---|---|---|---|---|---|---|
| ka (106M−1s−1) | kd (10−4s−1) | KD (pM) | KD (mut)/KD (wt) | t1/2 (min) | ||
| WT | 12 ± 4 | 3.4 ± 0.6 | 29 ± 11 | 34 | 44 ± 2 | |
| E17R | 15 ± 3 | 4 ± 1 | 26 ± 12 | 0.9 | 29 | NDc |
| E60R | 7 ± 1 | 4 ± 1 | 57 ± 18 | 2 | 29 | ND |
| Y69R | 9 ± 1 | 140 ± 20 | 1,500 ± 300 | 52 | 0.8 | >1,000 |
| T84R | 9 ± 1 | 4.2 ± 0.4 | 49 ± 9 | 1.7 | 28 | ND |
| L125R | 22 ± 8 | 4 ± 1 | 18 ± 8 | 0.6 | 29 | ND |
| S134R | 6 ± 1 | 7.0 ± 0.8 | 130 ± 30 | 4 | 17 | 500 ± 30 |
| N136W | 5 ± 1 | 7 ± 1 | 140 ± 40 | 5 | 17 | 240 ± 20 |
| K138Y | 5 ± 1 | 1.3 ± 0.4 | 25 ± 8 | 0.9 | 89 | ND |
| S171W | 18 ± 3 | 130 ± 20 | 720 ± 160 | 24 | 0.9 | >1,000 |
| S171Y | 17 ± 2 | 390 ± 50 | 2,300 ± 400 | 79 | 0.3 | >1,000 |
| I173Y | 12 ± 2 | 310 ± 20 | 2,600 ± 500 | 90 | 0.4 | >1,000 |
| I173R | 3.5 ± 0.7 | 470 ± 60 | 13,000 ± 3,000 | 448 | 0.3 | >1,000 |
| Δ51-65 | 2.2 ± 0.6 | 230 ± 30 | 10,000 ± 3,000 | 344 | 0.5 | >1,000 |
Values for ka, kd, and KD are means ± standard deviations. KD = kd/ka; t1/2 = 0.693/kd.
KD from equilibrium binding analysis.
ND indicates measurements that were not determined.
FIG. 4.
Mutational analysis of EVM1 reveals the chemokine binding surface. (A) SPR analysis of hCCL3 binding to immobilized EVM1 variants. Three representative sensorgrams are shown for WT EVM1, the S134R mutant, which has a small effect on KD, and the S171R mutant, which has a more drastic effect. The experimental curves (gray lines) were globally fit using a 1:1 mass transport model (black lines) to determine the kinetics parameters presented in Table 3. (B) Ribbon (left) and surface (right) diagrams highlight the residues targeted during mutagenesis. The Δ51-65 β2-β4 loop mutation involves the deletion of residues 51 to 65, replaced by GSG. All other mutations are single-amino-acid substitutions. Residues are color coded according to the effect of each mutation on the affinity for hCCL3 relative to that for the WT. Green, no change in affinity; pink, mild decrease in affinity (4- to 5-fold); red, large decrease in affinity (24- to 448-fold).
WT EVM1 binds to human CCL3 with a rapid association rate and a relatively slow dissociation rate, resulting in an apparent affinity of 29 pM. Of all of the single-amino-acid mutations, I173R produced the largest change (448-fold) in the binding affinity of hCCL3. A different mutation in the same position (I173Y) had a less drastic effect (90-fold). These significant reductions in affinity are due almost entirely to an increased dissociation rate, especially for I173Y, which has an on rate identical to that of the WT. The I173R mutation slightly decreases the association rate, presumably by introducing a positive charge in an otherwise electronegative environment. This suggests that electrostatics could be important for establishing a rapid on rate. It is also possible that a charged or polar substitution at this position, the only surface-exposed hydrophobic residue on β sheet II, interferes with an important hydrophobic interaction in the complex, thereby destabilizing the complex and increasing the rate of dissociation.
