Abstract
Severe acute respiratory syndrome coronavirus (SCoV) 7a protein is one of the viral accessory proteins. In expressing cells, 7a protein exhibits a variety of biological activities, including induction of apoptosis, activation of the mitogen-activated protein kinase signaling pathway, inhibition of host protein translation, and suppression of cell growth progression. Analysis of SCoV particles that were purified by either sucrose gradient equilibrium centrifugation or a virus capture assay, in which intact SCoV particles were specifically immunoprecipitated by anti-S protein monoclonal antibody, demonstrated that 7a protein was associated with purified SCoV particles. Coexpression of 7a protein with SCoV S, M, N, and E proteins resulted in production of virus-like particles (VLPs) carrying 7a protein, while 7a protein was not released from cells expressing 7a protein alone. Although interaction between 7a protein and another SCoV accessory protein, 3a, has been reported, 3a protein was dispensable for assembly of 7a protein into VLPs. S protein was not required for the 7a protein incorporation into VLPs, and yet 7a protein interacted with S protein in coexpressing cells. These data established that, in addition to 3a protein, 7a protein was a SCoV accessory protein identified as a SCoV structural protein.
Human coronavirus infections typically cause only mild or moderate diseases (19, 43), so the revelation that the newly identified severe acute respiratory syndrome (SARS) coronavirus (SCoV) is the etiologic agent for the global outbreak of life-threatening SARS in the winter of 2002/2003 was surprising (7, 17, 18, 34). Bats appear to be natural reservoirs of SCoV and SCoV-like viruses (20, 21). Specific mutations in the viral peplomer protein, S protein, allowed a SCoV-like virus to cross the species barrier and become a highly infectious human pathogen (22, 35). SCoV is an enveloped, positive-sense RNA virus with a spherical shape of approximately 100 nm in diameter. Like other coronaviruses, the SCoV membrane contains three viral proteins, S, M, and E. The ∼30-kb SCoV genomic RNA is bound with N protein to form a nucleocapsid complex, which is surrounded by the viral membrane. The genome organization of SCoV is also similar to that of other coronaviruses; the 5′ two-thirds of the genome encodes the gene 1 proteins, whose primary functions are associated with viral RNA synthesis, and the 3′ one-third encodes all of the structural proteins and accessory proteins, including 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b (24, 36, 40). Expression of 3a, 6, and 7a proteins has been confirmed to occur in infected cells and patients (8, 10, 46, 48). SCoV 3a protein is a viral structural protein (15, 37) and is released from 3a-expressing cells and virus-infected cells in membranous structures (13). Although other coronaviruses also produce accessory proteins (19, 43), the amino acid sequences of all of the SCoV accessory proteins have no homology with those of any other known viral proteins or nonviral proteins. Deletion of open reading frames 3a, 3b, 6, and 7a, either alone or in combination, does not affect the virus replication significantly in cell culture (45), demonstrating that these SCoV accessory proteins are not essential for virus replication in cell culture. Past studies of other coronaviruses suggested that many of the accessory proteins are important for viral virulence in vivo (6, 32, 33, 41, 44). Likewise, SCoV accessory proteins may play roles in SCoV pathogenesis, but the biological functions of these SCoV-specific accessory proteins are largely unexplored.
The 122-amino-acid (aa)-long SCoV 7a protein (also known as X4 protein or U122 protein) is a type I transmembrane protein consisting of a 15-aa signal peptide sequence at its N terminus, an 81-aa luminal domain, a 21-aa transmembrane domain, and a short C-terminal tail (30). The 7a gene is conserved in all SCoV strains isolated from humans and animals (21). The crystal structure of the luminal domain of the 7a protein has been resolved, revealing an unexpected topology similar to that of members of the immunoglobulin (Ig) superfamily (30). There are controversies in the literature as to whether 7a protein is localized in the endoplasmic reticulum or Golgi compartment (8, 16, 30). Reported biological functions of 7a protein based on expression studies include induction of apoptosis in various cell lines through a caspase-dependent pathway (38), inhibition of cellular protein synthesis, activation of p38 mitogen-activated protein kinase (MAPK) (16), and suppression of cell cycle progression at the G0/G1 phase (47); yet, it is still unclear whether 7a protein exerts these functions in infected cells. Nevertheless, these intriguing results collectively suggest that 7a protein is likely involved in virus-host interactions.
To further understand the function and properties of SCoV 7a protein, we examined whether 7a protein was a SCoV structural protein. Our data indicated that, in addition to the 3a protein, the 7a protein was a viral structural protein. We have also established a SCoV virus-like particle (VLP)-producing system by expressing SCoV proteins without using exogenous viruses and confirmed that the 7a protein was assembled into SCoV VLPs.
MATERIALS AND METHODS
Cells and virus.
Human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's essential medium supplemented with 10% fetal bovine serum, l-glutamine (2 mM), nonessential amino acids (0.1 mM), and kanamycin (100 μg/ml). Human colon carcinoma epithelium Caco2 cells were maintained in minimum essential medium (Eagle) with 20% fetal bovine serum, l-glutamine (2 mM), nonessential amino acids (0.1 mM), and kanamycin (100 μg/ml). Cells were incubated at 37°C in 5% CO2. The Urbani strain of SCoV was propagated in a biosafety level 3 laboratory as described previously (15, 27).
