Abstract
Post-translational modification of proteins by small polypeptides, such as ubiquitin, has emerged as a common and important mechanism for regulating protein function. Small ubiquitin-like modifier (SUMO) is a small protein that is structurally related to but functionally different from ubiquitin. We report the identification and functional analysis of AtSUMO1, AtSUMO2, and AtSCE1a as components of the SUMO conjugation (sumoylation) pathway in Arabidopsis. In yeast-two hybrid assays, AtSUMO1/2 interacts specifically with a SUMO-conjugating enzyme but not with a ubiquitin-conjugating enzyme. AtSCE1a, the Arabidopsis SUMO-conjugating enzyme ortholog, conjugates SUMO to RanGAP in vitro. AtSUMO1/2 and AtSCE1a colocalize at the nucleus, and AtSUMO1/2 are conjugated to endogenous SUMO targets in vivo. Analysis of transgenic plants showed that overexpression of AtSUMO1/2 does not have any obvious effect in general plant development, but increased sumoylation levels attenuate abscisic acid (ABA)–mediated growth inhibition and amplify the induction of ABA- and stress-responsive genes such as RD29A. Reduction of AtSCE1a expression levels accentuates ABA-mediated growth inhibition. Our results suggest a role for SUMO in the modulation of the ABA signal transduction pathway.
INTRODUCTION
Eukaryotic protein function is regulated in part by the covalent attachment of ubiquitin (Ub) and ubiquitin-like (Ubl) proteins. These small polypeptides are attached to target proteins by an isopeptide bond between the C terminus of the modifier and the ɛ-amino group of a Lys residue in the substrate. Ub/Ubl are synthesized as precursor proteins that are processed to expose the commonly conserved Ub/Ubl C-terminal (Gly-Gly) motif. Ub/Ubl conjugation to target proteins involves the sequential action of activating (E1), conjugating (E2), and ligase (E3) enzymes (Hochstrasser, 2000; Pickart, 2001). Small ubiquitin-like modifier (SUMO) resembles ubiquitin in its structure and mechanism of ligation. In yeast and animals, a single heterodimeric E1 (AOS1/UBA2) and a single E2 (UBC9) are essential for sumoylation. SUMO generally is conjugated to a target Lys located within a ψKxE consensus sequence (where ψ is an aliphatic residue). In contrast to ubiquitination, the E1 and E2 activities are sufficient for SUMO conjugation in vitro, suggesting E3-independent mechanisms for SUMO conjugation under these conditions (Melchior, 2000; Müller et al., 2001). Several E3-like factors that enhance the accumulation of SUMO-conjugated substrates both in vivo and in vitro have been identified in yeast and mammalian cells, although none appears to alter UBC9 specificity (Johnson and Gupta, 2001; Kahyo et al., 2001; Sachdev et al., 2001; Takahashi et al., 2001; Kotaja et al., 2002; Pichler et al., 2002).
In mammalian systems, both subunits of the SUMO E1 activating enzyme localize predominantly in the nucleus (Azuma et al., 2001; Rodriguez et al., 2001). Moreover, UBC9 is present in a complex with SUMO1-modified RanGAP1 and RanBP2 at the nuclear pore complex (Saitoh et al., 1997, 1998; Lee et al., 1998; Zhang et al., 2002). The modification of substrates by SUMO is reversible, and desumoylation is mediated by a family of SUMO proteases (Li and Hochstrasser, 1999, 2000), several of which also localize at the nuclear pore complex (Takahashi et al., 2000; Hang and Dasso, 2002; Zhang et al., 2002).
Many ubiquitin-modified proteins are targeted for degradation by the 26S proteasome. Meanwhile, the general consequence for SUMO modification remains unclear and may vary depending on the target (Müller et al., 2001). Sumoylation competes with ubiquitination for the same Lys within IκBα and proliferating cell nuclear antigen, protecting the SUMO-modified proteins from proteasome degradation (Desterro et al., 1998; Hoege et al., 2002). Sumoylation of mammalian RanGAP1 relocalizes it from the cytoplasm to the nuclear pore complex (Matunis et al., 1996; Mahajan et al., 1997). SUMO also is involved in the nuclear localization of promyelocytic leukemia (Kamitani et al., 1998), centromere segregation (Tanaka et al., 1999), and septin ring formation (Johnson and Blobel, 1999; Takahashi et al., 1999), among other phenomena. In addition, HsSUMO1 and HsSUMO2/3 are conjugated to different substrates, and HsSUMO2/3 conjugation has been proposed to constitute a new pathway of acute and reversible stress response (Saitoh and Hinchey, 2000).
Ubiquitin is the best-studied protein modifier in plants and plays an important regulatory role in the auxin signal transduction pathway (Estelle, 2001; Leyser, 2002; Xie et al., 2002). By contrast, little is known about the function of protein modification by SUMO. Two different studies have suggested that SUMO plays an important role in pathogen defense responses. The tomato SUMO ortholog (LeSUMO) interacts with the effector ethylene-inducing xylanase (EIX) from the fungus Trichoderma viridae in yeast two-hybrid assays. The expression of LeSUMO in tobacco transgenic plants suppressed the induction of the defense response by EIX, suggesting that LeSUMO may act as a repressor of the plant defense pathway (Hanania et al., 1999). AvrBsT, a virulence factor from the plant pathogen Xanthomonas campestris, possesses SUMO protease activity. Mutations in the predicted AvrBsT catalytic domain prevented the elicitation of the hypersensitive response in tobacco leaves (Orth et al., 2000).
To elucidate the role of SUMO conjugation in vivo, we used Arabidopsis as a model system. We show here that Arabidopsis contains four genes that code for SUMO paralogs and a single gene that encodes a SUMO-conjugating enzyme (AtSCE1a). AtSCE1a interacted specifically with SUMO in yeast two-hybrid assays and was able to conjugate HsSUMO1 to HsRanGAP1 in vitro. Analysis of transgenic plants with altered sumoylation levels suggested that SUMO modulates the signaling of the hormone abscisic acid (ABA), which mediates plant responses to environmental stresses such as cold, drought, and high salinity (Leung and Giraudat, 1998). During vegetative growth, endogenous ABA levels increased in response to these adverse environmental stimuli, and histochemical studies of root tissues supported the notion of an ABA-mediated inhibition of cell elongation and an arrest in mitotic cell activity (Himmelbach et al., 1998). Plants with increased sumoylation levels showed an attenuation of ABA-mediated growth inhibition, and the opposite effect was observed when AtSCE1a levels were reduced. In addition, the inducibility of some ABA- and stress-responsive genes was amplified in plants that overexpressed SUMO. These results suggest a regulatory role for SUMO in the ABA signaling pathway.
RESULTS
SUMO Is Encoded by a Multigene Family, and AtSCE1a Is Encoded by a Single-Copy Gene
To identify Arabidopsis SUMO orthologs, we performed protein BLASTA (Basic Local Alignment Search Tool A) searches against nonredundant databases of the National Center for Biotechnology Information using HsSUMO1 as a query. Two SUMO orthologs were identified with significant sequence identity to HsSUMO1, and these sequences were used to identify four other SUMO paralogs in the Arabidopsis translated genome database (ATH1_seq database) at TAIR (The Arabidopsis Information Resource).