Mutation of the nearby amino acid S171 to tyrosine produced the third-largest reduction (79-fold) in the interaction with hCCL3, even though the substitution maintained the polar and uncharged nature of the residue. When a large hydrophobic tryptophan residue was substituted in this position, the reduction in affinity was less striking (24-fold) than that for the WT, suggesting that the hydrophobic nature of the bulky residue helped to stabilize the complex. Mutation Y69R on the β4 strand is another point mutation with a dramatically reduced affinity (52-fold) compared to that for the WT. Although a positively charged arginine was substituted in this position, the on rate was not affected. A common feature of these five key mutations (I173R/Y, S177Y/W, and Y69R) is that their decreased affinities for CCL3 are a result of increased dissociation rates, such that the half-lives for these mutant complexes are all less than 1 min, compared to 34 min for the WT.
Additional mutations that had a lesser effect on hCCL3 binding include S134R and N136W. These substitutions displayed fourfold and fivefold reductions in affinity, respectively, which is a function of decreased on rates and increased off rates. Although the differences in affinity are minor, the half-lives for these mutations (17 min) are half of that of the WT, implying that these residues are nevertheless important determinants of the chemokine binding interface.
The boundaries of the chemokine binding surface on EVM1 can be more clearly defined by those mutations that had little effect on hCCL3 binding. Mutations E17R and L125R, located near the N terminus of EVM1, had no significant effect on the measured rate constants or binding affinity, suggesting that this conserved region of the molecule is not critically important for chemokine sequestration. Based on our results, the multiple signal peptide cleavage sites that we have identified at the N terminus of EVM1 should not be near the binding site and therefore are unlikely to affect chemokine binding. Similarly, the affinity of hCCL3 for EVM1 variants T84R, E60R, and K138Y, which surround the putative binding site, did not change relative to that for the WT. Although the mutation K138Y does not change the overall affinity for hCCL3, it does increase the off rate to 89 min, almost three times that of the WT.
The deletion of the entire β2-β4 loop (Δ51-65) resulted in the second-greatest decrease in affinity (344-fold) for hCCL3. Similar to the kinetics for the I173R mutation, the association rate is decreased to the lowest for any mutant (ka = 2.2 × 106 M−1 s−1) and the dissociation rate is increased such that the half-life is less than a minute. We surmise that sequence-independent interactions involving the loop are potentially important for chemokine binding, as the main chain has considerable solvent exposure, and mutation of a single invariant glutamate residue in the loop (E30R) had no discernible binding effect. Further, the β2-β4 loop does not appear to be responsible for CC-chemokine selectivity, since the loop deletion mutant did not acquire the capacity to bind other chemokine classes, for example, CXCL8 (interleukin-8) or XCL1 (lymphotactin) (data not shown).
In order to assess the role of the EVM1 binding site residues in a more general chemokine binding context, we measured the interaction of human CCL2 with the EVM1 variants. Equilibrium binding experiments were used to determine affinities due to difficulties obtaining accurate kinetics fits to the mutant binding data. This alternative method revealed a WT affinity (KD = 44 ± 2 nM) about twofold lower than the affinity calculated from the kinetics analysis (KD = 20 ± 4 nM). In agreement with the CCL3 results, the EVM1 S134R and N136W mutations had significantly decreased affinities for CCL2, with KDs of 500 ± 30 nM and 240 ± 20 nM, respectively (Table 3). Interestingly, the scales of deviation from the WT for the S134R and N136W mutants are comparable for both chemokines; CCL3 shows a 4- to 5-fold decrease in affinity, while CCL2 shows a 5- to 11-fold decrease. The affinities of the other EVM1 variants we tested (Y69R, S171W, S171Y, I173Y, I173R, and Δ51-65) were all too low to be accurately measured. Nevertheless, we estimate that the affinities of all of these variants for CCL2 are at least 2 orders of magnitude weaker than the affinity of the WT for CCL2.
