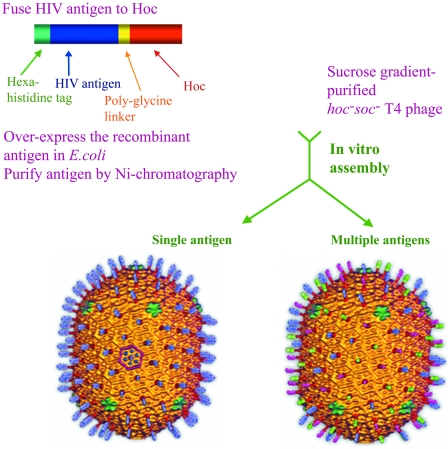
Abstract
Bacteriophage T4 capsid is an elongated icosahedron decorated with 155 copies of Hoc, a nonessential highly antigenic outer capsid protein. One Hoc monomer is present in the center of each major capsid protein (gp23*) hexon. We describe an in vitro assembly system which allows display of HIV antigens, p24-gag, Nef, and an engineered gp41 C-peptide trimer, on phage T4 capsid surface through Hoc-capsid interactions. In-frame fusions were constructed by splicing the human immunodeficiency virus (HIV) genes to the 5′ or 3′ end of the Hoc gene. The Hoc fusion proteins were expressed, purified, and displayed on hoc− phage particles in a defined in vitro system. Single or multiple antigens were efficiently displayed, leading to saturation of all available capsid binding sites. The displayed p24 was highly immunogenic in mice in the absence of any external adjuvant, eliciting strong p24-specific antibodies, as well as Th1 and Th2 cellular responses with a bias toward the Th2 response. The phage T4 system offers new direction and insights for HIV vaccine development with the potential to increase the breadth of both cellular and humoral immune responses.
A collaborative global effort is under way to develop an efficacious human immunodeficiency virus (HIV) vaccine (2, 5, 18). Considering the complexity of HIV replication and pathogenesis, it is widely recognized that an HIV vaccine, in order to be effective, should include multiple antigens and generate strong and broad neutralizing antibodies, as well as cell-mediated immune responses (2, 6, 18). Many strategies have been used to develop multicomponent HIV vaccine formulations. These include expression systems such as vaccinia virus and adenovirus, multiple antigenic peptides, and DNA prime-boost strategies (6, 30, 31). Although some of these approaches have been successful in rodent or nonhuman primate models, these methods have not yet translated effectively into a vaccine for humans (9). Recombinant platforms that allow construction of multicomponent vaccines eliciting both humoral and cellular immune responses are particularly attractive for advancing HIV vaccine development.
Bacteriophage T4 possesses unique features that lend itself to the development of a multicomponent vaccine platform. Phage T4 capsid is a prolate icosahedron (T=20) with precise dimensions: width of 86 nm and length of 119.5 nm (7, 11) (Fig. 1). It comprises 930 copies (155 hexamers) of the major capsid protein, gp23* (49 kDa; gold/blue protrusions, Fig. 1), 55 copies (11 pentamers) of the vertex protein gp24* (46 kDa; green subunits), 12 copies (one dodecamer) of the portal vertex protein gp20 (61 kDa; capsid-proximal ring at the base of the capsid), and two outer capsid proteins, Hoc (39 kDa) and Soc (10 kDa) (7). (The asterisk represents the cleaved form of the capsid protein following T4 capsid assembly-dependent maturation cleavages.) Hoc (for highly antigenic outer capsid protein; red spikes, Fig. 1) is present up to 155 copies per capsid particle, with each monomer occupying the center of the gp23* hexon. Soc (for small outer capsid protein; gold/purple subunits, Fig. 1) is present up to 810 copies per capsid particle, with each monomer bridging two gp23 monomers of adjacent hexamers (7, 13). Hoc and Soc are nonessential and bind to the outer capsid surface after the completion of capsid assembly (12). We and others have shown that foreign antigens fused to Hoc and Soc can be expressed in Escherichia coli and displayed on T4 capsid in vivo as part of phage morphogenesis and that the T4-displayed antigens are immunogenic in mice (14, 25).
FIG. 1.
Schematic of the phage T4 in vitro assembly system. Phage T4 capsid cryo-electron microscopy reconstructions (7) are shown with antigen “spikes” artificially fused to Hoc subunits. The left reconstruction shows blue spikes representing the display of single antigen (p24-gag in the present study), and the right reconstruction shows blue, green, and pink spikes representing the display of three antigens (p24-gag, Nef, and gp41 C-trimer in the present study). Hoc subunits are shown in dark red; the hexameric gp23* protrusions and the Soc subunits bridging the gp23* subunits form the capsid shell shown in gold; in one of the hexagons of the icosahedral face (left reconstruction), the gp23* and Soc subunits are shown in blue and purple, respectively, to distinguish these subunits on the capsid lattice. The vertex at the base of the capsid represents the unique portal vertex to which the neck and tail attach (not shown).
We hypothesized that Hoc/Soc-fused antigens can be assembled on T4 capsid in vitro, as long as the integrity of the capsid binding site is not compromised. Using purified phage and functionally well-characterized antigens, the binding process can be tightly controlled in vitro and made very efficient. Such a system would also be free of limitations that are inherent in the classic in vivo phage display systems such as M13, λ, T3/T7, and T4 (10, 28), which require intracellular expression and assembly of the antigen. Problems such as (i) a low and variable copy number of displayed antigen; (ii) nonspecific proteolysis leading to loss of critical epitopes; and (iii) structural heterogeneity due to aggregation, insolubility, and improper folding can be bypassed by in vitro binding. Immunization with the in vitro-assembled particulate antigens should stimulate strong immune responses, which can be further boosted by displaying additional targeting and/or enhancing molecules. (The term “assembly” here refers to the binding of Hoc fusion protein molecules to the symmetrically arranged binding sites on the T4 capsid surface. “Assembly,” “binding,” and “display” are interchangeably used to refer this process.) We have tested this hypothesis in two separate studies using two sets of antigens: anthrax protective antigen (27) and HIV antigens.
We report here a defined in vitro assembly system that allows high-density display of single as well as multiple HIV antigens on phage T4 capsid surface (Fig. 1). The basic system involves fusion of the HIV antigen to the N terminus of Hoc, expression in E. coli, preparation of highly purified antigen, and attachment in vitro of the fusion protein to Hoc-deficient phage through specific interactions with the capsid binding sites. The displayed HIV antigen would decorate the T4 capsid by occupying the center of gp23* hexons. More than one HIV antigen can be assembled on the same capsid, generating a multicomponent vaccine in a single binding step (Fig. 1), without incorporating any HIV genome sequences into the immunogen.
The major capsid protein of HIV virion, p24-gag, was chosen as a model antigen to develop the system and determine the immunogenicity of the T4-displayed p24. p24 is an important target for HIV vaccine development because of its high degree of sequence conservation among HIV isolates (8). Declining p24 antibody titers correlate with clinical deterioration and progression into AIDS (26). We show that the T4-displayed p24 was highly immunogenic in mice, eliciting strong humoral and cellular immune responses. In addition, other HIV antigens, Nef and a novel gp41C-peptide trimer were also targeted for display to assess the multivalent vaccine concept and the broad applicability of the system. The data show the efficient display of both single and multiple HIV antigens on phage T4 capsid and offer insights for designing novel particulate HIV vaccines that have not been demonstrated by other vector systems.
