Abstract
Lv1/TRIM5α (tripartite motif 5α) has recently emerged as an important factor influencing species-specific permissivity to retroviral infection in a range of primates, including humans. Old World monkey TRIM5α blocks human immunodeficiency virus type 1 (HIV-1) infectivity, and the human and New World monkey TRIM5α proteins are inactive against HIV-1 but active against divergent murine (N-tropic murine leukemia virus [MLV-N]) and simian (simian immunodeficiency virus from rhesus macaque [SIVmac]) retroviruses, respectively. Here we demonstrate antiviral activity of the first nonprimate TRIM protein, from cattle, active against divergent retroviruses, including HIV-1. The number of closely related human TRIM sequences makes assignment of the bovine sequence as a TRIM5α ortholog uncertain, and we therefore refer to it as bovine Lv1. Bovine Lv1 is closely related to primate TRIM5α proteins in the N-terminal RING and B-box 2 domains but significantly less homologous in the C-terminal B30.2 domain, particularly in the region shown to influence antiviral specificity. Intriguingly, some viruses restricted by bovine Lv1, including HIV-1 and MLV-N, are unable to synthesize viral DNA by reverse transcription, whereas restricted HIV-2 makes normal amounts of DNA. The data support the conclusion that TRIM protein-mediated restriction of retroviral infection is a more common attribute of mammals than previously appreciated.
Retroviruses rely on host cell biology to complete their life cycle, and their success throughout mammalian evolution and their ability to jump between divergent species imply the use of conserved pathways. The identification and characterization of these host-virus interactions are expected to enable improvement in animal models of infection, including human immunodeficiency virus (HIV)-AIDS, yield novel antiviral strategies, and facilitate the use of retroviruses for therapeutic gene delivery. A successful approach has been to characterize examples of species-specific or cell-type-specific retroviral infectivity. This has revealed that, in many cases, poor infection is due to the presence of dominant inhibitors of retroviral replication, as opposed to the nonpermissive cells lacking specific activities required by the virus (3, 7, 10, 20, 34, 39, 40, 46). Evidence for dominant antiviral factors encouraged the genetic screens that led to the discovery of the restriction factors APOBEC3G (32), tripartite motif 5α (TRIM5α) (34), and TRIM-Cyp (22, 30).
The prototype restriction factor is the murine Fv1 protein, which is derived from an endogenous retroviral gag sequence (5, 18). The gag-like Fv1 protein targets incoming murine leukemia virus (MLV) capsids (CAs) and blocks infection after viral DNA synthesis but before the formation of a provirus (16). There is also evidence to suggest that there may be dominant antiviral factors that restrict the late stage of the viral life cycle. The HIV-1 vpu protein has been shown to overcome a dominant block to viral egress that exists in human but not simian cells (8, 43). APOBEC3 proteins, particularly APOBEC3G, have also been shown to be able to effectively block retroviral infectivity (32, 47).
TRIM5α was first identified as a restriction factor active against HIV-1 in rhesus monkey cells (36) and later shown to encode the factors previously described as Ref1 in humans and Lv1 in monkeys, which are active against a broad range of divergent retroviruses in a species-specific way (12, 17, 26, 44). TRIM5 contains a TRIM comprising a RING domain, a B-box 2 domain, and a coiled coil. Multiple splice forms of TRIM5 have been detected that have been predicted to generate a series of proteins, each shorter from the C terminus (27). The α splice variant additionally encodes a C-terminal B30.2 (SPRY, RFP-like) domain which is essential for antiviral activity. Mutations in the B30.2 domain affect antiviral specificity and can broaden antiviral activity to previously insensitive retroviruses (21, 24, 36, 45). The viral determinant for sensitivity to TRIM5α is in the CA gene (7, 10, 17, 26, 44, 46). The simplest model of restriction is that TRIM5α interacts with incoming viral cores via B30.2-CA interactions leading to perturbation of the subsequent CA-dependent events in the viral life cycle. Indeed, a B30.2 domain-dependent TRIM5-N-tropic MLV (MLV-N) interaction has been demonstrated in vitro (31, 35). This interaction appears to lead to premature uncoating of incoming cores, leading to a loss of infectivity (35). In most cases, restriction by TRIM5α leads to a strong block to viral reverse transcription (17, 26, 34), but in the case of squirrel monkey TRIM5α restriction of simian immunodeficiency virus from rhesus macaque (SIVmac), the restricted virus reverse transcribes normally (46).