Construction of plasmids and transient protein expression.
Total intracellular RNA was extracted from SCoV-infected Vero E6 cells with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. SCoV S, M, N, E, 3a, and 7a genes were amplified from total RNA prepared from SCoV-infected cellsbyreverse transcription-PCR. The PCR products were cloned into a mammalian expression vector, pCAGGS, resulting in pCAGGS-S, pCAGGS-M, pCAGGS-N, pCAGGS-E, pCAGGS-3a, and pCAGGS-7a, respectively. All of the plasmids were confirmed using sequence analysis. The 7a gene was also cloned into pcDNA-myc plasmid (Invitrogen) to generate pcDNA-7a-myc, in which a myc tag was attached to the C terminus of the 7a protein. Cultures of 293T cells grown on 100-mm dishes were transfected with expression plasmids, as indicated in each experiment, by using TransIT-293 reagent (Mirus, Madison, WI) according to the manufacturer's instructions. At 48 h posttransfection, culture media were collected by centrifugation at 1,500 × g for 10 min at 4°C. The media were further clarified by filtration through 0.45-μm syringe filters and partially purified by centrifugation through 20% sucrose cushions at 26,000 rpm for 3 h in a Beckman SW28 rotor. Samples were resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 50 mM dithiothreitol). Cells were washed once with ice-cold phosphate-buffered saline (PBS). Total cell lysates were prepared by adding 1× SDS-PAGE loading buffer. All of the samples were incubated at 37°C for 30 min and subjected to Western blot analysis.
Antibodies and Western blot analysis.
Production of rabbit anti-SCoV 7a protein antibody and rabbit anti-SCoV 3a protein antibody was reported previously (13, 15). Rabbit anti-SCoV S polyclonal antibody (IGM-541) was obtained from IMGENEX, San Diego, CA. Rabbit anti-SCoV M antibody (AP6008b) was purchased from Abgent, San Diego, CA. Mouse anti-SCoV N antiserum was provided by Xiao-Hua Li at The University of Texas Southwestern Medical Center at Dallas. Mouse anti-SCoV S protein monoclonal antibody (NR-617) was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID. Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG, HRP-conjugated goat anti-mouse IgG, and HRP-conjugated donkey anti-goat IgG were purchased from Santa Cruz Biotechnology, CA. Protein samples were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad) with a Trans-Blot SD semidry transfer apparatus (Bio-Rad). The membranes were blotted with primary antibodies diluted 1:1,000 in PBS containing 5% nonfat milk overnight at 4°C. Membranes were washed and incubated with secondary antibodies diluted 1:3,000 for 1 h at room temperature. The protein bands were visualized with ECL reagent (Amersham Biosciences) by following the manufacturer's instructions.
Ultracentrifugation of SCoV samples on continuous sucrose gradients.
Supernatants from SCoV-infected Caco2 cells were inactivated by irradiation with 2 × 106 rads from a Gammacell 60Co source (model 109A; J. L. Shepherd and Associates, San Fernando, CA) as described previously (15). Inactivation of virus infectivity was confirmed by tissue culture assay. After centrifugation at 1,500 × g for 10 min at 4°C, samples were further clarified by filtration through 0.45-μm syringe filters. The clarified samples were partially purified by centrifugation through 20% sucrose cushions at 26,000 rpm for 3 h in a Beckman SW28 rotor and resuspended in NTE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA). Then, the samples were applied onto a 20 to 60% continuous sucrose gradient and subjected to centrifugation at 26,000 rpm for 18 h in an SW28 rotor. Twelve fractions were collected from the bottom of the gradient and diluted in NTE buffer. The SCoV particles in the fractions were pelleted through a 20% sucrose cushion at 38,000 rpm for 2 h using a Beckman SW41 rotor. The pellets were dissolved in 1× SDS-PAGE loading buffer and used for Western blot analysis.
Generation and analysis of SCoV VLPs.
Mixtures of plasmids containing pCAGGS-S, pCAGGS-M, pCAGGS-N, pCAGGS-E, pCAGGS-3a, or pCAGGS-7a in various combinations, as indicated in each experiment, were transfected into subconfluent 293T cells grown on 100-mm cell culture dishes. Culture media were harvested and analyzed using Western blotting as described above. In metabolic label experiments, the transfected cells were labeled with 100 μCi/ml Trans35S-Label (MP Biomedicals, CA) from 42 h to 48 h after transfection. The culture media were collected, clarified as described above, and centrifuged at 26,000 rpm for 3 h on a discontinuous sucrose gradient consisting of 60, 50, 30, and 20% sucrose by using an SW28 rotor. The VLP samples at the interface of 30 and 50% sucrose were collected, diluted with NTE buffer, and further loaded on the top of a 20 to 60% continuous sucrose gradient. After 18 h of centrifugation at 26,000 rpm in an SW28 rotor, 10 fractions were collected from the bottom of the gradient and measured for sucrose densities. Samples were diluted and recovered by centrifugation through a 20% sucrose cushion in a Beckman SW41 rotor. The pellets were dissolved in 1× SDS-PAGE loading buffer, applied to a 10% SDS-PAGE gel, and analyzed using autoradiography.
