Abstract
Hepatitis C viruses (HCVs) display a high level of sequence diversity and are currently classified into six genotypes and an increasing number of subtypes. Most likely, this heterogeneity is caused by genetic drift; evidence for recombination is scarce. To study the molecular heterogeneity of HCV in Vietnam, we analyzed 58 HCV RNA-positive sera from Vietnamese blood donors by sequence analysis of the CORE and NS5B regions. Phylogenetic analyses revealed the presence of genotype 1 (38%), genotype 2 (10.3%), and genotype 6 viruses (51.7%). All samples showed concordant results except for two (D3 and D54). Sample D54 was a mixed infection of genotype 2i and 6h viruses. Whole-genome analysis and bootscan analysis of sample D3, on the other hand, revealed a recombinant virus with genotype 2i and genotype 6p sequences at the 5′ and 3′ ends, respectively. The crossover point was located between nucleotide positions 3405 to 3464 (numbering according to prototype strain HCV-H, M67463) at the NS2/NS3 junction. The identification of this naturally occurring recombinant virus strengthens the concept that recombination may play a role in HCV epidemiology and evolution. Furthermore, the location of the recombination breakpoint may be relevant for constructing infectious chimeric viruses.
Hepatitis C virus (HCV), an important causative agent of acute and chronic hepatitis, is an enveloped plus-strand RNA virus that belongs to the family Flaviviridae (33). Its genome, approximately 9.4 kb in length, encodes both structural (Core, E1, and E2) and nonstructural (p7, NS2, NS3, NS4a/b, and NS5a/b) proteins in a single open reading frame (6). Short conserved untranslated regions (UTRs) located at the 5′ and 3′ ends of the genome are required for viral replication (9, 10). An internal ribosomal entry site in the 5′UTR is involved in protein translation (46).
Because of its heterogeneity, HCV is classified into six major genotypes and a large number of subtypes (36-38). Different genotypes display up to 30% sequence diversity, whereas subtypes vary by more than 20% (37, 38). The variability is distributed unequally across the genome, with regions such as E1 and E2 displaying most sequence diversity, whereas 5′UTR and CORE sequences are more conserved (39). Several methods were developed to determine the HCV genotype, such as serological genotyping, reverse transcription-PCR (RT-PCR) amplification with genotype specific primers, restriction fragment length polymorphism analysis, reverse hybridization assay, and sequence analysis (28). Of these methods, sequence analysis of phylogenetically informative regions is more reliable for genotype and subtype identification. Genotyping of HCV is important for prediction of the responses to and determining the duration of antiviral therapy (23, 28). Moreover, assessment of the distribution of genotypes in different parts of the world may help to understand the epidemiology and evolution of HCV.
The geographic distribution of HCV relates to different epidemic histories and routes of transmission. Some genotypes, such as 1a/b, 2, and 3a, are widely distributed in Western countries, the United States, and Japan as a result of transmission via blood transfusion and contaminated needles between intravenous drug users (2). These strains typically have limited sequence diversity, resulting from the recent introduction of a few strains from areas of endemicity (36). On the other hand, a more complex viral heterogeneity is observed in parts of Africa and Southeast Asia. In Western Africa, HCV infection is caused predominantly by genotype 2, whereas genotype 1 and 4 are most prevalent in Central Africa (4, 24). In both areas, a notable divergence of subtypes, especially broad genotype 2 variety in Ghana, was reported (5). Similar observations were made in Asia, where genotype 1, 3, and 6 dominate (33), and a large variety of genotype 6 was demonstrated in Vietnam, Thailand, Myanmar, China, and Hong Kong (8, 25, 34). This large sequence divergence points to a long-term presence of HCV infection among local populations through a variety of routes, including vertical, sexual, and household contact transmission (35, 41). The sequence diversity of HCV is supposedly caused by a high mutation rate of the RNA-dependent RNA-polymerase during replication (41). Interestingly, recombination, commonly seen among RNA viruses, including other members of the family Flaviviridae (14, 20, 47, 51), is not thought to play a major role (39, 40, 49). The fact that recombinant forms of HCV have been observed in nature, such as the St. Petersburg strains containing sequences of both genotype 2k and 1b (18) and a possible 1a/1b recombinant virus in Peru (7), would suggest that all requirements for genetic exchanges could be met. However, only a few chimeric HCV genomes were described, which may be due to the detection limits of currently used methods that are not suited for discovering recombinant strains. It is therefore possible that the true frequency of HCV recombination is underestimated (7, 18, 35), especially in areas of endemicity with a high prevalence of different HCV genotypes.
In the present study, we analyzed sera from 58 HCV RNA-positive blood donors from Ho Chi Minh City, Vietnam, in order to survey the molecular heterogeneity of HCV in this area. The CORE and NS5B genes of each sample were sequenced and phylogenetic analysis was performed. Two samples gave discordant results and were analyzed by full-length genome sequencing. Our results revealed one mixed genotype infection, whereas the other sample contained an intergenotypic recombinant virus.
MATERIALS AND METHODS
Serum samples.
Fifty-eight previously identified HCV-positive sera from the Pasteur Institute Ho Chi Minh City were collected. All samples were obtained from blood donors between 2000 and 2002. Samples were kept at −80°C until further analysis.