Here we identify a bovine TRIM protein, closely related to human and simian TRIM5α sequences, with strong antiviral activity against a range of divergent retroviruses. We refer to this protein as bovine Lv1. Our data indicate that antiretroviral activity of TRIM proteins is not limited to primates and suggest that such activities may be widespread in mammals.
MATERIALS AND METHODS
Cloning of bovine Lv1.
Bovine Lv1 was PCR amplified from cDNA prepared from Madin-Darby bovine kidney (MDBK) cells as previously described (46), with forward primer LY34 (5′-CAGACTGATCACCACCATGGCTTCAGGAATCCTG-3′ [Bcl1 restriction site underlined and Kozak sequence and ATG start codon in bold]) and reverse primer LY34 (5′-CTTCGATTCGAACTCAACAGCTTGGTGAGC-3′ [Csp45I site underlined]). Primers were designed on the basis of the bovine expressed sequence tag most closely related to human TRIM5. All of the PCR products amplified from MDBK cDNA and sequenced were derived from the gene described herein. The PCR product was cloned into MLV expression vector pCXCR between the Bc11 and Csp45I sites such that it was expressed under the control of the MLV long terminal repeat promoter in infected cells. CXCR also encodes red fluorescent protein (RFP), which is also expressed in infected cells. Lv1- and RFP-encoding vectors were prepared by 293T transfection with an NB-tropic Moloney MLV packaging construct (CMVintron) and the vesicular stomatitis virus G envelope protein (VSV-G) as previously described (17). CRFK cells were transduced with a vector encoding TRIM5α and RFP. Single-cell clones were isolated by limiting dilution, and positive clones were identified by red fluorescence microscopy. Positive clones were infected with green fluorescent protein (GFP)-encoding virus and, 48 h after that, assayed for green fluorescence by fluorescence-activated cell sorter (FACS) as previously described (17). The bovine Lv1 sequence was determined by sequencing three independent clones. DNA sequence analysis was performed with DNA Dynamo software (Blue Tractor Software, North Wales, United Kingdom).
Phylogenetic analysis.
Amino acid sequences were aligned in ClustalW (38), and the resulting alignment was manually adjusted. Uninformative regions and regions of low homology were removed to create a final alignment that was 487 amino acids in length. Bootstrapped phylogenies were constructed with 1,000 replicates of the neighbor-joining algorithm as implemented in PAUP 4.0 (37) and the Blosum62 amino acid transition matrix.
Viral vector preparation and infectivity assays.
VSV-G-pseudotyped, GFP-encoding retroviral vectors were prepared as previously described (3, 46), by triple transfection of 293T cells. Virus infectivity was measured by titrating serially diluted virus onto 105 cells per well in six-well plates. Infected cells were enumerated 48 h later by measuring GFP expression by FACS (BD Biosciences).
Disruption of Lv1 expression with small interfering RNA (siRNA).
Disruption of bovine Lv1 expression with siRNA was performed by transfecting 2.5 × 104 MDBK cells with siRNA oligonucleotides (QIAGEN) by using Oligofectamine (Invitrogen) according to the manufacturer's instructions in 24-well plates. The siRNA oligonucleotide sequence for bovine Lv1 was AUGCGAAUGUCAUCAUAAA. The irrelevant control oligonucleotide sequence was AGAGUUUAAUCAGCUACGA. Forty-eight hours later, cells were split 1:6 and incubated overnight. Cells were infected with serial dilutions of GFP-encoding MLV-N, B-tropic MLV (MLV-B), HIV-1, HIV-2, SIVmac, or EIAV prepared as previously described (3, 17, 46). Infected cells were enumerated by analysis of GFP expression by FACS (BD Bioscience). Titers of lentiviruses were calculated in infectious units per nanogram of reverse transcriptase (RT), determined by enzyme-linked immunosorbent assay (ELISA; CavidiTech, Uppsala, Sweden). Doses of MLV-N and MLV-B were equalized on murine SC1 cells, and titers are calculated as numbers of infectious units per 104 SC1 infectious units.
Quantitative PCR.
