Abstract
Higher plants possess medium-sized gene families that encode plasma membrane–localized sucrose transporters. For several plant species, it has been shown that at least one of these genes (e.g., AtSUC3 in Arabidopsis and LeSUT2 in tomato) differs from all other family members in several features, such as the length of the open reading frame, the number of introns, and the codon usage bias. For these reasons, and because two of these proteins did not rescue a yeast mutant defective in sucrose utilization, it had been speculated that this subgroup of transporters might have sensor functions. Here, we describe the detailed functional characterization and cellular localization of PmSUC3, the orthologous transporter from the Plantago major transporter family. The PmSUC3 protein is localized in the sieve elements of the Plantago phloem and mediates the energy-dependent transport of sucrose and maltose. In contrast to the situation in solanaceous plants, PmSUC3 is not colocalized with PmSUC2, the source-specific, phloem-loading sucrose transporter of Plantago. Moreover, PmSUC3 also was identified in sieve elements of sink leaves and in several nonphloem cells and tissues. Arguments for and against a potential sensor function for this type of sucrose transporter are presented, and the role of this type of transporter in the regulation of sucrose fluxes is discussed.
INTRODUCTION
In most higher plants, sucrose is the main type or even the only form of carbohydrate that is partitioned between the different sinks after its synthesis in the mature source leaves and its subsequent loading into the sieve element–companion cell complex. After the cloning of the first sucrose carrier cDNA from spinach, ps21 (Riesmeier et al., 1992), it was soon recognized that the original idea of a single higher plant sucrose transporter gene was wrong and that plants have at least two sucrose carrier genes (AtSUC1 and AtSUC2: Sauer and Stolz, 1994; PmSUC2: Gahrtz et al., 1994; PmSUC1: Gahrtz et al., 1996). Eventually, the initiation of genome and EST projects in Arabidopsis (Arabidopsis Genome Initiative, 2000) and other plants (Yu et al., 2002) revealed even larger numbers of sucrose transporter genes.
In Arabidopsis, the final number of sucrose carriers is nine (Arabidopsis Genome Initiative, 2000). Still growing families of three to five members were identified in tomato (Barker et al., 2000) and rice (Yu et al., 2002) and several other plant species. The identification of that many sucrose transporters raised the question of the physiological functions of the encoded proteins in carbohydrate metabolism and allocation. At least one of these proteins was expected to be localized to the phloem and to catalyze sucrose loading into the sieve element–companion cell complex; in fact, the promoters of StSUT1 (Riesmeier et al., 1993), PmSUC2 (Gahrtz et al., 1994), and AtSUC2 (Truernit and Sauer, 1995) were shown to be active in this tissue.
Anatomical analyses of minor vein structures in the phloem of numerous plants had shown that typically small sieve elements are surrounded by large companion cells oriented toward the adjacent phloem parenchyma or mesophyll (Behnke, 1989). Thus, it was expected that phloem loading should occur primarily via these companion cells. Immunohistochemical identification of the phloem-specific transporters PmSUC2 from Plantago and AtSUC2 from Arabidopsis in the companion cells confirmed this hypothesis (Stadler et al., 1995; Stadler and Sauer, 1996). However, despite a quite similar phloem anatomy, all phloem-specific transporters in solanaceous plants (potato, tomato, and tobacco) were found in sieve elements (SUT1: Kühn et al., 1997, 2003; SUT2: Barker et al., 2000; SUT4: Weise et al., 2000).
Further analyses revealed that sucrose transporters are responsible not only for phloem loading. PmSUC1 from Plantago, AtSUC1 from Arabidopsis, and NtSUT3 from tobacco were shown to be expressed exclusively or primarily in floral tissues, and a role for these transporters in phloem loading could be excluded (Gahrtz et al., 1996; Lemoine et al., 1999; Stadler et al., 1999).
A detailed comparison of genomic and cDNA sequences of all members of the Arabidopsis sucrose transporter family revealed that one of these genes, AtSUT2/AtSUC3, differed from all others (AtSUT2 was used by Barker et al., 2000; AtSUC3 was used by Meyer et al., 2000). First, the genomic sequence of AtSUT2/AtSUC3 is interrupted by 13 introns rather than by 2 to 4 introns, as in the other Arabidopsis sucrose transporter genes. Second, the protein has a higher apparent molecular mass, resulting from insertions of 30 and 50 amino acids in the first and ninth exons of the gene. Third, it has a low codon bias and low similarity to the Kozak consensus (Kozak, 1996), which is interpreted as an indication of low translation efficiency, as proposed for the amino acid sensor in yeast (Iraqui et al., 1999). Finally, Barker et al. (2000) reported that AtSUT2/AtSUC3 does not complement the yeast mutant SUSY7/ura3 (Riesmeier et al., 1992) and suggested that this protein may cause toxicity and be nonfunctional in yeast. Based on these results and on the observation that the well-characterized hexose sensors Snf3p and Rgt2p from bakers' yeast possess C-terminal extensions that trigger hexose transport in yeast (Özcan et al., 1996, 1998; Vagnoli et al., 1998), it was suggested that AtSUT2/AtSUC3 and related proteins from other species (e.g., LeSUT2 from tomato) might be sucrose sensors rather than transporters (Barker et al., 2000). However, Meyer et al. (2000) were able to show that AtSUT2/AtSUC3 can mediate sucrose transport in yeast, a result that was confirmed later by Schulze et al. (2000). Therefore, the sensor theory was modified, and it was proposed that SUT2/SUC3-type transporters might represent flux sensors that measure the transport rates of sucrose across the membrane (Schulze et al., 2000).
In solanaceous plants, these transporters (or flux sensors) were colocalized with the other sucrose transporters (e.g., LeSUT1 and LeSUT4) in phloem sieve elements (Barker et al., 2000). This led to the model that all sucrose transporters might interact to form heterooligomers, and indirect evidence for this postulated interaction was obtained using the split-ubiquitin system in bakers' yeast (Reinders et al., 2002). This possible interaction was discussed as a regulatory mechanism of SUT2/SUC3-type transporters for the rapid modulation of transport rates (Reinders et al., 2002).
Here, we further analyzed the hypothesis of SUT2/SUC3-type proteins being sensors that interact with other sucrose transporters to modulate transport efficiency rather than being sucrose transporters themselves (Barker et al., 2000; Reinders et al., 2002). To this end, we used the well-established system of Plantago major (common plantain), in which companion cell–specific transporters for sucrose (PmSUC2; Stadler et al., 1995) and sorbitol (PmPLT1 and PmPLT2; N. Sauer and M. Ramsperger-Gleixner, unpublished data) have been identified and characterized fully. Here, we show that in Plantago, PmSUC3 is not colocalized in companion cells with the high-affinity sucrose transporter PmSUC2. This finding differs from the situation in solanaceous plants and contradicts the model of SUT2/SUC3 as a sucrose sensor that interacts with other transporters. Moreover, PmSUC3 was localized in sieve elements of sink phloem, in which PmSUC2 is absent, and in several other sink tissues. The possible role of PmSUC3 in phloem loading or transport and the implications of this finding for the postulated sensor function are discussed.