Amplification of HCV CORE and NS5B regions.
Viral RNA was extracted from 100 μl of serum by using the High-Pure RNA isolation kit (Roche Diagnostics GmbH) and eluted with 30 μl of water. cDNA was synthesized from 10 μl of extracted RNA with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and random hexamer primers according to the instructions of the manufacturers. Core sequences (Fig. 1A, nucleotides 339 to 695, according to the numbering system for reference strain HCV-H GenBank accession number M67463) were amplified by nested PCR with the oligonucleotides s17, 410, 953, and 951 (Table 1), as described previously (24). NS5b sequences (Fig. 1A, nucleotides 8283 to 8624) were amplified by nested PCR with the primers Pr3, Pr4, Pr1, and Pr2 (Table 1) using conditions described in previous reports (32) with modifications. Briefly, in the first PCR, performed with Pr3 and Pr4, 1.5 mM MgCl2 was used in the reaction mixture, along with the following thermal profile: initial denaturation at 94°C for 90 s; 5 cycles of denaturation at 94°C for 30 s, annealing at 64°C for 45 s, and elongation at 72°C for 1 min, followed immediately by 30 cycles at 94°C for 30 s, 64°C with a drop of −0.5°C between each cycle for 45 S, and elongation at 72°C for 1 min. The last five cycles were performed at 94°C for 30 s, 48°C for 45 s, and 72°C for 1 min. A final elongation at 72°C for 10 min was also included. The nested PCR, performed with primers Pr1 and Pr2, was carried out on 2 μl of first PCR product with the following thermal profile: 95°C for 7 min, followed by 50 cycles at 95°C for 30 s, 63°C for 30 s, and 72°C for 30 s, with a final elongation step at 72°C for 10 min. The amplified products were gel purified and sequenced.
FIG. 1.
HCV genome organization and a schematic outline of the strategies used for RT-PCR amplification of CORE and NS5B regions (A) and overlapping fragments (B) covering the entire genome. Boxes represent the coding regions for the Core protein (C), envelope 1 and 2 (E1 and E2) proteins, p7, and nonstructural proteins 1 to 5 (NS1 to NS5). Also indicated are the 5′- and 3′untranslated regions (5′UTR and 3′UTR). Black lines and dotted lines indicate the first- and second-round PCR products, respectively. Forward and reverse primers are presented at the 5′ and 3′ ends of the PCR products. The nucleotide positions and genetic organization are according to the numbering system for the prototype strain HCV-H (accession number M67463).
TABLE 1.
Primers used for amplification and sequencing of HCV
| Primera | Sequence (5′ to 3′) | Position | Polarity | Source or reference |
|---|---|---|---|---|
| 16† | GRGGCGACACTCCACCAT | 16-33 | Forward | This study |
| s17† | ACCATAGATCACTCCCCTGT | 29-48 | Forward | 31 |
| 953 | AGGTCTCGTAGACCGTGCATCATG | 321-344 | Forward | 24 |
| 951 | CAYGTRAGGGTATCGATGAC | 697-716 | Reverse | 24 |
| 410 | ATGTACCCCATGAGGTCGGC | 731-750 | Reverse | 24 |
| 693 | AAYTTGGGTAARGTCATCG | 693-711 | Forward | This study |
| 845 | CCYGGTTGCTCYTTYTCTAT | 849-868 | Forward | This study |
| 1298 | CTGGGAYATGATGATGAA | 1298-1315 | Forward | This study |
| 1992† | GGNTGYACVTGGATGAA | 1992-2008 | Forward | This study |
| 2085 | TTSCKGAARCARTCVGTRGG | 2085-2104 | Reverse | This study |
| 3277F‡ | ATGGAGAAGAARVTYATCAC | 3277-3296 | Forward | This study |
| 3277R† | CTRATRABYTTVDTCTCCAT | 3277-3296 | Reverse | This study |
| 3636 | TGGTCYACATTGGTRTACATYTG | 3636-3658 | Reverse | This study |
| 4039ঠ| GGCAGYGGBAARAGYAC | 4039-4955 | Forward | This study |
| 4662† | CRCCYGTRTASCCRGTCAT | 4662-4680 | Reverse | This study |
| 4863 | CCSTCNGGHATGTTYGA | 4863-4879 | Forward | This study |
| A5310 | CCCAGGTGCTAGTGACGAC | 5304-5322 | Reverse | 31 |
| 5460† | TAGMRCAYTCYTCCATCTC | 5460-5478 | Reverse | This study |
| 5930‡ | AGTSGCYTTYAARRTCATG | 5930-5948 | Forward | This study |
| 6085 | GCTATBAGYCKGTTCATCCA | 6084-6103 | Reverse | This study |
| 6800 | TCCCBTGYGARCCBGAG | 6820-6836 | Forward | This study |
| 7039 | TGGMGGCARGARATGGG | 7038-7054 | Forward | This study |
| 7100‡ | AAGMGGVTCRAARGAGT | 7102-7118 | Reverse | This study |
| 7600 | TGVCGRATKAGRGAGTTGC | 7681-7699 | Reverse | This study |
| 8170 | ATGGGRTTCTCCTAYGAYAC | 8244-8263 | Forward | This study |
| Pr1 | TGGGGATCCCGTATGATACCCGCTGCTTTGA | 8245-8275 | Forward | 32 |
| Pr3¶ | TATGAYACCCGCTGYTTTGACTC | 8256-8278 | Forward | 32 |
| Pr2‡ | GGCGGAATTCCTGGTCATAGCCTCCGTGAA | 8616-8645 | Reverse | 32 |
| Pr4 | GCNGARTAYCTVGTCATAGCCTC | 8622-8644 | Reverse | 32 |
| 8625‡ | GARTACCTRGTCATAGC | 8625-8641 | Reverse | This study |
| 9325¶ | CTACRGTAAGTAGGAGTAGGC | 9325-9345 | Reverse | This study |
| 3′UTR¶ | AAAAAAAAAAAAAAAAAA | 9418-9435 | Reverse | This study |
†, Primer used for amplification of cDNA fragment nucleotides 17 to 5460; ‡, primer used for amplification of cDNA fragment nucleotides 3227 to 8625; ¶, primer used for amplification of cDNA fragment nucleotides 4039 to 9435.