Sequence identity to the mammalian SUMO proteins was insufficient to definitively classify the plant SUMO paralogs into the analogous HsSUMO1, HsSUMO2, or HsSUMO3 families. We proceeded to designate the Arabidopsis SUMO paralogs according to the nomenclature of Kurepa et al. (2003) (work published while this manuscript was under revision), although it does not reflect any phylogenetic or functional relationship to the human SUMOs. AtSUMO2, AtSUMO3, AtSUMO4, AtSUMO5, and AtSUMO6 share 87%, 54%, 38%, 34%, and 31% of amino acid sequence identity with AtSUMO1, respectively. AtSUMO1 and AtSUMO2 are highly related, and each shares 45% and 46% amino acid sequence identity with HsSUMO1, 53% and 54% with HsSUMO2, and 54% and 56% with HsSUMO3 respectively (Figure 1A). Despite being previously designated by Kurepa et al. (2003) as SUMO orthologs, it is not clear whether AtSUMO4 and AtSUMO6 are active members of the SUMO family because the signature C-terminal GG motif required for SUMO conjugation and deconjugation is absent from these proteins.
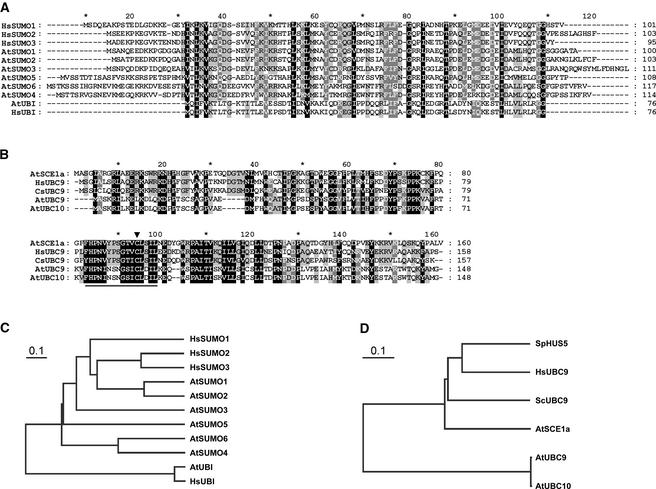
Figure 1.
Sequence Comparison of SUMO and SUMO-Conjugating Enzyme Orthologs in Arabidopsis.
(A) Amino acid sequence alignment of SUMO orthologs. HsSUMO1, HsSUMO2, HsSUMO3, AtUBI, HsUBI, and the six most significantly conserved Arabidopsis sequences retrieved from protein BLAST searches against the translated genome database at TAIR are shown. Black background and white letters corresponds to 100% conservation, gray background and white letters corresponds to 80% conservation, and gray background and black letters corresponds to 60% conservation.
(B) Amino acid sequence alignment of the SUMO-conjugating enzyme orthologs HsUBC9, ScUBC9, AtUBC9, AtUBC10, and AtSCE1a. The conserved catalytic Cys is indicated by an arrowhead, and the ubiquitin-conjugating enzyme signature (PDOC00163) present in all sequences is underlined.
(C) and (D) Phylogenetic tree representation of amino acid sequence distance among SUMO orthologs (C) and SUMO-conjugating enzyme orthologs (D).
In yeast and mammals, and in contrast to ubiquitin, the SUMO conjugation pathway has only one known E2 conjugating enzyme (UBC9) that is specific for SUMO and does not conjugate ubiquitin (Desterro et al., 1997; Gong et al., 1997; Johnson and Blobel, 1997; Schwarz et al., 1998). From protein BLASTA searches at the National Center for Biotechnology Information, we retrieved AtSCE1a as the Arabidopsis UBC9 ortholog. AtSCE1a was annotated previously as the Arabidopsis ortholog of Schizosaccharomyces pombe HUS5, a ubiquitin-conjugating enzyme required for normal mitosis (al-Khodairy et al., 1995). Because the name UBC9 is already assigned to a ubiquitin-conjugating enzyme in Arabidopsis (Girod et al., 1993), we used the name AtSCE1a to designate the SUMO-conjugating enzyme ortholog. AtSCE1a is 64% identical to HsUBC9, 59% identical to SpHUS5, and 58% identical to ScUBC9. Similarly, SpHUS5 is 66 and 61% identical to HsUBC9 and ScUBC9, respectively. AtSCE1a is 33% identical to the ubiquitin-conjugating enzymes AtUBC9 and AtUBC10, in contrast to the 98% sequence identity shared between AtUBC9 and AtUBC10 (Figure 1B). None of the Arabidopsis ubiquitin-conjugating enzymes annotated in the Arabidopsis translated genome database at TAIR shares >35% sequence identity with AtSCE1a, suggesting that a single-copy gene encodes the putative SUMO-conjugating enzyme. Phylogenetic trees representing the distances between groups of sequences were built using a cluster algorithm (Figures 1C and 1D).
Specific Interaction between AtSUMO1/2 and AtSCE1a
Comparative structural studies of SUMO-specific UBC9 with ubiquitin-specific conjugating enzymes revealed important differences that are believed to play a role in modifier discrimination (Giraud et al., 1998; Liu et al., 1999). Recently, in vitro experiments resulted in the identification of a region in ScUBC9 involved in noncovalent binding to ScSUMO (SMT3) (Bencsath et al., 2002). Because that study reported noncovalent interactions between SUMO and its conjugating enzyme, we assessed the ability of AtSCE1a to interact with AtSUMO1/2 in yeast two-hybrid assays. In these experiments, the capacity of the yeast strain HF7c to grow in the absence of His was used as a marker for the interaction between proteins. We found that His auxotrophy was restored only when AtSCE1a was cotransformed with AtSUMO1, AtSUMO2, or HsSUMO1 but not with an Arabidopsis ubiquitin, AtUBI. The opposite result was observed when AtSCE1a was substituted for an Arabidopsis ubiquitin-conjugating enzyme, AtUBC10 (Figure 2A).
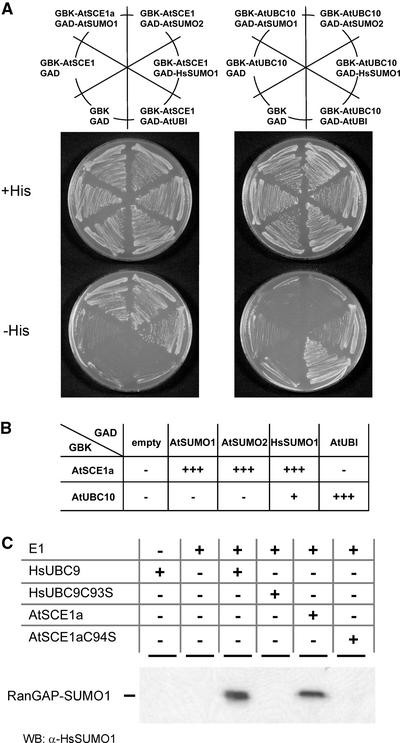
Figure 2.
AtSCE1a Interacts Specifically with SUMO in Yeast and Displays SUMO-Conjugating Activity in Vitro.