MATERIALS AND METHODS
Bacteriophage, bacteria, and DNAs.
The hoc−soc− mutant phage (hocQ21am; soc.del) was constructed in the laboratory (27). E. coli P301 (Δsup) was used to prepare wild-type and hoc−soc− phage stocks. E. coli XL-10 Gold cells (Stratagene) were used for initial transformation of the recombinant plasmids and stable maintenance of constructs for the long-term. The clones were transferred into the expression strains, E. coli BL21(DE3)/pLys-S or BL21(DE3)/RIL, for IPTG (isopropyl-β-d-thiogalactopyranoside)-induced overexpression of Hoc fusion proteins (29). The T7 expression vectors pET-15b and pET-28b (Ampr) (Novagen) were used for the cloning experiments. The HIV DNA corresponding to the full-length p24-gag sequence of HIV-1 su10 isolate was amplified from the pDAB72 plasmid; the HIV-nef DNA was amplified from pT7consnefhis6 plasmid (AIDS reagent program). The gp41 C-peptide sequence used for the synthetic gene construction corresponds to the consensus sequence based on HIV-1 HXB2 gp160 variant. Purified phage T4 DNA was used as a template for amplification of Hoc gene.
Construction of gene fusions.
A PCR-based splicing-by-overlap-extension strategy was used to construct various gene fusions (24). For example, the p24-hoc gene fusion was constructed by using the following four primers: p24-5′-forward primer (5′-CGC GGA TCC GCC TAT AGT GCA GAA CAT CCA GGG GC), p24 reverse fusion primer (5′-GGA GTT ATA TCA ACT GTA AAA GTC ATT CCA CCA GGC AAA ACT CTT GCC TTA TGG CC-3′), hoc forward fusion primer (5′-GGC CAT AAG GCA AGA GTT TTG CCT GGT GGA ATG ACT TTT ACA GTT GAT ATA ACT CC-3′), and hoc reverse primer (5′-CGC GGA TCC TTA TGG ATA GGT ATA GAT GAT ACC-3′) (italicized nucleotides represent the tag sequence added to the 5′ end for efficient digestion at the adjacent BamHI sequence that is underlined). The p24 reverse fusion primer consisted of nucleotide sequences corresponding to the 3′ end of p24 gene (boldface), Pro-Gly-Gly linker (boldface and underlined) and the 5′ end of Hoc gene; the hoc forward fusion primer had the complementary sequence. The p24 and Hoc genes were amplified separately using p24 forward and reverse fusion primers and hoc forward fusion and reverse primers, respectively. This created a 56-nucleotide overlap between the 3′ end of p24 and the 5′ end of hoc. In the second-stage PCR, the DNA fragments were “stitched” together by forced overlap extension, thus creating p24-Hoc gene fusion, which was then amplified by using the end primers (p24 forward and hoc reverse primers).
Hoc-p24 and Nef-Hoc were constructed by using the same strategy and appropriate primers. For construction of gp41 C-trimer-Hoc, the C-peptide trimer gene was first chemically synthesized. The nucleotide sequence corresponding to gp41 C-peptide (amino acid sequence WMEWDREINNYTSLIHSLIEESQNQQEKNEQEELLELDKWASLWNWF628-673 consisting of 2F5 and T-helper epitopes [19]) was repeated three times with a five-amino-acid linker, GGSGG, separating the repeats. A codon-optimized nucleotide sequence was generated by using GeneOptimizer software and assembled with the aid of GeneAssembler platform (Geneart, Germany). The C-trimer gene was fused to Hoc by the splicing-by-overlap-extension strategy using appropriate primers.
All amplifications were performed by using high-fidelity Precision-Taq DNA polymerase (Stratagene) for 25 to 30 cycles, with each cycle consisting of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 to 2 min. The stitched DNAs were digested with BamHI and purified by agarose gel electrophoresis (QIAGEN). The purified DNA was inserted into the BamHI site of pET-15b/pET-28b and transformed into E. coli XL-10 Gold cells. The primers were designed such that insertion in the right orientation would result in in-frame fusion of the construct with the upstream vector sequence corresponding to a 25-amino-acid peptide consisting of a hexahistidine sequence. Clones in the right orientation were sequenced for accuracy of the fusion and transformed into either BL21(DE3)/pLys-S or codon plus BL21(DE3)/RIL for protein expression.
Purification of recombinant proteins.
The native Hoc and HIV-Hoc recombinant proteins—p24-Hoc, linker-less p24-Hoc, Hoc-p24, Nef-Hoc, and gp41 C-trimer-Hoc—were overexpressed and purified using the procedures described below. Briefly, a 1-liter E. coli culture was induced at 30°C for 2.5 h with 1 mM IPTG. Cells were harvested by centrifugation at 3,000 rpm for 10 min, and the pellets were resuspended in 1/50 the volume of buffer 1 (50 mM phosphate buffer [pH 8.0], 300 mM NaCl, 20 mM imidazole) and lysed by French press treatment (1,000-lb pressure; Aminco French press). The homogenate was centrifuged at 15,000 rpm for 30 min. All of the recombinant proteins except gp41 C-trimer-Hoc partitioned into the supernatant, and each was purified by affinity chromatography on a Histrap column (AKTA-Prime; GE Healthcare). In the case of gp41 C-trimer-Hoc, the inclusion bodies were solubilized with 6 M urea and loaded onto a Histrap column preequilibrated with buffer 1 and washed with 50 mM imidazole. The bound protein was eluted with 150 ml of 6 to 0 M urea gradient. Peak fractions were pooled and dialyzed against buffer 2 (50 mM phosphate buffer [pH 7.0], 300 mM NaCl, 10 mM MgSO4) and concentrated (Amicon; 10,000 cutoff). In many cases, the proteins were further purified by fast-performance liquid chromatography gel filtration on a Hi-Load Superdex 200 column (16 mm by 60 cm; GE Healthcare). About 8 to 10 mg of protein with >95% purity was obtained from 1 liter of culture.
In vitro assembly on phage
T4 capsid. hoc−soc− phage was purified by velocity sucrose gradient centrifugation. About 2 × 1010 PFU of purified hoc−soc− phage were centrifuged in 1.5 ml of LowBind Eppendorf tubes at 13,000 rpm (4°C) for 1 h. The pellets were resuspended in 10 μl of buffer 2. One or more purified HIV-Hoc fusion proteins were added at the desired concentration, and the reaction mixture (100 μl) was incubated at 37°C for 45 min. Phage was sedimented by centrifugation as described above, and the pellets were washed twice with 1 ml of buffer 2 and resuspended in 10 to 20 μl of the same buffer. The sample was transferred to a fresh Eppendorf tube and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For immunizations, the phage were resuspended in an appropriate volume of phosphate-buffered saline (PBS) and used the next day.
Quantification of copy number.