RESULTS
Cloning of the PmSUC3 cDNA
cDNA sequences of AtSUC3 (Meyer et al., 2000), VvSUC12 (Davies et al., 1999), and LeSUT2 (Barker et al., 2000) were aligned to identify conserved sequence motifs, allowing the specific amplification of SUT2/SUC3-type cDNA and genomic fragments from other plant species by PCR. According to this alignment, we designed a pair of degenerate primers corresponding to nucleotides 876 to 896 and 1181 to 1159 in the open reading frame of the AtSUC3 cDNA (Meyer et al., 2000). Using these primers, we were able to amplify a 468-bp Plantago genomic fragment. Sequence analyses revealed that this fragment was derived from a Plantago SUT2/SUC3-type gene. This finding was supported by the observation that the fragment contained an intron that corresponded in position precisely to the ninth intron of the Arabidopsis AtSUC3 gene (Meyer et al., 2000). Therefore, the identified Plantago gene was named PmSUC3. The missing 5′ and 3′ parts of the PmSUC3 cDNA were obtained by rapid amplification of cDNA ends.
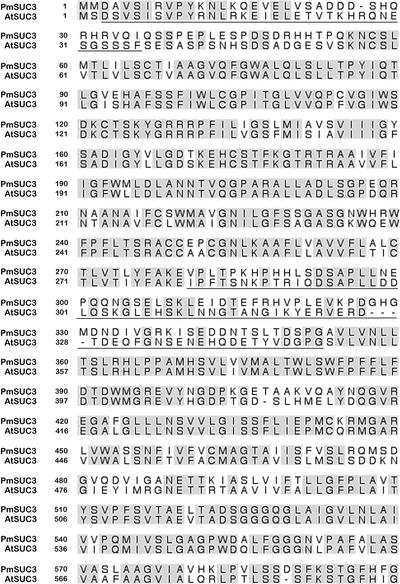
Figure 1 compares the deduced PmSUC3 protein sequence with the sequence of the previously characterized sucrose transporter AtSUC3 (Meyer et al., 2000). The PmSUC3 protein is 599 amino acids long and has a calculated molecular mass of 64.57 kD. It has similar extensions at the N terminus (∼40 amino acids) and in the central loop (∼60 amino acids) as the other transporters of the SUT2/SUC3 subfamily. These insertions are underlined in Figure 1. As can be seen, the degree of sequence conservation within the central insertion is much lower than in the residual protein.
Figure 1.
Comparison of the Deduced Protein Sequences of PmSUC3 and the Homologous AtSUC3 Protein from Arabidopsis.
Sequences were aligned with the program SeqVu (James Gardner, Garvan Institute of Medical Research, Sydney, Australia), and residues identical in both sequences are highlighted. Regions corresponding to the N-terminal and central extensions in SUT2/SUC3-type proteins are underlined.
Another characteristic of SUT2/SUC3-type transporters is their low pI, which is typically 3 to 4 units below those of all other sucrose transporters. This also is the case for PmSUC3, which has a pI of 6.5 (Table 1). The physiological role of this reproducibly found difference is unclear.
Table 1.
Comparison of Three SUT2/SUC3-Type Sucrose Transporters (PmSUC3, AtSUC3, and VvSUC12) from Three Different Species with Six Other Sucrose Transporters from the Same Plants
|
Plantago major
|
Arabidopsis thaliana
|
Vitis vinifera
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | SUC1 | SUC2 | SUC3 | SUC1 | SUC2 | SUC3 | SUC11 | SUC12 | SUC27 |
| MWapp | 53.39 | 54.02 | 64.57 | 54.86 | 54.53 | 63.97 | 53.94 | 65.25 | 53.97 |
| N | 503 | 510 | 599 | 513 | 512 | 594 | 501 | 608 | 505 |
| NGC | 288 | 291 | 234 | 238 | 244 | 226 | 222 | 202 | 326 |
| GC3 | 0.573 | 0.571 | 0.391 | 0.464 | 0.477 | 0.380 | 0.443 | 0.332 | 0.645 |
| pI | 8.60 | 10.02 | 6.50 | 9.25 | 9.55 | 5.81 | 9.47 | 6.50 | 10.23 |
MWapp, apparent molecular mass (kD); N, number of codons in the open reading frame; NGC, number of codons ending with C or G; CG3, CG3 index. The CG3 index is calculated as follows: the sum of informative codons whose third position is C or G (NGC) is divided by the total number of informative codons (N) in the coding sequence of the cDNA (Chiapello et al., 1998).
Functional Expression of PmSUC3 in Yeast
Barker et al. (2000) claimed that AtSUT2/AtSUC3 and LeSUT2 do not complement the yeast mutant SUSY7/ura3 (Riesmeier et al., 1992). Later, however, it was shown that recombinant AtSUT2/AtSUC3 does catalyze the uptake of 14C-labeled sucrose in yeast (Meyer et al., 2000; Schulze et al., 2000).
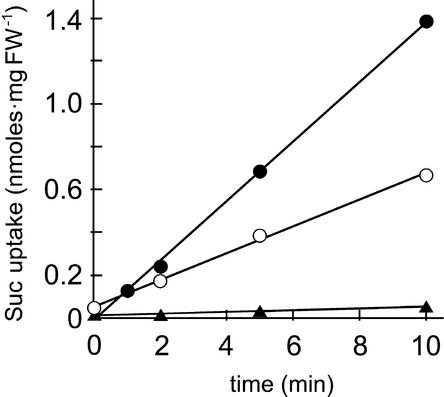
We studied the transport properties of recombinant PmSUC3 after expression of its cDNA in bakers' yeast. NotI restriction sites were introduced on both ends of the PmSUC3 cDNA, which then was cloned into the Escherichia coli/Saccharomyces cerevisiae shuttle vector NEV-N (Sauer and Stolz, 1994). Figure 2 shows that PmSUC3 can transport 14C-labeled sucrose across the plasma membrane of the PmSUC3-expressing yeast strain IBY20s and that the transport rate is enhanced significantly in the presence of d-glucose. This activation by glucose also has been described for other sucrose transporters expressed in yeast (Riesmeier et al., 1992; Gahrtz et al., 1994; Sauer and Stolz, 1994), and the assumption is that glucose metabolism provides energy for active transport and/or activates the plasma membrane H+-ATPase. No significant sucrose transport was observed in strain IBY20as, which expressed PmSUC3 in the antisense orientation (Figure 2).
Figure 2.
PmSUC3 Can Be Expressed in Yeast Cells and Catalyzes the Uptake of 14C-Sucrose.
Transport rates for 14C-sucrose were determined with the transgenic yeast cells expressing PmSUC3 in the sense (open circles; strain IBY20s) or the antisense (closed triangles; strain IBY20as) orientation. The initial concentration of 14C-sucrose was 1 mM in all experiments. The addition of d-glucose (closed circles; final concentration of 10 mM) clearly enhances the transport rates of PmSUC3. FW, fresh weight.