Thus, our mutational analysis indicates that the same EVM1 binding interface residues are employed for both CCL2 and CCL3 engagement despite the fact that CCL3 binds with affinity approximately 3 orders of magnitude higher and that these two CC chemokines share only 40% sequence identity. Mapping of our mutational analysis to the surface of the EVM1 structure reveals a cluster of five residues (I173, S171, S134, N136, and Y69) on the face of β sheet II that, along with the flexible β2-β4 loop, compose the general chemokine interaction surface of the decoy receptor (Fig. 4B).
DISCUSSION
Chemokines are crucial mediators of the immune response, and their role in viral infection is underscored by the many strategies that viruses have evolved to interfere with their functions. The large DNA viruses, such as poxviruses and herpesviruses, have acquired and modified host genes for chemokines and their GPCRs but also encode novel soluble proteins that broadly inhibit chemokines in the extracellular milieu. Analyses of decoy receptor interactions with chemokines have provided useful insights into structural aspects of viral immune evasion and chemokine biology. In addition, such studies could facilitate the design of therapeutics for the treatment of inflammatory diseases. In the present study, we have defined the chemokine binding surface of EVM1, a member of a novel family of poxvirus chemokine decoy receptors. Structural determination and site-directed mutagenesis were employed to determine the interaction site of EVM1 with the human CC chemokines CCL2 and CCL3. In total, mutations of five clustered residues in the conserved, surface-accessible patch on β sheet II led to diminished chemokine binding (Fig. 4B). The binding interface we have mapped is dominated by three residues: I173 and S171 on the β11 strand and Y69 on β4. Located between these three key amino acids are residues of significant but lesser importance for the interaction with CCL2 and CCL3: S134 and N136 on the β9 strand. An additional element necessary for high-affinity binding is the β2-β4 loop, which flanks the critical determinants on β sheet II.
An intriguing feature of the vCKBP family of proteins is their complete lack of sequence or structural similarity with known chemokine receptors (11). Despite these differences, vCKBPs and the cellular receptors are able to bind overlapping surfaces by engaging the same key residues on chemokines. The most extensive mutagenesis studies have used human CCL2, but much of what has been learned can be translated to other chemokines. CCL2 interacts with its cellular receptor CCR2b via two clusters of mostly basic residues (R24, K35, K38, K49, and Y13) (17). The key residues in this interaction, R24 and Y13, are located at opposite ends of CCL2 and are separated by a hydrophobic groove. In the proposed model, the receptor N terminus of CCR2b lies along the hydrophobic groove of CCL2 in an extended fashion. This arrangement places the DYDY motif of CCR2b near the basic cluster involving R24 and K49 of CCL2 and orients Y13 and the N terminus for productive signaling by the receptor (17).
Two subsequent studies determined that CCL2 residues R18, R24, and Y13 are also critical for binding to VV vCKBP (6, 36), while many other residues, both basic and hydrophobic, make less substantial contributions. The R24A mutation produced a large (17-fold) reduction in interaction with VV vCKBP; however, a charge reversal at the same position (R24E) was found to virtually abolish binding, suggesting that R24 forms a critical interaction with a negatively charged region of vCKBP (36). Based on the results of our study, the key arginine residues (R18 and R24) of CCL2 could easily interact with multiple acidic residues surrounding the patch of conserved residues at the binding site. The lack of conservation of specific acidic sites suggests that different acidic residues may be used by different chemokines, whose exact placements of basic residues also vary.
Studies of protein-protein interfaces have revealed the critical importance of buried hydrophobic residues in establishing long-lived stable complexes (41). Our mutational analysis indicates that this may well be the case for the EVM1/CCL3 interaction, since the I173 mutation has the single greatest effect on complex half-life. Among Orthopoxvirus vCKBPs, the equivalent position is invariantly isoleucine, whereas the similarly hydrophobic leucine residue is found in Leporipoxvirus vCKBPs. Perhaps analogous to the hydrophobic residues at the N termini of host chemokine receptors, I173 may be important for interacting with the hydrophobic groove of chemokines. In addition, immediately adjacent to I173, EVM1 has its own highly conserved hydrophobic pocket along the interface of the two β sheets. This region is probably involved with ligand binding; however, our mutagenesis approach cannot effectively address this because binding is likely to include backbone atoms and hydrophobic interactions with structurally important apolar residues within the cleft.