A laser densitometer (PDSI) and ImageQuant software (Molecular Dynamics, GE Healthcare) were used to quantify the density volumes of T4 gp23*, p24-Hoc, and other Hoc fusion bands after SDS-PAGE. The system was calibrated with bovine serum albumin (BSA) as the protein standard, and the antigen density measurements were well within the linear range established, with an R2 value of 0.99 (99% confidence level). In some experiments, the p24-Hoc band(s) migrated very close to gp18 (71 kDa), the tail-sheath protein of T4. (For reasons unknown, native Hoc or HIV-Hoc fusion proteins showed two closely migrating bands, which separated better under certain gel conditions; see, for example, Fig. 4C.) In such cases, the density corresponding to the same area in the control wild-type T4 lane was subtracted from that of the displayed phage lane. The copy number of the displayed antigen was determined by normalizing the density with the control gp23* band for which the precise copy number was established to be 930 per phage particle (7).
FIG. 4.
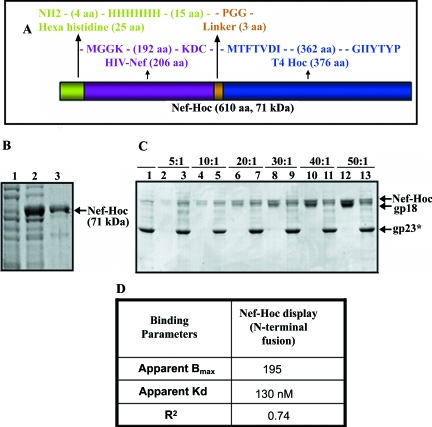
Display of HIV Nef. Nef sequence was fused in frame to the N terminus of Hoc (A) and overexpressed in E. coli and purified (B). Lanes: 1 and 2, Nef-Hoc clones analyzed by SDS-10% PAGE before (0 h) or after (3 h) IPTG induction, respectively; 3, purified Nef-Hoc protein after Ni-affinity chromatography. (C) The hoc−soc− particles (1010 PFU) were incubated with the purified Nef-Hoc at various ratios of Nef-Hoc to Hoc binding sites under standard in vitro assembly conditions. The samples were electrophoresed on an SDS-4 to 20% PAGE gel and stained with Coomassie blue. Lanes: 1, control hoc−soc− phage; 2, 4, 6, 8, 10, and 12, Nef-Hoc at the ratios shown at the top; 3, 5, 7, 9, 11, and 13, phage showing displayed Nef-Hoc. (D) Table showing the binding parameters for Nef-Hoc calculated from C, as described in the legend to Fig. 3.
Immunizations.
Six- to eight-week-old female BALB/c mice (Jackson Laboratory, Bar Harbor, MA) were immunized with three intramuscular injections (six mice/group, 50 μl/mouse) at an interval of 3 weeks. The antigen doses were as follows: ca. 16 μg each of Hoc-p24-T4 or p24-Hoc-T4 containing 100 ng each of displayed p24 or 10 μg of E. coli-expressed p24 (soluble p24). Each group consisted of six mice. One week after the boost, spleens and lymph nodes were obtained from three immunized and three naive mice and then processed. Mice were bled before immunization and then every 2 weeks. Blood was collected from each of the six mice up to week 7 and thereafter from the three remaining mice. In a separate experiment, six mice were immunized with 100 ng of p24-Hoc-T4 on weeks 0, 3, and 6. Blood was collected from all six mice on weeks 5, 8, and 10 and thereafter from the three remaining mice on weeks 12, 18, 20, and 37. All of the sera were stored at −20°C until assayed.
p24-specific IgG antibody responses.
Individual sera were analyzed for the presence of p24-specific immunoglobulin G (IgG) antibodies by a solid-phase enzyme-linked immunosorbent assay (ELISA) as described previously (23). The baculovirus expressed p24 (100 μl per well of a 1-μg/ml solution) was used as the coating antigen. The data are expressed as endpoint titers, with the titer being defined as the highest dilution that yielded an optical density (A405) reading of greater than or equal to twice the background values. The titers were calculated after subtracting the mean absorbance of triplicate wells lacking antigen from the absorbance of triplicate wells containing antigen at each serum dilution.
p24-specific IgG subclass antibody responses.
Antigen-specific IgG antibody subclass levels were also determined by ELISA as previously described (23). Immulon 2HB flat-bottom microtiter plates (96 well; Dynatech, Chantilly, VA) were coated with p24 (0.1 μg of antigen per well) for test sera or goat anti-mouse IgG for IgG subclass standard curves. The antigen-coated plates were blocked and test sera or IgG1, IgG2a, IgG2b, or IgG3 standards diluted in PBS-BSA-casein buffer were added to the plates, followed by incubation at room temperature for at least 2 h. The plates were washed three times with wash buffer and incubated 1 h at room temperature with peroxidase-labeled secondary antibody (diluted 1:1,000 in PBS-BSA-casein buffer; sheep anti-mouse IgG1, IgG2a, IgG2b, and IgG3), followed by the addition of ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate. After 30 min, the reaction was stopped by the addition of 100 μl per well of 1% SDS, and the plates were read immediately at 405 nm. The concentrations (in micrograms/milliliter) of antigen-specific IgG subclass antibodies in the test sera were determined from the respective standard curves run on each plate.
T-cell proliferation.
One week after the boost, lymphocytes obtained from the spleens and lymph nodes from three naive and three immunized mice were cultured in RPMI 1640-EHAA medium (BioWhittaker, Walkersville, MD) at 1:1 (vol/vol) containing 0.5% normal mouse serum, 8 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 100 μM 2-mercaptoethanol. Cultures were set up in 96-well plates in triplicate at a concentration of 2.5 × 105 to 5 × 105 cells/ml in the presence or absence of the antigen. Lymphocytes were cultured for 5 days with different concentrations of p24, medium alone, or with concanavalin A (as a positive control) or ovalbumin (as a nonspecific antigen/negative control). During the last 16 h of the culture period, cells were pulsed with 1 μCi of [3H]thymidine per well (23). Cells were then harvested onto glass fiber filters. The data are expressed as stimulation indices ([3H]thymidine incorporation in the presence of antigen divided by the same in the absence of antigen) plus or minus standard error.
ELISPOT assay.
Spleen and lymph node cells secreting gamma interferon (IFN-γ) and interleukin-4 (IL-4) were analyzed by enzyme-linked immunospot (ELISPOT) assay as previously described (22). Single-cell suspensions (106 cells/well) prepared from the spleens and lymph nodes (three mice/group) of naive and immunized mice were plated on anti-IFN-γ (clone RMGG-1; Biosource)- or anti-IL-4 antibody (BVD4-1D11; Pharmingen)-coated 96-well nitrocellulose-backed MultiScreen-IP sterile plates. Cells were incubated with or without p24 antigen (10 μg/ml) or the cytotoxic-T-lymphocyte (CTL) epitope of p24 (10 μg/ml) for 18 h at 37°C in a humidified CO2 incubator. Plates were washed and overlaid with biotinylated anti-IFN-γ (clone XMG 1.2) or biotinylated anti-IL-4 (clone BVD6-24G2; Pharmingen), followed by the addition of avidin-conjugated alkaline phosphatase. The plates were washed, and bound IFN-γ or IL-4 was detected by the addition of BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium. The plates were washed with water, and the individual spots were visualized and counted the next day using a stereo binocular microscope. The average number of spots/number of cells plated was calculated.