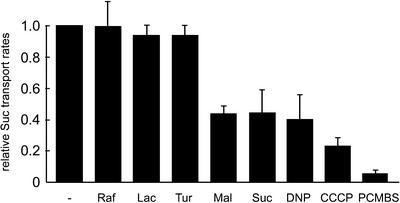
Further transport analyses were performed in the presence of various unlabeled compounds representing potential substrates of PmSUC3 or in the presence of inhibitors, such as the uncouplers carbonyl cyanide-m-chlorophenylhydrazone (CCCP) and dinitrophenol (DNP), or the sulfhydryl group inhibitor p-(chloromercuri)benzene sulfonic acid (PCMBS; Figure 3). Of the tested competitors, only maltose and sucrose competed with the uptake of 14C-labeled sucrose. No inhibition was observed for turanose, lactose, or raffinose. Uncouplers, such as DNP and CCCP, strongly reduced the PmSUC3-dependent transport rates, indicating that sucrose transport by PmSUC3 might be an energized H+ symport. Transport rates also were reduced by PCMBS, a classic inhibitor of plant sucrose transporters (Ludwig et al., 2000). Both the substrate specificity and the observed sensitivity to inhibitors are typical properties of higher plant sucrose transporters.
Figure 3.
Transport Properties of PmSUC3 in Yeast Cells.
Transport of 14C-sucrose (1 mM) by strain IBY20s was analyzed in the presence of uncouplers (DNP or CCCP) or in the presence of the sulfhydryl group inhibitor PCMBS. Inhibitors were added to a final concentration of 50 μM. Transport of 14C-sucrose also was analyzed in the presence of other potential substrates added at a 10-fold excess (final concentrations of 10 mM). Each bar represents results from at least three independent analyses. Lac, lactose; Mal, maltose; Raf, raffinose; Suc, sucrose; Tur, turanose.
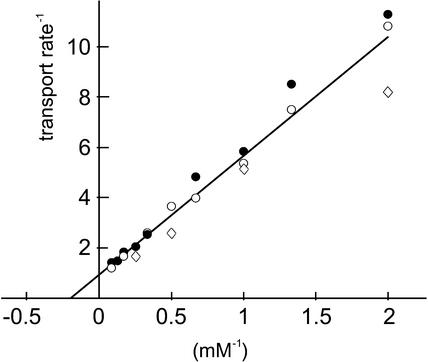
The affinity of PmSUC3 for its physiological substrate sucrose was determined in yeast strain IBY20s. The data obtained in three independent analyses are shown in Figure 4. The Km value for sucrose of PmSUC3 was 5.5 ± 1.1 mM, which was approximately fivefold higher than the Km value for sucrose of PmSUC2, the companion cell–specific phloem loader.
Figure 4.
Km Value of PmSUC3 for Sucrose Transport in Transgenic Yeast Cells.
The Lineweaver-Burk diagram shown contains data from three independent experiments represented by three different symbols. From the individual values of the three analyses, a Km value of 5.5 ± 1.1 mM was calculated.
Immunolocalization of PmSUC3 in Plantago Phloem
Barker et al. (2000) identified LeSUT2 in sieve elements, the same cell type in which all other phloem-specific sucrose transporters had been found previously in solanaceous plants. We were interested to determine whether PmSUC3 also is colocalized with the phloem loader PmSUC2 in the companion cells of Plantago (Gahrtz et al., 1994; Stadler et al., 1995) or whether PmSUC3 is localized in a different cell type. Therefore, we raised antibodies against a peptide of 15 amino acids corresponding to residues 337 to 351 in the PmSUC3 protein sequence shown in Figure 1. This region is part of the central loop found only in this subfamily of transporters. It is not conserved between the different SUT2/SUC3-type transporters (Figure 1) and thus should recognize PmSUC3 with high specificity.
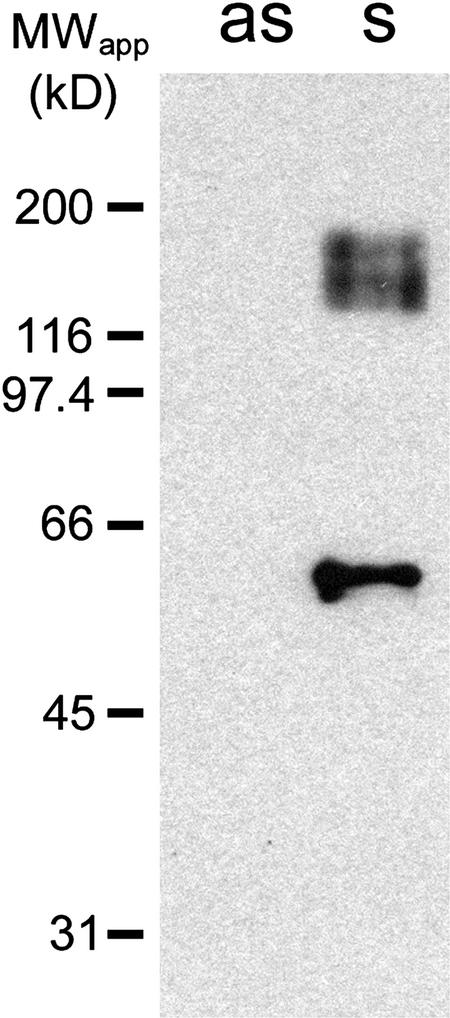
In Figure 5, the quality of this anti-PmSUC3 antiserum was tested on plasma membrane proteins from yeast strains IBY20s and IBY20as. On protein gel blots, the anti-PmSUC3 antiserum reacted only with protein extracts from the PmSUC3-expressing strain IBY20s and not with extracts from the antisense yeast strain. Moreover, the apparent molecular mass of the monomer was ∼56 kD, which was slightly lower than the molecular mass deduced from the cDNA sequence (64.6 kD). This migration at a lower apparent molecular mass, as well as the appearance of aggregates at ∼150 kD, is typical for highly lipophilic proteins (Beyreuther et al., 1980; Gahrtz et al., 1994).
Figure 5.
Identification of PmSUC3 in Transgenic Yeast Cells.
SDS-solubilized plasma membrane proteins (4 μg/lane) from yeast cells expressing PmSUC3 in the sense (s = IBY20s) or the antisense (as = IBY20as) orientation were separated on a polyacrylamide gel, transferred to a nitrocellulose filter, and incubated with anti-PmSUC3 antiserum at a dilution of 1:500. Binding of antibodies to PmSUC3 was detected with anti-rabbit IgG antiserum conjugated to peroxidase. MWapp, apparent molecular mass.