SCoV virus capture assay.
A SCoV virus capture assay was performed as reported previously (13). Briefly, supernatants from SCoV-infected Caco2 cells or transfected cells were partially purified by centrifugation through a 20% sucrose cushion. The pellet was suspended in NTE buffer containing a protease inhibitor cocktail (Sigma) with bovine serum albumin (BSA) at a concentration of 0.3%. Samples were precleared by incubation with 40 μl of protein A/G Sepharose 4 Fast Flow beads (Amersham Biosciences) at 4°C for 1 h. Then, samples were collected and mixed with 5 μg of mouse anti-SCoV S protein monoclonal antibody (NR-617) or mouse anti-H2KkDk (H2K) monoclonal antibody (29). After incubation at 4°C overnight, 5 μg of goat anti-mouse IgG (Fcγ fragment specific; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was added and incubated at 4°C for 3 h. Then, 40 μl of protein A/G Sepharose Fast Flow beads was added and incubated for another 3 h at 4°C. After centrifugation at 750 × g for 5 min, the supernatant was collected. The pellets were washed with NTE buffer five times. Finally, the collected supernatant and the pellets were suspended in SDS-PAGE loading buffer and used for SDS-PAGE and Western blot analysis.
Coimmunoprecipitation analysis.
Subconfluent 293T cells were transfected with plasmids by means of TransIT-293 reagent. At 24 h posttransfection, cells were harvested and washed once with ice-cold PBS. Cell lysates were prepared in radioimmunoprecipitation assay lysis buffer (1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, and protease inhibitor cocktail in PBS) on ice for 30 min and briefly centrifuged using an Eppendorf microcentrifuge at maximum speed to remove cell debris. Supernatants were collected and precleared by incubation with 40 μl of protein A/G Sepharose Fast Flow beads for 1 h at 4°C. Then, 1 μg of rabbit anti-7a protein antibody, mouse anti-myc monoclonal antibody (9E10; Santa Cruz), or mouse anti-SCoV S protein monoclonal antibody (NR-617) was added, as indicated in each experiment, to precipitate the 7a protein, 7a-myc protein, or S protein at 4°C overnight. The samples were incubated further with 40 μl of protein A/G Sepharose Fast Flow beads for an additional 3 h at 4°C. After centrifugation at 750 × g for 5 min, precipitated immune complexes were washed three times with lysis buffer and twice with PBS. Finally, the immunoprecipitated proteins were dissolved in 2× SDS-PAGE loading buffer and analyzed via Western blot assay.
RESULTS
SCoV 7a protein is a SCoV structural protein.
To test whether SCoV 7a protein was a viral structural protein, SCoV that was released from infected Caco2 cells was purified using a 20 to 60% sucrose gradient centrifugation. Twelve fractions were collected from the bottom of the gradient, and the presence of SCoV proteins in each fraction was analyzed using Western blot analysis. Consistent with our past studies (13), the strongest S, N, and M protein signals were detected in fractions 5 (density, 1.20 g/ml) and 6 (density, 1.18 g/ml) (Fig. 1), showing that the buoyant density of SCoV was between 1.18 and 1.20 g/ml. Western blot analysis clearly demonstrated that the peak of SCoV 7a protein was also in fractions 5 and 6, suggesting that the 7a protein was a SCoV structural protein.
FIG. 1.
Association of SCoV 7a protein with purified SCoV particles. Supernatant from SCoV-infected Caco2 cells was clarified, and SCoV particles were pelleted down through a 20% sucrose cushion. The pellet was suspended in NTE buffer and purified by ultracentrifugation on a continuous 20 to 60% sucrose gradient. Twelve fractions were taken from the bottom. The purified viruses in each fraction were pelleted by centrifugation through a 20% sucrose cushion. Samples were dissolved in 1× SDS-PAGE loading buffer. Western blot analysis using anti-7a antibody (7a), anti-SCoV S protein antibody (S), anti-SCoV N protein antibody (N), and anti-SCoV M protein antibody (M) was performed to detect virion 7a, S, N, and M proteins, respectively.
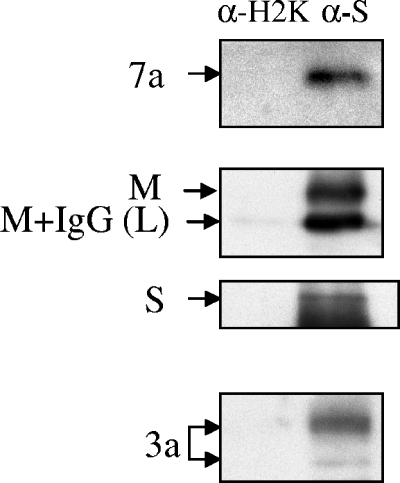
To further confirm the presence of 7a protein in SCoV particles, a virus capture assay using anti-SCoV S protein monoclonal antibody was conducted. This method allows purification of intact SCoV from the culture media of virus-infected cells with high specificity and efficacy (13, 31). Partially purified SCoV was incubated with anti-SCoV S monoclonal antibody and subsequently captured using immunoprecipitation. Our results demonstrated that anti-SCoV S monoclonal antibody successfully precipitated intact SCoV particles carrying M, S, and 3a proteins (Fig. 2). SCoV 7a protein was also precipitated by anti-SCoV S monoclonal antibody. In contrast, an unrelated H2K monoclonal antibody failed to precipitate any of these viral proteins, exhibiting the specificity of the virus capture assay. The virus capture assay was quite effective for precipitating intact SCoV particles, as neither the 7a protein nor the M protein, the most abundant viral structural protein, was detected in the supernatant of the immunoprecipitated samples (data not shown). Because 7a protein was identified in the SCoV samples purified with two different purification methods, we concluded that 7a protein was a novel SCoV structural protein.