RESULTS AND DISCUSSION
In vitro assembly of HIV p24 on bacteriophage T4 capsid.
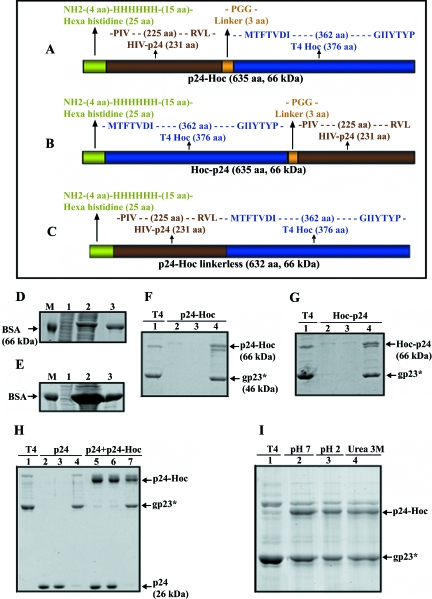
Three p24 fusions were constructed: p24-Hoc (p24 fused to the N terminus of Hoc; Fig. 2A), Hoc-p24 (p24 fused to the C terminus of Hoc; Fig. 2B), and linker-less p24-Hoc (same as p24-Hoc but without the intervening Pro-Gly-Gly linker; Fig. 2C), each with an N-terminal hexahistidine tag. The fusion proteins were expressed in E. coli up to >10% of the total cell protein (Fig. 2D and E, lanes 1 and 2; data for linker-less p24-Hoc are not shown since they are very similar to p24-Hoc) and ca. 90% of the protein remained in the soluble form. The proteins were purified to >90% purity (Fig. 2D and E; lanes 3) with a yield of about 8 to 10 mg per 1 liter of culture. Quantitative ELISA showed that the E. coli-produced p24 showed reactivity comparable to that of the baculovirus-p24 using p24 antibodies to baculovirus-p24 and vice versa.
FIG. 2.
In vitro display of HIV p24 on phage T4. p24 was fused in frame to the N terminus (A) or C terminus (B) of Hoc or to the N terminus of Hoc without the Pro-Gly-Gly linker (C) (see Materials and Methods). (D and E) Expression and purification of p24-Hoc (D) and Hoc-p24 (E). Lanes: 1 and 2, E. coli samples analyzed by SDS-10% PAGE before (0 h) or after (3 h) IPTG induction, respectively; 3, purified protein after Ni-affinity chromatography; M, BSA (66 kDa) used as a molecular mass standard. (F and G) In vitro assembly of p24-Hoc (F) and Hoc-p24 (G) on hoc−soc− phage (see Materials and Methods). Lanes: 1, control hoc−soc− phage; 2, starting p24-Hoc or Hoc-p24; 3, supernatant containing the unbound protein; 4, phage pellet containing the displayed protein. The samples were electrophoresed by SDS-4 to 20% PAGE and stained with Coomassie blue. (On a 4 to 20% gradient SDS-PAGE gel, p24-Hoc migrates slightly slower than gp18 [see, for example, panels F and G], whereas on an SDS-10% PAGE gel, it migrates faster [see, for example, Fig. 3A]). This anomalous behavior of Hoc is linked to the C-terminal domain of Hoc, although the reasons are unknown (unpublished results).(H) Specificity of in vitro binding. hoc−soc− particles (lane 1) were incubated with either p24 (lanes 2 to 4) or a mixture of p24 and p24-Hoc (lanes 5 to 7) under standard assembly conditions, and the unbound and bound fractions were analyzed. A 25:1 ratio of p24 (lane 2) or a mixture of p24 and p24-Hoc (lane 5) to capsid binding sites was used. Lanes: 2 and 5, starting p24, or p24 and p24-Hoc mixture; 3 and 6, supernatants containing the unbound proteins; 4 and 7, phage pellet containing the displayed protein. (I) Stability of displayed p24-T4 phage. The displayed p24-T4 particles were treated with control pH 7 buffer (lane 2), pH 2 buffer (lane 3), or 3 M urea (lane 4). Lane 1 (T4) represents control hoc−soc− phage. The samples in panels H and I were electrophoresed on a SDS-10% PAGE gel and stained with Coomassie blue.
To test for in vitro assembly, sucrose gradient-purified hoc−soc− phage particles were incubated with purified p24-Hoc or Hoc-p24, and the phage were sedimented by high-speed centrifugation. After two washes to remove any nonspecifically bound protein, the phage pellet was subjected to SDS-PAGE. In numerous experiments, a new band corresponding to the fusion protein consistently appeared with the phage particles, suggesting that the HIV p24 is now transferred to the phage capsid (Fig. 2F and G, compare lanes 4 to lanes 1).
The interaction between p24-Hoc and capsid is specific and stable. Free p24, when not fused to Hoc, did not bind to hoc−soc− phage significantly (Fig. 2H, lanes 2 to 4). When both p24 and p24-Hoc were added to the reaction mixture, only p24-Hoc preferentially assembled onto the hoc−soc− phage (lanes 5 to 7). Treatment of p24-T4 particles with acidic buffer (pH 2.0) (Fig. 2I, lane 3) or 3 M urea (lane 4) did not cause significant disassociation of the displayed p24 protein, indicating a strong interaction between Hoc and gp23*. (Some disruption and loss of T4 phage occurred at pH 2, although the Hoc-capsid interaction remained intact.)
Quantitative analysis of p24-Hoc binding to phage T4 capsid.
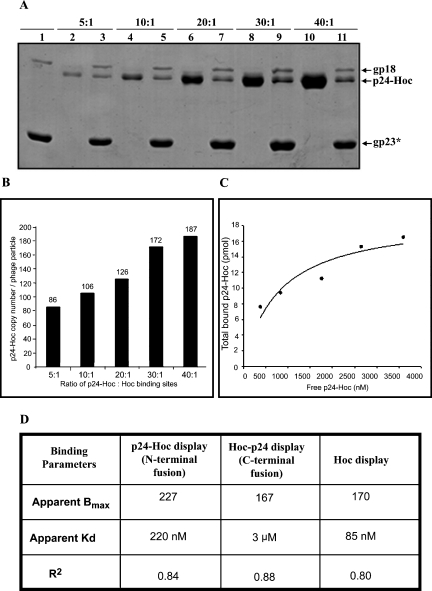
The ratio of p24-Hoc molecules to capsid binding sites was varied, and the number of bound p24-Hoc molecules per particle was determined by laser densitometry. The Coomassie blue-stained p24-Hoc band was compared to the gp23* band for which the copy number is known (930 copies per particle). As shown in Fig. 3A, the copy number of bound p24-Hoc increased with increasing ratio and reached near saturation at a ratio of 40:1, at which point the copy number was 187 (Fig. 3B). This is consistent with the predicted binding of one p24-Hoc per gp23* hexon; a preliminary cryo-electron microscopy reconstruction of p24-T4 indeed showed Hoc in the center of each gp23* hexon (A. Fokine, P. Chipman, and M. Rossmann, unpublished data).