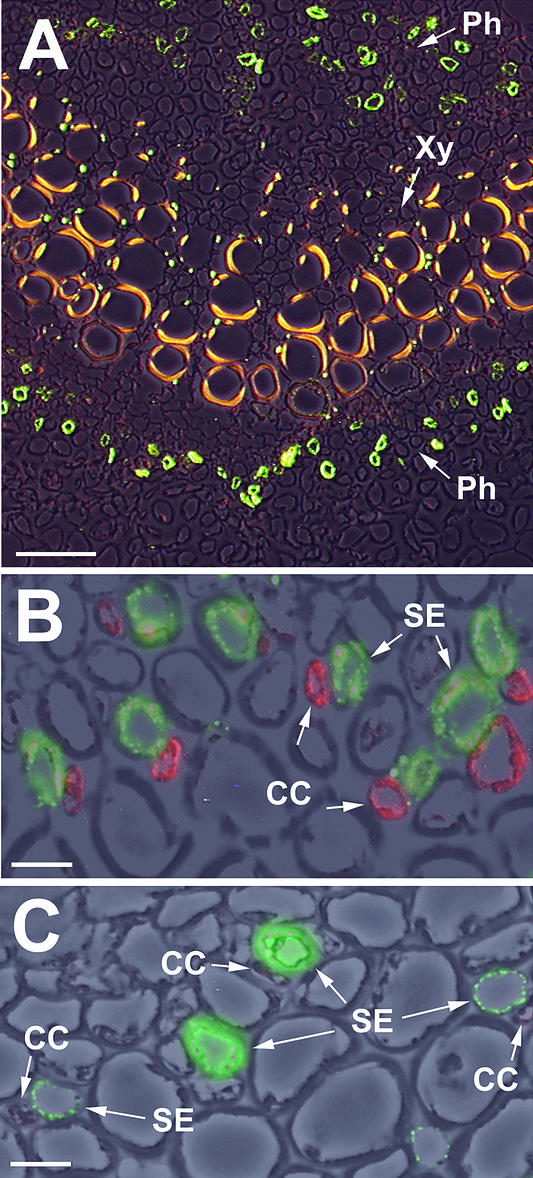
When this specific anti-PmSUC3 antiserum was used for immunohistochemical analyses of Plantago leaf sections, a single cell type showed fluorescence after treatment with a dye-coupled second antibody (Figure 6A). The labeled cells were located exclusively within the bicollateral phloem (in fully developed vascular tissue) on both sides of the xylem. The identity of these cells was determined by double labeling of sections with anti-PmSUC3 antiserum and with the monoclonal anti-PmSUC2 antibody 1A2 (Stolz et al., 1999). Binding of these antibodies was analyzed with anti-mouse IgG and anti-rabbit IgG second antibodies coupled to different fluorescent dyes. Figure 6B shows that this treatment results in the labeling of cell pairs, with one cell showing red fluorescence (indicating binding of anti-PmSUC2 antibody) and one cell showing green fluorescence (indicating binding of anti-PmSUC3 antibody). Because PmSUC2 (red fluorescence) has been characterized as a companion cell–specific sucrose transporter by immunodetection (Stadler et al., 1995), it can be used as an internal standard for the Plantago vascular system. Thus, PmSUC3 is located in the sieve elements of the Plantago phloem. The specificity of all labeling reactions was confirmed by titrating the signal with the PmSUC3-specific peptide that had been used for the production of anti-PmSUC3 antiserum (see Methods). No PmSUC3 signals were found, if the antiserum was preincubated with peptide conjugated to ovalbumin. Incubation with ovalbumin alone had no effect (data not shown).
Figure 6.
Immunolocalization of PmSUC3 in Sieve Elements of the Plantago Phloem.
(A) Cross-section through the vascular bundle of a Plantago leaf treated with anti-PmSUC3/fluorescein isothiocyanate–conjugated anti-rabbit antiserum (green fluorescence in the phloem [Ph]). A photograph taken under white light and a photograph taken under excitation light were superimposed. The yellow fluorescence of xylem vessels in the center (Xy) results from phenolic compounds in the walls of these cells. Bar = 50 μm.
(B) Cross-section through a medium-sized vascular bundle of a Plantago source leaf double stained with anti-PmSUC3/Alexa Fluor 488 goat anti-rabbit IgG (green fluorescence) and anti-PmSUC2/Alexa Fluor 546 goat anti-mouse IgG (red fluorescence). Three photographs (one taken under white light and two taken under excitation light) were superimposed. CC, companion cell; SE, sieve element. Bar = 1 μm.
(C) Cross-section through a medium-sized vascular bundle of a Plantago sink leaf double stained with anti-PmSUC3/Alexa Fluor 488 goat anti-rabbit IgG (green fluorescence) and anti-PmSUC2/Alexa Fluor 546 goat anti-mouse IgG (red fluorescence, which was not detectable in sinks). Three photographs (one taken under white light and two taken under excitation light) were superimposed. Bar = 1 μm.
Most importantly, there was a clear difference in the expression patterns of PmSUC2 and PmSUC3 in sink and source tissues. Expression of PmSUC2 is known to be source specific (Truernit and Sauer, 1995; Imlau et al., 1999). This was confirmed by the analyses shown in Figure 6C, in which the localizations of PmSUC2 and PmSUC3 were analyzed in sink leaves. The red PmSUC2-specific fluorescence is completely absent in the vascular bundles of sink leaves, whereas the green PmSUC3-specific fluorescence is seen in the sieve elements of both sink and source leaves.
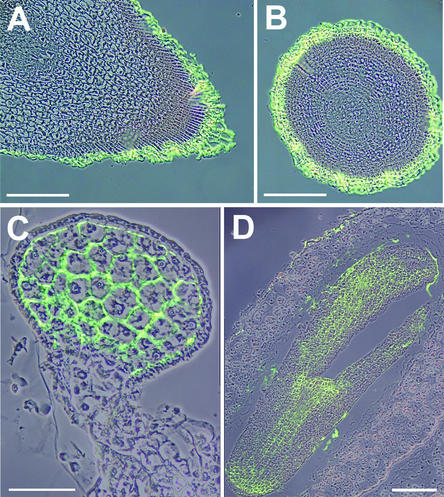
Identification of PmSUC3 in Nonphloem Tissues
Using the anti-PmSUC3 antiserum, PmSUC3 protein also was detected in other tissues. Figures 7A and 7B show strong signals in the root tips (calyptra), and Figures 7C and 7D show strong immunosignals in the embryo at different developmental stages. As in the sieve elements, all of this signal could be titrated away by pretreatment of the affinity-purified anti-PmSUC3 antiserum with ovalbumin-bound anti-PmSUC3 peptide (data not shown).
Figure 7.
Identification of PmSUC3 in the Root Tips of Plantago.
(A) Longitudinal section through a Plantago root tip. Two photographs were superposed, one taken under white light and one taken under excitation light to visualize Alexa Fluor 488–decorated PmSUC3.
(B) Cross-section through a Plantago root tip presented as described for (A).
(C) Longitudinal section through a Plantago embryo in the globular state presented as described for (A).
(D) Longitudinal section through a Plantago embryo in the torpedo state presented as described for (A).
Bars = 50 μm for (C) and 100 μm for (A), (B), and (D).
