Abstract
The varicella-zoster virus major transactivator, IE62, can activate expression from homologous and heterologous promoters. High levels of IE62-mediated activation appear to involve synergy with cellular transcription factors. The work presented here focuses on functional interactions of IE62 with the ubiquitously expressed cellular factor USF. We have found that USF can synergize with IE62 to a similar extent on model minimal promoters and the complex native ORF28/29 regulatory element, neither of which contains a consensus IE62 binding site. Using Gal4 fusion constructs, we have found that the activation domain of USF1 is necessary and sufficient for synergistic activation with IE62. We have mapped the regions of USF and IE62 required for direct physical interaction. Deletion of the required region within IE62 does not ablate synergistic activation but does influence its efficiency depending on promoter architecture. Both proteins stabilize/increase binding of TATA binding protein/TFIID to promoter elements. These findings suggest a novel mechanism for the observed synergistic activation which requires neither site-specific IE62 binding to the promoter nor a direct physical interaction with USF.
The varicella-zoster virus (VZV) major transcriptional activator, commonly designated the immediate-early 62 protein or IE62, is a potent and promiscuous transactivator of both homologous and heterologous promoters (reviewed in reference 38). The IE62 protein contains 1,310 amino acids (aa), and the major functional domains within the protein with respect to transactivation are an N-terminal acidic activation domain (AD) and a DNA binding domain. The IE62 activation domain (aa 1 to 86) is compositionally similar to other acidic activation domains found in herpes simplex virus VP16 and the pseudorabiesvirus major transactivator but shows little homology to those domains at the individual amino acid level (4, 34).
The DNA binding domain of IE62 is contained within aa 468 to 640 of the IE62 sequence. Previous studies showed that bacterially expressed fragments of IE62 containing this region are capable of binding to a variety of VZV and non-VZV promoter elements (2, 50, 51, 53). The ability of IE62 to interact directly with DNA has been shown to be an important aspect of its mechanism of activation since mutations in IE62 which ablate DNA binding also abrogate transactivation (49). However, it is unclear if IE62 is required to bind to a specific sequence. DNase protection studies by Wu and Wilcox (53) identified a consensus sequence (-ATCGT-) to which a recombinant fusion protein containing the IE62 DNA binding domain bound tightly. Other work, however, indicated that IE62 is capable of binding numerous sequences within promoters (2, 22, 50). Finally, an extensive analysis by Perera (32) showed that IE62 is capable of transactivation of minimal promoters containing only a TATA box and lacking all known or permuted IE62 binding sites.
The mechanism(s) of IE62 activation is largely unknown. Perera (32) showed that IE62 was able to achieve differential levels of transcriptional activation of model promoters depending on the nature of the TATA motif. Direct physical interaction between a fragment of IE62 (aa 273 to 724) and TATA binding protein (TBP) and TFIIB was also demonstrated in that study. Although a TATA element alone is able to mediate IE62 activation as evidenced on artificial minimal reporters and the VZV ORF21 promoter (5, 32), studies of a number of other VZV promoters indicate that an upstream cellular factor binding site, in addition to a TATA element, is required for significant IE62-mediated activation (23, 24, 30, 55). The essential role of one such factor, USF, in mediating IE62 activation of the VZV ORF28/29 regulatory element has been extensively documented, and a direct physical interaction between IE62 and USF has been demonstrated (23-25, 37).