FIG. 2.
SCoV capture assay. SCoV particles in clarified supernatants from SCoV-infected Caco2 cells were pelleted down by centrifugation on a 20% sucrose cushion. The suspended pellets were immunoprecipitated by a nonspecific mouse anti-H2K monoclonal antibody (α-H2K) or by a mouse anti-SCoV S monoclonal antibody (α-S) to capture virions. Immunoprecipitated virions were separated by SDS-PAGE and subsequently analyzed by Western blotting using rabbit anti-SCoV 7a antibody (7a), rabbit anti-SCoV M antibody (M), rabbit anti-SCoV S protein (S), and rabbit anti-3a protein (3a) to detect viral proteins in the captured virions. M+IgG (L) represents a mixture of a minute IgG light chain signal (see left lane) and a fast-migrating major M protein signal.
SCoV 3a protein is dispensable for incorporation of 7a protein into virions.
A past study reporting the interaction between expressed 3a and 7a proteins (39) led us to examine the possibility that SCoV 7a protein was incorporated into SCoV particles through an interaction between proteins 3a and 7a. To this end, we have established a SCoV VLP system, in which SCoV VLPs were produced from the cells coexpressing viral structural proteins. SCoV genes were cloned downstream of the chicken β-actin promoter in the pCAGGS mammalian expression plasmid for the expression of viral proteins. Subconfluent 293T cells were cotransfected with pCAGGS-S, expressing SCoV S protein, pCAGGS-M, expressing SCoV M protein, pCAGGS-N, expressing SCoV N protein, and pCAGGS-E, expressing SCoV E protein. All of these proteins are common coronavirus structural proteins. Cells were metabolically labeled with Tran[35S] for 6 h from 42 to 48 h posttransfection. At 48 h posttransfection, culture media were collected and clarified. Released VLPs were partially purified through ultracentrifugation on a discontinuous sucrose gradient for 3 h and subsequently on a continuous sucrose gradient overnight. Ten fractions were collected from the bottom, and the VLPs in each fraction were pelleted through 20% sucrose cushions. Samples were examined using SDS-PAGE and autoradiography. SCoV VLPs containing S, M, and N proteins were detected, and most of them were found in fraction 6 (sucrose density, 1.167 g/ml) (Fig. 3). We were unable to detect the E protein by separating SCoV VLP proteins in a higher concentration of a gel, suggesting a low abundance of E protein in SCoV VLPs.
FIG. 3.
Production and purification of SCoV VLPs. Cultures of 293T cells were transfected with 14 μg of pCAGGS-S, 1.4 μg of pCAGGS-M, 2.8 μg of pCAGGS-N, and 9 μg of pCAGGS-E. The cells were labeled with Tran[35S] for 6 h from 42 to 48 h posttransfection. The culture media were clarified, and VLPs were purified by centrifugation on a discontinuous sucrose gradient and on a subsequent continuous sucrose gradient. Ten fractions were collected from the bottom of the gradient. VLP samples in each fraction were pelleted by centrifugation through 20% sucrose cushions. The pellets were dissolved in 1× SDS-PAGE loading buffer and analyzed using SDS-PAGE and autoradiography.
The role of 3a protein in the assembly of 7a protein into SCoV particles was studied by examining incorporation of 7a protein into VLPs in the presence and absence of SCoV 3a protein. Subconfluent 293T cells were transfected with a mixture of pCAGGS-S, pCAGGS-M, pCAGGS-N, pCAGGS-E, and pCAGGS-7a, which expresses 7a protein, either with or without pCAGGS-3a, which expresses 3a protein. For VLPs without 3a protein, pCAGGS plasmid was used instead of pCAGGS-3a. At 48 h after transfection, released VLPs in the clarified culture media were pelleted by using ultracentrifugation through a 20% sucrose cushion. Then, two SCoV VLP preparations, with or without 3a protein, were suspended in NTE buffer containing 0.3% of BSA and subjected to the virus capture assay with anti-SCoV S monoclonal antibody. As expected, Western blot analysis of the captured SCoV VLPs indicated that both VLPs contained N, S, and M proteins (Fig. 4). The captured VLPs from both preparations contained 7a protein. VLPs from the cells coexpressing 3a protein also contained 3a protein. Although the 7a protein levels in the VLPs from the 3a-expressing cells were slightly higher than levels from the cells not expressing 3a protein, the amounts of intracellular 7a protein were also similarly larger in the former (Fig. 4). Two S protein signals were detected in the cell extracts, and we suspect that the fast-migrating signal represented an unglycosylated form of S protein. The partially excised band above the N protein in the captured VLPs represented the 50-kDa IgG heavy chain, which migrated closely to the 45-kDa N protein. These data demonstrated that the expressed 3a protein and 7a protein were incorporated into SCoV VLPs yet that expression of 3a protein was dispensable for incorporation of 7a protein into SCoV VLPs. Our data also indicated that the major viral structural proteins, i.e., S protein, M protein, N protein, and E protein, were sufficient for the assembly of 7a protein into virus particles, while other viral proteins, including those proteins encoded by gene 1 and other SCoV accessory proteins, were not required for the assembly of 3a and 7a proteins into SCoV VLPs.