(A) Yeast two-hybrid assay. Yeast HF7c cells were cotransformed with plasmids expressing GAL4BD-AtSCE1a or GAL4B-AtUBC10 and GAL4AD alone or fused to AtSUMO1, AtSUMO2, HsSUMO1, or AtUBI. As a negative control, cells also were cotransformed with plasmids expressing GAL4BD and GAL4AD. The ability to grow in the absence of His indicates protein interaction. The plates were incubated for 2 days at 30°C, and the results were scored. GBK corresponds to GAL4BD and GAD corresponds to GAL4AD.
(B) Table representing the growth efficiency of each pair of plasmids. The number of plus signs is proportional to the growth rate, and minus signs indicate growth similar to that of the negative control.
(C) AtSCE1a displays E2 SUMO-conjugating activity in vitro. SUMO conjugation of RanGAP1 peptide (amino acids 420 to 589) was performed in the presence of human E1 and HsUB9, AtSCE1a, or mutants in the catalytic Cys (HsUBC9-C93S or AtSCE1a-C94S). Reaction products were resolved by SDS-PAGE, and conjugation of HsSUMO1 to RanGAP1 was examined by protein gel blot analysis using anti-HsSUMO1 antibodies. The reaction containing HsUBC9 was diluted 20 times before SDS-PAGE to compare the apparent molecular masses of the reaction products. WB, protein gel blot analysis.
AtSCE1a Has SUMO-Conjugating E2 Activity
The structural basis for E2-mediated SUMO conjugation was described within the context of a complex between HsUBC9 and RanGAP1 (Bernier-Villamor et al., 2002). All of the residues identified as being involved in the UBC9–substrate interaction are conserved in AtSCE1a. This observation led us to hypothesize that AtSCE1a may be able to conjugate HsSUMO1 to RanGAP1. Accordingly, we performed an in vitro assay using, as E2, AtSCE1a, HsUBC9, or their respective Cys-to-Ser substitutions within the conserved catalytic domain. Figure 2C shows that HsSUMO1 was conjugated to HsRanGAP1 only when native HsUBC9 or AtSCE1a was present in the reaction mixture. As expected, the mutant forms HsUBC9-C93S and AtSCE1a-C94S were inactive. Although SUMO conjugation was less efficient in the presence of AtSCE1a compared with HsUBC9, these results, together with the interaction specificity observed in the yeast two-hybrid assays, provide corroborating evidence that AtSCE1a is a SUMO-conjugating enzyme.
Intracellular Localization of AtSUMO1/2 and AtSCE1a
Most of the SUMO conjugates in mammalian cells are found in the nucleus, with a significant fraction being localized specifically to nuclear bodies. In HeLa cells, HsSUMO1, HsSUMO2, and HsSUMO3 are localized predominantly at the nuclear membrane, nuclear bodies, and cytoplasm, respectively (Su and Li, 2002). The SUMO-conjugating enzyme HsUBC9 is predominantly nuclear and partially concentrated at the nuclear envelope of mammalian cells in association with filaments of the nuclear pore complex (Lee et al., 1998; Zhang et al., 2002). In plants, the only study reported to date on SUMO localization was performed in tomato; that study suggested that tomato SUMO (LeSUMO) is a cytoplasmic protein (Hanania et al., 1999). Because green fluorescent protein (GFP) was fused to the LeSUMO C terminus for these experiments, the observed localization might be attributable to GFP released from LeSUMO:GFP as a result of C-terminal processing of LeSUMO by endogenous SUMO proteases (Li and Hochstrasser, 1999, 2000).
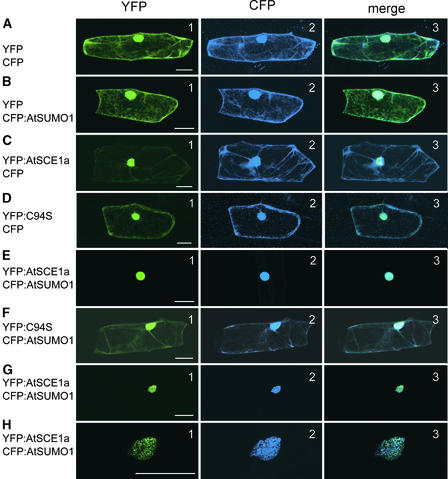
To analyze the localization of the Arabidopsis SUMO-conjugating system, fluorescent proteins were fused to the N terminus of AtSCE1a and AtSUMO1/2 to preserve SUMO:fluorescent protein fusions from SUMO-processing reactions (cyan fluorescent protein [CFP]:AtSUMO1, yellow fluorescent protein [YFP]:AtSCE1a, and YFP:AtSCE1a-C94S). Figures 3A2 and 3B2 show that AtSUMO1 was distributed widely in the cytoplasm and the nucleus in a manner similar to CFP, whereas AtSCE1a was localized preferentially in the nucleus (Figure 3C1). A point mutation in the AtSCE1a catalytic site, AtSCE1a-C94S, prevented efficient nuclear localization, suggesting a possible coupling of the catalytic activity to cellular localization (Figure 3D1). When AtSCE1a was coexpressed with AtSUMO1, both proteins colocalized strongly in the nucleus, with little signal detected in the cytoplasm (Figures 3E1 and 3E2). Again, colocalization was dependent on the integrity of the AtSCE1a catalytic Cys-94 (Figures 3F1 and 3F2). In some cells, AtSCE1a and AtSUMO1 colocalized to nuclear bodies (Figures 3G1, 3G2, 3H1, and 3H2). The same results were obtained when AtSUMO2 was used instead of AtSUMO1 (data not shown).
Figure 3.
AtSCE1a Colocalizes with AtSUMO1/2 in the Nucleus.
Epidermal onion cells were transformed transiently with vectors expressing the following fluorescent proteins: YFP + CFP (A), YFP + CFP:AtSUMO1 (B), YFP:AtSCE1a + CFP (C), YFP:AtSCE1a-C94S + CFP (D), YFP:AtSCE1a + CFP:AtSUMO1 ([E] to [H]), and YFP:AtSCE1a-C94S + CFP:AtSUMO1 (F). Row (H) is a higher magnification of row (G). Signal from YFP, CFP, and the merge of both signals are shown in the left (1), middle (2), and right (3) columns, respectively. Bars = 50 μm.
(His)6:AtSUMO1 and (His)6:AtSUMO2 Are Conjugated to Endogenous SUMO Target Proteins
To analyze endogenous SUMO conjugates in Arabidopsis, we performed protein gel blot analyses using anti-AtSUMO1 antibody. Because SUMO may be deconjugated rapidly from its targets by SUMO-specific proteases that are inhibited by N-ethylmaleimide (NEM) (Li and Hochstrasser, 1999), we tested the effect of NEM on the proteins detected by the anti-AtSUMO1 antibody. A ladder of proteins with an apparent molecular mass of >60 kD was detected only when NEM was included in the extraction buffer, suggesting that the SUMO conjugates protected by the NEM treatment are mainly high molecular mass proteins (Figure 4A).