A TaqMan PCR to measure viral DNA synthesis was performed with primer and probe sequences specific to GFP as previously described (3). Cells (105) were infected in six-well plates in triplicate with equivalent doses of DNase-treated virus. Six hours after infection, total DNA was extracted from two samples with a DNeasy kit (QIAGEN, Chatsworth, CA). The third sample was subjected to FACS analysis 48 h after infection to enumerate infected cells. DNA (100 ng) was subjected to TaqMan quantitative PCR as previously described (3, 42).
Nucleotide sequence accession number.
The Lv1 sequence described here has been submitted to GenBank and assigned accession no. DQ380509.
RESULTS
MDBK cells are differentially permissive to a range of VSV-G-pseudotyped retroviral vectors.
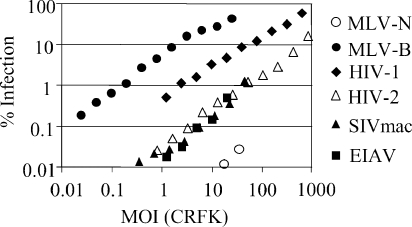
MDBK cells strongly restrict infection by MLV-N after target cell entry and before reverse transcription (4, 39). They have also been shown to be poorly permissive to HIV type 1 (HIV-1) (13). We therefore sought to examine the permissivity of MDBK cells to infection with other retroviruses and to identify the antiviral host factors involved. In order to compare the permissivity of a particular cell line to that of a series of distantly related viruses, the dose of each virus must be standardized. Primate lentiviruses are often standardized by measurement of RT activity with an ELISA to measure a synthetic product of viral RT (9, 11, 14). Such an assay cannot be reliably used to compare doses of viruses as unrelated as HIV-1, equine infectious anemia virus (EIAV), and the gamma retroviruses MLV-N and MLV-B as their RT enzymes are not likely to be equally active in the assay. We therefore standardized the viral dose by titration in permissive Crandell-Reese feline kidney (CRFK) cells. Standardization of the virus dose for any cell line is likely to be influenced to some degree by its inevitable differential permissivity to each virus. Interpretation of such standardization data must therefore be cautious. However, CRFK cells have been shown to be particularly permissive to a range of viruses (13). Moreover, closely related viruses such as MLV-N and MLV-B (39) or SIVmac and HIV-2 (46) have similar titers on these cells. Viral doses are therefore plotted as multiplicities of infection (MOIs) on CRFK cells. For example, a multiplicity of 1 of MLV-B will infect around 66% of CRFK cells and around 10% of MDBK cells (Fig. 1).
FIG. 1.
MDBK cells are differentially permissive to a range of VSV-G-pseudotyped retroviral vectors. Serial dilutions of VSV-G-pseudotyped, GFP-encoding vectors from MLV-N (○) MLV-B (•), HIV-1 (⧫), HIV-2 (▵), SIVmac (▴), and EIAV (▪) were used to infect MDBK cells. The percentage of infected cells was measured by FACS 48 h after infection. Viral doses were determined by titration onto CRFK cells, and MOIs were calculated. For example, an MOI of 0.1 infects 10% of the CRFK cells. This experiment is representative of three replicates with independently prepared virus preparations.
We prepared VSV-G-pseudotyped, GFP-encoding retroviral vectors from MLV-N, MLV-B, HIV-1, HIV-2, EIAV, and SIVmac and determined their infectious titers on CRFK cells as previously described (3). Serial dilutions of each virus were then titrated onto MDBK cells (Fig. 1). MLV-B is high titer on MDBK cells, and MLV-N is around 3 orders of magnitude less infectious, as previously described (39). HIV-1 is the most infectious of the lentiviral vectors, and HIV-2, SIVmac, and the horse lentivirus EIAV all have similar low infectivities on these cells, in comparison to CRFK cells.
Identification of a bovine Lv1 gene.