HCV full-length genome amplification.
Total RNA was isolated from 100 μl of serum by proteinase K digestion and phenol-chloroform extraction as previously described (31) and dissolved in 20 μl of sterile water. To generate a long fragment of cDNA, RT was performed on 10 μl of RNA with Expand reverse transcriptase (Roche Diagnostics GmbH) and primer 5460, 8625, or 3′UTR (Table 1 and Fig. 1B), according to the manufacturer's instructions, in a total volume of 30 μl at 42°C for 2 h. We amplified 10-μl volumes of cDNA with 2.5 U of Expand High-Fidelity enzyme mixture (Roche Diagnostics GmbH), 1× Expand PCR buffer, 0.4 mM concentrations of each dideoxynucleotide, and 0.4 μM concentrations of outer primer pairs (Table 1), with a thermal profile that was described previously (31). First-round amplification from each cDNA reaction resulted in three overlapping PCR fragments (nucleotides 16 to 5460, 3227 to 8625, and 4039 to 9416; Fig. 1B). Next, 2-μl volumes of amplified product were subjected to nested PCR with inner primers to generate overlapping genome fragments (nucleotides 17 to 3277, 1992 to 4662, 4039 to 7100, 5930 to 8645, and 8256 to 9325; Fig. 1B) under conditions similar to those used for the first PCR. Amplified products were gel purified with a Qiaquick gel extraction kit (QIAGEN) and ligated directly into plasmid pCR2.1 (TA Cloning Kit; Invitrogen). For each amplicon, five positive clones were selected and sequenced.
DNA sequencing and sequence analysis.
Sequence reactions on PCR products and plasmids were performed by using a BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems) and an ABI Prism 3100 autosequencer (Applied Biosystems). Sequencing primers are shown in Table 1. Multiple sequence alignments were generated with the BioEdit program (version 7.0.1). Phylogenetic analyses were conducted by using the Kimura two-parameter model and neighbor-joining methods. Bootstrapping (1,000 replicates) was tested, and consensus trees were produced by using MEGA version 3.0 (19).
Reference strains used in the present study were obtained from GenBank (accession number): genotypes 1a (M67463), 1b (D90208), 2a (AF169005, AB047644), 2b (AF238486), 2c (D50409), 2f (D49754), 2k (AB031663), 2i (L48499, X76411), 3a (D28917), 3b (D49374), 3k (10a, 0D63821), 4a (Y11604), 5a (Y13184), 6a (Y12083), 6b (D84262), 6c (7d, D37857 and D37843), 6d (7b, D84263), 6e (7a, D87365 and D88478), 6f (7c, D38078 and D38079), 6g (11a, D63822), 6 h (9a, D84265), 6i (9b, D37864 and D37850), 6j (9c, D37862 and D37848), 6k (8b, D84264), 6l (8a, D87359 and D88472), and 6p (L38340 and L38380). The sequence of hepatitis G-B virus (U22304) was used as an outgroup.
Similarity and bootscanning analyses of whole-genome sequences were performed by using the Simplot program (version 2.5 for Windows 95/NT; available from the author, Stuart C. Ray [http://sray.med.som.jhmi.edu/RaySoft/SimPlot/]).
Nucleotide sequence accession numbers.
New sequences reported in the present study have been submitted to GenBank and have been assigned accession numbers DQ155444 to DQ155501 (CORE sequences), DQ155502 to DQ155559 (NS5B sequences), DQ155560 (D3 complete genome sequence), DQ155561 (D54 genotype 2i complete genome sequence), and DQ155562 and DQ155566 (D54 5′UTR-to-CORE sequences).
RESULTS
Genotyping of HCV isolates from Ho Chi Minh City, Vietnam.
