Abstract
Coronaviruses can infect a variety of animals including poultry, livestock, and humans and are currently classified into three groups. The interspecies transmissions of coronaviruses between different hosts form a complex ecosystem of which little is known. The outbreak of severe acute respiratory syndrome (SARS) and the recent identification of new coronaviruses have highlighted the necessity for further investigation of coronavirus ecology, in particular the role of bats and other wild animals. In this study, we sampled bat populations in 15 provinces of China and reveal that approximately 6.5% of the bats, from diverse species distributed throughout the region, harbor coronaviruses. Full genomes of four coronavirues from bats were sequenced and analyzed. Phylogenetic analyses of the spike, envelope, membrane, and nucleoprotein structural proteins and the two conserved replicase domains, putative RNA-dependent RNA polymerase and RNA helicase, revealed that bat coronaviruses cluster in three different groups: group 1, another group that includes all SARS and SARS-like coronaviruses (putative group 4), and an independent bat coronavirus group (putative group 5). Further genetic analyses showed that different species of bats maintain coronaviruses from different groups and that a single bat species from different geographic locations supports similar coronaviruses. Thus, the findings of this study suggest that bats may play an integral role in the ecology and evolution of coronaviruses.
Coronaviruses (CoVs) are enveloped, single-stranded, positive-sense RNA viruses (4). Based on serological and genetic features, coronaviruses are classified into three groups (14). These viruses infect various species of poultry, livestock, and pets and also humans, causing acute and chronic respiratory, enteric, hepatic, and central nervous system diseases (45). Prior to the outbreak of severe acute respiratory syndrome (SARS), most of our knowledge regarding coronaviruses resulted from investigations associated with animal health. As such, until recently, the evolutionary and ecological aspects of coronavirus had not been extensively studied.
In March 2003, after the outbreak of SARS, a novel coronavirus (SARS-CoV) was identified as the etiologic agent responsible for human infection (20, 31). The identification of SARS-like coronavirus in Himalayan palm civets and raccoon dogs in live-animal markets in southern China suggested that SARS was a possible zoonosis (10). Further virological surveillance confirmed that the infectious source of SARS was from those live-animal markets and confirmed its zoonotic origin (11). However, subsequent studies suggested that those market animals were intermediate hosts rather than the natural reservoirs of SARS-CoV, as extensive surveillance studies did not detect the virus in either farmed or wild animals of the same species (17).
Recent studies have suggested horseshoe bats (Rhinolophus spp.) as possible natural reservoirs of SARS-like coronavirus (23, 25). However, genome sequence comparison of the spike (S) genes from bat SARS-like coronavirus and civet SARS-like coronavirus revealed only 64% genetic homology, suggesting that the evolutionary pathway of SARS-CoV remains to be fully described. Given the high biodiversity of bats, along with significant population size, broad geographical distribution, and the ability to migrate and along with the detection of many emerging viruses (1, 7), it is reasonable to consider that bats may contain the direct progenitor of SARS-CoV. Moreover, a growing number of novel coronaviruses have recently been identified, such as HCoV-NL63 (42) and HCoV-HKU1 (47) from humans and some avian infectious bronchitis virus (IBV)-like coronaviruses from different avian species (16, 26). These accumulated findings suggest that coronaviruses may have a much wider distribution in the animal kingdom than previously thought.
To explore the natural distribution of the virus in bat populations and also to understand the possible role of bats in coronavirus ecology, we conducted a virological surveillance study in China. Genetic analysis revealed that bat coronaviruses mainly clustered into three different groups: group 1, a group including all SARS and SARS-like coronaviruses from different hosts (putative group 4), and an independent bat coronavirus group (putative group 5). Further characterization of bat coronaviruses revealed high genetic diversity across a large geographic distribution and revealed that different species of bats maintain coronaviruses from different groups and that the same species of bat from different geographic locations can also contain the same type of coronavirus. Thus, the findings of this study suggest that bats may play an integral role in the ecology and evolution of coronaviruses.
MATERIALS AND METHODS
Sampling.
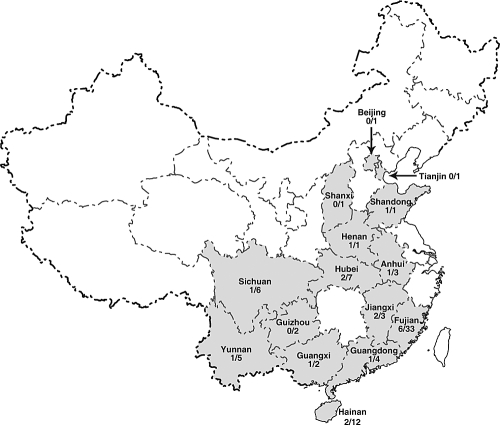
From November 2004 to March 2006, 985 bats from 35 species (belonging to 14 genera in three families) were captured and sampled from their natural habitats at 82 locations in 15 provinces throughout China (Fig. 1; Table 1). Bat identification in the field was initially determined by morphology (1, 44), and identifications were confirmed by sequence analysis of the mitochondrial cytochrome b DNA as previously described (43). Identification of species in the genus Myotis could not be readily made, and these specimens were recorded as Myotis sp. Most bats were captured in the wild from natural roosts; however, some were also sampled from populated areas. Oropharyngeal and anal swabs from each bat were taken, placed in transport medium, kept in liquid nitrogen for transportation to the laboratory, and then stored at −80°C. All captured bats were released after samples were taken.
FIG. 1.
Map of China showing 15 provinces where coronavirus surveillance in bats was conducted. Numbers indicate number of sites positive over the total number of sites sampled in each province.
TABLE 1.