FIG. 4.
Assembly of SCoV 7a protein into VLPs. Initially, 3 μg of pCAGGS-S, 3 μg of pCAGGS-M, 3 μg of pCAGGS-N, 0.5 μg of pCAGGS-E, and 2 μg of pCAGGS-7a with 1.5 μg of pCAGGS-3a (3a+) or 1.5 μg of pCAGGS empty plasmid (3a−) were transfected into subconfluent 293T cells. At 48 h posttransfection, media were harvested and clarified and the released VLPs were pelleted by centrifugation through a 20% sucrose cushion. Cell extracts were also prepared at 48 h posttransfection. SCoV VLPs were resuspended in NTE buffer containing 0.3% of BSA and immunoprecipitated with mouse anti-SCoV S monoclonal antibody. The captured SCoV VLPs (VLP) and cell lysates (Cell lysate) were analyzed using Western blot analysis with anti-7a antibody (7a), anti-S protein antibody (S), anti-M protein antibody (M), anti-N protein antibody (N), and anti-3a antibody (3a).
Analysis of 7a protein assembly into SCoV VLPs.
The SCoV VLP system was used to further characterize the role of SCoV viral proteins in incorporation of 7a protein into SCoV VLPs. Cultures of 293T cells were transfected with different combinations of plasmids, as shown in Fig. 5; pCAGGS plasmid was added to adjust the total amount of DNAs to the same level in all of the different samples. The released VLPs were recovered by ultracentrifugation through a 20% sucrose cushion and examined on Western blots (Fig. 5). Consistent with our previous study (13), 7a protein was not released from 7a protein-expressing cells. A low level of 7a protein was detected in the pellets of the culture media prepared from the cells cotransfected with pCAGGS-E and pCAGGS-7a, suggesting that coexpression of E protein and 7a protein resulted in the release of a low level of 7a protein into culture fluid. Because expression of mouse hepatitis virus (MHV) E protein alone (23) or infectious bronchitis virus E protein alone (5) results in release of membrane vesicles containing E protein, the 7a protein might have been released into putative SCoV E protein membrane vesicles. A substantial increase in the amount of 7a protein in culture media was observed to occur when 7a protein was coexpressed with E and M proteins (Fig. 5). Western blot analysis also revealed the presence of M protein in the same sample. Past studies of coronavirus assembly demonstrated that coexpression of M and E proteins results in VLP production and that S protein is dispensable for VLP production (42). Accordingly, our data suggested that VLPs containing 7a protein were released from the cells expressing M, E, and 7a proteins. The amounts of 7a protein in the released VLPs did not change substantially when N and S proteins were coexpressed with M, E, and 7a proteins, although a tendency towards increased amounts of M protein in the presence of N protein was seen. In fact, when a reduced amount of pCAGGS-M was used, cotransfection of pCAGGS-N with pCAGGS-M and pCAGGS-E substantially increased VLP production compared with cotransfection of the latter two plasmids only (unpublished data), suggesting that SCoV N protein expression somehow promoted SCoV VLP production. These data implied that 7a protein was assembled into VLPs containing M and E proteins.
FIG. 5.
Release of SCoV 7a protein in the presence of other viral structural proteins. 7a protein was coexpressed with S protein, M protein, N protein, and E protein in different combinations, as indicated, in 293T cells. Plasmid quantities for this analysis were 1.5 μg of pCAGGS-S, 1.5 μg of pCAGGS-M, 1.5 μg of pCAGGS-N, 0.25 μg of pCAGGS-E, and 1 μg of pCAGGS-7a. Total DNA levels were adjusted by adding pCAGGS. At 48 h after transfection, media were harvested, and released VLPs and/or membrane vesicles were pelleted through a 20% sucrose cushion. Media (Media) and cell lysates (Cell) were analyzed for N protein, M protein, and 7a protein by Western blotting.
7a protein interacts with S protein.
To understand the mechanism of 7a protein assembly into SCoV VLPs, we next used coimmunoprecipitation to determine if 7a protein interacts with M protein in cells expressing both proteins. We were unable, however, to demonstrate the expected interaction (data not shown). We also tested whether anti-7a antibody could coimmunoprecipitate E protein along with 7a protein in cells coexpressing 7a and E proteins. The anti-7a antibody pulled down the 7a protein successfully, but we failed to detect coimmunoprecipitated E protein (data not shown). A reciprocal experiment using anti-E protein antibody was not performed, due to the lack of an antibody suitable for immunoprecipitation.