The cellular transcription factor USF (upstream stimulatory factor) is a member of the helix-loop-helix (HLH) family of regulatory proteins. USF binds to a symmetrical DNA sequence (5′-GGTCACGTGACC-3′) first identified in the adenovirus major late promoter (ADMLP) (12, 41). Purified human USF is composed of 43- and 44-kDa polypeptides, designated USF1 and USF2, respectively, which coexist as both homo- and heterodimers, with the heterodimer as the major species (10, 41, 42). USF1 and USF2 are conserved in their C-terminal regions, which contain the bHLH-Zip domains involved in DNA binding and dimerization (44, 45), and in the USF-specific region (USR), which is essential for activation of initiator element-mediated transcription (20). Their N-terminal regions containing transcriptional activation domains show considerable diversity (15, 20). As part of its mechanism of action, USF is believed to stabilize TBP binding and facilitate the formation of transcription preinitiation complexes (41, 52). Consensus binding sites for USF have been found in nearly one-fourth of the putative VZV promoter elements controlling expression of the 71 viral open reading frames (39), indicating that USF likely plays an important role in VZV replication. This has recently been confirmed in a study showing that VZV replication was significantly impaired in a cell line expressing a dominant-negative form of USF (37). The demonstration of a direct physical interaction between IE62 and USF in the same study suggests that IE62 may target promoters through interactions with this cellular factor.
The specific part played by IE62 in cooperation with USF for promoter activation is not fully understood. In one study, IE62 was shown to be able to activate the ORF28/29 regulatory element in an osteosarcoma cell line (Saos-2) in which USF is expressed but lacks transcriptional activity (36). It was therefore suggested that IE62 could substitute for a cellular coactivator that is required for USF activation but which is absent in the Saos-2 cells. However, the ability of IE62 to activate promoters containing USF binding sites in other cell lines where USF is active (23, 55) indicates that IE62 plays a role other than, or in addition to, that of the cellular USF coactivator.
In the work presented here, we found that IE62 and USF individually stimulated expression from model promoters and the VZV ORF28/29 regulatory element. However, the level of activation observed when both were present was 20- to 30-fold greater than the sum of the individual contributions. Further experiments showed that the activation domain of USF1 was both necessary and sufficient to mediate IE62 activation and that TBP/TFIID was captured in protein pull-down experiments using a glutathione S-transferase (GST) fusion protein containing the USF1 activation domain. The regions involved in the direct physical interaction between USF1 and IE62 were mapped to the DNA binding domain of USF and to amino acids 238 to 258 of IE62, respectively. Deletion of this 20-amino-acid sequence in IE62 resulted in no significant difference in the ability of the protein to transactivate both model and native promoters. However, alteration of the architecture of the ORF28/29 regulatory element resulted in less efficient activation by IE62 carrying the deletion than by that carrying the wild-type protein. Finally while both IE62 and USF increased the level of TBP/TFIID binding to DNA fragments containing specific promoter elements, they showed no ability to increase each other's binding. Thus, it is possible that TBP/TFIID may be part of a molecular bridge between USF and IE62. These findings suggest a novel mechanism for the observed synergistic activation which requires neither site-specific IE62 binding to the promoter nor a direct physical interaction with USF.
MATERIALS AND METHODS
Reporter constructs.
The dual luciferase bidirectional reporter vectors pRFL/WT and pRFL/USFm have been described previously (55). The chimeric TA-Luc reporter vectors were constructed based on the pGL-2 basic vector (Promega, Madison, WI). The TATA element and flanking sequence derived from the adenovirus major late promoter were inserted between the XhoI and HindIII sites of the pGL-2 vector to generate the pTALuc minimal reporter vector. The flanking region was mutated in order to eliminate a functional Maz/Sp1 site immediately downstream of the TATA element. The sequence inserted was 5′-GGCTATAAAAGGAAGCTCGGAGCCGTTCGTCCTC-3′. The USF binding site (5′-GTAATCACGTGATTTGT-3′) derived from the VZV ORF28/29 regulatory element was inserted between the KpnI and MluI sites in the pTALuc vector, 25 bp upstream of the TATA box. The TATA element and the core consensus USF site are underlined. The pUSFmTALuc, pUSFTAmLuc, and pUSFmTAm reporter vectors were generated by site-specific mutation of pUSFTALuc at the USF site, TATA element, or both, respectively. The base substitutions in the USF site in these vectors are identical to those in the pRFL/USFm reporter vector (5′-GTAATCACGCTCTTTGT-3′; the mutant USF site is underlined, and the specific base substitutions are in boldface type). In the vectors carrying the TATA element mutation the sequence 5′-TATAAA-3′ was replaced with 5′-AACGCTT-3′. The pG1TALuc vector was constructed by inserting a single copy of the Gal4 binding motif into the MluI site in the pTALuc vector. The Gal4 binding motif is the 17-mer palindrome consensus sequence CGGAGGACAGTACTCCG (7).