Coronavirus distribution in different bat species and locations
| Family and species of bat | Common name | No. sampled (no. positive)
|
Group(s) | |
|---|---|---|---|---|
| Sites | Bats | |||
| Rhinolophidae | ||||
| Rhinolophus pusillus | Least horseshoe bat | 27 | 101 | |
| Rhinolophus malayanus | Malayan horseshoe bat | 2 | 15 | |
| Rhinolophus affinis | Intermediate horseshoe bat | 9 | 60 | |
| Rhinolophus ferrumequinum | Greater horseshoe bat | 11 (3) | 41 (4) | 1, 4, and 5 |
| Rhinolophus thomasi | Thomas's horseshoe bat | 5 | 12 | |
| Rhinolophus sinicus | Chinese horseshoe bat | 16 (1) | 66 (1) | 4 |
| Rhinolophus pearsoni | Pearson's horseshoe bat | 10 (1) | 48 (1) | 1 |
| Rhinolophus macrotis | Big-eared horseshoe bat | 11 (1) | 38 (1) | 4 |
| Rhinolophus rex | King horseshoe bat | 2 | 2 | |
| Rhinolophus luctus | Woolly horseshoe bat | 4 | 4 | |
| Rhinolophus osgoodi | Osgood's horseshoe bat | 1 | 1 | |
| Hipposideros armiger | Great leaf-nosed bat | 13 | 58 | |
| Hipposideros larvatus | Intermediate leaf-nosed bat | 1 | 3 | |
| Hipposideros pratti | Pratt's leaf-nosed bat | 2 | 9 | |
| Hipposideros pomona | Pomona leaf-nosed bat | 1 | 1 | |
| Coelops frithi | East Asian tailless leaf-nosed bat | 2 | 6 | |
| Aselliscus stoliczkanus | Stoliczka's Asian trident bat | 1 | 7 | |
| Vespertilionidae | ||||
| Pipistrellus pipistrellus | Common pipistrelle | 4 (1) | 27 (6) | 5 |
| Pipstrellus abramus | Japanese pipistrelle | 8 (3) | 41 (14) | 5 |
| Scotophilus kuhlii | Lesser Asiatic yellow house bat | 2 (1) | 43 (5) | 1 |
| Myotis daubentonii | Daubenton's bat | 4 | 41 | |
| Myotis mystacinus | Whiskered bat | 1 | 1 | |
| Myotis ricketti | Rickett's big-footed bat | 8 (4) | 53 (13) | 1 |
| Myotis chinensis | Large Myotis | 2 | 3 | |
| Myotis sp.a | 9 | 80 | ||
| Nyctalus aviator | Birdlike noctule | 2 | 6 | |
| Nyctalus noctula | Noctule bat | 3 | 17 | |
| Scotomanes ornatus | Harlequin bat | 1 | 1 | |
| Barbastella leucomelas | Eastern barbastelle | 1 | 1 | |
| Tylonycteris pachypus | Lesser bamboo bat | 1 (1) | 14 (2) | 5 |
| Ia io | Great evening bat | 1 | 8 | |
| Murina leucogaster | Greater tube-nosed bat | 1 | 5 | |
| Miniopterus schreibersi | Schreiber's long-fingered bat | 15 (3) | 135 (17) | 1 |
| Pteropodidae | ||||
| Cynopterus sphinx | Greater short-nosed fruit bat | 2 | 6 | |
| Rousettus leschenaulti | Leschenault's Rousette | 1 | 31 | |
| Total | 35 | 82 (19) | 985 (64) | |
Identification of many Myotis specimens was possible only to generic level.
Viral detection and isolation.
Viral RNA was extracted from oropharyngeal and anal swabs with the QIAamp viral RNA minikit (QIAGEN, Westburg, The Netherlands) and used as the template for reverse transcription-PCR (RT-PCR) detection of the coronavirus RNA-dependent RNA polymerase (RdRp) gene as previously described (10). Primers conserved for all known coronaviruses were designed for RT-PCR detection (39). Primer sequences are available upon request. The RdRp PCR products were gel purified using the QIAquick PCR purification kit (QIAGEN) and sequenced to confirm virus identification (see below). Virus isolation was attempted using several cell lines (FRHK4, Vero E6, and CV1) for five of the PCR-positive samples; however, no cytopathic effect was observed in any of the cell lines. PCR detection confirmed that no viruses had grown in cell culture.
As many coronaviruses have been recently identified in different animals from different regions, to avoid confusion the nomenclature of bat coronaviruses from this study is given in the following format: host, geographic location of sampling, sample number, and year, e.g., BtCoV/Rhinolophus ferrumequinum/Hubei/273/2004 (abbreviation, BtCoV/273/04).
Genome analysis.
The nucleotide data obtained from diagnostic sequencing of the RdRp fragment were analyzed with available coronavirus sequences in GenBank and used to determine the diversity of the detected coronaviruses and to select representative strains for full genome sequencing. Four viruses in two new coronavirus lineages were selected for complete genome sequencing.
RNA extraction was done using the viral RNA kit from QIAGEN, and cDNA synthesis was conducted with random hexamer, gene-specific, and oligo(dT) primers. Degenerate primers for cDNA amplification and sequencing were designed from multiple alignments of GenBank sequence data using the program CODEHOP (35). Conventional PCR using Platinum Taq DNA high-fidelity polymerase (Invitrogen) and gene-specific primers was then used for filling gaps between the CODEHOP-amplified regions. Shotgun sequencing (38) with the Zero Blunt PCR cloning kit (Invitrogen) was conducted for large PCR fragments generated from specific primers between the CODEHOP-amplified regions (35). For regions that could not be amplified using CODEHOP, we used the method of rapid amplification of cDNA ends with second-generation 5′/3′ kits for rapid amplification of cDNA ends (Roche). Sequencing was performed by using the BigDye Terminator version 3.1 cycle sequencing kit on an ABI PRISM 3700 DNA analyzer (Applied Biosystems) following the manufacturer's instructions. All primer sequences are available upon request.
The open reading frames (ORFs) of each of the four complete genomes were identified and mapped using the program SeqBuilder (Lasergene version 6.1; DNAStar, Madison, WI) and confirmed using Z-Curve (8). Homology searches of identified ORFs against other known coronaviruses were conducted in the GenBank and Pfam databases (2). Protein precursors produced by ORF1ab were predicted using the program Z-Curve (8). Prediction of transmembrane (TM) domains was performed using TMpred (12).
Sequence similarity.
Full-length amino acid alignments of each of the major gene products were used to calculate the similarity (p distances) within and between the different coronavirus groups, including putative groups 4 and 5, using MEGA3 (21). The virus sequences used in this analysis are the same as those in the phylogenetic trees.