We next tested whether 7a protein interacted with S protein. Subconfluent 293T cells were transfected with pCAGGS-7a alone, pCAGGS-S alone, or a mixture of pCAGGS-7a and pCAGGS-S. At 24 h posttransfection, clarified cell extracts were incubated with mouse anti-SCoV S monoclonal antibody. Western blot analysis of the precipitated immunocomplex demonstrated that anti-S antibody coimmunoprecipitated 7a protein along with S protein (Fig. 6A). The anti-S antibody did not immunoprecipitate 7a protein from the extracts of pCAGGS-7a-transfected cells (Fig. 6A), indicating it did not cross-react with 7a protein. Western blot analysis of the cell extracts also showed that coexpression of 7a protein and S protein did not affect the accumulation of either protein. A complementary experiment, looking at whether anti-7a antibody coimmunoprecipitated S protein, showed that anti-7a antibody did not coimmunoprecipitate the S protein from coexpressing cells (data not shown). To circumvent this problem, we coexpressed 7a-myc protein, in which a myc tag was attached at the C terminus of 7a protein, and S protein in 293T cells and performed the immunoprecipitation experiment using anti-myc antibody (Fig. 6B). Anti-myc antibody coprecipitated S protein along with 7a-myc protein. Again, cotransfection of two plasmids did not affect the intracellular accumulation of S and 7a proteins. These data revealed the interaction of 7a protein with S protein in coexpressing cells.
FIG. 6.
Interaction of SCoV 7a protein with S protein in coexpressing cells. Five micrograms of pCAGGS-7a plasmid and 5 μg of pCAGGS-S plasmid, either alone or in combination, as indicated (A), or 5 μg of pcDNA-7a-myc and 5 μg of pCAGGS-S, either alone or in combination, as indicated (B), were transfected into 293T cells. At 24 h after transfection, cell lysates were prepared in radioimmunoprecipitation assay lysis buffer, and expressed proteins were immunoprecipitated with mouse anti-SCoV S monoclonal antibody (A) or anti-myc antibody (9E10) (B). Precipitated immune complexes were examined using Western blot analysis with rabbit anti-S antibody and anti-7a antibody, as indicated. Levels of 7a protein, 7a-myc protein, and S protein in cell lysates (Input) were also examined by Western blot assay. IP, immunoprecipitation; IB, immunoblotting.
DISCUSSION
The present study unambiguously demonstrated that SCoV 7a protein was a SCoV structural protein by detecting SCoV 7a protein in purified SCoV both by a sucrose gradient purification method and by a SCoV capture assay using anti-S antibody. To understand the mechanism of the assembly of 7a protein into SCoV, a SCoV VLP production system that used a eukaryotic expression vector for expression of viral structural proteins was developed. SCoV VLPs containing 7a protein were produced from cells coexpressing 7a, S, M, E, and N proteins. Although 3a protein and 7a protein have been shown to interact with each other in coexpressing cells (39) and 3a protein is a SCoV structural protein (15), 3a protein was not necessary for assembly of 7a into SCoV VLPs. Consistent with our previous study (13), 7a protein was not released into culture fluid from the cells expressing 7a protein alone, while coexpression of 7a and E proteins resulted in the release of 7a protein into culture medium. SCoV 7a protein was released more efficiently from the cells coexpressing 7a, E, and M proteins, most probably as a form of SCoV VLPs containing M, E, and 7a protein. Coimmunoprecipitation analysis demonstrated that 7a protein interacted with SCoV S protein in coexpressing cells; however, the biological significance of this interaction is unclear.
SCoV 3a protein was the first SCoV accessory protein identified as a viral structural protein (15, 37). Here, we further determined that SCoV 7a protein was the second SCoV accessory protein identified as a viral structural protein. The structural status of accessory proteins of other coronaviruses has not been studied extensively; only MHV I protein is known to be assembled into virus particles (9). The presence of two SCoV accessory proteins in the SCoV virion implies that some of the accessory proteins of other coronaviruses may also be viral structural proteins. In addition to being a structural protein, SCoV 3a protein is released from SCoV-infected cells and 3a-expressing cells in membranous structures (13). In contrast, the present study (Fig. 5) and our previous study (13) both demonstrated that 7a protein was not released from 7a-expressing cells. Analysis of SCoV samples purified by equilibrium ultracentrifugation on sucrose gradients showed that most of the 7a protein was detected in the fractions containing abundant SCoV S, N, and M proteins (Fig. 1). Also, SCoV 7a protein was detected only in SCoV particles that were captured by anti-S antibody in the SCoV capture assay (Fig. 2) and not in the postimmunoprecipitation supernatant (data not shown). These data demonstrated that, unlike SCoV 3a protein, 7a protein was released only from SCoV-infected cells in association with virus particles.