Comparison of EVM1 to M3, the chemokine binding protein encoded by gammaherpesvirus 68, can provide insight into common features of chemokine sequestration by viruses, in addition to highlighting novel mechanisms exploited by EVM1. Currently, M3 is the only structure of a viral chemokine binding protein bound to a chemokine, human CCL2 (4). M3 exists in solution as a head-to-tail dimer; the N-terminal domain of one monomer interacts with the C-terminal domain of the second monomer, forming of a cleft between the domains. One chemokine can bind to each cleft, resulting in a 2:2 complex. Unlike EVM1, M3 binds and inhibits chemokines from all four structural classes. This promiscuity is attributed to conformational plasticity (i.e., structural rearrangements between the domains) and the use of flexible loops as primary contact regions. Instead of using flexible loops on an oligomeric framework for ligand binding, EVM1 uses a rigid scaffold secured by two conserved disulfides that hold the strands of β sheet II together. The intrinsic rigidity of the binding interface may preclude EVM1 from binding to multiple classes of chemokines, a trade-off that may be necessary for developing high-affinity, lasting interactions with select members of the CC class.
The C-terminal domain of M3 forms a β-strand interaction with CCL2 that is analogous to the interface of the CCL2 homodimer and consequently contacts Y13, a key residue at the binding interface of CCL2 with CCR2b and EVM1 (4, 17, 36). M3 has a proline residue (P272) that packs up against the invariant disulfide bond of chemokines, thereby mimicking the interaction of P8 at the CCL2 dimer interface. In addition to several other CC chemokines, CCL3 has a proline at a similar position, which may be involved in dimerization. EVM1 does not have such a surface-exposed proline, so we presume that it is not utilizing this same mimicry strategy but has come up with a distinctive mechanism for chemokine engagement.
The N-terminal domain of M3 is especially acidic and binds to the most basic patch of CCL2 involving R18, K19, R24 and K49, while also making extensive hydrophobic contacts with the hydrophobic groove of CCL2 (4). Interestingly, our structural alignment demonstrated that the surface of the M3 N-terminal domain that interacts with chemokines is analogous to β sheet II of EVM1, which we now know contains the chemokine binding site. While M3 primarily uses the acidic s2b-s3 loop of the N-terminal domain to contact CCL2, the corresponding loops on the bottom of the EVM1 structure (e.g., the β11-β12 loop [Fig. 2A]) are not expected to be important for chemokine binding. In addition, the acidic β2-β4 loop of EVM1 that is critical for chemokine binding is equivalent to the s5-s6a loop in M3, which is buried at the dimer interface. Although acidic loops play a prominent role in chemokine sequestration by both decoy receptors, the positions of the loops on the β-sandwich structures are distinct.
Electrostatics play an important role in molecular recognition by enhancing the association rates of proteins and by permitting complexes to attain proper binding orientations (37). Like M3, EVM1 is an extremely acidic protein (pI ∼4.6), with a significant cluster of acidic residues around the binding site on β sheet II. Both decoy receptors appear to target the basic patches of residues that chemokines have on their surfaces to mediate receptor interactions (4, 36). We speculate that this electrostatic complementarity at the binding interface mediates the extremely rapid association rates that are observed for both M3 and EVM1 with chemokines; however, this hypothesis has not yet been experimentally addressed.