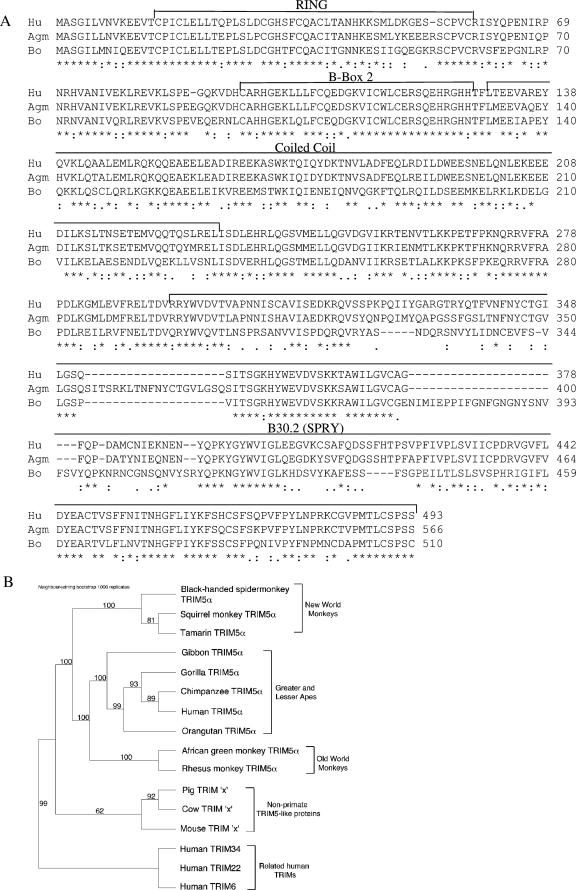
The block to MLV-N in MDBK cells is after viral entry and before viral reverse transcription and is characteristic of blocks to retroviral infection mediated by primate Lv1/TRIM5α proteins. We therefore hypothesized that an Lv1-like activity exists in MDBK cells, despite the fact that TRIM5α had previously been observed only in primates. Consistent with this hypothesis, we were able to clone and sequence a gene closely related to human TRIM5α expressed in MDBK cells. Alignment of bovine, human, and African green monkey (Agm) sequences reveals that the bovine protein is highly homologous to human and simian proteins, particularly in the N-terminal RING and B-box 2 domains and in certain parts of the B30.2 domain (Fig. 2A). Notably, the region of the B30.2 domain previously shown to determine the antiviral specificity of primate TRIM5α proteins, around residues 320 to 350 (21, 24, 29, 36, 45), is not conserved. Furthermore, an insertion in the bovine sequence, residues 349 to 375, is reminiscent of a 20-amino-acid insertion in the Agm B30.2 domain, relative to the human and rhesus sequences, although there is no homology between the Agm and bovine insertions. Interestingly, the inserted sequence in Agm has been implicated in its antiviral specificity (21).
FIG. 2.
Identification of a gene for bovine Lv1. (A) Bovine Lv1 sequence (Bo) aligned with the human (Hu) and Agm TRIM5α sequences. Domains are according to reference 34. (B) Phylogenetic tree showing the relationship of bovine Lv1 to 10 primate TRIM5α genes, to related TRIM-encoding genes in humans (those for TRIM6, TRIM22, and TRIM34), and to TRIM5α-like proteins identified in mice and domestic pigs. Bootstrap values, based on 1,000 neighbor-joining replicates, are shown for each node where support was greater than 60%. Asterisk, identical residue; colon, conserved substitution; period, semiconserved substitution; gap, no conservation.
We further examined the evolutionary relationship of the bovine Lv1 sequence to primate TRIM5α sequences by constructing a phylogenetic tree (Fig. 2B). We also included three further human TRIM sequences (TRIM6, TRIM22, and TRIM34) in this analysis as they are closely related to TRIM5 in humans, are arranged in tandem on chromosome 11, and on the basis of their close homology, are likely to represent duplications of an ancient TRIM sequence. Finally, we included the murine and porcine expressed sequence tags most closely related to primate TRIM5α genes. As expected, primate TRIM5α sequences cluster according to primate phylogeny into three groups representing New World and Old World monkeys and apes. The TRIM sequences from cattle and pigs appear to be relatively closely related, but the relationship of these proteins to the larger subset of TRIM5α-like genes is more difficult to discern, and it is not clear from this analysis whether the TRIM sequences from pigs and cows are orthologous with TRIM5α or with another closely related TRIM protein in humans.
Reduction of bovine Lv1 levels in MDBK cells increases their permissivity to viral infection.