FIG. 3.
Quantification of p24-Hoc displayed on hoc−soc− phage. (A) The hoc−soc− particles (1010 PFU) were incubated with purified p24-Hoc at various ratios of p24-Hoc molecules to Hoc binding sites, and in vitro assembly was performed under standard conditions. Lanes: 1, control hoc−soc− phage; 2, 4, 6, 8, and 10, p24-Hoc at the ratios indicated at the top; 3, 5, 7, 9, and 11, phage with displayed p24-Hoc. The samples were electrophoresed on a SDS-10% PAGE gel and stained with Coomassie blue. The positions of gp23*, p24-Hoc, and gp18 (tail-sheath protein of phage T4; 71 kDa) are labeled with arrows. (B and C) The density volumes of the bound and unbound p24-Hoc and gp23* were quantified by laser densitometry and normalized to that of the gp23* band in the respective lane (930 copies per particle), and the copy number of p24-Hoc per particle was determined (B). The data were fitted by nonlinear regression using the GraphPad Prism-4 software (C) and the binding parameters, apparent Bmax (defined as maximum number of p24-Hoc molecules per phage particle), Kd (association constant), and R2 (degree of confidence) were calculated (D). All of these analyses were also performed for the display of Hoc-p24 and native Hoc (data not shown), and the corresponding values are shown in the table.
It should however be noted that, when >40:1 ratios were used, the copy number of p24-Hoc increased further, reaching 347 at a ratio of 100:1 (data not shown). This was equivalent to about twice the expected copy number and likely due to oligomerization of p24-Hoc and display of oligomers. The p24-gag, being the major capsid protein of the HIV virion, tends to form oligomers in solution; the E. coli-overexpressed p24 has been reported to form dimers to dodecamers in a concentration-dependent manner (4).
The apparent Bmax (maximum copy number per particle) and Kd (association constant) for p24-Hoc binding, as calculated from the binding curve, are 227 and 220 nM, respectively (Fig. 3C and D). The corresponding values for the binding of “native” Hoc (i.e., recombinant Hoc with a hexahistidine tag at the N terminus, which was constructed and purified similar to the strategy described in Fig. 2) are 170 and 85 nM, respectively (Fig. 3D). Thus, the binding affinity of Hoc is lowered by ∼2.6-fold upon fusion to HIV p24. The linker-less p24-Hoc showed binding characteristics very similar to those of p24-Hoc, suggesting that the linker as such did not affect the binding affinity of p24-Hoc. On the other hand, Hoc-p24 showed a lower Bmax of 167 and a high Kd (3 μM) compared to p24-Hoc (Fig. 3D). A 35-fold decrease in the affinity for Hoc-p24 binding suggests that fusion to the C terminus of Hoc interfered with the Hoc-capsid interactions. This is consistent with our recent mutagenesis data, which maps the capsid binding site to the C terminus of Hoc (T. Sathaliyawala and V. Rao, unpublished data). Thus, fusion to the C terminus distorted the capsid-binding site, resulting in greatly reduced affinity, and is not a desirable option for antigen fusion.
Display of HIV Nef.
The general ability of the in vitro system to display HIV antigens was tested using HIV Nef and gp41 proteins. Nef is a conserved 31-kDa multifunctional protein and exerts pleiotropic effects through modulation of cellular gene expression and signaling pathways, thereby contributing to the progression of HIV infection into AIDS (15). Individuals infected with Nef-defective HIV mutants show slow progression to AIDS symptoms and, in macaques, the disease progression is attenuated (21). Being a frequent target for CTL responses, Nef is considered to be a strong candidate for inclusion in multicomponent HIV vaccine formulations (17).
Full-length Nef was fused to the N terminus of Hoc, overexpressed and purified by the schemes described above (Fig. 4A and B). Purified Nef-Hoc was efficiently displayed on T4 phage by the in vitro assembly system (Fig. 4C). Nef, unlike p24-gag, exists as a monomer in solution; the apparent Bmax of 195 (Fig. 4D) is consistent with the binding of one Nef-Hoc monomer per gp23* hexon. The apparent Kd of 130 nM for Nef-Hoc binding is similar to that for native Hoc binding.
Display of an engineered HIV gp41 C-peptide trimer.
gp41, the membrane-anchored subunit of trimeric HIV envelope receptor complex, consists of two conserved heptad repeats (N-peptide and C-peptide), which are transiently exposed during fusion of the HIV envelope to the host cell membrane (32). The pre-hairpin structural intermediate consisting of a triple stranded coiled-coil N-peptide with extended C-peptide helices is a vulnerable target for antibody interaction (19). Neutralizing human monoclonal antibodies to the C-peptide epitopes—2F5, 4E10, and Z13—arrest HIV infection of both laboratory-adapted and primary HIV isolates (2, 3, 20). Thus, this region has been the focus of vaccine design and development.
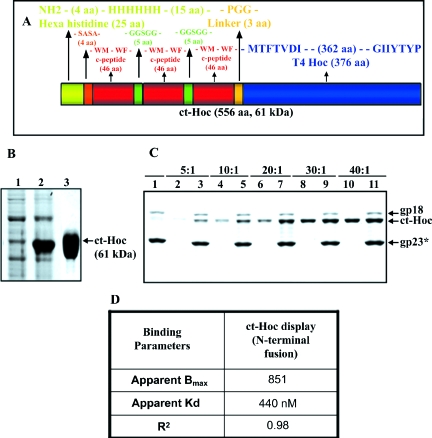
A synthetic gene consisting of three C-peptide repeats (hereafter referred to as “C-trimer” or “ct” in the figures) was constructed and fused to the N terminus of Hoc (Fig. 5A). Repeats of C-peptide are expected to increase the density of neutralizing epitopes on the T4 capsid surface. The 61-kDa C-trimer-Hoc was overexpressed in E. coli up to >10% of total cell protein and purified by urea denaturation and renaturation (Fig. 5B). Gel filtration and native polyacrylamide gel analyses showed that the purified C-trimer-Hoc exists as oligomers in solution (data not shown).
FIG. 5.
Display of HIV gp41 C-trimer. The synthetic nucleotide sequence of gp41 C-trimer (ct) was fused in-frame to the N terminus of Hoc (A; see Materials and Methods for details) and overexpressed in E. coli and purified (B). Lanes: 1 and 2, ct-Hoc clones analyzed by SDS-10% PAGE before (0 h) or after (3 h) IPTG induction, respectively; 3, purified ct-Hoc protein after Ni-affinity chromatography. (C) The hoc−soc− particles (1010 PFU) were incubated with the purified ct-Hoc at various ratios of ct-Hoc to Hoc binding sites under standard in vitro assembly conditions. The samples were electrophoresed on SDS-4 to 20% PAGE gel and stained with Coomassie blue. Lanes: 1, control hoc−soc− phage; 2, 4, 6, 8, and 10, ct-Hoc at the ratios shown on top; 3, 5, 7, 9, and 11, phage showing displayed ct-Hoc. (D) Table showing the binding parameters for ct-Hoc calculated from panel C as described in the legend to Fig. 3.