Phylogenetic studies.
For the structural proteins, spike (S), membrane (M), envelope (E), and nucleocapsid (N), only full-length sequences were included in the analyses. For the replicase domains, the conserved sequence regions of the RdRp and helicase (HEL) were used. Multiple alignments of bat coronaviruses with other known coronaviruses were conducted with the programs TransAlign (3) and ClustalW (41) and manually optimized with Se-Al (33). Phylogenetic trees were constructed using the neighbor-joining criterion with the Jukes-Cantor model (JC69) in the programs MEGA3 (21) and PAUP* version 4.0b (40). Gaps were treated as missing data in all analyses.
Recombination analysis.
Sliding window analysis was used to detect recombination within the RdRp, S, and N genes. The same multiple alignments used for phylogenetic tree reconstruction, with the outgroup excluded, were analyzed using the difference of the sum of squares method (window size, 300, with steps of 100 amino acids) in the program Topali (29). The RDP method, as implemented in program RDP version 2 (28), was also used for recombination detection with the percentage of identity for recombinant sequences set from 0 to 100.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in GenBank under accession numbers DQ648786 to DQ648797 and DQ648799 to DQ648858.
RESULTS
Surveillance and prevalence.
To understand the prevalence and distribution of coronavirus in bats, 985 individuals belonging to 35 species, from three bat families, were sampled at 82 sites in 15 provinces of China (Fig. 1; Table 1). All samples were collected from apparently healthy individuals and tested for the presence of coronavirus by RT-PCR detection of a 440-bp RdRp gene fragment. A total of 64 (6.5%) samples tested positive in 19 of the 82 sites, located in 12 provinces (Fig. 1). Ten of the 35 species tested were found to harbor coronavirus; 57 (89%) positive samples were detected from six species of the family Vespertilionidae, and the rest were from four species of the Rhinolophidae. The Vespertilionidae and Rhinolophidae accounted for 48% and 47% of the samples, respectively (Table 1).
Two colonies of bats, from different sampling sites, had much higher positive rates than average. One Miniopterus schreibersi colony had a 55% (11/20) positive rate, while a Pipistrellus abramus colony had a 35% (11/31) positive rate. All positive samples were from anal swabs, and none from throat swabs, suggesting that the gastrointestinal tract is the principal replication site of coronavirus infection in those bats.
There were also some species of bats that had high sample numbers, but in which all individuals were negative for coronavirus: 84 individuals of the genus Hipposideros (58 from Hipposideros armiger), 101 specimens of Rhinolophus pusillus, and 37 samples from two genera of the Pteropodidae.
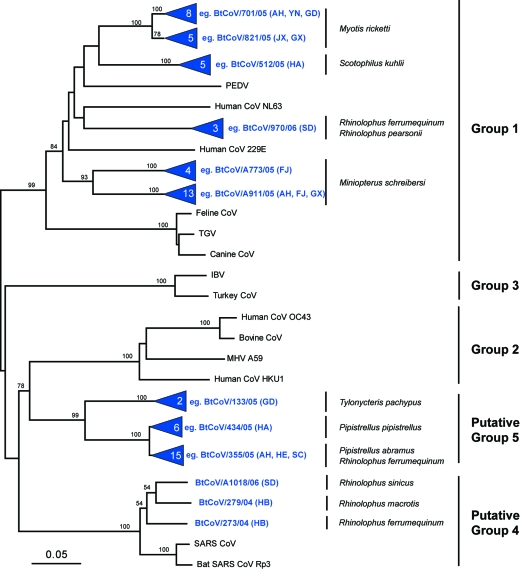
To determine the overall diversity of coronaviruses that were isolated from bats, preliminary phylogenetic analysis of the RdRp fragment obtained from RT-PCR detection revealed that all viruses characterized fell within the previously recognized coronavirus groups, including the SARS-CoV group. Of the 65 viruses, only three bat coronaviruses were closely related to SARS-CoV (putative group 4) and 40 clustered with group 1 viruses, while the remaining 22 viruses form a separate group that is most closely related to group 2 viruses (putative group 5); however, there was no statistical support for this relationship (Fig. 2). None of the coronaviruses characterized in this study were phylogenetically related to group 3.
FIG. 2.
Phylogenetic relationships of 64 coronaviruses isolated from bats in China. The tree was generated based on 440 nucleotides of the RNA-dependent RNA polymerase region by the neighbor-joining method in the MEGA program. Numbers above branches indicate neighbor-joining bootstrap values (percent) calculated from 1,000 bootstrap replicates. Terminal nodes containing bat coronaviruses isolated in this study are collapsed and represented by a blue triangle with the number of viruses indicated within. The tree was rooted to Breda virus (AY427798). Scale bar, 0.05 substitution per site. Red text indicates provinces from where viruses were isolated. Abbreviations: AH, Anhui; FJ, Fujian; GD, Guangdong; GX, Guangxi; HA, Hainan; HB, Hubei; HE, Henan; JX, Jiangxi; SC, Sichuan; SD, Shandong; YN, Yunnan.
Genetic analysis revealed the presence of species-specific host restriction of coronavirus in bats. For all species, but one, that were sampled and found to harbor coronavirus, those viruses from a single species all clustered together with high bootstrap support (Fig. 2). The one exception was R. ferrumequinum, which tested positive for group 1, 4, and 5 viruses. Furthermore, in instances where the same bat species was sampled in different provinces, those species were found to harbor coronaviruses that clustered together (Fig. 2). Species specificity was also evident when two bat species from the same cave in Guangxi, Miniopterus schreibersi and Myotis ricketti, were positive for group 1 coronaviruses, represented by BtCoV/A911/05 and BtCoV/821/05, respectively, but the viruses from each species did not cluster together in the phylogenetic analysis (Fig. 2).
These findings suggest that genetically divergent coronaviruses are commonly present in, and specific to, different species of bats in China.
Genome organization.