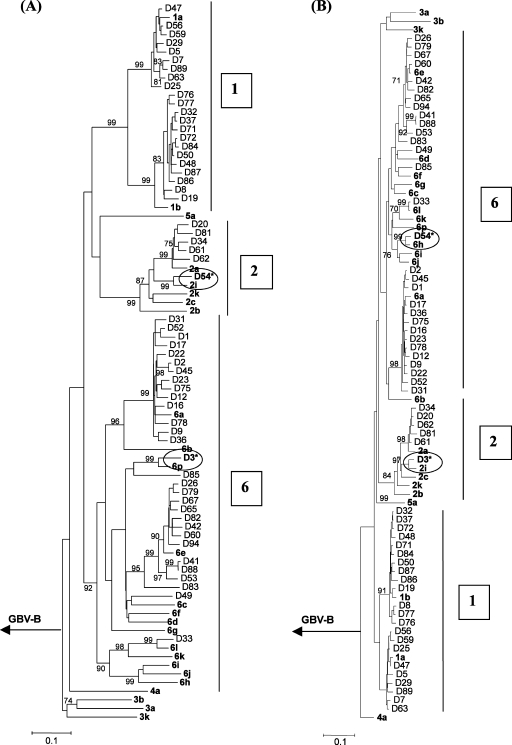
We obtained 58 serum samples from Vietnamese blood donors, which were previously found to be HCV antibody positive. Viral RNA served as a template for RT-PCR amplification of the Core and NS5B sequences (Fig. 1). To study HCV diversity, a phylogram, based on 333 nucleotides of the NS5B isolated from these 58 samples and 26 prototype strains was constructed (Fig. 2A). This analysis revealed the presence of genotype 1 (38%), genotype 2 (10.3%), and genotype 6 (51.7%) viruses. All Vietnamese 1b isolates subclustered together with a bootstrap value of 80%, while most of the subtype 1a strains were grouped with the 1a reference sequence (Fig. 2A). Five samples were characterized as subtype 2a, and one strain (D54) was identified as subtype 2i. High levels of sequence divergence were observed among the genotype 6 viruses; 4 HCVs (D41, D53, D83, and D88) exhibited separate branches from prototype strains, supported by bootstrap values. These data are a showpiece example of why it is so difficult to subtype genotype 6 variants based on reference sequences.
FIG. 2.
Rooted neighbor-joining trees depicting the phylogenetic relationships among HCV field variants and prototype strains. Trees were constructed for a 333-nucleotide segment of the NS5B gene (A) and a 357-nucleotide segment of the CORE gene (B), using the Kimura two-parameter model. Confidence values (>70%) calculated by bootstrap analysis (1,000 replicates) are indicated at the major branching points. Branch lengths are drawn to scale. The prototype HCV strains obtained from GenBank are indicated by boldface letters and hepatitis G-B virus (U22304) was used as an outgroup sequence. Samples that gave discordant results in the phylogenetic trees are specified by asterisks and grouped to the related strain with circular mark.
To confirm the consistency of genotyping, we constructed a phylogram based on the 357 nucleotides of the CORE sequence (Fig. 2B). Although the HCV CORE sequences of Vietnamese genotype 1 and 2 strains showed less divergence compared to prototypes than in the NS5B sequence, the genotype 6 strains showed considerable variety. Overall, 56 of 58 samples showed concordant genotyping results. Interestingly, sample D3 was characterized as genotype 2i based on the CORE region but typed as a genotype 6p after phylogenetic analysis of the NS5B sequence. Sample D54 on the other hand was classified as genotype 6h by analysis of the CORE sequence but grouped with genotype 2i after NS5B analysis.
Characterization of sample D54: mixed infection with HCV genotypes 2 and 6.
Conflicting phylogenetic data, such as were obtained for the CORE and NS5B sequences in samples D3 and D54, are indicative of the presence of either a mixed infection with two different genotypes or an intergenotypic recombinant virus. Upon analysis with the 5′UTR reverse-hybridization line probe assay (INNO-LiPA HCV II), HCV from sample D3 was identified as genotype 2 and patient D54 was shown to have a mixed infection with HCVs of genotype 2 and genotype 1 or 6 variant (data not shown).
To further characterize sample D54, the 5′-UTR-to-CORE region (nucleotides 29 to 752) was amplified, cloned, and sequenced. The alignment of CORE sequences obtained from sample D54 with prototype strains indicated that this patient had been doubly infected with HCV genotype 2i (clone 20) and 6h (clone 1) viruses (Fig. 3). Complete genome sequences of both genotypes from sample D54 were determined following the strategy indicated in Fig. 1. No evidence for recombination was found. Phylogenetic analysis of CORE, E1, and NS5B sequences of HCV from sample D54 confirmed that one of the full-length sequences was of genotype 2i, whereas the other was of genotype 6 h (data not shown).
FIG. 3.
Alignment of CORE sequences obtained from sample D54. Two representative clones, clone 1 (C1) and clone 20 (C20), were aligned with genotype 2a, 2k, 2i, 6a, 6d, and 6 h prototype strains. The D54 sequences are indicated in boldface. The accession numbers of references strains are given in Materials and Methods. Residues identical to the major sequence are indicated by dashes, and numbering of the nucleotides starts at the first codon of the CORE gene.
Characterization of sample D3: HCV genotype 2/6 recombinant.