As shown in Fig. 5C, the C-trimer-Hoc is efficiently displayed on T4 capsid by the in vitro assembly system. Binding reached close to saturation at a relatively low C-trimer-Hoc/binding site ratio of 20:1. The apparent Kd for binding is 440 nM, and the Bmax is 851 copies per capsid (Fig. 5C and D). The 5.6 times greater Bmax is consistent with the observation that the C-trimer-Hoc forms oligomers in solution, possibly leading to variable affinities for the display of different C-trimer oligomers on T4 capsid. We have also observed various C-trimer oligomerization patterns depending on the length of storage; however, display as such was not affected by the state of oligomerization, as all preparations were efficiently displayed in saturation binding experiments.
Multicomponent display.
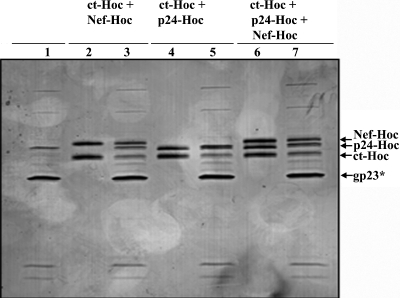
Combinations of two or three antigens were added to the in vitro binding mixture to test for the display of multiple HIV antigens on T4 capsid (Fig. 6). The data from numerous experiments showed that two (Nef-Hoc and C-timer-Hoc, lane 3; p24-Hoc and C-trimer-Hoc; lane 5) or three antigens (Nef-Hoc, p24-Hoc, and C-trimer-Hoc; lane 7) could be efficiently displayed. Typically, the copy number for each antigen in the multicomponent display was lower than when each was individually displayed. This is to be expected due to competition among the antigens for the same number of binding sites on the capsid.
FIG. 6.
Multiple HIV antigen display. The hoc−soc− particles (1010 PFU) were incubated with two HIV antigens (ct-Hoc and Nef-Hoc [lanes 2 and 3] and ct-Hoc and p24-Hoc [lanes 4 and 5]) or three HIV antigens (ct-Hoc, p24-Hoc, and Nef-Hoc [lanes 6 and 7]) under standard in vitro assembly conditions. The samples were electrophoresed on a SDS-13% PAGE gel and stained with Coomassie blue. Lanes: 1, control hoc−soc− phage; 2, 4, and 6, combinations of the proteins as indicated at the top; 3, 5 and 7: phage showing the displayed antigens. The positions of gp23*, ct-Hoc, p24-Hoc, and nef-Hoc are labeled with arrows.
Humoral responses elicited by T4-displayed p24.
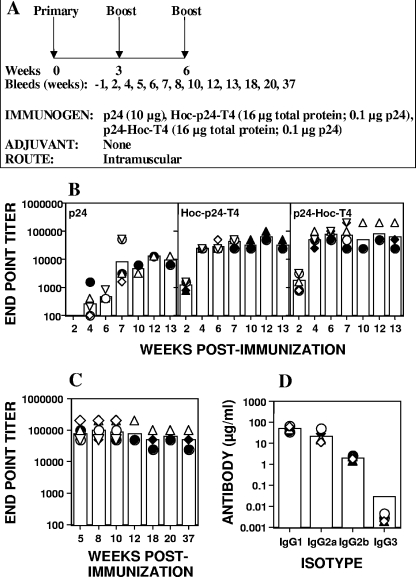
The immunogenicity of displayed antigen was tested in a murine model with p24 as a model antigen (Fig. 7A). Individual mouse serum samples were analyzed by ELISA for p24-specific IgG antibodies using baculovirus-expressed p24 as the capture antigen. This assay specifically determined antibody titers against native p24, but at the same time ruled out false positives against any contaminants present in the E. coli-produced p24. Mice immunized with soluble p24 antigen (10 μg) induced poor antibody responses, with endpoint titers in the range of 4,000 to 9,000 (Fig. 7B). In contrast, mice immunized with about 100 ng of T4-displayed p24 elicited strong and long-lasting anti-p24 IgG titers; the geometric mean endpoint titers ranged from 64,000 to 72,000 at week 13 postimmunization (Fig. 7B). In a repeat experiment, the p24-specific IgG titers were sustained even after 37 weeks postimmunization, with a mean endpoint titer of about 50,000 (Fig. 7C). Similar anti-p24 titers have been reported in other studies using DNA prime-boost and vaccinia virus vectors or adenovirus prime-boost strategies (30). However, unlike these other studies wherein 10 μg of p24 was used, our studies used 100-fold less (about 100 ng) T4-displayed p24 antigen.
FIG. 7.
Immunogenicity of T4-displayed HIV p24. (A) Experimental design for immunization. Female BALB/c mice (six/group) were immunized by intramuscular injections at weeks 0, 3, and 6 with the antigen doses as indicated. Three mice from each group were euthanized at week 7 for analysis of the cellular responses. (B) In the first experiment, blood was collected from each of the six mice at weeks 2, 4, 6, and 7 and thereafter from the three remaining mice at weeks 10, 12, and 13. (C) In the second experiment, blood was collected from all six mice at weeks 5, 8, and 10 and thereafter from the three remaining mice at weeks 12, 18, 20, and 37. (B) Display of p24 on phage T4 enhances antibody responses to p24. BALB/c mice were immunized with p24 (10 μg), Hoc-p24-T4 (16 μg of phage protein displaying 100 ng of p24), and p24-Hoc-T4 (16 μg of phage protein displaying 100 ng of p24). Individual serum samples were analyzed in triplicate by ELISA for the presence of p24-specific IgG antibodies using baculovirus-expressed p24 as the coating antigen. Each bar represents the geometric mean endpoint titers for each group. Each symbol represents the endpoint titer for an individual mouse. (C) Display of p24 on phage T4 induces long-lasting p24-specific IgG antibodies. The experimental design is the same as in panel B except that sera were collected for up to 37 weeks and analyzed for p24-specific IgG antibodies by ELISA. (D) Subclass analysis was performed on individual serum samples collected at week 8 from p24-Hoc-T4-immunized mice (see panel C) using peroxidase-conjugated goat anti-mouse subclass specific IgG as the secondary antibody. The absorbance (A405) was measured at the 30-min endpoint. The amount of antibody in each sample was calculated from a standard graph. Each bar represents the geometric mean for each subclass, and individual serum samples from six mice are represented by the various symbols.
Analysis of subclass specificity showed that both IgG1 and IgG2a antibodies were induced, indicating that a mixed Th1/Th2 response was generated (Fig. 7D). The responses, however, were skewed more toward a Th2 response since the induction of IgG1 antibodies was higher than that of IgG2a. This is consistent with the induction of IL-4-producing cells in the spleen and lymph nodes (Table 1).
TABLE 1.