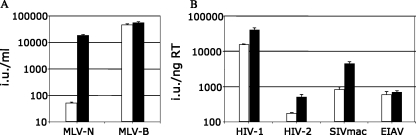
We directly examined the role of bovine Lv1 in the poor permissivity of bovine cells by reducing its expression with siRNA and measuring viral infectivity. Treatment with siRNA directed against bovine Lv1 specifically increased MLV-N, HIV-1, HIV-2, and SIVmac infectivity but not MLV-B or EIAV infectivity (Fig. 3A and B). Transfection of an irrelevant siRNA had no effect on the infectivity of any of the viruses (data not shown). siRNA to Lv1 did not rescue MLV-N infectivity to the level of MLV-B infectivity, suggesting an incomplete reduction of bovine Lv1 expression, consistent with the fact that MDBK cells are difficult to transfect with siRNA (data not shown). These data suggest that bovine Lv1 has a broad antiviral effect and is largely responsible for the poor permissivity of bovine cells to retroviral infection.
FIG. 3.
Reduction of Lv1 levels in MDBK cells increases their permissivity to viral infection. (A and B) Titers of MLV-N, MLV-B, HIV-1 HIV-2, SIVmac, and EIAV on MDBK cells after transfection of bovine TRIM5α-specific siRNA (black bars) or untreated MDBK cells (white bars). The viral doses used were chosen to infect between 0.5 and 5% of the cells in the absence of siRNA. MLV-N and MLV-B doses were equalized by infection of permissive murine SC1 cells and are plotted as infectious units (i.u.) per milliliter on MDBK cells. RT activity of HIV-1, HIV-2, SIVmac, and EIAV was measured by ELISA, and titers are plotted as infectious units per nanogram of RT. Each error bar indicates the standard error of the mean of three repetitions.
Expression of bovine Lv1 in permissive feline cells renders them able to restrict a range of divergent retroviruses.
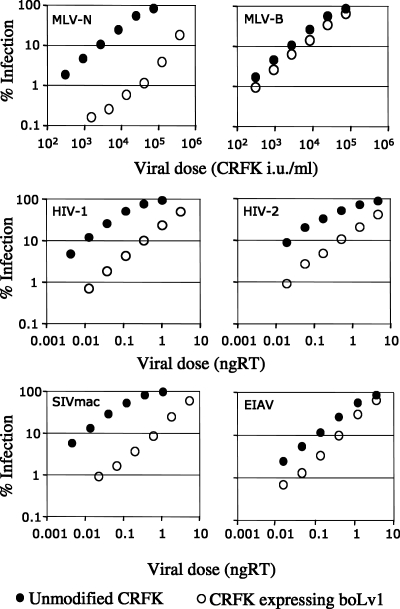
To further examine the ability of bovine Lv1 to restrict viral infection, we expressed the protein in CRFK cells by using an MLV vector as previously described (17). In these experiments, the feline cells expressing the bovine Lv1 protein are clonal and coexpress a fluorescent marker gene. We called these cells CRFKboLv1. We then compared the permissivity of CRFKboLv1 and unmodified CRFK cells to infection by a range of retroviruses (Fig. 4). Expression of bovine Lv1 renders CRFK cells almost 2 orders of magnitude less permissive to MLV-N. As expected, the infectivity of MLV-B is not significantly affected by bovine Lv1 expression. These data are reminiscent of the strong restriction of MLV-N by the human and Agm TRIM5α proteins and the insensitivity of MLV-B to all of the TRIM5α proteins tested thus far. We also tested the permissivity of CRFKboLv1 to HIV-1, HIV-2, SIVmac, and EIAV GFP-encoding vectors (Fig. 4). HIV-1, HIV-2, and SIVmac are all restricted by around 10-fold in CRFKboLv1 compared to unmodified CRFK cells. EIAV is weakly restricted by overexpression of bovine Lv1. However, the lack of an effect on EIAV infectivity after transfection of bovine Lv1-specific siRNA (Fig. 3) suggests that bovine Lv1 does not significantly contribute to the poor infectivity of EIAV in MDBK cells and this small inhibition is likely to be due to the expansion of antiviral specificity often seen when factors are overexpressed (6, 23, 46). Data in Fig. 4 support the notion that much of the poor permissivity of bovine cells to retroviral infectivity is due to the expression of a bovine TRIM5α-like Lv1 protein with broad antiviral specificity.
FIG. 4.