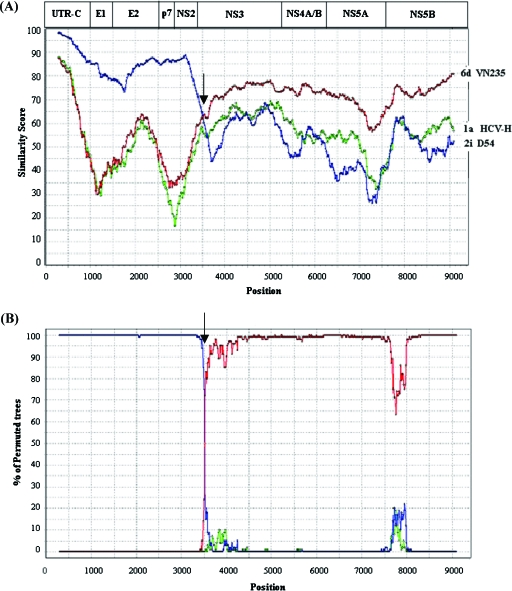
Sequence analysis of 10 individual clones of 5′UTR-CORE sequences from sample D3, confirmed the data from the INNO-LiPA HCV II line probe assay, suggesting the presence of a viral strain belonging to genotype 2i (data not shown). Because the data depicted in Fig. 2 strongly indicate the presence of both genotype 2i and 6p sequences, we suspected the presence of a recombinant virus. The full-length genome sequence of sample D3 was determined. In order to identify recombination breakpoints, we used similarity plot and bootscanning analysis (Fig. 4). Since complete sequences of genotype 2i and genotype 6p were not found in the sequence databases, we used genotype 6d (VN235, D84236) and the genotype 2i strain from sample D54 as reference sequences. Genotype 1a (HCV-H, M67463) was included as an outgroup sequence. Figure 4A shows the genetic distances between the reference strains and the potential recombinant HCV strain of sample D3. The 5′UTR-to-NS2 region of the HCV strain of sample D3 is more related to subtype 2i sequences, whereas the NS3-to-NS5B region is more closely related to subtype 6d. Thus, the Simplot analysis (Fig. 4) corroborates the results from the phylogenetic trees (Fig. 2), providing evidence that the HCV strain in sample D3 is a recombinant virus. It is of note that the recombinant virus of sample D3 shows ∼25% sequence divergence compared to genotype 6d viruses in its C-terminal part of the genome.
FIG. 4.
(A) Similarity plots of the intergenotypic recombinant D3 strain and HCV genotype 1a (HCV-H), 2i (D54), and 6d (VN235) based on the entire genome using a window size of 600 bp, a step size of 20 bp, and the maximum-likelihood parameter. Gaps were ignored, and 1,000 bootstrap replicates were used. (B) Bootscan plots showing the likelihood of clustering of the putative intergenotypic recombinant of sample D3 with reference strains, with 1,000 bootstrap replicates and neighbor-joining tree analysis. As a reference, the genomic organization of HCV, drawn to scale, is depicted at the top. Arrows indicate the recombination breakpoints. The nucleotide positions are numbered as described in the legend of Fig. 1.
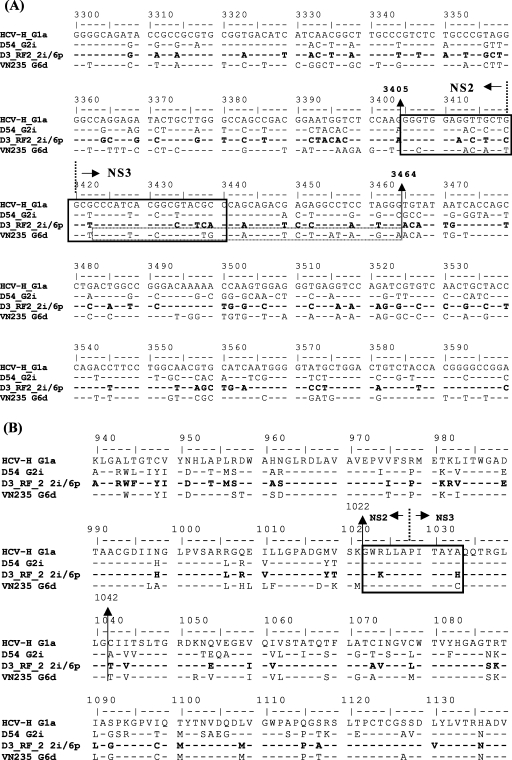
Bootscan analysis of the D3 HCV genome revealed the existence of only one recombination breakpoint, located within the NS2-NS3 region (Fig. 4B), between nucleotides 3405 and 3464 (Fig. 5A) and amino acids 1022 and 1042 (Fig. 5B). Interestingly, the recombination site is a conserved region at the amino acid level (Fig. 5B), which is cleaved by the NS2-NS3 protease (15, 43). Almost all cases of genetic exchanges in RNA viruses are thought to occur via copy-choice RNA recombination (26, 51). Template switching is supposedly aided by homology between the donor and acceptor strand and secondary RNA structures (1, 26). According to the proposed mechanism of HCV recombinant formation described by Kalinina et al. (17), the secondary RNA structure around the recombination site may enhance genetic exchanges in vivo. We could identify a stable hairpin structure (nucleotides 3423 to 3464) in the positive strand of the genotype 6d strain VN235 that is not present in genotype 2i and that may have played a role in the creation of our recombinant genotype 2i/6 virus (data not shown).