Identification of T-DNA Insertion Mutants of AtSUC3
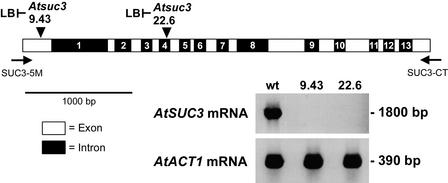
If SUT2/SUC3-type genes encode transporters with an additional sensor function, and if these proteins interact with other sucrose transporters to regulate their activity, a severe phenotype might be expected in mutant plants that lack this gene. Therefore, we screened the collection of Arabidopsis T-DNA insertion mutants generated at the University of Wisconsin (Krysan et al., 1999) and identified two independent insertion lines, Atsuc3-9.43 and Atsuc3-22.6, that carry mutations in the AtSUC3 gene. Figure 8 shows the positions of the T-DNA insertions in these two lines (after nucleotide 154 in Atsuc3-9.43 and after nucleotide 1331 in Atsuc3-22.6; in both lines, the precise insertion could be determined only for the left border of the T-DNA) and the reverse transcriptase–PCR (RT-PCR) results that confirmed the absence of AtSUC3 mRNA in these mutant lines.
Figure 8.
Identification of T-DNA Insertions in the Arabidopsis AtSUC3 Gene.
The positions of the T-DNA insertions in the Arabidopsis lines Atsuc3-9.43 (first exon) and Atsuc3-22.6 (fourth intron) are indicated by arrowheads, and binding sites for the PCR primers SUC3-5M and SUC3-CT are represented by arrows. The orientations of the identified left borders (LB) of the T-DNA insertions are indicated. The indicated primers were used to perform RT-PCR on total RNA isolated from Arabidopsis wild-type (wt) plants or from the two different T-DNA insertion lines. RT-PCR products were identified only with RNA from wild-type plants. In control reactions, fragments of the Arabidopsis ACT1 mRNA (An et al., 1996) were amplified from all RNA preparations.
When the phenotypes of the mutant lines and of the corresponding wild-type plants (Arabidopsis ecotype Wassilewskija) were compared, we observed an altered floral morphology for most flowers in Atsuc3-9.43 mutant plants. This phenotype looked exactly like that described for apetala3 mutants and is observed in ∼10% of the Wisconsin lines (http://www.biotech.wisc.edu/Arabidopsis/) as a result of the presence of an AP3 fragment in the T-DNA insertions used. Therefore, we concentrated on the Atsuc3-22.6 insertion line that also had no AtSUC3 mRNA (Figure 8). At ambient CO2 concentrations and under the growth conditions described in Methods, we were unable to detect any difference between plants of the Atsuc3-22.6 mutant line and Wassilewskija wild-type plants.
DISCUSSION
Here, we describe the functional properties of the PmSUC3 protein from Plantago, a new member of the SUT2/SUC3-type sucrose transporter subfamily. Using a specific anti-PmSUC3 antiserum, PmSUC3 was identified in the sieve elements of the Plantago phloem and in nonphloem cells of several other tissues. The protein showed all of the characteristics described for other members of this subfamily, such as an N-terminal extension, an enlarged central loop, and a lower pI than other higher plant sucrose transporter genes (Table 1). The important results are (1) that PmSUC3 is localized in phloem sieve elements and (2) that it can mediate the transport of sucrose (Km = 5 mM) across yeast plasma membranes at high rates. Sieve element specificity also has been shown for the LeSUT2 sucrose transporter (Barker et al., 2000). However, in tomato, LeSUT2 is colocalized with the high-affinity sucrose transporter LeSUT1 (Kühn et al., 1997) and the low-affinity sucrose transporter LeSUT4 (Weise et al., 2000). For LeSUT2, it was suggested that this type of transporter might be nonfunctional (Barker et al., 2000) or have little transport activity (Schulze et al., 2000) and that it could interact with LeSUT1 and LeSUT4 to modulate their transport activity (Reinders et al., 2002). Here, we show that the situation is different for the Plantago PmSUC3 protein.
Plantago Has Different Sucrose Transporters in Sieve Elements and in Companion Cells
The newly characterized sucrose transporter PmSUC3 is located in the sieve elements (Figure 6), whereas the high-affinity phloem loader PmSUC2 (Gahrtz et al., 1994) is located in the companion cells of the Plantago phloem (Stadler et al., 1995). PmSUC2 and all other phloem loaders were shown to be expressed primarily in source leaves or during the sink-source transition of expanding leaves (Gahrtz et al., 1994; Truernit and Sauer, 1995; Imlau et al., 1999). The essential roles of the companion cell–specific SUC2 sucrose transporters (described in Plantago and Arabidopsis) and of the sieve element–specific SUT1 sucrose transporters (described in solanaceous plants) in phloem loading have been well documented in analyses of suc2 knockout plants (Gottwald et al., 2000) and SUT1 antisense plants (Kühn et al., 1996).
Our data have major implications for the predicted functions of SUT2/SUC3-type transporters in planta. For LeSUT2, it was speculated that it may be a sensor that interacts with the colocalized phloem loaders to modulate sucrose import into the phloem (Reinders et al., 2002). Such an interaction cannot occur between PmSUC2 and PmSUC3 in the phloem of Plantago, in which the different transporters are located in different cell types (Figure 6B) and the genes are expressed at different developmental stages (Figure 6C).
Do SUT2/SUC3-Type Transporters Have a Function as Sucrose Sensors?
One argument in the discussion about a potential function of SUT2/SUC3 transporters in sucrose sensing concerned the low codon bias of their genes (Barker et al., 2000). Such a low codon bias had been observed in bakers' yeast for the genes of the transporter-like sensor proteins Ssy1p, Rgt2p, and Snf3p, in which it correlates with a low efficiency of mRNA translation (Iraqui et al., 1999). In Table 1, the CG content of the codons in the PmSUC3 open reading frame and in two other SUT2/SUC3-type transporter cDNAs from Arabidopsis (AtSUC3) and grapevine (VvSUC12) are presented and compared with the CG contents of other sucrose transporter cDNAs of these plants. The frequency of codons using C or G at the third position is given in absolute numbers and as CG3 index.
CG3 indices of a large number of Arabidopsis genes have been determined by Chiapello and co-workers (1998). In these analyses, it has been found that genes with a high CG3 index (up to 0.6) often encode ubiquitous, highly abundant housekeeping and photosynthetic genes (e.g., the gene for the photosystem II type-I chlorophyll a/b binding protein LHB1B1, with a CG3 index of 0.618). Genes with a low CG3 index (down to 0.25) frequently show tissue-specific expression, often in relation to stress conditions, and may encode proteins abundant under particular physiological conditions (e.g., the gene of the G-box binding factor GBF3 [CG3 = 0.277]). Additionally, the genes of several highly abundant proteins were found to have low CG3 values, such as the hexokinase gene HK1 (CG3 = 0.355) and the Gly-rich protein gene GFP-7 (CG3 = 0.267).