Production of IFN-γ and IL-4 in spleen and lymph node cellsa
| Immunogen | No. of spots/106 cells
|
|||||
|---|---|---|---|---|---|---|
| Spleen
|
Lymph node
|
|||||
| p24, IFN-γ | Peptide, IFN-γ | p24, IL-4 | p24, IFN-γ | Peptide, IFN-γ | p24, IL-4 | |
| p24-Hoc-T4 | 231 | 14 | 163 | 70 | 22 | 228 |
| Hoc-p24-T4 | 50 | 4 | 161 | 12 | 0 | 100 |
| p24 | 21 | 9 | 53 | 4 | 0 | 36 |
| Naive | 2 | 2 | 12 | 0 | 0 | 0 |
Immune and naive spleen and lymph node cells were pooled separately from three mice per group and stimulated in vitro with p24 (10 μg) or with the p24-CTL (10 μg) epitope for 18 h. The number of cells activated to secrete IFN-γ or IL-4 was determined by an ELISPOT assay as described in Materials and Methods. The data are representative of three independent experiments.
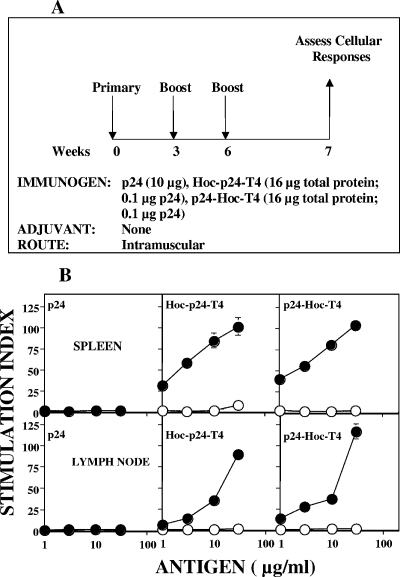
Cellular responses.
Antigen-specific CD4+ T cells are essential for the induction of both CD8+ T-cell and memory T-cell responses (22). Analysis of p24-T4-immunized mice (Fig. 8A) showed a robust T-cell proliferative recall response when the spleen (Fig. 8B, top panels) and lymph node cells (bottom panels) were cultured in vitro with baculovirus-p24. Spleen cells from mice immunized with T4-displayed p24 gave a threefold-greater proliferative response than that obtained with spleen cells from mice immunized with soluble p24. Lymph node cells gave slightly lower responses than spleen cells, and the amount of p24 antigen required for proliferation was ∼10-fold higher. The negative control antigen, ovalbumin, and naive spleen cells did not induce any proliferative responses to either p24 or ovalbumin, thus demonstrating the specificity of the measured responses.
FIG. 8.
Display of p24 on phage T4 enhances T-cell proliferative responses to p24. (A) Experimental design for immunization. Female BALB/c mice (six/group) were immunized by intramuscular injections at weeks 0, 3, and 6 with the antigens as described in Fig. 7. One week after the final boost, the mice were euthanized to assess the cellular responses. (B) Lymphocytes from the spleen (top panels) and lymph nodes (bottom panels) of mice were collected 1 week after the boost, pooled, and cultured in triplicate either in the presence or in the absence of various concentrations of baculovirus-expressed p24 (•) or ovalbumin (○). T-cell proliferation was determined by measuring [3H]thymidine incorporation. The data are depicted as stimulation indices ([3H]thymidine incorporation in the presence of antigen divided by [3H]thymidine incorporation in the absence of antigen ± the standard error).
The cytokine-producing cell repertoire (Table 1) also showed a mixed Th1 and Th2 immune response, again with a bias toward a Th2 response. While a strong CD4+ T-cell response, as shown by IFN-γ- and IL-4-producing cells in response to in vitro stimulation with soluble p24, was induced from immune spleen and lymph node cells, the CD8+ T-cell responses were less strongly induced, as shown by the ELISPOT data with immune cells in response to the p24 CTL peptide (Table 1). p24 CTL peptide-specific CD8+ T cells secreting IFN-γ were identified in both spleen and lymph node cells, with the lymph node cells showing a more pronounced effect. The large number of IFN-γ-secreting cells seen with p24 could be due to the presence of more than one CTL epitope in p24. However, only a single CTL epitope, and not overlapping peptides, was tested in the ELISPOT assay. It is possible that multiple epitopes were recognized, suggesting a broad-based immune response. The major difference in immunogenicity between Hoc-p24-T4 (C-terminal Hoc fusion) and p24-Hoc-T4 (N-terminal Hoc fusion) was seen in the induction of number of IFN-γ-secreting cells. Although immunization with Hoc-p24-T4 induced a similar number of IL-4-secreting cells in the spleen in response to in vitro stimulation with soluble p24, there was a 5-fold reduction in the number of p24-specific IFN-γ-secreting cells in both spleen and lymph node cells (Fig. 8B). The peptide-specific CD8+ T cells were also greatly diminished in the spleen, and none were detected in the lymph nodes. The poor CD8+ T-cell response seen with the T4 displayed protein implies that the displayed antigen was channeled to the major histocompatibility complex (MHC) class II pathway and was not efficiently cross presented through the class I pathway.
Exogenous antigens do not normally induce CD8+ T-cell responses because they are internalized by the endosomes or lysosomes and enter the MHC class II pathway. However, exogenous antigens can enter the MHC class I pathway by a phenomenon known as cross-presentation (1). The lack of a CD8+ T-cell response seen after immunization with the C-terminally fused Hoc-p24 displayed protein compared to the response obtained after immunization with p24-Hoc-T4 could be due to differences in the exposed epitopes leading to reduced cross presentation. Display of a targeting and/or enhancing molecule (CD40 ligand, cytokine, etc.) and inclusion of adjuvants in the antigen preparation could further boost the immune responses, in particular the CD8+ T-cell responses, by channeling the antigen more efficiently to the cytoplasmic compartment of the antigen-presenting cell for processing through the MHC class I pathway, which in turn would enhance the immunogenicity of the displayed antigen and generate a strong CD8+ T-cell immune response.
In conclusion, we have developed a defined bacteriophage T4 in vitro display system that can efficiently display HIV antigens on T4 capsid. Three highly purified candidate HIV antigens—p24, Nef, and gp41 C-peptide trimer—which differ in structure, biological function, and oligomeric state, were displayed either individually or in combination up to the full capacity of available binding sites. The displayed p24 was highly immunogenic, eliciting strong antibody and cellular immune responses upon immunization with very small amounts of antigen. In separate experiments, anthrax antigens in the range of 85 to 90 kDa were efficiently displayed either individually (27) or in combinations (S. B. Shivachandra et al., unpublished data). The displayed anthrax antigens were highly immunogenic in mice in the absence of any externally added adjuvant, generating strong anthrax-specific antibodies, as well as lethal toxin-neutralizing antibodies (27). The in vitro approach utilized in the present study has been further extended to Soc fusion, allowing the display of 965 copies of multiple antigens per phage particle using both Hoc and Soc, the maximum capacity of the T4 capsid (Q. Li et al., unpublished data).
These successes of the in vitro approach using a broad range of full-length antigens offer insights for the design of novel particulate HIV antigens, which can potentially elicit broader and stronger humoral and cellular immune responses. For instance, it is possible to construct hybrid T4-HIV particles displaying multiple gp41 peptide mimics through Hoc display and trimeric gp120 complexes through Soc display, which might stimulate strong neutralizing antibodies (16). Since the capsid-assembled Soc forms trimers, display of “native” trimeric HIV gp120/gp160 complexes using glycosylated gp120-Soc produced in mammalian expression systems would allow the preparation of a stabilized complex. Furthermore, targeting of the particles to specific immune cells can be attempted by codisplay of a particular targeting ligand, or a combined prime-boost strategy can be designed by cloning the DNAs inside and displaying the proteins outside.