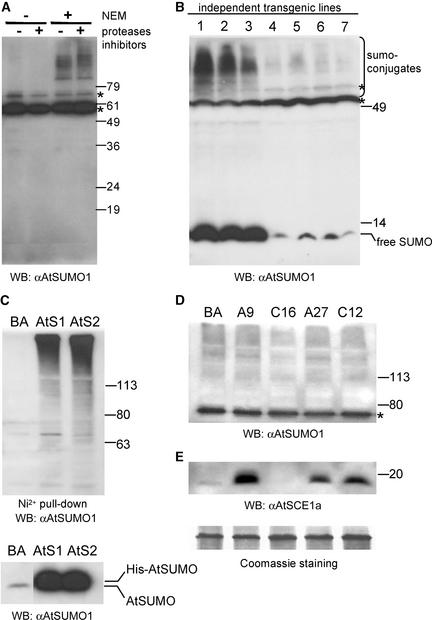
Figure 4.
SUMO Conjugates in Arabidopsis.
(A) Detection of endogenous SUMO conjugates. Protein extracts were prepared in the presence or absence of general protease inhibitors and in the presence or absence of NEM, a Cys protease inhibitor. Endogenous SUMO conjugates were detected by protein gel blot analysis (WB) using anti-AtSUMO1 polyclonal antibody.
(B) Analysis of independent T1 transgenic lines for His-AtSUMO1 expression. Protein extracts from leaves excised from 3-week-old transgenic plants were prepared in Laemmli (1970) buffer and detected by protein gel blot analysis as described for (A).
(C) Purification of His-AtSUMO1/2 conjugates. Two T2 transgenic lines expressing similar levels of free His-AtSUMO1 (AtS1) and free His-AtSUMO2 (AtS2) (bottom gel) were used to purify proteins modified by the His-tagged SUMOs using Ni2+ affinity. As a negative control, purification also was performed on protein extracts from a transgenic plant carrying an empty vector (pBA002 [BA]). Purified proteins were detected by protein gel blot analysis as for (A).
(D) Analysis of endogenous SUMO conjugates in T2 transgenic plants expressing AtSCE1a or AtSCE1a-C94S. Transgenic lines expressing AtSCE1a-His (A9 and A27) and AtSCE1a-C94S-His (C16 and C12) were detected by protein gel blot analysis as described for (A).
(E) Analysis of AtSCE1a protein levels. Expression levels of AtSCE1a were analyzed using anti-AtSCE1a polyclonal antibody in the same protein samples shown in (D). A portion of the membrane stained with Coomassie Brilliant Blue R 250 is shown as a loading control.
Asterisks indicate nonspecific bands. Numerals indicate migration positions of molecular mass markers in kilodaltons.
To study the physiological role of sumoylation in Arabidopsis, we generated several independent transgenic plants that expressed (His)6:AtSUMO1 (Figure 4B) and (His)6:AtSUMO2 (data not shown). SUMO conjugation levels in these transgenic lines correlated with the levels of free SUMO as detected by protein gel blot analysis. Interestingly, the apparent molecular mass of SUMO conjugates detected in these transgenic plants was similar to that in wild-type plants, suggesting that the recombinant (His)6:AtSUMO1/2 modified physiological SUMO targets. Affinity purification of the His-tagged proteins confirmed that His-AtSUMO1/2 were conjugated to high molecular mass proteins (Figure 4C).
We also generated transgenic plants that expressed AtSCE1a:(His)6 and AtSCE1a-C94S:(His)6. From the 20 independent lines analyzed for each construct, one line (C16) transformed with 35S-AtSCE1a-C94S:(His)6 showed diminished levels of the endogenous AtSCE1a, probably as a result of cosuppression (Figure 4E). A twofold reduction of AtSCE1a mRNA levels in this line was detected by RNA gel blot analysis (data not shown). Protein gel blot analysis using anti-AtSUMO1 antibody revealed that plants that overexpressed AtSCE1a:(His)6 showed no detectable differences in endogenous sumoylated protein levels compared with control plants. By contrast, a mild reduction of sumoylation levels was observed in the transgenic line that exhibited cosuppression of the endogenous AtSCE1a protein (Figure 4D). Sumoylation levels were more affected by AtSCE1a cosuppression than by AtSCE1a-C94S mutant expression (Figure 4D).
SUMO Modulates the ABA Signal Transduction Pathway
Transgenic plants that exhibited increased or decreased sumoylation levels from AtSUMO1/2-overexpressing or AtSCE1a- cosuppressed lines, respectively, showed no impairment in general development. Consequently, we analyzed whether hormonal and abiotic stress responses were affected in these plants.
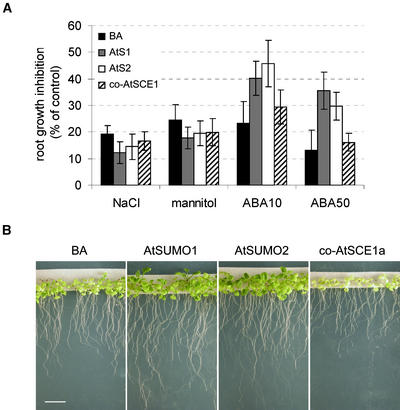
Figure 5A shows that ABA-mediated root growth inhibition was reduced significantly in plants that overexpressed either AtSUMO1 or AtSUMO2 compared with empty vector control plants, but no significant differences were observed in plants that exhibited cosuppression of AtSCE1a. Transgenic plants that expressed 35S-AtSUMO1/2 displayed wild-type responses to auxin (0.1 and 1 μM 1-naphthylacetic acid), ethylene (0.2 and 2 μM 1-aminocyclopropane-1-carboxylic acid), or cytokinin (0.5 μM benzyladenine) with respect to root growth (data not shown). Similarly, the root growth of transgenic plants was inhibited by 200 mM NaCl or 400 mM mannitol to the same extent as that of vector control plants.
Figure 5.
Sumoylation Modulates ABA-Induced Growth Inhibition.
(A) ABA-mediated root growth inhibition is attenuated in SUMO-overexpressing plants. T2 transgenic plants expressing His-AtSUMO1/2 (AtS1 and AtS2), AtSCE1a cosuppression (co-AtSCE1a), or carrying the empty vector (pBA002 [BA]) were germinated on medium A plates for 3 days, transferred to medium A control or medium A supplemented with 200 mM NaCl, 400 mM mannitol, 10 μM ABA (ABA10), or 50 μM ABA (ABA50), and incubated for 4 additional days before root growth was scored (n = 20). Root length obtained in control medium A was taken to be 100%, and growth in the other conditions was expressed with respect to the control value. Error bars are shown.
(B) ABA-induced growth inhibition is attenuated in SUMO-overexpressing plants and accentuated in plants showing AtSCE1a cosuppression. Three-day-old T2 seedlings were transferred to medium A supplemented with 5 μM ABA and incubated for 24 days before photographs were taken. Bar = 1 cm.
Interestingly, when plants were incubated for 24 days in 5 μM ABA, transgenic plants with AtSCE1a cosuppression showed the highest growth inhibition and were highly chlorotic. ABA-mediated growth inhibition was less acute in control plants, whereas transgenic plants that overexpressed AtSUMO1/2 showed the best growth capability under these conditions (Figure 5B). Control experiments showed no growth differences between transgenic plants grown on medium A (data not shown).