In fact, the CG3 index of the SUT2/SUC3-type transporter genes shown in Table 1 is reproducibly lower than that of the other sucrose transporter cDNAs. However, the values of 0.380 for AtSUC3 and 0.391 for PmSUC3 are more in the middle range of CG3 indices and are comparable to those of other Arabidopsis membrane transporter genes. Examples are the K+-channel gene KAT1-1 (CG3 = 0.388; Anderson et al., 1992), the plasma membrane H+-ATPase gene AHA10 (CG3 = 0.346; Harper et al., 1994), and the vacuolar H+-ATPase gene AVA-P (CG3 = 0.395; Perera et al., 1995). These values show that the codon bias of SUT2/SUC3-type transporters cannot be used to support the sensor function of SUT2/SUC3-type transporters.
Another argument concerned the low transport rate of SUT2 in transgenic bakers' yeast (Barker et al., 2000; Schulze et al., 2000). However, we were able to show that both AtSUC3 (Meyer et al., 2000) and PmSUC3 (this study) can transport sucrose at significant rates across the plasma membranes of AtSUC3- or PmSUC3-expressing yeast cells. This finding demonstrates that a negative result, such as the inability of a cDNA to complement a mutation in a heterologous expression system, cannot be used as an argument for a potential sensor function in planta. N. Sauer and M. Ramsperger-Gleixner (unpublished data) clearly showed that the successful expression of Plantago sorbitol transporters in yeast and the observed transport rates depend strongly on the 5′ flanking sequences of the expressed cDNAs. Optimization of these sequences for the yeast translation machinery will strongly enhance the efficiency of expression.
A third argument for the sensor function of SUT2/SUC3-type transporters concerned the observed extensions of ∼30 amino acids at the N terminus and ∼50 amino acids in the central loop (Figure 1). In fact, this difference could be used as an argument for a transport-independent function of these extensions, and experimental evidence supporting this model was presented by Meyer and co-workers (2000). Those authors showed that the extension of the central loop can be removed from the AtSUC3 protein with little effect on the sucrose transport capacity of the protein. In further analyses (S. Meyer and N. Sauer, unpublished data), a similar result was obtained with an N-terminally deleted AtSUC3 protein. However, these observations do not provide any information about the physiological functions of the deleted extensions. They might be involved in protein–protein interaction for signaling, but they also could be necessary for the modulation of transport activity in planta, for a switch from import to export, or for another yet unknown function.
Recently, a draft sequence of the rice genome was published that covered >90% of the gene-containing regions (Yu et al., 2002). In this sequence, the putative sucrose transporter genes OsSUT1 to OsSUT5 were identified (Aoki et al., 2003). Two of these genes (OsSUT1 and OsSUT4) were shown to possess the identical number of introns (13) as the SUT2/SUC3-type transporters from dicots, and two genes have quite similar intron numbers (OsSUT3: 9 introns; OsSUT5: 12 introns). Surprisingly, the deduced amino acid sequences of these four rice sucrose transporters cluster together with the SUT2/SUC3-type transporters from dicots, such as AtSUC3 and PmSUC3 (Aoki et al., 2003). Moreover, this cluster clearly is separated from the clusters that contain the typical dicot transporters, such as AtSUC1, PmSUC2, AtSUT4, and LeSUT1 (Aoki et al., 2003). However, only one of these four rice transporters has N-terminal and central loop extensions similar to those of the dicot SUT2/SUC3 transporters (OsSUT4: 595 amino acids). This finding demonstrates that monocot plants have significantly larger numbers of SUT2/SUC3-type transporters. However, as in dicots, only one of these transporters has the additional cytoplasmic extensions found in PmSUC3 and AtSUT2/AtSUC3.
Apparently, every higher plant species has one gene that encodes this special type of sucrose transporter. Therefore, one would predict that SUT2/SUC3-type transporters are essential for plant growth and development. However, homozygous knockout mutants of the Arabidopsis T-DNA insertion line Atsuc3-22.6 (Figure 8) showed no recognizable change in phenotype under standard growth conditions. The possibility cannot be excluded that a phenotype appears only if plants are grown under different, more challenging conditions.
What Is the Physiological Function of the PmSUC3 Sucrose Transporter?
PmSUC3 is localized in sieve elements and can transport sucrose at a high rate. Its Km value for sucrose is approximately fivefold higher than that of the companion cell–specific PmSUC2 phloem loader. In all of the plants analyzed, the companion cells are considerably larger in the regions of phloem loading than in the transport phloem or in sink tissues, emphasizing the important role of companion cells in phloem loading. Conversely, the diameter of sieve elements increases strongly after the loading zone in the minor veins (Behnke, 1989), supporting their role in long-distance transport. Thus, it is unlikely that the sieve element–specific PmSUC3 transporter is involved in the regulation of phloem loading. This notion is supported by the observation that PmSUC3 is also found in sieve elements of the phloem in sink leaves, in which no phloem loading occurred and expression of PmSUC2 was missing (Figure 6). Unloading of sucrose in sink leaves was shown to occur symplastically (Imlau et al., 1999). This and the lower substrate affinity (5 mM) suggests a role for PmSUC3 in sucrose retrieval along the phloem path. The higher Km value may be important to make this retrieval less efficient and to leave part of the sucrose for an apoplastic supply to lateral sinks along the phloem.
PmSUC3 also was identified in nonphloem cells and tissues, such as in the calyptra, in the embryo (Figure 7), and in pollen tubes (data not shown). The signals in roots and in embryos could be titrated away completely with ovalbumin-bound PmSUC3 peptide that had been used to raise the antiserum. The titration worked less efficiently for the pollen tubes. However, we found activity of the AtSUC3 promoter and signals with anti-AtSUC3 antiserum in the same tissues of Arabidopsis (S. Meyer, C. Lauterbach, and N. Sauer, unpublished data), supporting the described PmSUC3 localizations in Plantago. All PmSUC3-expressing tissues outside of the phloem represent sinks and depend on a massive supply of organic carbon for biosynthetic purposes and growth. Thus, PmSUC3 might act as a sucrose importer into these tissues.
METHODS
Strains and Growth Conditions
Plantago major wild-type plants were grown in potting soil in a greenhouse under ambient conditions. Arabidopsis thaliana (ecotype Wassilewskija) wild-type and mutant plants were grown at 21°C under short-day conditions (8 h of light/16 h of dark) and transferred to long-day conditions (16 h of light/8 h of dark) after bolting. For cloning in Escherichia coli, we used strain DH5α (Hanahan, 1983); yeast expression was performed in Saccharomyces cerevisiae strain SEY2102 (Emr et al., 1983).