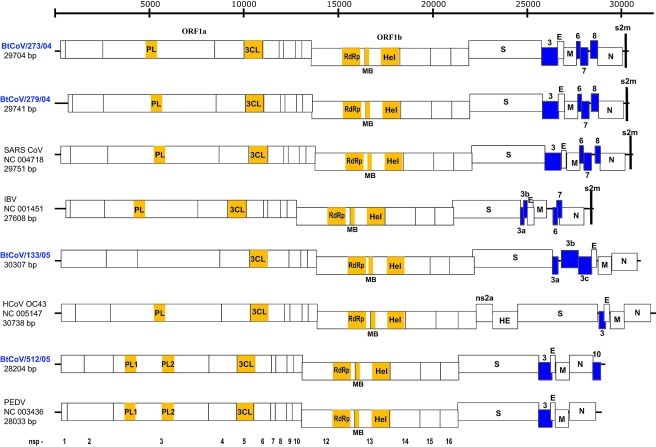
Based on preliminary phylogenetic analysis of the RdRp gene (Fig. 2), four strains, representing the diversity of bat coronaviruses isolated in this study, were selected for full genome sequencing: BtCoV/Tylonycteris pachypus/Guangdong/133/2005 (BtCoV/133/05), BtCoV/Rhinolophus ferrumequinum/Hubei/273/2004 (BtCoV/273/04), BtCoV/R. macrotis/Hubei/279/2004 (BtCoV/279/04), and BtCoV/Scotophilus kuhlii/Hainan/512/2005 (BtCoV/512/05). An additional five viruses were selected for partial sequencing of the RdRp, HEL, and S genes: BtCoV/S. kuhlii/Hainan/515/2005 (BtCoV/515/05), BtCoV/S. kuhlii/Hainan/527/2005 (BtCoV/527/05), BtCoV/Pipistrellus pipistrellus/Hainan/434/05 (BtCoV/434/05), BtCoV/P. abramus/Sichuan/355/2005 (BtCoV/355/05), and BtCoV/Myotis ricketti/Yunnan/701/2005 (BtCoV/701/05). Sequences generated in this study were analyzed with all available coronavirus sequence data in public databases. Comparison of the genome organization of bat coronaviruses with that of representative strains of other coronavirus is presented in Fig. 3 and Table 2.
FIG. 3.
Linear representation of the ORFs of the bat coronaviruses and representative known coronaviruses from each group. Conserved functional domains in ORF1a and ORF1b are indicated by yellow boxes. The following predicted domains are shown: pepain-like proteases 1 and 2 (PL1 and PL2), 3C-like protease (3CL), RdRp, metal ion-binding domain (MB), and helicase (Hel). Putative ORFs are indicated by blue boxes and numbered according to their order in the genome: BtCoV/R. ferrumequinum/Hubei/273/04 (BtCoV/273/04), BtCoV/R. macrotis/Hubei/279/04 (BtCoV/279/04), BtCoV/T. pachypus/Guangdong/133/05 (BtCoV/133/05), BtCoV/S. kuhlii/Hainan/512/05 (BtCoV/512/05), SARS-CoV, PEDV, avian IBV, and human coronavirus OC43 (HCoV-OC43).
TABLE 2.
Comparison of coronavirus genome structuresa
| Feature | BtCoV/273/04 (29,704b)
|
BtCoV/279/04 (29,741)
|
SARS-CoV (29,751)
|
IBV (27,608)
|
BtCoV/133/05 (30,307)
|
HCoV-OC43 (30,738)
|
BtCoV/512/05 (28,204)
|
PEDV (28,033)
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start (bp) | End (bp) | No. of aa | Start (bp) | End (bp) | No. of aa | Start (bp) | End (bp) | No. of aa | Start (bp) | End (bp) | No. of aa | Start (bp) | End (bp) | No. of aa | Start (bp) | End (bp) | No. of aa | Start (bp) | End (bp) | No. of aa | Start (bp) | End (bp) | No. of aa | |
| ORF1a | 261 | 13,382 | 4,373 | 897 | 13,415 | 4,173 | 265 | 13,398 | 4,378 | 529 | 12,354 | 3,942 | 260 | 13,564 | 4,435 | 211 | 13,341 | 4,377 | 294 | 12,650 | 4,119 | 297 | 12,620 | 4,108 |
| nsp1 | 261 | 797 | 179 | 897 | 1,157 | 87 | 265 | 801 | 179 | 529 | 753 | 75 | 260 | 2,800 | 847 | 211 | 948 | 246 | 294 | 1,514 | 407 | 297 | 626 | 110 |
| nsp2 | 798 | 2,717 | 640 | 1,158 | 2,717 | 520 | 802 | 2,718 | 639 | 754 | 2,547 | 598 | 2,801 | 4,420 | 540 | 949 | 2,763 | 605 | 1,515 | 2,984 | 490 | 627 | 2,918 | 764 |
| nsp3 (PL pro) | 2,718 | 8,468 | 1,917 | 2,718 | 8,501 | 1,928 | 2,719 | 8,484 | 1,922 | 2,548 | 7,323 | 1,592 | 4,421 | 8,632 | 1,404 | 2,764 | 8,460 | 1,899 | 2,985 | 7,886 | 1,634 | 2,982 | 7,847 | 1,622 |
| nsp5 (3CL) | 9,969 | 10,886 | 306 | 10,002 | 10,919 | 306 | 9,985 | 10,902 | 306 | 8,866 | 9,786 | 307 | 10,154 | 11,071 | 306 | 9,949 | 10,857 | 303 | 9,330 | 10,235 | 302 | 9,288 | 10,193 | 302 |
| ORF1b | 13,382 | 21,469 | 2,695 | 13,415 | 21,502 | 2,695 | 13,398 | 21,485 | 2,695 | 12,354 | 20,417 | 2,687 | 13,564 | 21,639 | 2,691 | 13,341 | 21,497 | 2,718 | 12,650 | 20,674 | 2,674 | 12,620 | 20,641 | 2,673 |
| nsp12 (RdRp) | 13,356 | 16,150 | 932 | 13,389 | 16,183 | 932 | 13,372 | 16,166 | 932 | 12,313 | 15,131 | 940 | 13,541 | 16,341 | 934 | 13,318 | 16,100 | 928 | 12,627 | 15,406 | 927 | 12,597 | 15,376 | 927 |
| nsp13 (HEL) | 16,151 | 17,953 | 601 | 16,184 | 17,986 | 601 | 16,167 | 17,969 | 601 | 15,132 | 16,931 | 600 | 16,342 | 18,135 | 598 | 16,101 | 17,909 | 603 | 15,407 | 17,197 | 597 | 15,377 | 17,167 | 597 |
| Ns2 | 21,507 | 22,343 | 279 | |||||||||||||||||||||
| HE | 22,355 | 23,629 | 425 | |||||||||||||||||||||
| S | 21,476 | 25,201 | 1,241 | 21,509 | 25,234 | 1,241 | 21,492 | 25,259 | 1,255 | 20,368 | 23,856 | 1,162 | 21,584 | 25,636 | 1,350 | 23,644 | 27,729 | 1,361 | 20,671 | 24,786 | 1,371 | 20,638 | 24,789 | 1,383 |
| ORF3/3a | 25,211 | 26,035 | 274 | 25,244 | 26,068 | 274 | 25,268 | 26,092 | 274 | 23,856 | 24,029 | 57 | 25,663 | 25,938 | 91 | 27,817 | 28,146 | 109 | 24,786 | 25,460 | 224 | 24,789 | 25,463 | 224 |