Although the β2-β4 loop is the most divergent region of EVM1 compared to related vCKBPs, it appears to be critical for both rapid complex formation and slow dissociation. We speculate that the concentration of charge and the flexibility of the loop allow it to lure in chemokines via electrostatic effects, resulting in a very rapid on rate. Once bound, the loop may wrap around the ligand to neutralize some of its surface charge, thereby stabilizing the complex and ensuring a long half-life. Thus, the vCKBP β2-β4 loop can perhaps be thought of as a thumb on a hand, gripping chemokines to the β-sheet II palm where we have mapped the other critical determinants for chemokine sequestration.
The identification of a binding surface on EVM1 has implications for this entire family of chemokine binding proteins. We envision that the initial interaction of chemokines with EVM1 involves a rapid nonspecific electrostatic attraction. Once a favorable electrostatic environment is formed, the interactions necessary to form a stable complex will select for CC chemokines, although the determinants of CC-chemokine specificity remain unclear. One possible scenario involves binding of the chemokine N loop across β sheet II such that the N terminus is near the EVM1 β strand and the hydrophobic groove of the chemokine interacts with the surface of the β sheet. The β2-β4 loop could contact the chemokine β1 strand, while acidic residues around the β11 strand at the edge the β sandwich could interact with key basic residues (18 and 24) on the chemokine. Alternatively, our data would support a model in which the N-terminal region of the chemokine N loop forms an antiparallel β-strand interaction with the EVM1 β11 strand, reminiscent of the chemokine interaction with the C-terminal domain of M3 (4). In this case, the functionally important chemokine residue 13, an aromatic or hydrophobic amino acid, would be situated in the hydrophobic pocket at the edge of the EVM1 β sandwich and the invariant dicysteine residues would be positioned directly adjacent to the functionally critical I173 of EMV1. The N loop could extend across the top of β sheet II, allowing the key basic residues, R18 and R24 of CCL2, to interact with acidic residues near the α1 helix and on the β2-β4 loop, respectively. In agreement with past mutagenesis studies (6, 36), both scenarios allow the N loop, the β3 strand, and the hydrophobic groove of chemokines to make extensive contacts with the surface of EMV1 β sheet II.
Members of the vCKBP family possess the unique ability to bind selectively to CC chemokines with high affinity. The results presented here represent the only study thus far that examines the structural determinants for chemokine sequestration by a member of this novel family, EVM1. The identification of the EVM1 binding site will facilitate the design of more structure-based and library-based EVM1 variants, perhaps with altered specificities, which can be used as reagents to examine the roles of individual chemokines in specific experimental systems or to dissect the vast chemokine networks in more-complex models of viral infection and disease. Additionally, in a model of allergic airway hyperreactivity, VV vCKBP effectively ameliorated some aspects of the disease (14). Therefore, chemokine inhibitors based on the newly defined features of the EVM1 binding surface could be useful in the design of therapeutics for the treatment of similar inflammatory conditions.
Acknowledgments
We thank Grant Nybakken and Vess Mitaksov for comments on the manuscript.
This work was supported by Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE) grant U54-AI05716003 and NIH grant R01-AI05142604 to D.H.F.