Molecular Cloning of a PmSUC3 cDNA
A partial genomic fragment was obtained by PCR with Plantago genomic DNA using the degenerate primers Suc3c876f (5′-AGAYTCTGCNCCTYTNTTGGA-3′) and Suc3c1181r (5′-TCNCKNCCCATCCARTCNGTRTC-3′) that were deduced from alignments of AtSUC3 (Meyer et al., 2000), VvSUC12 (Davies et al., 1999), and LeSUT2 (Barker et al., 2000) cDNA sequences. PmSUC3-specific primers were synthesized according to the obtained genomic sequence and were used to generate the complete coding sequence of the PmSUC3 cDNA using the 5′/3′-RACE Kit from Roche Molecular Biochemicals (Mannheim, Germany). For 5′ rapid amplification of cDNA ends (RACE), we used the primers PmSUC3ML-Tail1r (5′-CAATCCTATCATTATCCATGCCATGCCC-3′), PmSUC3ML-Tail2r (5′-CGGGCTTAACTTCTAATGGGACATG-3′), and PmSUC3ML-Tail3r (5′-CAGATCCATTTTGCTGAGGTTCATCC-3′), and for 3′ RACE, we used the primers PmSUC3-ML1f (5′-TTGGATGAACCTCAGCAAAATGGATCTG-3′) and PmSUC3-ML421f (5′-GTCCTGGTTCCCCTTTTTCCTC-3′). From the sequences obtained by the 5′ RACE and 3′ RACE, the primers PmSUC3-5 (5′-GAATCTGAGCGGCCGCAAATTCAAATCACCGACATG-3′) and PmSUC3-3 (5′-GAATCTGAGCGGCCGCTCAGCCAAAATGGAATCCAG-3′) were designed that allowed the amplification of the entire PmSUC3 open reading frame from the Plantago mRNA preparation. These primers also introduced NotI cloning sites at both ends of the cDNA, which were used to clone the PCR fragment into the E. coli/S. cerevisiae shuttle vector NEV-N (Sauer and Stolz, 1994). All constructs were sequenced. The resulting clones were named pIB20s (insert in the sense orientation) and pIB20as (insert in the antisense orientation).
Expression in Bakers' Yeast and Transport Analyses
The plasmids pIB20s and pIB20as were used to transform yeast strain SEY2102 (Emr et al., 1983). The resulting strains were named IBY20s and IBY20as. Transport of 14C-sucrose was analyzed in Na-phosphate buffer, pH 5.0, in the presence of 10 mM d-glucose as described (Sauer et al., 1990).
Isolation of Yeast Plasma Membranes
Plasma membranes were isolated according to the protocol of Stolz et al. (1994). Membrane proteins were solubilized with SDS and separated onto SDS–polyacrylamide gels as described by Laemmli (1970). Protein transfers to nitrocellulose and protein gel blot analyses were performed according to the protocol of Dunn (1986). The nitrocellulose was incubated overnight in affinity-purified anti-PmSUC3 antiserum (diluted 1:500 in blocking buffer). Binding of antibody to PmSUC3 protein was detected by treatment of the filter with anti-rabbit IgG antiserum-peroxidase conjugate (diluted 1:4000 in blocking buffer [50 mM Tris-HCI, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, and 1% skim milk powder]) followed by incubation with Lumi-Light Western Blotting Substrate (Roche Diagnostics, Mannheim, Germany).
Immunohistochemical Techniques
Anti-PmSUC3 antisera were raised in rabbits (Pineda-Antikörper-Service, Berlin, Germany) against a synthetic peptide (RKISEDDNTSLTDSP) coupled to a carrier protein. The sequence was derived from a peptide located in the predicted central loop of the protein.
Plantago tissue was prepared, fixed in methacrylate, sectioned, and transferred to adhesion microscope slides (Linaris, Wertheim, Germany) as described previously (Stadler and Sauer, 1996). Root tips (Figure 7) were isolated from soil-grown plants, rinsed with water for a few seconds, and fixed immediately. Methacrylate was removed by incubation of the slides for 3 min in 100% acetone. Sections were rehydrated by sequential incubation in ethanol of decreasing concentrations (100, 95, 80, 60, and 30%) and blocked for 1 h (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% skim milk powder). After overnight incubation with affinity-purified anti-PmSUC3 antiserum (for phloem location: diluted 1:10 in blocking buffer; for nonphloem location: diluted 1:60 in blocking buffer) and/or monoclonal anti-PmSUC2 antiserum (diluted 1:4; Stolz et al., 1999), sections were washed five times with blocking buffer. For the detection of anti-PmSUC3 antiserum, sections were incubated for 1 h with a 1:300 dilution of Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes, Leiden, The Netherlands) or with anti-rabbit IgG–fluorescein isothiocyanate–isomer 1 conjugate (Sigma, Taufkirchen, Germany). For double stainings of sections with polyclonal anti-PmSUC3 antibody and monoclonal anti-PmSUC2 antibody 1A2, Alexa Fluor 546 goat anti-mouse IgG (Molecular Probes) was added in addition (diluted 1:100) to visualize antibody binding to PmSUC2. After five final washes with blocking buffer, the slides were rinsed with water and mounted using the ProLong Antifade Kit (Molecular Probes). Photographs were taken on a fluorescence microscope (Zeiss, Göttingen, Germany) with appropriate excitation light.
For antibody-peptide competition experiments, a conjugate of the specific peptide with ovalbumin was used. Before immunolocalization, the affinity-purified antiserum was incubated for 2 to 3 h at room temperature with 200 μg/mL conjugate or pure ovalbumin.
Identification of Arabidopsis T-DNA Insertion Lines
T-DNA insertion lines from the collection described by Krysan and co-workers (1999) were identified by reverse transcriptase–PCR (http://www.biotech.wisc.edu/Arabidopsis/). Total RNA from Arabidopsis plants was isolated according to standard protocols.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the genes and proteins mentioned in this article are as follows: PmSUC3 cDNA (AJ534442), LHB1B1 (X64459), GBF3 (U17891), HK1 (U18754), GFP-7 (Z11868), OsSUT1 gene (AF280050), OsSUT2 mRNA (AB091672), OsSUT3 gene (AF419298), OsSUT4 mRNA (AB091673), and OsSUT5 mRNA (AB091674).
Acknowledgments
We thank Robert T. Furbank (Commonwealth Scientific and Industrial Research Organization Plant Industry, Canberra, Australia) for allowing us to use unpublished data on newly cloned rice sucrose transporter genes and cDNAs. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to N.S. (SPP1108 - Sa 382/12-1).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010967.
References
- An, Y.Q., Huang, S., McDowell, J.M., McKinney, E.C., and Meagher, R.B. (1996). Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell 8, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.A., Huprikar, S.S., Kochian, L.V., Lucas, W.J., and Gaber, R.F. (1992). Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89, 3736–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, N., Hirose, T., Scofield, G.N., Whitfeld, P.R., and Furbank, R.T. (2003). The sucrose transporter gene family in rice. Plant Cell Physiol. 44, 223–232. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Barker, L., Kühn, C., Weise, A., Schulz, A., Gebhardt, C., Hirner, B., Hellmann, H., Schulze, W., Ward, J.M., and Frommer, W.B. (2000). SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke, H.-D. (1989). Structure of the phloem. In Transport of Photoassimilates, D.A. Baker, and J.A. Milburn, eds (Harlow, UK: Longman Scientific & Technical), pp. 79–137.