FIG. 5.
Alignment of nucleotide (A) and amino acid (B) sequences of the NS2/NS3 region of the intergenotypic HCV recombinant of sample D3 and reference strains. The recombinant sequence(s) is shown in boldface. Boxes with solid lines indicate the cleavage site for the NS2/NS3 protease. The dotted box indicates the sequence forming the postulated hairpin proposed to promote recombination. The broken lines indicate the protease cleavage site of the NS2-NS3 protein. Vertical arrows overlap nucleotides 3405 to 3464 or amino acids 1022 to 1042 and represent the recombination breakpoint. The nucleotide and amino acid positions are according to the numbering system described in the legend of Fig. 1.
To rule out the possibility of RT and PCR artifacts, we used a different reverse primer (4662; Table 1) to generate a cDNA fragment of HCV from sample D3 and amplified a short PCR product covering the NS2/NS3 region with primer 3277F and 4662 (positions 3277 to 4662) (Fig. 1). The results confirmed the recombination breakpoint.
DISCUSSION
Until recently, there was no evidence for recombination in HCVs. Therefore, it has been implicitly assumed that HCV diversity was generated through genetic drift. Recently, however, both intergenotypic and intragenotypic recombinant HCVs have been identified (7, 18). In the present study, we have identified a second intergenotypic recombinant form of HCV.
Molecular heterogeneity of HCV in 58 HCV-positive serum samples from blood donors in Ho Chi Minh City were analyzed. Phylogenetic analyses revealed the presence of genotype 1 (38%), genotype 2 (10.3%), and genotype 6 (51.7%) viruses. The Vietnamese genotype 1a and 1b viruses in our study formed separate clusters and showed lower diversity than the genotype 6 strains. This suggests that genotype 1 viruses were introduced in Vietnam relatively recently and were spread by intravenous drug users and through the use of contaminated blood products (35, 41). The higher heterogeneity of genotype 6 viruses would indicate, on the other hand, that these strains have circulated in Vietnam for a longer period of time (29, 41). The Vietnamese genotype 2a viruses clustered together and are related to subtype 2a from Japan. Interestingly, one of the strains that infected patient D54 was identified as a relatively rare subtype 2i, based on comparison with the CORE, E1, and NS5B sequences of prototype 2i strain HN4. Subtype 2i was first identified in France in 1994 (30); the origin of the Vietnamese 2i virus is unknown. It is not unlikely that it was introduced in Vietnam by French citizens.
The most striking observation in our study is the identification of an HCV recombinant in Vietnam; the second intergenotypic recombinant that has been described thus far. This virus contains sequences from subtype 2i and genotype 6. The genotype 6 region in this virus is probably most closely related to a new subtype 6p, of which recently a short NS5B sequence was reported (42). The new HCV recombinant thus can be classified as a candidate RF2_2i/6p strain. The recombination breakpoint is located between nucleotides 3405 and 3464. In the HCV polyprotein, this region is cleaved by the NS2-NS3 protease and is reasonably conserved among different HCV genotypes (43). The copy choice model, originally proposed in the case of poliovirus recombination (20, 45), supports almost all possible mechanisms of RNA viruses described thus far. In this case, hybrid RNAs might be formed when the viral RNA-dependent RNA polymerase complex switches during replication. Our data suggest that both the high level of homology between donor and acceptor strand and the presence of a hairpin structure in the region where the genetic exchange occurred facilitated the generation of this recombinant.
It is worth noting that the recombination breakpoints in both identified intergenotypic HCV recombinants are located in the NS2/NS3 region of the genome (18). This suggests that this region of the genome is especially suitable for creating viable intergenotypic recombinants in vivo. Moreover, this observation may aid researchers in designing chimeric HCV genomes for in vitro experiments, using the recently constructed infectious clone of HCV (21, 50, 54).
The frequency of successful recombination events is determined among others by (i) the properties inherent to the viral replicase, (ii) the odds of the double infection of single cells, and (iii) the viability of the recombinant progeny and the increase in the fitness of the recombinant virus compared to its nonrecombinant parents. Overall, there is ample evidence that the generation of HCV recombinants is a rare event (35, 40, 49). On the one hand, most recombinants generated may not be stable, which is illustrated by the fact that chimeric HCV replicons, even constructed from closely related HCV subtypes 1a and 1b, often fail to replicate in cell culture (11, 13). Recent studies indicated that optimal HCV replication complexes composed of the various NS proteins, cis-acting RNA elements, and cellular factors are required for HCV replication (12, 22, 52). Possibly, recombinants that maintain successful replication complexes are scarce, and they may not be able to compete with parental viruses. However, even if new replication-competent recombinant viruses are generated, they may be hampered in infectious virus particle formation, as suggested by recent experiments with infectious full-length genotype 1a/2a chimeras (21).