REFERENCES
- 1.Alcami, A. 2003. Structural basis of the herpesvirus M3-chemokine interaction. Trends Microbiol. 11:191-192. [DOI] [PubMed] [Google Scholar]
- 2.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcami, A., J. A. Symons, P. D. Collins, T. J. Williams, and G. L. Smith. 1998. Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J. Immunol. 160:624-633. [PubMed] [Google Scholar]
- 4.Alexander, J. M., C. A. Nelson, V. van Berkel, E. K. Lau, J. M. Studts, T. J. Brett, S. H. Speck, T. M. Handel, H. W. Virgin, and D. H. Fremont. 2002. Structural basis of chemokine sequestration by a herpesvirus decoy receptor. Cell 111:343-356. [DOI] [PubMed] [Google Scholar]
- 5.Barton, G. J. 1993. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6:37-40. [DOI] [PubMed] [Google Scholar]
- 6.Beck, C. G., C. Studer, J. F. Zuber, B. J. Demange, U. Manning, and R. Urfer. 2001. The viral CC chemokine-binding protein vCCI inhibits monocyte chemoattractant protein-1 activity by masking its CCR2B-binding site. J. Biol. Chem. 276:43270-43276. [DOI] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Blaszczyk, J., E. V. Coillie, P. Proost, J. V. Damme, G. Opdenakker, G. D. Bujacz, J. M. Wang, and X. Ji. 2000. Complete crystal structure of monocyte chemotactic protein-2, a CC chemokine that interacts with multiple receptors. Biochemistry 39:14075-14081. [DOI] [PubMed] [Google Scholar]
- 9.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 10.Buller, R. M., and G. J. Palumbo. 1991. Poxvirus pathogenesis. Microbiol. Rev. 55:80-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carfi, A., C. A. Smith, P. J. Smolak, J. McGrew, and D. C. Wiley. 1999. Structure of a soluble secreted chemokine inhibitor vCCI (p35) from cowpox virus. Proc. Natl. Acad. Sci. USA 96:12379-12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carson, M. 1997. Ribbons. Methods Enzymol. 277:493-505. [PubMed] [Google Scholar]
- 13.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 14.Dabbagh, K., Y. Xiao, C. Smith, P. Stepick-Biek, S. G. Kim, W. J. Lamm, D. H. Liggitt, and D. B. Lewis. 2000. Local blockade of allergic airway hyperreactivity and inflammation by the poxvirus-derived pan-CC-chemokine inhibitor vCCI. J. Immunol. 165:3418-3422. [DOI] [PubMed] [Google Scholar]
- 15.Esteban, D. J., and R. M. Buller. 2005. Ectromelia virus: the causative agent of mousepox. J. Gen. Virol. 86:2645-2659. [DOI] [PubMed] [Google Scholar]
- 16.Graham, K. A., A. S. Lalani, J. L. Macen, T. L. Ness, M. Barry, L. Y. Liu, A. Lucas, I. Clark-Lewis, R. W. Moyer, and G. McFadden. 1997. The T1/35kDa family of poxvirus-secreted proteins bind chemokines and modulate leukocyte influx into virus-infected tissues. Virology 229:12-24. [DOI] [PubMed] [Google Scholar]
- 17.Hemmerich, S., C. Paavola, A. Bloom, S. Bhakta, R. Freedman, D. Grunberger, J. Krstenansky, S. Lee, D. McCarley, M. Mulkins, B. Wong, J. Pease, L. Mizoue, T. Mirzadegan, I. Polsky, K. Thompson, T. M. Handel, and K. Jarnagin. 1999. Identification of residues in the monocyte chemotactic protein-1 that contact the MCP-1 receptor, CCR2. Biochemistry 38:13013-13025. [DOI] [PubMed] [Google Scholar]
- 18.Holm, L., and C. Sander. 1996. Mapping the protein universe. Science 273:595-603. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard, S. J., S. F. Campbell, and J. M. Thornton. 1991. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 220:507-530. [DOI] [PubMed] [Google Scholar]
- 20.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 21.Karupiah, G., T. N. Fredrickson, K. L. Holmes, L. H. Khairallah, and R. M. Buller. 1993. Importance of interferons in recovery from mousepox. J. Virol. 67:4214-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lalani, A. S., K. Graham, K. Mossman, K. Rajarathnam, I. Clark-Lewis, D. Kelvin, and G. McFadden. 1997. The purified myxoma virus gamma interferon receptor homolog M-T7 interacts with the heparin-binding domains of chemokines. J. Virol. 71:4356-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lalani, A. S., J. Masters, K. Graham, L. Liu, A. Lucas, and G. McFadden. 1999. Role of the myxoma virus soluble CC-chemokine inhibitor glycoprotein, M-T1, during myxoma virus pathogenesis. Virology 256:233-245. [DOI] [PubMed] [Google Scholar]