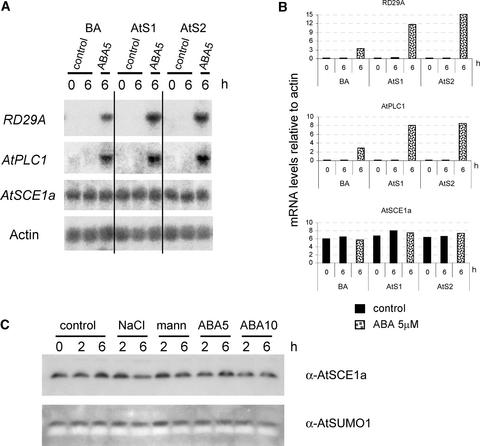
Analysis of mRNA levels of ABA- and stress-responsive genes upon ABA treatment revealed that RD29A and AtPLC1 expression levels were upregulated in AtSUMO1- and AtSUMO2-overexpressing plants compared with vector control plants (Figures 6A and 6B). Other analyzed genes—KIN2, COR47, RD22, and AtMyb—showed minor or no induction differences (data not shown). When plants that exhibited AtSCE1a cosuppression were treated with ABA, we did not observe significant differences in the RD29A mRNA levels compared with control plants (data not shown). This result was not completely surprising because, in these plants, more severe ABA-mediated growth inhibition was observed only after prolonged incubation in ABA (Figure 4D).
Figure 6.
AtSUMO Regulates the Expression of Stress-Responsive Genes.
Twelve-day-old T2 transgenic plants expressing His-AtSUMO1/2 or plants harboring the empty vector (pBA002 [BA]) were treated with or without 5 μM ABA.
(A) Expression levels of stress-responsive genes are modified in AtSUMO-overexpressing plants. Actin mRNA levels were used as loading controls.
(B) Graphs representing relative expression levels upon ABA treatment. Values are expressed relative to actin mRNA levels.
(C) Analysis of AtSCE1a and SUMO protein levels under ABA and stress treatments. Protein extracts from empty vector control plants were examined by protein gel blot analysis using anti-AtSCE1a or anti-AtSUMO1 polyclonal antibodies. mann, mannitol.
We performed protein gel blot analyses on protein extracts from control plants treated with NaCl, mannitol, and ABA and found that free AtSUMO and AtSCE1a protein levels were not altered significantly (Figure 6C). Similar results were obtained with AtSCE1a (Figure 6A) and AtSUMO1/2 (data not shown) mRNA levels. These results are consistent with the observation that levels of SUMO protein conjugates were not altered visibly by any treatment (data not shown).
DISCUSSION
We have confirmed the identity of AtSUMO1, AtSUMO2, and AtSCE1a as components of the sumoylation pathway in Arabidopsis using yeast two-hybrid and in vitro sumoylation assays and analyzed their biological functions. The Arabidopsis SUMO-conjugating enzyme AtSCE1a appears to be encoded by a single-copy gene, like UBC9, which in yeast and mammals is encoded by a single gene (Melchior, 2000). By contrast, 37 genes are predicted to encode ubiquitin-conjugating enzymes in Arabidopsis, although the biochemical functions of approximately half of them have not been determined (Bachmair et al., 2001). In yeast two-hybrid assays, AtSCE1a interacted specifically with AtSUMO1/2 and HsSUMO1 but not with AtUBI, and AtSUMO1/2 did not interact with a ubiquitin-conjugating enzyme, AtUBC10. In vitro, AtSCE1a was able to conjugate HsSUMO1 to RanGAP1, suggesting that it is both competent for SUMO transfer from E1 and capable of conjugating SUMO to a consensus receptor Lys. The observation that AtSCE1a conjugated HsSUMO less efficiently that HsUBC9 could be the result of a poor interaction between AtSCE1a and either human E1 or RanGAP1 or both, although comparative analysis between HsUBC9 and AtSCE1a has shown that all residues described previously to be important in HsUBC9-RanGAP binding (Bernier-Villamor et al., 2002) are conserved in AtSCE1a. These results strongly suggest that the genes identified by homology searches, AtSUMO1/2 and AtSCE1a, are in fact components of the Arabidopsis sumoylation apparatus and not components of the ubiquitin-conjugating or any other ubiquitin-like conjugating system.
Among the roles assigned to sumoylation, the regulation of protein localization is reported most commonly. A recent study showed that in HeLa cells, HsSUMO1, HsSUMO2, and HsSUMO3 localize predominantly to the nuclear membrane, nuclear bodies, and cytoplasm, respectively (Su and Li, 2002). In yeast, a predominantly SUMO nuclear localization also has been observed in all stages of the cell cycle, in addition to the bud-neck localization during mitosis (Johnson and Blobel, 1999). Although we cannot conclude that the signal in our localization experiments corresponds to conjugated or free SUMO, the observation that both AtSUMOs are distributed in the nucleus and cytoplasm suggests that SUMO it is not related directly to nuclear targeting in Arabidopsis.
We found that the SUMO-conjugating enzyme AtSCE1a was localized mainly in the nucleus, although not specifically at the nuclear envelope, as in other organisms. In mammals, a fraction of UBC9 interacts with the nuclear pore complex, where it associates with SUMO1-modified RanGAP1 and RanBP2 (Mahajan et al., 1998; Saitoh et al., 1998; Zhang et al., 2002). By contrast, Arabidopsis RanGAP1 does not contain the homologous domain for sumoylation, and its localization at the nuclear envelope is mediated by the WPP motif that is considered to be unique to plants (Rose and Meier, 2001). Similarly, the SUMO attachment domain is not present in Rna1p, the Saccharomyces cerevisiae and Schizosaccharomyces pombe RanGAP homolog (Hillig et al., 1999). These differences may explain the absence of AtSCE1a-specific nuclear envelope localization that we observed.
When AtSUMO1 or AtSUMO2 was coexpressed with AtSCE1a, colocalization was observed predominantly in the nucleus and was dependent on the integrity of the catalytic Cys in AtSCE1a. Although several possibilities could explain this result, the most likely cause of this colocalization is that AtSUMO1/2 are conjugated to nuclear targets more efficiently in the presence of AtSCE1a. The identification of SUMO targets in Arabidopsis should help to clarify their subcellular localization.
Among the transgenic plants generated, only plants that overexpressed AtSUMO1/2 or plants with AtSCE1a cosuppression showed increased or diminished levels of endogenous SUMO conjugates, respectively. It is noteworthy that SUMO conjugates in transgenic plants that overexpressed AtSUMO1/2 accumulated in a similar high molecular mass pattern, although more efficiently, compared with endogenous SUMO conjugates, as determined by protein gel blot analysis. This finding suggests that overexpression of AtSUMO1/2 does not significantly alter the selection of substrates for sumoylation. Plants with increased levels of SUMO conjugates showed no obvious impairment of general growth. It is possible that these plants display physiological adaptation to the higher levels of sumoylation or that the overmodified SUMO targets are not involved in any basic growth process. Similar considerations apply to transgenic plants with diminished levels of SUMO conjugates as a consequence of endogenous AtSCE1a cosuppression. By contrast, RNA interference experiments with UBC9 performed in Caenorhabditis elegans resulted in embryonic arrest after gastrulation, pleiotropic defects in larval development, or, in a small percentage, sterility (Jones et al., 2001). It remains to be determined if higher AtSCE1a cosuppression levels would result in important growth defects.