Expression of bovine Lv1 in permissive feline cells renders them able to restrict a range of retroviruses. Serial dilutions of the indicated VSV-G-pseudotyped, GFP-encoding viruses were titrated onto cloned CRFK cells expressing bovine Lv1 (boLv1; open symbols) or unmodified CRFK cells as a control (solid symbols). Percentages of infected cells were measured by FACS 48 h after infection. RT activities of HIV-1, HIV-2, SIVmac, and EIAV were measured by ELISA in nanograms (ngRT). Results are representative of two independent experiments performed with two independent virus preparations and two clones of CRFKboLv1 cells. i.u., infectious units.
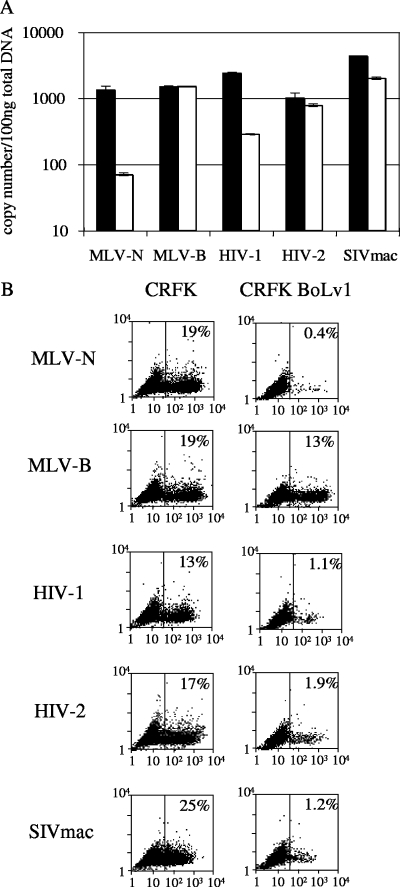
Bovine Lv1 restriction of viral infection can be before or after reverse transcription.
TRIM5α usually restricts retroviral infectivity very early after target cell entry, before significant viral reverse transcription (17, 26, 34). An exception is TRIM5α from New World squirrel monkeys, which strongly restricts SIVmac infectivity, despite normal levels of SIVmac DNA synthesis (46). In order to examine whether bovine Lv1 is able to block viral reverse transcription, we infected CRFK cells or CRFK cells expressing bovine Lv1 and measured viral DNA by TaqMan quantitative PCR 6 h after exposure to virus as previously described (3) (Fig. 5A). Cells were infected in triplicate, and 6 h after infection two samples were DNA extracted for PCR. The third sample was subjected to FACS analysis 48 h after infection to measure infected cells (Fig. 5B). We used a fixed dose of virus equivalent to a MOI of around 0.2 on unmodified CRFK cells. Expression of bovine Lv1 led to a significant reduction in MLV-N DNA synthesis, compared to MLV-B, as it does in bovine cells (4). HIV-1 DNA synthesis is also reduced by around 10-fold, which corresponds to a 10-fold reduction in infectivity on the bovine Lv1-expressing cells. In contrast, there was moderate inhibition of viral DNA synthesis by SIVmac and no inhibition for HIV-2, despite the observed 10-fold reduction in infectivity. The consequence of restriction by bovine Lv1 therefore depends on the virus rather than on the Lv1 protein, and bovine Lv1 is able to restrict virus before or after reverse transcription.
FIG. 5.
Restriction of viral infection by bovine Lv1 can be before or after reverse transcription. (A) Measurement of viral cDNA synthesis after bovine Lv1-restricted and unrestricted infection by TaqMan quantitative PCR. CRFK cells (black bars) or CRFK cells expressing bovine Lv1 were infected in triplicate with MOIs of viruses of around 0.2, determined on unmodified CRFK cells. At 6 h after infection, DNA was extracted from two samples and subjected to quantitative PCR in duplicate with primers and a probe specific for GFP. (B) The percentage of infected cells in the third sample was measured by FACS 48 h after infection. Errors are standard errors of the means. Results are representative of two independent experiments performed with two different CRFKboLv1 clones.