| ORF3b | 24,029 | 24,223 | 64 | 26,119 | 26,976 | 285 | ||||||||||||||||||
| ORF3c | 26,992 | 27,675 | 227 | |||||||||||||||||||||
| E | 26,060 | 26,290 | 76 | 26,093 | 26,323 | 76 | 26,117 | 26,347 | 76 | 24,207 | 24,533 | 108 | 27,745 | 27,993 | 82 | 28,133 | 28,387 | 84 | 25,441 | 25,671 | 76 | 25,444 | 25,674 | 76 |
| M | 26,337 | 27,002 | 221 | 26,374 | 27,039 | 221 | 26,398 | 27,063 | 221 | 24,505 | 25,182 | 225 | 28,008 | 28,667 | 219 | 28,402 | 29,094 | 230 | 25,678 | 26,361 | 227 | 25,682 | 26,362 | 226 |
| ORF6 | 27,013 | 27,204 | 63 | 27,050 | 27,241 | 63 | 27,074 | 27,265 | 63 | 25,488 | 25,685 | 65 | ||||||||||||
| ORF7 | 27,212 | 27,580 | 122 | 27,249 | 27,617 | 122 | 27,273 | 27,641 | 122 | 25,682 | 25,930 | 82 | ||||||||||||
| ORF8 | 27,718 | 28,086 | 122 | 27,755 | 28,120 | 122 | 27,864 | 28,118 | 84 | |||||||||||||||
| N | 28,088 | 29,353 | 421 | 28,135 | 29,397 | 420 | 28,120 | 29,388 | 422 | 25,873 | 27,102 | 409 | 28,705 | 29,979 | 424 | 29,104 | 30,450 | 448 | 26,372 | 27,556 | 394 | 26,374 | 27,699 | 441 |
| ORF10 | 27,571 | 27,960 | 129 | |||||||||||||||||||||
| s2m | 29,555 | 29,586 | 29,599 | 29,630 | 29,590 | 29,621 | 27,477 | 27,508 | ||||||||||||||||
For blank cells the corresponding ORF was either not present or not identified.
Numbers in parentheses after the virus names are genome sizes in base pairs.
All four bat coronaviruses had classic coronavirus genome organization in which the replicase gene and structural protein genes are arranged in the order 5′-ORF1a and ORF1b, S, E, M, and N (Fig. 3). The genome size of these bat coronaviruses varied: the longest was 30.3 kb, for BtCoV/133/05, and the shortest was 28.2 kb, for BtCoV/512/05.
Putative ORFs coding for nonstructural proteins or accessory proteins were deduced and analyzed if transcription-regulating sequences (TRSs) were present close to, and upstream of, potential initiating methionine residues. The ORFs of nonstructural proteins vary significantly among different bat coronaviruses. The genome organization of BtCoV/273/04 and that of BtCoV/279/04 were essentially the same and were similar to that of SARS-CoV. The genome organization of BtCoV/512/05 is most similar to that of porcine epidemic diarrhea virus (PEDV), while the genome of BtCoV/133/05 is unlike that of all known coronaviruses (Fig. 3).
In the genome of all coronaviruses, approximately the first two-thirds of the genome is composed of the two large replicase ORFs ORF1a and ORF1b, which encode virus replicase polyproteins pp1a and pp1ab (14). Proteolytic processing end products and putative functional domains of the replicase polyproteins were identified. The nonstructural proteins nsp1 and nsp2 were the most variable among these bat coronaviruses, while papain-like protease (PL), 3C-like protease (3CL), RdRp, metal binding (MB), and HEL functional domains were conserved in all genomes, except that of BtCoV/133/05 (Fig. 3). Coronaviruses generally employ two papain-like proteases, PL1 and PL2, to process the N-proximal regions of the replicative polyproteins. PL1 and PL2 were identified in BtCoV/512/05; however, only one PL domain was identified in BtCoV/273/04 and BtCoV/279/04. It is noteworthy that in BtCoV/133/05 both nsp1 and nsp2 were highly divergent from other coronaviruses and that the PL domain could not be identified in any of the nonstructural proteins (Fig. 3; Table 2).
ORFs located between the S and E genes and between the M and N genes were predicted and are numbered according to their order in the genome (Fig. 3; Table 2). In viruses BtCoV/273/04, BtCoV/279/04, and BtCoV/512/05, there is a single ORF between the S and E genes (ORF3). In BtCoV/273/04 and BtCoV/279/04 ORF3 is predicted to encode a similar protein of 274 amino acids (aa) with two predicted TM helices in the N-terminal sequence. BLAST and Pfam searches failed to identify any sequences similar to this protein. In BtCoV/512/05 ORF3 encodes a predicted 224-aa protein also with two predicted TM domains in the N-terminal sequence.
The region between the S and E genes in BtCoV/133/05 is the longest among all known coronaviruses, at 2,013 bp (Fig. 3). Furthermore, in BtCoV/133/05 this region contains three predicted ORFs (ORF3a, ORF3b, and ORF3c), with predicted proteins of 91, 285, and 227 aa, respectively. Each of these ORFs has a conserved TRS upstream of the ORFs: UUAACGAACUU (9 nucleotides) AUG for OFR3a and UUAACGAACUU AUG for ORF3b and ORF3c. The ORF3c-encoded protein contains three TM domains, but no matching proteins could be identified.