Coronavirus VLPs have been used to study the mechanisms of virus assembly (5, 23, 42). Most previous studies used exogenous viruses to drive expression of viral proteins (2, 23, 28, 42). Studies of SCoV using recombinant baculovirus expression systems found that expression of SCoV M and E proteins was sufficient for VLP formation (11, 26). If S protein is expressed with M and E proteins, VLPs showing typical coronavirus-like morphology and size are produced (11, 26). In addition, SCoV VLP production by use of a recombinant vaccinia virus has been reported (12). Huang et al. demonstrated that the eukaryotic expression vector-mediated expression of SCoV M and N proteins results in accumulation of intracellular SCoV pseudoparticles (14). Although they did not show the release of pseudoparticles (or VLPs) from plasmid-transfected cells, their results were distinctly different from those of previous studies that have demonstrated the importance of M and E proteins for efficient coronavirus VLP production (42). The difference in the methods of expressing viral protein might have led to the different results. Otherwise, the mechanism of SCoV assembly may be distinctly different from that known for other coronaviruses. The present study showed that coexpression of SCoV S, M, E, and N proteins resulted in efficient production and release of SCoV VLPs (Fig. 3). Furthermore, our data suggested that coexpression of E, M, and 7a proteins resulted in efficient SCoV VLP production (Fig. 5). Our SCoV VLP system not only eliminated possible unwanted effects caused by exogenous viruses in the study of coronavirus assembly but also yielded VLP samples free of these exogenous viruses. Furthermore, these particles could serve as sources of viral antigens for diagnostic and immunization purposes as well as for detailed analyses of coronavirus structure that would not entail handling a highly pathogenic coronavirus (e.g., SCoV) in a biosafety level 3 laboratory.
Release of 7a protein was detected from cells coexpressing 7a and E proteins but not from cells expressing 7a protein alone (Fig. 5), suggesting that E protein induced the release of 7a protein. MHV E protein is released in membrane vesicles from E protein-expressing cells and MHV-infected cells (23), and infectious bronchitis virus E protein is released from E protein-expressing cells (5); likewise, SCoV E protein may be released in membrane vesicles from E protein-expressing cells. If this is the case, coexpressed 7a protein might be included in the putative E protein-containing vesicles. Coexpression of 7a, E, and M proteins resulted in efficient release of 7a protein into culture media (Fig. 5). M protein was also detected in the culture media, and the possibility is very likely that SCoV VLPs containing M, E, and 7a proteins were released from the coexpressing cells. We suspect that E or M protein interacted with 7a protein to facilitate the assembly of 7a protein into VLPs; investigating possible 7a-E and/or 7a-M interactions is another topic of study. Although we showed that 7a protein interacted with S protein in coexpressing cells (Fig. 6), it appears that this interaction was not essential for the assembly of 7a protein into virus particles (Fig. 5).
To estimate the abundance of SCoV 3a and 7a proteins in SCoV particles, the gradient-purified SCoV particles were applied to an SDS-PAGE gel, and the gel was stained with colloidal Coomassie blue (Bio-Rad). In addition to major viral structural S, N, and M proteins, multiple bands, which probably represented contaminated host proteins and/or partially degraded viral proteins, were detected (data not shown). These bands interfered with the positive identification of the 3a and 7a proteins. Although Western blot analysis clearly showed the presence of 3a and 7a proteins in SCoV VLPs from the cells coexpressing S, M, E, N, 3a, and 7a proteins (Fig. 4), we were unable to demonstrate the presence of 3a and 7a proteins in the purified radiolabeled SCoV VLPs from the cells coexpressing the same proteins (data not shown), suggesting the presence of low abundances of 3a and 7a proteins in SCoV VLPs.
Although 7a protein is not essential for SCoV replication in cell culture (45), its assembly into SCoV implies the importance of 7a protein in the viral replication circle; the presence of 7a protein in SCoV particles indicates that virion-associated 7a protein may exert its function early in infection. The incoming 7a protein might alter the intracellular environment immediately after infection, by itself or by binding a putative cellular ligand(s), in a way that might be beneficial for virus replication. Previous studies demonstrated that expressed 7a protein affects host cells by inducing apoptosis (38), inhibiting translation of cellular protein (16), activating p38 MAPK (16), and blocking cell cycle progression at G0/G1 (47). In fact, activation of p38 MAPK occurs in both SCoV infection and MHV infection (1, 25). In the case of MHV, activated p38 MAPK promotes virus replication through virus-induced translation initiation factor 4E activation (1). In MHV-infected cells, the cell cycle is arrested at the G0/G1 phase, which may benefit MHV replication and MHV pathogenesis (4). In contrast, MHV replication is suppressed in the cells that undergo apoptosis (3). Accordingly, some of these reported functions of SCoV 7a protein from overexpression studies appear to be beneficial for SCoV replication, whereas some do not. Further studies using SCoV, its 7a gene deletion mutant (45), SCoV VLPs carrying 7a protein, and those lacking 7a protein will be useful to determine the biological functions of virion-associated 7a protein.