SUMO has also been shown to play a role in stress responses. In mammalian cells, sumoylation by HsSUMO2 and HsSUMO3 increased upon oxidative and heat stresses (Saitoh and Hinchey, 2000), whereas LeSUMO has been reported to be involved in biotic stress responses in tomato (Hanania et al., 1999). Similarly, during the revision of this manuscript, Kurepa et al. (2003) reported that the levels of AtSUMO1 and 2 conjugates, but not AtSUMO3 conjugates, increased in response to abiotic stress such as heat shock, H2O2, ethanol, and the amino acid analog canavanine. The hormone ABA mediates plant responses to environmental stresses such as cold, drought, and high salinity (Leung and Giraudat, 1998). We have observed that ABA-mediated root growth inhibition was reduced in SUMO-overexpressing plants compared with wild-type plants after exposure to 10 and 50 μM ABA for 4 days. Under these conditions, plants showing AtSCE1a cosuppression had wild-type responses. In addition, after incubation for 3 weeks in the presence of 5 μM ABA, SUMO-expressing plants continued to develop more efficiently than wild-type plants according to plant size and greening degree. Consistently, plants showing cosuppression of the SUMO-conjugating enzyme AtSCE1a displayed a more severe growth inhibition and were highly chlorotic compared with wild type. These results suggest that this mild reduction of AtSCE1a levels is not sufficient to affect short-term responses to ABA, but instead it seems to have an effect on growth processes under continued exposure to 5 μM ABA. It is conceivable that plants showing higher reduction of AtSCE1a levels would display an accentuated hypersensitivity to ABA-mediated growth inhibition.
RNA gel blot analysis showed that in response to ABA, RD29A and AtPLC1 mRNA levels were upregulated in AtSUMO1/2-overexpressing plants compared with wild-type plants. Differences in the expression of other stress- and/or ABA-responsive genes were less dramatic, suggesting a specialized role of SUMO in ABA signaling modulation. None of the investigated genes was expressed in the absence of ABA treatment, probably because the SUMO targets involved in the upregulation of these genes are not present or accessible to the sumoylation apparatus under normal conditions or because the mechanisms involved in the recognition of these sumoylated proteins are not fully functional in the absence of ABA.
Numerous genes are upregulated under stress conditions in vegetative tissues and their products are required for ABA signaling, such as AtPLC1, or are considered to play a protective role in stress tolerance, such as RD/COR/KIN/LTI (Zhu, 2001). AtPLC1, which encodes phospholipase C1, is required for secondary responses to ABA signals (Sánchez and Chua, 2001). In AtSUMO-overexpressing plants, the amplified induction of AtPLC1 likely results in increased inositol 1,4,5-triphosphate levels, which could contribute to the upregulation of other stress-responsive genes such as RD29A, as we have observed. In AtSUMO-overexpressing plants, upregulation of RD29A is not sufficient to confer increased tolerance to abiotic stresses, as was similarly reported for the fry1 mutant (Xiong et al., 2001). fry1 shows increased 1,4,5-triphosphate levels and upregulation of RD29A but is more sensitive to salt and drought stresses. Moreover, the developmental phenotype of AtSUMO-overexpressing plants indicates a reduced ABA response; likewise in the hos15 mutant, RD29A upregulation in response to ABA did not correlate with an ABA-hypersensitive phenotype (Alabadi et al., 2002). Thus, it is possible that alternative branches of ABA signaling lead to RD29A expression, ABA-mediated growth inhibition, and abiotic stress tolerance.
Our results suggest that SUMO plays a dual role in ABA signaling. On the one hand, sumoylation positively modulates the induction of ABA-responsive genes; on the other hand, sumoylation contributes to an attenuation of the ABA signaling branch, leading to growth inhibition. A similar situation was described for the rpn12a-1 mutant (Smalle et al., 2002). RPN12 encodes a subunit of the proteasome, and a T-DNA insertion mutant affected in RPN12a showed decreased cytokinin responses, whereas transcript levels of cytokinin-inducible genes were amplified. In this case, it was proposed that an inhibitory feedback loop that requires RPN12a might be responsible for desensitizing the plants in response to cytokinins. In AtSUMO1/2-overexpressing plants, gene upregulation in response to ABA could lead to the synthesis of factors required for the attenuation of ABA-mediated growth inhibition.
Ubiquitin has been implicated in the modulation of auxin as well as ABA (Frugis and Chua, 2002; López-Molina et al., 2003) signal transduction via controlled proteolysis. We show that sumoylation also contributes to the regulation of ABA signaling. The identification of SUMO paralogs and the SUMO-conjugating enzyme from Arabidopsis will enable the isolation and study of SUMO conjugates associated with the modulation of ABA responses in plants.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia was used in this study. Seeds were surface-sterilized by incubation in 50% bleach and 0.01% Triton X-100 and washed three times with water. Seeds were kept for 3 days in the dark at 4°C, plated on medium A, and transferred to a tissue culture room under constant white fluorescent light (27 μmol·m−2·s−1) at 22°C. Medium A contains full-strength Murashige and Skoog (1962) salts (JRH Biosciences, Lenexa, KS), pH 5.7, 0.8% BactoAgar (Difco), and 1% sucrose. For abscisic acid and stress treatments, 3-day-old seedlings were transferred to medium A supplemented with abscisic acid (mixed isomers; Sigma), 200 mM NaCl, or 400 mM mannitol. For RNA and protein gel blot analysis, 10-day-old seedlings were removed from the plates and grown for 2 days in liquid medium A. The medium was replaced by liquid medium A (control) or liquid medium A supplemented with 5 μM ABA, 10 μM ABA, 200 mM NaCl, or 400 mM mannitol. Plant samples were collected at 0, 2, and 6 h, and the samples were frozen.
Plasmid Constructs
AtSUMO1 and AtSUMO2 cDNAs were amplified by PCR from a cDNA library generated previously (Ballesteros et al., 2001). A first PCR was performed using the primers AP1 and LML4 or LML5 to amplify AtSUMO1 and AtSUMO2, respectively. The PCR product was diluted 1:25, and nested PCR was performed to replace the primer AP1 with the primer AP2 in the reaction mixture. Primers AP1 and AP2 (Marathon cDNA adaptor primers; Clontech, Palo Alto, CA) are complementary to the adaptor that was ligated to the cDNA during library construction. Primers LML4 (5′-GACGGTACCGCGGGATGTCTGCTACTCCGGAAGAAG-3′) and LML5 (5′-GACGGTACCGCGGGATGTCTGCAAACCAGGAGGAAG-3′) are complementary to the 3′ untranslated region of AtSUMO1 and AtSUMO2, respectively, and they also contain KpnI and SacII restriction sites. AtSCE1a cDNA was obtained from the ABRC (Ohio State University, Columbus). Appropriate restriction sites were introduced by PCR according to the sites contained in each expression vector. PCR was performed using Pfu polymerase (Stratagene), and PCR products were verified by DNA sequencing. The binary vector pBA002, a BASTA resistance marker (Kost et al., 1998), was used for the 35S promoter constructs. Plasmids were introduced into Agrobacterium tumefaciens strain ABI, and Arabidopsis Columbia plants were used for in planta transformation (Clough and Bent, 1998). HsSUMO clones were obtained from Mark Hochstrasser (Yale University, New Haven, CT; Li and Hochstrasser, 1999). HsE1, HsUBC9, HsUBC8-C93S, and HsSUMO1 proteins were expressed and purified as described by Bernier-Villamor et al. (2002).