In BtCoV/273/04 and BtCoV/279/04 the region between the M and N genes is a 1,085- and a 1,095-bp sequence, respectively, that contains three ORFs (ORF6, ORF7, and ORF8) of 63, 122, and 122 aa, respectively (Fig. 3). ORF7 is predicted to have two TM domains, in both the N- and C-terminal sequences, while for ORF8 one TM helix is predicted. BLAST and Pfam searches failed to identify sequences similar to any of the three predicted proteins. This region between the M and N genes is absent in BtCoV/133/05 and BtCoV/512/05 (Fig. 3). The sequence region between the M and N genes of BtCoV/273/04 and BtCoV/279/04 and other SARS-like CoVs showed a gene organization similar to that of IBV (22, 46). Analysis of this region in a representative IBV (NC_001451) revealed a much shorter region (692 bp) also with two ORFs (ORF6 and ORF7) predicted to encode proteins of 65 and 82 aa, respectively. However, unlike BtCoV/273/04 and BtCoV/279/04, in IBV no conserved TRSs were identified upstream of the three ORFs.
Downstream of the N gene in BtCoV/512/05, there is a 387-bp sequence (ORF10) that is predicted to encode a 129-aa protein with a putative signal peptide at the N-terminal region and three TM domains. This sequence region is absent in all known coronaviruses including BtCoV/133/05, BtCoV/273/04, and BtCoV/279/04 (Fig. 3). No matching protein was identified in GenBank or Pfam.
The hemagglutinin esterase protein, which is present in group 2 coronaviruses (6) and presumably obtained by horizontal gene transfer from influenza C virus (48), was not present in any of the bat coronaviruses analyzed in this study. In the 3′ untranslated region a stem-loop II-like (s2m) motif (15) was recognized in BtCoV/273/04 and BtCoV/279/04 but not in BtCoV/133/05 and BtCoV/512/05 (Fig. 3). This motif is also present in group 3 coronaviruses and SARS-CoV but not in other coronaviruses (34, 37).
Sequence similarity.
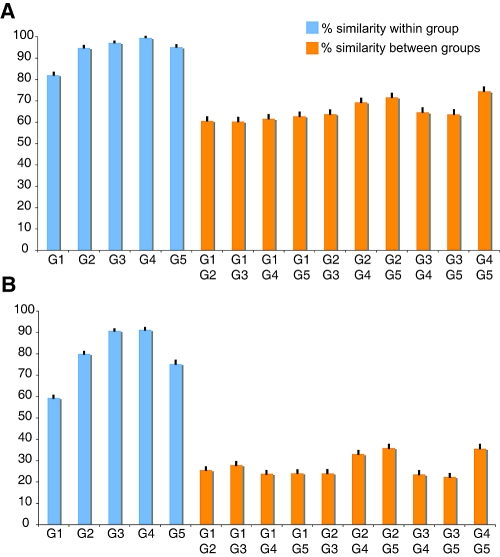
To understand the interrelationship between the BtCoVs and the other known coronaviruses, similarity analysis within and between groups was conducted (9). Analysis of the RdRp amino acid sequence showed that, within groups, the similarity ranged from 82 to 99%, while between different groups, including the putative groups 4 and 5 in the present study, the similarity range was 60 to 74% (Fig. 4A). In contrast, within-group similarities of the S protein were from 59 to 91% and between-group similarities were from 22 to 36% (Fig. 4B). Similar patterns were observed for the remaining major gene products: more-conserved genes usually had higher similarity between different groups, and less-conserved genes had lower similarity between groups (data not shown).
FIG. 4.
Similarity histogram of RdRp (A) and spike (B) genes based on alignments from the program TransAlign.
Phylogenetic analysis.
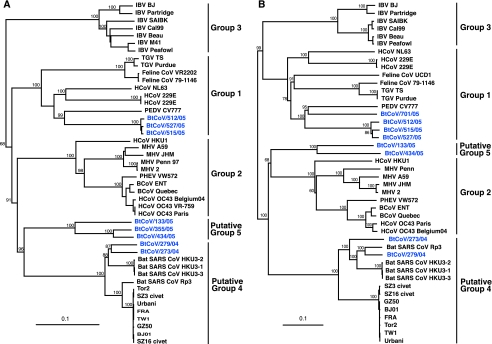
To further define the evolutionary pathway of those novel BtCoVs, each of the major genes was phylogenetically analyzed. In all genes analyzed, represented by the HEL and S gene trees, the bat coronaviruses did not form a single group (Fig. 5). As in the preliminary analysis, five groups, all with 100% bootstrap support, were apparent (Fig. 2 and 5). The same relationships were apparent in all genes analyzed, with the exception of group 1 bat CoVs (BtCoV/512/05, BtCoV/515/05, and BtCoV/527/05) and putative group 5 viruses (represented by BtCoV/133/05).
FIG. 5.
Phylogenetic relationships of the helicase (A) and spike (B) genes of representative coronaviruses isolated from bats in China. Trees were generated by the neighbor-joining method in the PAUP program. Numbers above branches indicate neighbor-joining bootstrap values (percent) calculated from 1,000 bootstrap replicates. Analyses were based on 1,833 nucleotides for the helicase gene and 3,510 nucleotides for the spike gene. The trees were rooted to Breda virus (AY427798). Scale bar, 0.1 substitution per site.
In the HEL, N, and E gene phylogenies, putative group 5 viruses fall as the sister group to the SARS and SARS-like CoV group (putative group 4), which also contains two bat coronaviruses from this study (BtCoV/273/04 and BtCoV/279/04). However, in the S, M, and RdRp gene analyses, group 5 viruses are most closely related to group 2 coronaviruses.