However, the fact that recombinant HCV viruses have been found (7, 18) suggests that in principal the viral replicase is allowing the generation of chimeric genomes. The overlap in genotype distributions in many parts of the world would in theory increase the chance of double infections and thus the likelihood of recombination, especially in cases of frequent repeated needle sharing over short time intervals by infected drug users and the use of contaminated blood products. Although most of the HCV recombinant strains might be selected out by natural selection (35, 49), the fact that recombinant viruses have been identified would suggest that at least some recombinants are viable.
Based on these findings we may currently underestimate the true frequency of HCV recombination. Recombination events in HCV are difficult to detect if they occurred between variants of the same subtype or between highly diverse subtypes. Even though there are some reports describing different genotype and subtype sequences of different regions obtained from single sample (27, 53), there are always doubts due to the possibility of mixed infections or contaminated samples (27). Moreover, most studies of HCV variability are based on analysis of single, short genomic regions, making the detection of potential recombinants unlikely. Therefore, the current methodology of sequencing short genomic regions as markers for entire viral genomes is challenged now, since it may not produce accurate results of all genetic characteristics. Sequence analysis of whole HCV genomes would possibly increase the detection of natural HCV recombinants.
Recombination has been well documented for many RNA viruses (51), including other members of the family Flaviviridae (3, 16, 44, 47, 48, 51), of which evolution was for long supposed to be clonal, with diversity generated by the accumulation of mutations. The mere fact that recombinant HCV viruses have been identified (7, 18; the present study), suggests that the assumption that HCV diversity is caused only via genetic drift is no longer tenable and that the role recombination plays in HCV evolution and biology demands serious consideration. Clearly, the fact that recombination occurs between subtypes and genotypes of HCV not only presents a serious impediment to vaccine development but also to prediction of the response to antiviral therapy.
Acknowledgments
The Commission of the European Community, HECSA Project (ICA4-CT-1999-10009), supported this study.
We thank Vu Thuy Yen, Jan Brouwer, and Paula van Luijt for technical assistance and Byron Martina for valuable discussions.
REFERENCES
- 1.Alejska, M., A. Kurzyniska-Kokorniak, M. Broda, R. Kierzek, and M. Figlerowicz. 2001. How RNA viruses exchange their genetic material. Acta Biochim. Pol. 48:391-407. [PubMed] [Google Scholar]
- 2.Alter, M. J. 1999. Hepatitis C virus infection in the United States. J. Hepatol. 31(Suppl. 1):88-91. [DOI] [PubMed] [Google Scholar]
- 3.Becher, P., M. Orlich, and H. J. Thiel. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J. Virol. 75:6256-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candotti, D., J. Temple, F. Sarkodie, and J. P. Allain. 2003. Frequent recovery and broad genotype 2 diversity characterize hepatitis C virus infection in Ghana, West Africa. J. Virol. 77:7914-7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colina, R., D. Casane, S. Vasquez, L. Garcia-Aguirre, A. Chunga, H. Romero, B. Khan, and J. Cristina. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31-37. [DOI] [PubMed] [Google Scholar]
- 8.Doi, H., C. Apichartpiyakul, K. I. Ohba, M. Mizokami, and H. Hotta. 1996. Hepatitis C virus (HCV) subtype prevalence in Chiang Mai, Thailand, and identification of novel subtypes of HCV major type 6. J. Clin. Microbiol. 34:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gates, A. T., R. T. Sarisky, and B. Gu. 2004. Sequence requirements for the development of a chimeric HCV replicon system. Virus Res. 100:213-222. [DOI] [PubMed] [Google Scholar]
- 12.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu, B., A. T. Gates, O. Isken, S. E. Behrens, and R. T. Sarisky. 2003. Replication studies using genotype 1a subgenomic hepatitis C virus replicons. J. Virol. 77:5352-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, E. C., M. Worobey, and A. Rambaut. 1999. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 16:405-409. [DOI] [PubMed] [Google Scholar]
- 17.Kalinina, O., H. Norder, and L. O. Magnius. 2004. Full-length open reading frame of a recombinant hepatitis C virus strain from St Petersburg: proposed mechanism for its formation. J. Gen. Virol. 85:1853-1857. [DOI] [PubMed] [Google Scholar]
- 18.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 20.Lai, M. M. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 22.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 24.Mellor, J., E. C. Holmes, L. M. Jarvis, P. L. Yap, P. Simmonds, et al. 1995. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J. Gen. Virol. 76(Pt. 10):2493-2507. [DOI] [PubMed] [Google Scholar]