Acknowledgments
We thank Elaine Morrison for technical assistance in performing all of the immunizations and bleedings and Steven McQuinn for assisting with the design of the T4 capsid images in Fig. 1.
All research was conducted in compliance with the Animal Welfare Act and other Federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the NRC Publication Guide for the Care and Use of Laboratory Animals.
The information contained herein reflects the views of the authors and should not be construed to represent those of the Department of the Army or the Department of Defense.
REFERENCES
- 1.Brode, S., and P. A. Macary. 2004. Cross-presentation: dendritic cells and macrophages bite off more than they can chew! Immunology 112:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 3.Conley, A. J., J. A. Kessler, II, L. J. Boots, J. S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esparza, J. 2005. The global HIV vaccine enterprise. Int. Microbiol. 8:93-101. [PubMed] [Google Scholar]
- 6.Excler, J. L. 2005. AIDS vaccine development: perspectives, challenges, and hopes. Indian J. Med. Res. 121:568-581. [PubMed] [Google Scholar]
- 7.Fokine, A., P. R. Chipman, P. G. Leiman, V. V. Mesyanzhinov, V. B. Rao, and M. G. Rossmann. 2004. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA 101:6003-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. St John, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert, P. B., Y. L. Chiu, M. Allen, D. N. Lawrence, C. Chapdu, H. Israel, D. Holman, M. C. Keefer, M. Wolff, and S. E. Frey. 2003. Long-term safety analysis of preventive HIV-1 vaccines evaluated in AIDS vaccine evaluation group NIAID-sponsored phase I and II clinical trials. Vaccine 21:2933-2947. [DOI] [PubMed] [Google Scholar]
- 10.Hoess, R. H. 2002. Bacteriophage lambda as a vehicle for peptide and protein display. Curr. Pharm. Biotechnol. 3:23-28. [DOI] [PubMed] [Google Scholar]
- 11.Ishii, T., and M. Yanagida. 1975. Molecular organization of the shell of the Teven bacteriophage head. J. Mol. Biol. 97:655-660. [DOI] [PubMed] [Google Scholar]
- 12.Ishii, T., and M. Yanagida. 1977. The two dispensable structural proteins (Soc and Hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 109:487-514. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki, K., B. L. Trus, P. T. Wingfield, N. Cheng, G. Campusano, V. B. Rao, and A. C. Steven. 2000. Molecular architecture of bacteriophage T4 capsid: vertex structure and bimodal binding of the stabilizing accessory protein, Soc. Virology 271:321-333. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, J., L. Abu-Shilbayeh, and V. B. Rao. 1997. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 65:4770-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistance: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M., Q. Z. Montefiori, D. C. Haynes, B. F. Reinherz, and H. X. Liao. 2005. Comparison of HIV type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res. Hum. Retrovir. 21:58-67. [DOI] [PubMed] [Google Scholar]
- 17.Kong, W. P., Y. Huang, Z. Y. Yang, B. K. Chakrabarti, Z. Moodie, and G. J. Nabel. 2003. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J. Virol. 77:12764-12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letvin, N. L. 2005. Progress toward an HIV vaccine. Annu. Rev. Med. 56:213-223. [DOI] [PubMed] [Google Scholar]
- 19.McGaughey, G. B., G. Barbato, E. Bianchi, R. M. Freidinger, V. M. Garsky, W. M. Hurni, J. G. Joyce, X. Liang, M. D. Miller, A. Pessi, J. W. Shiver, and M. J. Bogusky. 2004. Progress toward the development of a HIV-1 gp41-directed vaccine. Curr. HIV. Res. 2:193-204. [DOI] [PubMed] [Google Scholar]
- 20.Mehandru, S., J. Galovich, G. Stiegler, B. Vcelar, A. Hurley, C. Hogan, S. Vasan, H. Katinger, C. J. Petropoulos, and M. Markowitz. 2004. Neutralization profiles of newly transmitted human immunodeficiency virus type 1 by monoclonal antibodies 2G12, 2F5, and 4E10. J. Virol. 78:14039-14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piguet, V., and D. Trono. 1999. The Nef protein of primate lentiviruses. Rev. Med. Virol. 9:111-120. [DOI] [PubMed] [Google Scholar]
- 22.Rao, M., M. Bray, C. R. Alving, P. Jahrling, and G. R. Matyas. 2002. Induction of immune responses in mice and monkeys to Ebola virus after immunization with liposome-encapsulated irradiated Ebola virus: protection in mice requires CD4+ T cells. J. Virol. 76:9176-9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao, M., G. R. Matyas, T. C. Vancott, D. L. Birx, and C. R. Alving. 2004. Immunostimulatory CpG motifs induce CTL responses to HIV type I oligomeric gp140 envelope protein. Immunol. Cell Biol. 82:523-530. [DOI] [PubMed] [Google Scholar]
- 24.Rao, V. B., and M. S. Mitchell. 2001. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J. Mol. Biol. 314:401-411. [DOI] [PubMed] [Google Scholar]
- 25.Ren, Z., and L. W. Black. 1998. Phage T4 SOC and HOC display of biologically active, full-length proteins on the viral capsid. Gene 215:439-444. [DOI] [PubMed] [Google Scholar]
- 26.Sei, Y., P. H. Tsang, F. N. Chu, I. Wallace, J. P. Roboz, P. S. Sarin, and J. G. Bekesi. 1989. Inverse relationship between HIV-1 p24 antigenemia, anti-p24 antibody and neutralizing antibody response in all stages of HIV-1 infection. Immunol. Lett. 20:223-230. [DOI] [PubMed] [Google Scholar]
- 27.Shivachandra, S. B., M. Rao, L. Janosi, T. Sathaliyawala, G. R. Matyas, C. R. Alving, S. H. Leppla, and V. B. Rao. 2006. In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc-capsid interactions: a strategy for efficient display of large full-length proteins. Virology 345:190-198. [DOI] [PubMed] [Google Scholar]
- 28.Smith, G. P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315-1317. [DOI] [PubMed] [Google Scholar]
- 29.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 30.Tritel, M., A. M. Stoddard, B. J. Flynn, P. A. Darrah, C. Y. Wu, U. Wille, J. A. Shah, Y. Huang, L. Xu, M. R. Betts, G. J. Nabel, and R. A. Seder. 2003. Prime-boost vaccination with HIV-1 Gag protein and cytosine phosphate guanosine oligodeoxynucleotide, followed by adenovirus, induces sustained and robust humoral and cellular immune responses. J. Immunol. 171:2538-2547. [DOI] [PubMed] [Google Scholar]
- 31.Wong, S. B., and R. F. Siliciano. 2005. Contribution of virus-like particles to the immunogenicity of human immunodeficiency virus type 1 Gag-derived vaccines in mice. J. Virol. 79:1701-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]