In all genes analyzed, except the S gene, group 1 bat coronaviruses are most closely related to PEDV (bootstrap support, 99%), and these viruses cluster with HCoV-NL63 and HCoV-229E (Fig. 5A). In the S gene tree, while group 1 bat coronaviruses still clustered together with PEDV, they were now most closely related to those coronaviruses from domestic animals (Fig. 5B). The relationship of group 1 bat coronaviruses to PEDV, transmissible gastroenteritis virus, and feline coronaviruses demonstrates that virus transmission may occur between bats, livestock, and companion animals, presenting a possible pathway for human infection.
None of the viruses sequenced in this study was the direct progenitor of SARS. It is noteworthy that within putative group 4 the SARS-like viruses from bats clustered together, away from SARS viruses from other mammalian hosts (Fig. 5), suggesting that other intermediate hosts or viruses were involved in the emergence of SARS.
Taken together, the above phylogenetic findings demonstrated that bats had a relatively high diversity of coronaviruses and harbor a distinct lineage (putative group 5) that may represent a novel coronavirus group. These relationships are in consensus with the results of the genomic and sequence similarity analyses.
Recombination analysis.
To evaluate if the different gene phylogenies for group 1 bat CoVs and putative group 5 viruses were due to recombination, a sliding window analysis was conducted. Results of this analysis indicated that while some areas of the RdRp, S, and N genes may be recombinant, there was no statistical support for this conclusion. Furthermore, those potentially recombinant areas were highly divergent and ambiguously aligned, and the different phylogenies were therefore likely due to variation in the rates of substitution and not recombination between coronaviruses (13, 30).
DISCUSSION
The recent identification of SARS-like and other coronaviruses in bats suggested that they may play an important role in the ecology of these viruses. In the present study we investigated 35 of the 120 bat species identified in China (44) and revealed that approximately 6% of bats, from 10 different species sampled in 12 provinces, were positive for coronavirus. Our findings indicate that bats with coronavirus infection are commonly observed in this region.
Phylogenetic analyses of the present study revealed high genetic diversity of coronaviruses in bats from this region. Except for SARS-like viruses, many bat coronaviruses clustered with existing group 1 viruses; while others formed a separate lineage that included only viruses from bats (putative group 5). Within group 1, the bat CoVs did not form a single group but were highly divergent and related to coronaviruses previously identified from different domestic animals.
Our findings also revealed that within the SARS and SARS-like CoV group (putative group 4) the S gene and other genes clustered into two subgroups, one of bat CoVs and another of SARS viruses from humans and other mammalian hosts. As the similarity of the S genes between those two subgroups is only approximately 80% and since coronaviruses usually have low mutation rates (24), it seems unlikely that these viruses have diverged due to host adaptation within such a short time period. Therefore, the direct progenitor of the SARS-CoV from civets in the animal markets of southern China and the ecological and evolutionary pathway that led to the emergence of SARS have still not been fully determined.
The association between almost all of the coronaviruses that we sequenced and a single bat species demonstrates a high degree of host restriction for coronavirus in bat populations. For example, similar viruses were detected in Myotis ricketti from Anhui, Guangdong, and Yunnan, approximately 1,600 km distant, while two different bat species sampled in the same cave had different coronaviruses. This wide distribution may be associated with bat migration. It also appears that SARS-like CoVs from bats are restricted to different species of Rhinolophus. Furthermore, Hipposideros, which belongs to the same family as Rhinolophus, and all members of the Pteropodidae all tested negative for coronavirus, even though many individuals were sampled (Table 1). As such, these viruses may be restricted to just a few families and genera, and further information regarding which taxonomic groups of bats may host coronaviruses will provide an insight into the evolution and ecology of coronaviruses.
While there have been previous reports of recombination in coronaviruses as a major evolution pattern (18, 19), it is likely that at least some of this is due to those sequence areas being highly divergent. This study did not find any convincing evidence for recombination events in the bat coronaviruses tested. This information further supports the high degree of host specificity seen for bat coronaviruses, as two divergent viruses are unlikely to coinfect the same bat species, let alone the same individual. However, it must be noted that there was one instance of a single bat species being infected with coronaviruses from two different groups (Fig. 2; Table 1).
It is also possible that coronavirus may cause a persistent or long-term infection in bat species as observed for other coronaviruses in vivo and in vitro (5, 36). Each of the previous studies that have identified coronaviruses in bats has sampled at various times and in different areas of China, and all have successfully identified coronaviruses from the samples (23, 25, 32). In addition, the present study was conducted over 17 months in provinces throughout China, and positive samples were identified almost year-round.
In the present study, all bat coronaviruses tested had classical coronavirus genome organization (4). However, BtCoV/133/05 from putative group 5 had the longest genome characterized from bats, a large noncoding region at the start of the genome in which we were unable to identify the PL domain, and also three ORFs between the S and E genes.
The continued identification of novel coronaviruses from different hosts, especially bats, suggests that coronaviruses are more diverse than previously thought (14). Therefore, the classification of the group may need to be modified to match this increasing diversity. The results of this study suggest that many novel coronaviruses cannot be easily accommodated in the current classification, as antigenic data are not available in many cases due to difficulty in virus isolation (14, 23, 25, 32). Genetic data also indicate that some of these novel coronaviruses are intermediate strains that fall between the established groups. Therefore, based on phylogenetic relationships, low genetic similarity, and unique genome organization we propose a new putative coronavirus group (group 5) and also support the suggestion that SARS-like coronaviruses belong to group 4 (27). The proliferation of coronaviruses identified from different hosts has also led to confusion in naming the viruses. We have therefore used a standardized naming system based on the influenza A virus convention. While any changes in nomenclature and taxonomy must be arrived at through consensus in the scientific community, we believe that it is reasonable to consider these issues.
In considering the diversity of species and the habitats that they occupy, large population sizes and densities, and the ability to migrate, bats appear to be ideal candidates for the natural reservoirs of all coronaviruses (7). The current study revealed that coronaviruses in bats exhibit high genetic diversity and high prevalence across a wide geographical distribution, possibly with asymptomatic or persistent infection. However, as bats are a large order that account for approximately 20% of extant mammalian species (1), so far only a small proportion of the total species number have been investigated and those only from China (23, 25, 32). There is also a general lack of knowledge regarding the prevalence of coronaviruses in other animal groups, and it will be difficult to reach solid conclusions until more is known regarding the frequency and diversity of coronaviruses in other animals, especially those that share ecological space with bats.