DISCUSSION
We have identified a bovine TRIM protein showing significant homology to the primate restriction factor TRIM5α and demonstrated that it strongly influences the permissivity of bovine cells to retroviral infection. Although this function strongly suggests a relationship with TRIM5α proteins in primates, it is difficult to determine with absolute certainty whether the bovine gene represents an ortholog of primate TRIM5α. Phylogenetic analysis places the bovine sequence between primate TRIM5α sequences and the closely related human TRIM6, TRIM22, and TRIM34 proteins (Fig. 2B). However, TRIM proteins with antiviral function are likely to undergo rapid evolution because of strong positive selection (29), and consequently their evolutionary relationships across species may have been obscured. Furthermore, the chromosomal arrangement of TRIM5, TRIM6, TRIM22, and TRIM34 in tandem in humans suggests a history of duplication for TRIM genes that not only obscures synteny but may have differed across mammalian lineages, making a definition of orthology based on the identification of syntenous regions in different species difficult if not impossible. Nevertheless, such an assignment is largely semantic, particularly since sequence homology suggests that TRIM5α is likely to be derived from duplication of TRIM6 or vice versa. The overriding observation is that the bovine sequence is closely related to primate TRIM5α proteins and has a similar antiviral activity. On this basis, we refer to the bovine protein as bovine Lv1.
Alignment of bovine and primate sequences demonstrates a high level of conservation in the N-terminal RING and B-box 2 domains but lower homology in the C-terminal B30.2 domain (Fig. 2A). The role of the RING and B-box 2 domains in restriction remains unclear. High conservation of the RING domain suggests that it is important for TRIM5 function. RING domains are found in E3 ligases for proteins such as ubiquitin or SUMO, suggesting that modification of incoming cores may underlie the antiviral mechanism, although this remains controversial (15, 24, 25). Remarkably, an inactive allele of TRIM5α carrying the RING domain polymorphism H43Y has recently been described in the human population (28). The inactivity of this protein also supports a role for the RING domain in restriction, as well as a nonessential role for TRIM5α in humans. The bovine sequence has a conservative change to asparagine at this position (Fig. 2).
Analyses of primate TRIM5α B30.2 domain sequences reveal variable regions under selection in recent primate evolution, presumably under pressure from pathogenic endogenous or exogenous viruses (29, 33). Remarkably, both the bovine and Agm Lv1 proteins restrict HIV-1, HIV-2, SIVmac, EIAV, and MLV-N but not MLV-B (Fig. 1 and 4) (10). It therefore seems that Lv1 antiviral determinants unrelated at the sequence level can be closely related in terms of antiviral specificity. The fact that MDBK cells, and therefore bovine Lv1, restrict MLV-N but not MLV-B with specificity for amino acid 110 in the MLV CA (4) demonstrates that the viral restriction sensitivity determinant is in the CA protein, as it is for other Lv1/TRIM5α alleles (7, 10, 17, 26, 44, 46). These observations support the notion that Lv1/TRIM5α is an effective antiviral, targeting CA from a broad array of unrelated retroviruses to block infection. It remains unclear how HIV-1 has escaped restriction by human TRIM5α and also why MLV-B and Moloney MLV are insensitive to any of the TRIM5α alleles thus far tested. One possibility is that MLV's relative insensitivity to TRIM5α is related to the fact that the MLV CA does not have the exposed loop on its surface (19) that is targeted by TRIM5α (11, 14, 46), making it easier for MLV to escape TRIM5α activity.
Most TRIM5α-restricted viruses make very little viral DNA, indicating that the block to infection is very early after the virus has entered the target cell. However, in some cases, exemplified by restriction of SIVmac by squirrel monkey TRIM5α, the restricted virus is able to reverse transcribe normally (46). The murine gag-like restriction factor Fv1 is also able to block MLV infection after reverse transcription, and this is conserved when Fv1 is expressed in human cells (4). Despite the apparently later interaction between Fv1 and virus, Fv1 and TRIM5α can compete for incoming virus (23). This and the fact that bovine Lv1 restricts MLV-N and HIV-1 before reverse transcription, whereas restricted HIV-2 is able to synthesize normal levels of DNA (Fig. 5), suggest that the timing of the block is dependent on both the factor and the virus. It seems likely, therefore, that restriction factors interact with incoming virus very early after entry and it is the consequences of these interactions that vary. In some cases, the restricted virus is unable to reverse transcribe whereas in other cases restricted virus is able to make normal levels of DNA despite the fact that it will ultimately be noninfectious. These observations are consistent with TRIM5α interacting with incoming CA and perturbing the normal rearrangement or uncoating events that are important during reverse transcription and trafficking.