- 24.Lalani, A. S., T. L. Ness, R. Singh, J. K. Harrison, B. T. Seet, D. J. Kelvin, G. McFadden, and R. W. Moyer. 1998. Functional comparisons among members of the poxvirus T1/35kDa family of soluble CC-chemokine inhibitor glycoproteins. Virology 250:173-184. [DOI] [PubMed] [Google Scholar]
- 25.Lau, E. K., S. Allen, A. R. Hsu, and T. M. Handel. 2004. Chemokine-receptor interactions: GPCRs, glycosaminoglycans and viral chemokine binding proteins. Adv. Protein Chem. 68:351-391. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, P. M. 2001. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol. 2:116-122. [DOI] [PubMed] [Google Scholar]
- 27.Ng, A., D. C. Tscharke, P. C. Reading, and G. L. Smith. 2001. The vaccinia virus A41L protein is a soluble 30 kDa glycoprotein that affects virus virulence. J. Gen. Virol. 82:2095-2105. [DOI] [PubMed] [Google Scholar]
- 28.Nibbs, R. J., S. M. Wylie, I. B. Pragnell, and G. J. Graham. 1997. Cloning and characterization of a novel murine beta chemokine receptor, D6. Comparison to three other related macrophage inflammatory protein-1alpha receptors, CCR-1, CCR-3, and CCR-5. J. Biol. Chem. 272:12495-12504. [DOI] [PubMed] [Google Scholar]
- 29.Nicholls, A., K. A. Sharp, and B. Honig. 1993. The program grasp. Biophysical J. 64:166-170. [Google Scholar]
- 30.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-344. [DOI] [PubMed] [Google Scholar]
- 31.Parry, C. M., J. P. Simas, V. P. Smith, C. A. Stewart, A. C. Minson, S. Efstathiou, and A. Alcami. 2000. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raport, C. J., J. Gosling, V. L. Schweickart, P. W. Gray, and I. F. Charo. 1996. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J. Biol. Chem. 271:17161-17166. [DOI] [PubMed] [Google Scholar]
- 33.Reading, P. C., J. A. Symons, and G. L. Smith. 2003. A soluble chemokine-binding protein from vaccinia virus reduces virus virulence and the inflammatory response to infection. J. Immunol. 170:1435-1442. [DOI] [PubMed] [Google Scholar]
- 34.Rot, A., and U. H. von Andrian. 2004. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 22:891-928. [DOI] [PubMed] [Google Scholar]
- 35.Schuck, P. 1996. Kinetics of ligand binding to receptor immobilized in a polymer matrix, as detected with an evanescent wave biosensor. I. A computer simulation of the influence of mass transport. Biophys. J. 70:1230-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seet, B. T., R. Singh, C. Paavola, E. K. Lau, T. M. Handel, and G. McFadden. 2001. Molecular determinants for CC-chemokine recognition by a poxvirus CC-chemokine inhibitor. Proc. Natl. Acad. Sci. USA 98:9008-9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheinerman, F. B., R. Norel, and B. Honig. 2000. Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 10:153-159. [DOI] [PubMed] [Google Scholar]
- 38.Smith, C. A., T. D. Smith, P. J. Smolak, D. Friend, H. Hagen, M. Gerhart, L. Park, D. J. Pickup, D. Torrance, K. Mohler, K. Schooley, and R. G. Goodwin. 1997. Poxvirus genomes encode a secreted, soluble protein that preferentially inhibits beta chemokine activity yet lacks sequence homology to known chemokine receptors. Virology 236:316-327. [DOI] [PubMed] [Google Scholar]
- 39.Smith, V. P., and A. Alcami. 2000. Expression of secreted cytokine and chemokine inhibitors by ectromelia virus. J. Virol. 74:8460-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Berkel, V., J. Barrett, H. L. Tiffany, D. H. Fremont, P. M. Murphy, G. McFadden, S. H. Speck, and H. I. Virgin. 2000. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol. 74:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young, L., R. L. Jernigan, and D. G. Covell. 1994. A role for surface hydrophobicity in protein-protein recognition. Protein Sci. 3:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]