Bioinformatics
Sequence alignments were performed using the CLUSTAL W program of the EMBL-EBI (http://www.ebi.ac.uk/). Alignment shading was performed using GeneDoc version 2.6.001 (Karl Nicholas, PIttsburgh Supercomputing Center, www.psc.edu/biomed/genedoc). Color codes in the sequences are explained in the legend to Figure 1. Trees were predicted using the TreeTop Phylogenetic Prediction program at the GeneBee-Molecular Biology Server (http://www.genebee.msu.su/). The bars on the trees shown in Figure 1 represent the branch length equivalent of 0.1 amino acid changes per residue.
Yeast Two-Hybrid Experiments
Expression vectors pGBKT7 and pGADT7 (Clontech) were introduced into the yeast strain HF7c (Feilotter et al., 1994) by the lithium acetate method as described in the Clontech Yeast Protocols Handbook. Protein interactions were analyzed using His auxotrophy as a selective marker.
In Vitro Sumoylat10ion Assays
SUMO conjugation was assayed with RanGAP1 peptide (amino acids 420 to 589) as described by Bernier-Villamor et al. (2002). Reactions mixtures contained 2 μM glutathione S-transferase (GST)-RanGAP1, 0.3 μM human E1, 0.3 μM HsUBC9 or 3 μM AtSCE1a, and 8 μM HsSUMO1 in the reaction buffer (1 mM ATP, 50 mM NaCl, 20 mM Hepes, pH 7.5, 0.1% Tween 20, 5 mM MgCl2, and 0.1 mM DTT). After incubation at 37°C for 4 h, reactions were stopped by the addition of protein-loading buffer and the mixture was boiled for 5 min. Three microliters of each reaction mixture was resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, MA), and SUMO conjugation to GST-RanGAP was examined by protein gel blot analysis using anti-HsSUMO1 polyclonal antibody (diluted 1:1000; Alexis, San Diego, CA).
Transient Expression of Fluorescent Protein Fusions in Onion Cells
AtSUMO1, AtSUMO2, and AtSCE1a were fused in frame to the 3′ end of the coding sequences of yellow fluorescent protein (YFP) or cyan fluorescent protein (CFP). The fusion proteins were expressed from a 35S constitutive promoter. Onion epidermal cells were bombarded with 5 μg of DNA constructs using a helium biolistic gun (Kost et al., 1998). Treated epidermal cells were kept in the dark at room temperature for 16 h before analysis by confocal microscopy (LSM 510 microscope; Zeiss, Jena, Germany). YFP fluorescence was imaged using excitation with the 514-nm line of the argon laser and a 530-nm band-pass emission filter, and CFP fluorescence was imaged using excitation with the 458-nm line of the argon laser and a 475- to 525-nm band-pass emission filter. Imaging of YFP and CFP was performed sequentially. Samples were scanned with the Z-stack mode, and the projection of the image stacks was calculated with the LSM 510 microscope's three-dimensional functions. Signals from both channels corresponding to each image were compared using Adobe Photoshop 6.0 (Mountain View, CA), and the results shown in the merged columns indicate protein colocalization (Figure 3).
Antibody Production, Immunoblot Analysis, and Affinity Purification
AtSUMO1 and AtSCE1a coding regions were cloned into pET28 expression vectors (Novagen, Madison, WI). His-tagged AtSUMO1 and AtSCE1a expressed in Escherichia coli BL21 were purified using HisTrap columns (Amersham Pharmacia). Polyclonal antibodies were raised against purified His-AtSUMO1 and His-AtSCE1a in rabbits (Cocalico Biological, Reamstown, PA). Affinity purification of the antibodies was performed by coupling 0.5 mg of the appropriate recombinant protein (His-AtSUMO1 or His-AtSCE1a) to 1 mL of cyanogen bromide–activated Sepharose 6-MB (Amersham Pharmacia) according to the manufacturer's protocol.
Plant tissues used for immunoblot analysis were frozen and ground in liquid nitrogen. Protein extracts were prepared in PE buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 2 mM N-ethylmaleimide, and complete protease inhibitors; Roche, Indianapolis, IN) unless stated otherwise. Protein concentration was determined using the Bio-Rad DC Protein Assay, and 70 μg of total protein was resolved by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes and examined by protein gel blot analysis.
SUMO conjugates were affinity purified using nickel–nitrilotriacetic acid agarose (Ni-NTA) Superflow resins (Qiagen, Valencia, CA). Protein extracts were prepared by adding 1 volume of TBS (10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) containing 2% SDS to 1 volume of frozen tissue. Extracts were boiled for 5 min and centrifuged at 14,000 rpm for 20 min. One volume of the supernatant (1 mL) was diluted 1:4 with TBS containing 1% Triton X-100 and then incubated overnight at room temperature in the presence of 50 μL of Ni-NTA beads. The beads were washed five times with 1 mL of TBS containing 5 mM imidazole, and bound proteins were eluted by boiling the Ni-NTA beads for 5 min in Laemmli (1970) buffer. Purified proteins were examined by protein gel blot analysis using anti-AtSUMO1 polyclonal antibody.
RNA Isolation and RNA Gel Blot Analysis
Total RNA was extracted from frozen ground samples using the Qiagen RNeasy Plant Mini Kit. Total RNA (5 μg per lane) was separated under denaturing conditions on 1% agarose gels, and RNA was blotted onto Hybond-XL membranes (Amersham Pharmacia). Probes were synthesized with Amersham Ready-To-Go Labeling Beads (−dCTP), and hybridizations were performed with ULTRAhyb solution (Ambion, Austin, TX) according to the manufacturer's instructions. Probes are described by Sánchez and Chua (2001) and Urao et al. (1993). For the AtSCE1a gene, we used the first exon as a probe.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The protein sequences used for BLAST searches and homology analysis correspond to the following accession numbers: HsSUMO1 (U67122), HsSUMO2 (XM_036093), HsSUMO3 (XM_039809), AtSUMO1 (At4g26840), AtSUMO2 (At5g55160), AtSUMO3 (At5g55170), AtSUMO4 (At5g48710), AtSUMO5 (At2g32765), AtSUMO6 (At5g48700), AtUBI (At4g02890), HsUBI (GI:576323), HsUBC9 (U66867), ScUBC9 (NP_010219), AtUBC9 (NM_118934), AtUBC10 (At5g53300), and AtHUS5 (U44976).
Acknowledgments
We thank J.P. Sánchez for helpful discussions and Mark Hochstrasser for providing the HsSUMO1 clones. We also thank A. North from the Rockefeller University bioimaging center for technical support. L.M.L. was supported by a postdoctoral fellowship from the Spanish Ministerio de Ciencia y Tecnología. This work was supported by National Institutes of Health Grant GM65872 to C.D.L. and National Institutes of Health Grant GM44640 and a Bayer CropScience grant to N.-H.C.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009902.