Acknowledgments
This work was supported by the National Special Task Force Fund for Identification of the Animal Reservoir of SARS-CoV, National Institutes of Health grant AI95357, the Li Ka Shing Foundation, and the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government.
REFERENCES
- 1.Altringham, J. D. 1996. Bats: biology and behavior, p. 1-48. Oxford University Press, Oxford, United Kingdom.
- 2.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. Database Issue 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bininda-Emonds, O. R. P. 2005. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics 6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W., and R. S. Baric. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 70:3947-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton, B. T. 2001. Introduction to current focus on Hendra and Nipah viruses. Microbes Infect. 3:277-278. [DOI] [PubMed] [Google Scholar]
- 8.Gao, F., H. Y. Ou, L. L. Chen, W. X. Zheng, and C. T. Zhang. 2003. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 553:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjunes. 2003. A comparative sequence analysis to revise the current taxonomy of the family coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. M. Peiris, and L. L. M. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 11.Guan, Y., H. Field, G. J. D. Smith, and H. L. Chen. 2005. SARS coronavirus: an animal reservoir? p. 79-83. In M. Peiris, L. J. Anderson, A. D. M. E. Osterhaus, K. Stohr, and K. Y. Yuen (ed.), Severe acute respiratory syndrome. Blackwell Publishing, Malden, Mass.
- 12.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 13.Holmes, E. C., and A. Rambaut. 2004. Viral evolution and the emergence of SARS coronavirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:1059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes, K. V. 2001. Coronaviruses, p. 1187-1203. In D. M. Knipe, P. M. Howley, D. E. Griffin, et al (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 15.Jonassen, C. M., T. Jonassen, and B. Grinde. 1998. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen. Virol. 79:715-718. [DOI] [PubMed] [Google Scholar]
- 16.Jonassen, C. M., T. Kofstad, I. L. Larsen, A. Lovland, K. Handeland, A. Follestad, and A. Lillehaug. 2005. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos). J. Gen. Virol. 86:1597-1607. [DOI] [PubMed] [Google Scholar]
- 17.Kan, B., M. Wang, H. Jing, H. Xu, X. Jiang, M. Yan, W. Liang, H. Zheng, K. Wan, Q. Liu, B. Cui, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, X. Qi, K. Chen, L. Du, K. Gao, Y. T. Zhao, X. Z. Zou, Y. J. Feng, Y. F. Gao, R. Hai, Y. Du, Y. Guan, and J. Xu. 2005. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 79:11892-11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keck, J. G., L. H. Soe, S. Makino, S. A. Stohlman, and M. M. Lai. 1988. RNA recombination of murine coronaviruses: recombination between fusion-positive mouse hepatitis virus A59 and fusion-negative mouse hepatitis virus 2. J. Virol. 62:1989-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kottier, S. A., D. Cavanagh, and P. Britton. 1995. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology 213:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDue, W. J. Bellini, and J. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 22.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leparc-Goffart, I., S. T. Hingley, M. M. Chua, X. Jiang, E. Lavi, and S. R. Weiss. 1997. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology 239:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 26.Liu, S., J. Chen, J. Chen, X. Kong, Y. Shao, Z. Han, L. Feng, X. Cai, S. Gu, and M. Liu. 2005. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (pavo cristatus) and teal (Anas). J. Gen. Virol. 86:719-725. [DOI] [PubMed] [Google Scholar]
- 27.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petresue, A. G. Tobertson, J. E. Schein, S. Siddiqui, D. E. Smailus, J. M. Stott, and G. S. Yang. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 28.Martin, D., and E. Rybicki. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562-563. [DOI] [PubMed] [Google Scholar]
- 29.Milne, I., F. Wright, G. Rowe, D. F. Marshal, D. Husmeier, and G. McGuire. 2004. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 20:1806-1807. [DOI] [PubMed] [Google Scholar]
- 30.Moya, A., E. C. Holmes, and F. González-Candelas. 2004. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2:279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, R. W. Yung, T. K. Ng, K. Y. Yuen, and SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poon, L., D. K. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. Leung, S. K. Lau, P. C. Woo, K. Y. Suen, K. Y. Yeun, Y. Guan, and J. S. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rambaut, A. 1996. Se-Al: sequence alignment editor. http://evolve.zoo.ox.ac.uk/.
- 34.Robertson, M. P., G. Igel, R. Baertsch, D. Haussler, M. Ares, Jr., and W. G. Scott. 2005. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 3:86-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose, T. M., J. G. Henikoff, and S. Henikoff. 2003. CODEHOP (Consensus-Degenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schickli, J. H., B. D. Zelus, D. E. Wentworth, S. G. Sawicki, and K. V. Holmes. 1997. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 71:9499-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineages. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song, W. J., Q. W. Qin, J. Qiu, C. H. Huang, F. Wang, and C. L. Hew. 2004. Functional genomics analysis of Singapore grouper iridovirus: complete sequence determination and proteomic analysis. J. Virol. 78:12576-12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephensen, C. B., D. B. Casebolt, and N. N. Gangopadhyay. 1999. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 60:181-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swofford, D. L. 2001. PAUP*: phylogenetic analysis using parsimony (and other methods) 4.0 beta. Sinauer Associates, Sunderland, Mass.
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, H., B. Linag, J. Feng, L. Sheng, and S. Y. Zhang. 2003. Molecular phylogenetic of Hipposiderids (Chiroptera: Hipposideridae) and Rhinolophids (Chiroptera: Rhinolophidae) in China based on mitochondrial cytochrome b sequences. Folia Zool. 52:259-268. [Google Scholar]
- 44.Wang, Y. X. 2003. A complete checklist of mammal species and subspecies in China—a taxonomic and geographic reference. China Forestry Publishing House, Beijing, China. (In Chinese.)
- 45.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, A. K., L. Wang, L. W. Sneed, and E. W. Collison. 1993. Analysis of a hypervariable region in the 3′ non-coding end of the infectious bronchitis virus genome. Virus Res. 28:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, X. M., K. G. Kousoulas, and J. Storz. 1992. The hemagglutinin/esterase gene of human coronavirus strain OC43: phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology 186:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]