- Beyreuther, K., Bieseler, B., Ehring, R., Griesser, H.-W., Mieschendahl, M., Müller-Hill, B., and Triesch, I. (1980). Investigation of structure and function of lactose permease of Escherichia coli. Biochem. Soc. Trans. 8, 675–676. [DOI] [PubMed] [Google Scholar]
- Chiapello, H., Lisacek, F., Caboche, M., and Henaut, A. (1998). Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 209, GC1–GC38. [DOI] [PubMed] [Google Scholar]
- Davies, C., Wolf, T., and Robinson, S.P. (1999). Three putative sucrose transporters are differentially expressed in grapevine tissues. Plant Sci. 147, 93–100. [Google Scholar]
- Dunn, S.D. (1986). Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal. Biochem. 157, 144–153. [DOI] [PubMed] [Google Scholar]
- Emr, S.D., Scheckman, R., Flessel, M.C., and Thorner, J. (1983). An MFα1-SUC2 (σ-factor-invertase) gene fusion for study of protein localisation and gene expression in yeast. Proc. Natl. Acad. Sci. USA 80, 7080–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahrtz, M., Schmelzer, E., Stolz, J., and Sauer, N. (1996). Expression of the PmSUC1 sucrose carrier gene from Plantago major L. is induced during seed development. Plant J. 9, 93–100. [DOI] [PubMed] [Google Scholar]
- Gahrtz, M., Stolz, J., and Sauer, N. (1994). A phloem specific sucrose-H+ symporter from Plantago major L. supports the model of apoplastic phloem loading. Plant J. 6, 697–706. [DOI] [PubMed] [Google Scholar]
- Gottwald, J.R., Krysan, P.J., Young, J.C., Evert, R.F., and Sussman, M.R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 97, 13979–13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Harper, J.F., Manney, L., and Sussman, M.R. (1994). The plasma membrane H+-ATPase gene family in Arabidopsis: Genomic sequence of AHA10, which is expressed primarily in developing seeds. Mol. Gen. Genet. 244, 572–587. [DOI] [PubMed] [Google Scholar]
- Imlau, A., Truernit, E., and Sauer, N. (1999). Cell-to-cell and long distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui, I., Vissers, S., Bernard, F., De Craene, O.-J., Boles, E., Urrestarazu, A., and Andre, B. (1999). Amino acid signalling in Saccharomyces cerevisiae: A permease-like sensor of external amino acids and F-boc protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad specificity amino acid permease. Mol. Cell. Biol. 19, 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (1996). Interpreting cDNA sequences: Some insights from studies on translation. Mamm. Genome 7, 563–574. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, C., Franceschi, V.R., Schulz, A., Lemoine, R., and Frommer, W.B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300. [DOI] [PubMed] [Google Scholar]
- Kühn, C., Hajirezaei, M.-R., Fernie, A.R., Roessner-Tunali, U., Czechowski, T., Hirner, B., and Frommer, W.B. (2003). The sucrose transporter StSUT1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiol. 131, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, C., Quick, W.P., Schulz, A., Riesmeier, J.W., Sonnewald, U., and Frommer, W.B. (1996). Companion cell-specific inhibition of the potato sucrose transporter SUT1. Plant Cell Environ. 19, 1115–1123. [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lemoine, R., Bürkle, L., Barker, L., Sakr, S., Kühn, C., Regnacq, M., Gaillard, C., Delrot, S., and Frommer, W.B. (1999). Identification of a pollen-specific sucrose transporter-like protein NtSUT3 from tobacco. FEBS Lett. 454, 325–330. [DOI] [PubMed] [Google Scholar]
- Ludwig, A., Stolz, J., and Sauer, N. (2000). Plant sucrose-H+ symporters mediate the transport of vitamin H. Plant J. 24, 503–509. [DOI] [PubMed] [Google Scholar]
- Meyer, S., Truernit, E., Hümmer, C., Besenbeck, R., Stadler, R., and Sauer, N. (2000). AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. Plant J. 24, 869–882. [DOI] [PubMed] [Google Scholar]
- Özcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signalling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, S., Dover, J., Rosenwald, A.G., Woelfl, S., and Johnston, M. (1996). Two glucose transporters in S. cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93, 12428–12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, I.Y., Li, X., and Sze, H. (1995). Several distinct genes encode nearly identical to 16 kDa proteolipids of the vacuolar H+-ATPase from Arabidopsis thaliana. Plant Mol. Biol. 29, 227–244. [DOI] [PubMed] [Google Scholar]
- Reinders, A., Schulze, W., Kühn, C., Barker, L., Schulz, A., Ward, J.M., and Frommer, W.B. (2002). Protein–protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell 14, 1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J.W., Hirner, B., and Frommer, W.B. (1993). Potato sucrose transporter expression in minor veins indicates a role in phloem loading. Plant Cell 5, 1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J.W., Willmitzer, L., and Frommer, W.B. (1992). Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, N., Friedländer, K., and Gräml-Wicke, U. (1990). Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J. 9, 3045–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, N., and Stolz, J. (1994). SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana. Expression and characterization in baker's yeast and identification of the histidine tagged protein. Plant J. 6, 67–77. [DOI] [PubMed] [Google Scholar]
- Schulze, W., Weise, A., Frommer, W.B., and Ward, J.M. (2000). Function of the cytosolic N-terminus of sucrose transporter AtSUC2 in substrate affinity. FEBS Lett. 485, 189–194. [DOI] [PubMed] [Google Scholar]
- Stadler, R., Brandner, J., Schulz, A., Gahrtz, M., and Sauer, N. (1995). Phloem loading by the PmSUC2 sucrose carrier from Plantago major occurs into companion cells. Plant Cell 7, 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., and Sauer, N. (1996). The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta 109, 299–306. [Google Scholar]
- Stadler, R., Truernit, E., Gahrtz, M., and Sauer, N. (1999). The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 19, 269–278. [DOI] [PubMed] [Google Scholar]
- Stolz, J., Ludwig, A., Stadler, R., Biesgen, C., Hagemann, K., and Sauer, N. (1999). Structural analysis of a plant sucrose carrier using monoclonal antibodies and bacteriophage lambda surface display. FEBS Lett. 453, 375–379. [DOI] [PubMed] [Google Scholar]
- Stolz, J., Stadler, R., Opekarová, M., and Sauer, N. (1994). Functional reconstitution of the solubilized Arabidopsis thaliana STP1 monosaccharide-H+ symporter in lipid vesicles and purification of the histidine tagged protein from transgenic Saccharomyces cerevisiae. Plant J. 6, 225–233. [DOI] [PubMed] [Google Scholar]
- Truernit, E., and Sauer, N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196, 564–570. [DOI] [PubMed] [Google Scholar]
- Vagnoli, P., Coons, D.M., and Bisson, L.F. (1998). The C-terminal domain of Snf3p mediates glucose-responsive signal transduction in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 160, 31–36. [DOI] [PubMed] [Google Scholar]
- Weise, A., Barker, L., Kühn, C., Lalonde, S., Buschmann, H., Frommer, W.B., and Ward, J.M. (2000). A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity is localized in enucleate sieve elements of plants. Plant Cell 12, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]