References
- Alabadi, D., Devoto, A., and Eckardt, N. (2002). Arabidopsis research heats up in Seville. Plant Cell 14, 1987–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Khodairy, F., Enoch, T., Hagan, I.M., and Carr, A.M. (1995). The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J. Cell Sci. 108, 475–486. [DOI] [PubMed] [Google Scholar]
- Azuma, Y., Tan, S.H., Cavenagh, M.M., Ainsztein, A.M., Saitoh, H., and Dasso, M. (2001). Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 15, 1825–1827. [DOI] [PubMed] [Google Scholar]
- Bachmair, A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. (2001). Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 10, 463–470. [DOI] [PubMed] [Google Scholar]
- Ballesteros, M.L., Bolle, C., Lois, L.M., Moore, J.M., Vielle-Calzada, J.P., Grossniklaus, U., and Chua, N.H. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15, 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencsath, K.P., Podgorski, M.S., Pagala, V.R., Slaughter, C.A., and Schulman, B.A. (2002). Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 277, 47938–47945. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor, V., Sampson, D.A., Matunis, M.J., and Lima, C.D. (2002). Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Desterro, J.M., Rodriguez, M.S., and Hay, R.T. (1998). SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 2, 233–239. [DOI] [PubMed] [Google Scholar]
- Desterro, J.M., Thomson, J., and Hay, R.T. (1997). Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417, 297–300. [DOI] [PubMed] [Google Scholar]
- Estelle, M. (2001). Proteases and cellular regulation in plants. Curr. Opin. Plant Biol. 4, 254–260. [DOI] [PubMed] [Google Scholar]
- Feilotter, H.E., Hannon, G.J., Ruddell, C.J., and Beach, D. (1994). Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 22, 1502–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugis, G., and Chua, N.H. (2002). Ubiquitin-mediated proteolysis in plant hormone signal transduction. Trends Cell Biol. 12, 308–311. [DOI] [PubMed] [Google Scholar]
- Giraud, M.F., Desterro, J.M., and Naismith, J.H. (1998). Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for SUMO rather than ubiquitin. Acta Crystallogr. D Biol. Crystallogr. 54, 891–898. [DOI] [PubMed] [Google Scholar]
- Girod, P.A., Carpenter, T.B., van Nocker, S., Sullivan, M.L., and Vierstra, R.D. (1993). Homologs of the essential ubiquitin conjugating enzymes UBC1, 4, and 5 in yeast are encoded by a multigene family in Arabidopsis thaliana. Plant J. 3, 545–552. [DOI] [PubMed] [Google Scholar]
- Gong, L., Kamitani, T., Fujise, K., Caskey, L.S., and Yeh, E.T. (1997). Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272, 28198–28201. [DOI] [PubMed] [Google Scholar]
- Hanania, U., Furman-Matarasso, N., Ron, M., and Avni, A. (1999). Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 19, 533–541. [DOI] [PubMed] [Google Scholar]
- Hang, J., and Dasso, M. (2002). Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966. [DOI] [PubMed] [Google Scholar]
- Hillig, R.C., Renault, L., Vetter, I.R., Drell, T., IV, Wittinghofer, A., and Becker, J. (1999). The crystal structure of rna1p: A new fold for a GTPase-activating protein. Mol. Cell 3, 781–791. [DOI] [PubMed] [Google Scholar]
- Himmelbach, A., Iten, M., and Grill, E. (1998). Signalling of abscisic acid to regulate plant growth. Philos. Trans. R. Soc. Lond. B 353, 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M. (2000). Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 8, 153–157. [DOI] [PubMed] [Google Scholar]
- Hoege, C., Pfander, B., Moldovan, G.L., Pyrowolakis, G., and Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., and Blobel, G. (1997). Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272, 26799–26802. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., and Blobel, G. (1999). Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S., and Gupta, A.A. (2001). An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744. [DOI] [PubMed] [Google Scholar]
- Jones, D., Crowe, E., Stevens, T.A., and Candido, E.P. (2001). Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: Ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 3, Research0002.1–Research0002.15. [DOI] [PMC free article] [PubMed]
- Kahyo, T., Nishida, T., and Yasuda, H. (2001). Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718. [DOI] [PubMed] [Google Scholar]
- Kamitani, T., Kito, K., Nguyen, H.P., Wada, H., Fukuda-Kamitani, T., and Yeh, E.T. (1998). Identification of three major sentrinization sites in PML. J. Biol. Chem. 273, 26675–26682. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kotaja, N., Karvonen, U., Janne, O.A., and Palvimo, J.J. (2002). PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22, 5222–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa, J., Walker, J.M., Smalle, J., Gosink, M.M., Davis, S.J., Durham, T.L., Sung, D.Y., and Vierstra, R.D. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278, 6862–6872. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, G.W., Melchior, F., Matunis, M.J., Mahajan, R., Tian, Q., and Anderson, P. (1998). Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 273, 6503–6507. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2002). Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53, 377–398. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (1999). A new protease required for cell-cycle progression in yeast. Nature 398, 246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20, 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Jin, C., Liao, X., Shen, Z., Chen, D., and Chen, Y. (1999). The binding interface between an E2 (UBC9) and a ubiquitin homologue (UBL1). J. Biol. Chem. 274, 16979–16987. [DOI] [PubMed] [Google Scholar]
- López-Molina, L., Mongrand, S., Kinoshita, N., and Chua, N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, R., Delphin, C., Guan, T., Gerace, L., and Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., Gerace, L., and Melchior, F. (1998). Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Coutavas, E., and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior, F. (2000). SUMO-nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Müller, S., Hoege, C., Pyrowolakis, G., and Jentsch, S. (2001). SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell. Biol. 2, 202–210. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Orth, K., Xu, Z., Mudgett, M.B., Bao, Z.Q., Palmer, L.E., Bliska, J.B., Mangel, W.F., Staskawicz, B., and Dixon, J.E. (2000). Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Pichler, A., Gast, A., Seeler, J.S., Dejean, A., and Melchior, F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Ubiquitin enters the new millennium. Mol. Cell 8, 499–504. [DOI] [PubMed] [Google Scholar]
- Rodriguez, M.S., Dargemont, C., and Hay, R.T. (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659. [DOI] [PubMed] [Google Scholar]
- Rose, A., and Meier, I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98, 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F., and Grosschedl, R. (2001). PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, H., and Hinchey, J. (2000). Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252–6258. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., Pu, R.T., and Dasso, M. (1997). SUMO-1: Wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci. 22, 374–376. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., Sparrow, D.B., Shiomi, T., Pu, R.T., Nishimoto, T., Mohun, T.J., and Dasso, M. (1998). Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8, 121–124. [DOI] [PubMed] [Google Scholar]
- Sánchez, J.P., and Chua, N.H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13, 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, S.E., Matuschewski, K., Liakopoulos, D., Scheffner, M., and Jentsch, S. (1998). The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Babiychuk, E., Kushnir, S., Durski, A., and Vierstra, R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14, 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., and Li, S. (2002). Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 296, 65–73. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Iwase, M., Konishi, M., Tanaka, M., Toh-e, A., and Kikuchi, Y. (1999). Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem. Biophys. Res. Commun. 259, 582–587. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Kahyo, T., Toh-e, A., Yasuda, H., and Kikuchi, Y. (2001). Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276, 48973–48977. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Mizoi, J., Toh-e, A., and Kikuchi, Y. (2000). Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J. Biochem. 128, 723–725. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Nishide, J., Okazaki, K., Kato, H., Niwa, O., Nakagawa, T., Matsuda, H., Kawamukai, M., and Murakami, Y. (1999). Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell. Biol. 19, 8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao, T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Lee, B.H., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Saitoh, H., and Matunis, M.J. (2002). Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22, 6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2001). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406. [DOI] [PubMed] [Google Scholar]