The isolation of a broadly effective antiviral TRIM protein from cattle indicates that innate immune activity of TRIM proteins extends beyond primates, and it will be interesting to examine the antiviral activity of other TRIM molecules from divergent vertebrates. Our data suggest that antiviral TRIM proteins are likely to be widespread within mammals and represent a conserved mechanism of protection from pathogenic retroviruses. Furthermore, the ability of drugs such as arsenic trioxide (2, 17) or cyclosporine (1, 41) to manipulate viral sensitivity to host restriction factors suggests that these pathways might be a target for novel therapeutics.
Acknowledgments
This work was funded by the Wellcome Trust (G.T.), a UCL Graduate School scholarship and Bogue fellowship (Z.K.), and EU CASCADE (R.G.).
We thank Kevin Tennill for technical assistance and Sam Wilson for critical reading of the manuscript.
REFERENCES
- 1.Berthoux, L., S. Sebastian, E. Sokolskaja, and J. Luban. 2005. Cyclophilin A is required for TRIM5α-mediated resistance to HIV-1 in Old World monkey cells. Proc. Natl. Acad. Sci. USA 102:14849-14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthoux, L., G. Towers, C. Gurer, P. Salomoni, P. P. Pandolfi, and J. Luban. 2003. As2O3 enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J. Virol. 77:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besnier, C., L. Ylinen, B. Strange, A. Lister, Y. Takeuchi, S. P. Goff, and G. Towers. 2003. Characterization of murine leukemia virus restriction in mammals. J. Virol. 77:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 6.Bock, M., K. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottlinger, H. G., T. Dorfman, E. A. Cohen, and W. A. Haseltine. 1993. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. USA 90:7381-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatziioannou, T., S. Cowan, and P. D. Bieniasz. 2004. Capsid-dependent and -independent postentry restriction of primate lentivirus tropism in rodent cells. J. Virol. 78:1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatziioannou, T., S. Cowan, U. K. Von Schwedler, W. I. Sundquist, and P. D. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, Y., M. K. Collins, P. A. Radcliffe, K. A. Mitrophanous, and Y. Takeuchi. 2002. Gene transduction efficiency in cells of different species by HIV and EIAV vectors. Gene Ther. 9:932-938. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, Y., L. Ylinen, M. Kahar-Bador, and G. J. Towers. 2004. The influence of gag on HIV-1 species specific tropism. J. Virol. 78:11816-11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 16.Jolicoeur, P., and E. Rassart. 1980. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J. Virol. 33:183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilly, F. 1970. FV-2: identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J. Natl. Cancer Inst. 45:163-169. [PubMed] [Google Scholar]
- 19.Mortuza, G. B., L. F. Haire, A. Stevens, S. J. Smerdon, J. P. Stoye, and I. A. Taylor. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431:481-485. [DOI] [PubMed] [Google Scholar]
- 20.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama, E. E., H. Miyoshi, Y. Nagai, and T. Shioda. 2005. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5α determines species-specific restriction of Simian immunodeficiency virus SIVmac infection. J. Virol. 79:8870-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Passerini, L. D., Z. Keckesova, and G. J. Towers. 2006. Retroviral restriction factors Fv1 and TRIM5α act independently and can compete for incoming virus before reverse transcription. J. Virol. 80:2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Caballero, D., T. Hatziioannou, F. Zhang, S. Cowan, and P. D. Bieniasz. 2005. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 79:15567-15572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyer, S. L., L. I. Wu, J. M. Akey, M. Emerman, and H. S. Malik. 2006. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5α in humans. Curr. Biol. 16:95-100. [DOI] [PubMed] [Google Scholar]
- 29.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 31.Sebastian, S., and J. Luban. 2005. TRIM5α selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 33.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79:6111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 35.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stremlau, M., M. J. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (* and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref1 restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 42.Towers, G. J., D. Stockholm, V. Labrousse-Najburg, F. Carlier, O. Danos, and J. C. Pages. 1999. One step screening of retroviral producer clones by real time quantitative PCR. J. Gene Med. 1:352-359. [DOI] [PubMed] [Google Scholar]
- 43.Varthakavi, V., R. M. Smith, S. P. Bour, K. Strebel, and P. Spearman. 2003. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA 100:15154-15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 46.Ylinen, L., Z. Keckesova, S. J. Wilson, S. Ranasinghe, and G. J. Towers. 2005. Differential restriction of HIV-2 and SIVmac by TRIM5α alleles. J. Virol. 79:11580-11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]