- 25.Mellor, J., E. A. Walsh, L. E. Prescott, L. M. Jarvis, F. Davidson, P. L. Yap, and P. Simmonds. 1996. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core-based genotyping assay. J. Clin. Microbiol. 34:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 27.Ndjomou, J., O. G. Pybus, and B. Matz. 2003. Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J. Gen. Virol. 84:2333-2341. [DOI] [PubMed] [Google Scholar]
- 28.Nolte, F. S. 2001. Hepatitis C virus genotyping: clinical implications and methods. Mol. Diagn. 6:265-277. [DOI] [PubMed] [Google Scholar]
- 29.Pybus, O. G., M. A. Charleston, S. Gupta, A. Rambaut, E. C. Holmes, and P. H. Harvey. 2001. The epidemic behavior of the hepatitis C virus. Science 292:2323-2325. [DOI] [PubMed] [Google Scholar]
- 30.Qu, D., O. Hantz, M. Gouy, L. Vitvitski, J. S. Li, F. Berby, S. P. Tong, and C. Trepo. 1994. Heterogeneity of hepatitis C virus genotypes in France. J. Gen. Virol. 75 (Pt. 5):1063-1070. [DOI] [PubMed] [Google Scholar]
- 31.Rispeter, K., M. Lu, S. Lechner, A. Zibert, and M. Roggendorf. 1997. Cloning and characterization of a complete open reading frame of the hepatitis C virus genome in only two cDNA fragments. J. Gen. Virol. 78(Pt. 11):2751-2759. [DOI] [PubMed] [Google Scholar]
- 32.Sandres-Saune, K., P. Deny, C. Pasquier, V. Thibaut, G. Duverlie, and J. Izopet. 2003. Determining hepatitis C genotype by analyzing the sequence of the NS5b region. J. Virol. Methods 109:187-193. [DOI] [PubMed] [Google Scholar]
- 33.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 34.Shinji, T., Y. Y. Kyaw, K. Gokan, Y. Tanaka, K. Ochi, N. Kusano, T. Mizushima, S. Fujioka, H. Shiraha, A. A. Lwin, Y. Shiratori, M. Mizokami, M. Khin, M. Miyahara, S. Okada, and N. Koide. 2004. Analysis of HCV genotypes from blood donors shows three new HCV type 6 subgroups exist in Myanmar. Acta Med. Okayama 58:135-142. [DOI] [PubMed] [Google Scholar]
- 35.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 36.Simmonds, P. 1999. Viral heterogeneity of the hepatitis C virus. J. Hepatol. 31(Suppl. 1):54-60. [DOI] [PubMed] [Google Scholar]
- 37.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 38.Simmonds, P., E. C. Holmes, T. A. Cha, S. W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74(Pt. 11):2391-2399. [DOI] [PubMed] [Google Scholar]
- 39.Simmonds, P., D. B. Smith, F. McOmish, P. L. Yap, J. Kolberg, M. S. Urdea, and E. C. Holmes. 1994. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol. 75(Pt. 5):1053-1061. [DOI] [PubMed] [Google Scholar]
- 40.Smith, D. B., and P. Simmonds. 1997. Review: molecular epidemiology of hepatitis C virus. J. Gastroenterol. Hepatol. 12:522-527. [DOI] [PubMed] [Google Scholar]
- 41.Stumpf, M. P., and O. G. Pybus. 2002. Genetic diversity and models of viral evolution for the hepatitis C virus. FEMS Microbiol. Lett. 214:143-152. [DOI] [PubMed] [Google Scholar]
- 42.Stuyver, L., A. Wyseur, W. van Arnhem, F. Lunel, P. Laurent-Puig, J. M. Pawlotsky, B. Kleter, L. Bassit, J. Nkengasong, L. J. van Doorn, and G. Maertens. 1995. Hepatitis C virus genotyping by means of 5′-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 38:137-157. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, R., T. Suzuki, K. Ishii, Y. Matsuura, and T. Miyamura. 1999. Processing and functions of hepatitis C virus proteins. Intervirology 42:145-152. [DOI] [PubMed] [Google Scholar]
- 44.Tolou, H. J., P. Couissinier-Paris, J. P. Durand, V. Mercier, J. J. de Pina, P. de Micco, F. Billoir, R. N. Charrel, and X. de Lamballerie. 2001. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 82:1283-1290. [DOI] [PubMed] [Google Scholar]
- 45.Tolskaya, E. A., L. I. Romanova, V. M. Blinov, E. G. Viktorova, A. N. Sinyakov, M. S. Kolesnikova, and V. I. Agol. 1987. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology 161:54-61. [DOI] [PubMed] [Google Scholar]
- 46.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Twiddy, S. S., and E. C. Holmes. 2003. The extent of homologous recombination in members of the genus Flavivirus. J. Gen. Virol. 84:429-440. [DOI] [PubMed] [Google Scholar]
- 48.Uzcategui, N. Y., D. Camacho, G. Comach, R. Cuello de Uzcategui, E. C. Holmes, and E. A. Gould. 2001. Molecular epidemiology of dengue type 2 virus in Venezuela: evidence for in situ virus evolution and recombination. J. Gen. Virol. 82:2945-2953. [DOI] [PubMed] [Google Scholar]
- 49.Viazov, S., A. Widell, and E. Nordenfelt. 2000. Mixed infection with two types of hepatitis C virus is probably a rare event. Infection 28:21-25. [DOI] [PubMed] [Google Scholar]
- 50.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80(Pt. 10):2535-2543. [DOI] [PubMed] [Google Scholar]
- 52.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yun, Z., C. Lara, B. Johansson, I. Lorenzana de Rivera, and A. Sonnerborg. 1996. Discrepancy of hepatitis C virus genotypes as determined by phylogenetic analysis of partial NS5 and core sequences. J. Med. Virol. 49:155-160. [DOI] [PubMed] [Google Scholar]
- 54.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]