Acknowledgments
This work was supported by Public Health Service grant AI29984 to S.M. and NIH grant N01-AI-30039 via Respiratory Pathogen Research Unit, SARS, to C.-T.K.T. C.H. was supported by the James W. McLaughlin Endowment, and N.I. was supported by a fellowship for long-term overseas research for young investigators sponsored by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Banerjee, S., K. Narayanan, T. Mizutani, and S. Makino. 2002. Murine coronavirus replication-induced p38 mitogen-activated protein kinase activation promotes interleukin-6 production and virus replication in cultured cells. J. Virol. 76:5937-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. J., and S. Makino. 2002. Murine coronavirus-induced apoptosis in 17Cl-1 cells involves a mitochondria-mediated pathway and its downstream caspase-8 activation and bid cleavage. Virology 302:321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C.-J., and S. Makino. 2004. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 78:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 8.Fielding, B. C., Y. J. Tan, S. Shuo, T. H. Tan, E. E. Ooi, S. G. Lim, W. Hong, and P. Y. Goh. 2004. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 78:7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, F., D. Peng, S. T. Hingley, S. R. Weiss, and P. S. Masters. 1997. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 71:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng, H., Y. M. Liu, W. S. Chan, A. W. Lo, D. M. Au, M. M. Waye, and Y. Y. Ho. 2005. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 579:6763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, Y., P. H. Lin, C. Y. Liu, S. P. Lee, and Y. C. Chao. 2004. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 318:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh, P. K., S. C. Chang, C. C. Huang, T. T. Lee, C. W. Hsiao, Y. H. Kou, I. Y. Chen, C. K. Chang, T. H. Huang, and M. F. Chang. 2005. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 79:13848-13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, C., K. Narayanan, N. Ito, C. J. Peters, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. J. Virol. 80:210-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, Y., Z. Y. Yang, W. P. Kong, and G. J. Nabel. 2004. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 78:12557-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 79:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, and P. Palese. 2006. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 80:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 18.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviruses, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 20.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 22.Li, W., C. Zhang, J. Sui, J. H. Kuhn, M. J. Moore, S. Luo, S. K. Wong, I. C. Huang, K. Xu, N. Vasilieva, A. Murakami, Y. He, W. A. Marasco, Y. Guan, H. Choe, and M. Farzan. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda, J., A. Maeda, and S. Makino. 1999. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 25.Mizutani, T., S. Fukushi, M. Saijo, I. Kurane, and S. Morikawa. 2004. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 319:1228-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortola, E., and P. Roy. 2004. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 576:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossel, E. C., C. Huang, K. Narayanan, S. Makino, R. B. Tesh, and C. J. Peters. 2005. Exogenous ACE2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J. Virol. 79:3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan, K., C. J. Chen, J. Maeda, and S. Makino. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77:2922-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure (Cambridge) 13:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278:52347-52354. [DOI] [PubMed] [Google Scholar]
- 32.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balasch, J. Plana, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul, P. S., E. M. Vaughn, and P. G. Halbur. 1997. Pathogenicity and sequence analysis studies suggest potential role of gene 3 in virulence of swine enteric and respiratory coronaviruses. Adv. Exp. Med. Biol. 412:317-321. [DOI] [PubMed] [Google Scholar]
- 34.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu, X. X., P. Hao, X. J. Song, S. M. Jiang, Y. X. Liu, P. G. Wang, X. Rao, H. D. Song, S. Y. Wang, Y. Zuo, A. H. Zheng, M. Luo, H. L. Wang, F. Deng, H. Z. Wang, Z. H. Hu, M. X. Ding, G. P. Zhao, and H. K. Deng. 2005. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 280:29588-29595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 37.Shen, S., P. S. Lin, Y. C. Chao, A. Zhang, X. Yang, S. G. Lim, W. Hong, and Y. J. Tan. 2005. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 330:286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan, Y. J., B. C. Fielding, P. Y. Goh, S. Shen, T. H. Tan, S. G. Lim, and W. Hong. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 78:14043-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan, Y.-J., E. Teng, S. Shen, T. H. P. Tan, P.-Y. Goh, B. C. Fielding, E.-E. Ooi, H.-C. Tan, S. G. Lim, and W. Hong. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 78:6723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 41.Vaughn, E. M., P. G. Halbur, and P. S. Paul. 1995. Sequence comparison of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3-1 genes. J. Virol. 69:3176-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wesley, R. D., R. D. Woods, and A. K. Cheung. 1991. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J. Virol. 65:3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, C. J., Y. C. Chen, C. H. Hsiao, T. C. Kuo, S. C. Chang, C. Y. Lu, W. C. Wei, C. H. Lee, L. M. Huang, M. F. Chang, H. N. Ho, and F. J. Lee. 2004. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 565:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan, X., J. Wu, Y. Shan, Z. Yao, B. Dong, B. Chen, Z. Zhao, S. Wang, J. Chen, and Y. Cong. 2006. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology 346:74-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng, R., R. F. Yang, M. D. Shi, M. R. Jiang, Y. H. Xie, H. Q. Ruan, X. S. Jiang, L. Shi, H. Zhou, L. Zhang, X. D. Wu, Y. Lin, Y. Y. Ji, L. Xiong, Y. Jin, E. H. Dai, X. Y. Wang, B. Y. Si, J. Wang, H. X. Wang, C. E. Wang, Y. H. Gan, Y. C. Li, J. T. Cao, J. P. Zuo, S. F. Shan, E. Xie, S. H. Chen, Z. Q. Jiang, X. Zhang, Y. Wang, G. Pei, B. Sun, and J. R. Wu. 2004. Characterization of the 3a protein of SARS-associated coronavirus in infected Vero E6 cells and SARS patients. J. Mol. Biol. 341:271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]