Expression constructs for transient transfections.
The cloning of the pCMV62 plasmid expressing wild-type IE62 under the control of the cytomegalovirus immediate-early (IE) promoter was described previously (31, 33). The IE62 gene was derived from the EcoRI E fragment of the genome of the low-passage-number North American clinical isolate Scott (47). The pCMV62d20 plasmid was derived from the pCMV62 plasmid by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, Tex.) to delete aa 238 to 258 within IE62.
The DNA sequence encoding full-length USF1 (aa 1 to 310) was derived by reverse transcription-PCR using total RNA extracted from MeWo cells (C. Grose, University of Iowa) and then cloned into the pQE-tri vector between the EcoRI and XhoI sites to produce the pQE-USF1 vector. RBUSF1 and BUSF1, which contain aa 155 to 310 and aa 197 to 310 of USF1, respectively, were PCR amplified from full-length USF1 and cloned into the same restriction sites to create the pQE-RBUSF1 and pQE-BUSF1 vectors.
The pGU series of plasmids expressing Gal4-USF fusion proteins were constructed based on the pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA), within which the Gal4 DNA binding domain (aa 1 to 147) was inserted between the HindIII and BamHI sites and the USF1 fragments were inserted between the BamHI and EcoRI sites. pGU/FL contains the full-length USF1 coding sequence (aa 1 to 310). pGU/AD contains the sequence of the activation domain of USF1 (aa 1 to 156). pGU/AR contains the sequence of the activation domain and USR of USF1 (aa 1 to 197). pGU/RB contains the sequence of the USR and DNA binding domain of USF1 (aa 157 to 310), and pGU/B contains the sequence of the DNA binding domain of USF1 (aa 198 to 310). The structures of the USF fusion constructs are summarized in Table 1.
TABLE 1.
Summary of USF fusion constructs
| Construct | Composition (amino acids)
|
|
|---|---|---|
| N terminus | C terminus | |
| pQE-USF1 | Full-length USF1 1-310 | His tag |
| pQE-RBUSF1 | RBUSF1 (USR and bHLH-Zip) 155-310 | His tag |
| pQE-BUSF1 | BUSF1 (bHLH-Zip) 197-310 | His tag |
| pGU/FL | Gal4 DNA binding domain 1-147 | Full-length USF1 1-310 |
| pGU/AD | Gal4 DNA binding domain 1-147 | USF1 AD 1-156 |
| pGU/AR | Gal4 DNA binding domain 1-147 | USF1 AR (AD and USR) 1-197 |
| pGU/RB | Gal4 DNA binding domain 1-147 | RBUSF1 (USR and bHLH-Zip) 157-310 |
| pGU/B | Gal4 DNA binding domain 1-147 | BUSF1 (bHLH-Zip) 198-310 |
| pTrc-USF1/AD | USF1 1-155 | His tag |
| pTrc-USF1/AR | USF1 1-197 | His tag |
| pHF2 | His tag | USF1/ΔN 105-310 |
Expression vectors used in protein pull-down assays.
Full-length His-USF1 and His-USF-1/BD (197-310) were expressed from the pQE plasmids described above. The pTrcHis2C vector (Invitrogen, Carlsbad, CA) was used to express His-USF1 (1-155) and His-USF1 (1-197) by inserting the corresponding USF1 coding sequence between the NcoI and EcoRI sites present in the parental plasmid, resulting in the pTrc-USF1/AD and pTrc-USF1/AR constructs, respectively. The pHF2 plasmid expressing His-USF1/ΔN (aa 105 to 310) was the gift of Michele Sawadogo (M. D. Anderson Cancer Institute).
The pGEX-IE62 series was constructed using the pGEX-4T-3 plasmid (Amersham Biosciences, Piscataway, NJ) to encode the N-terminal portions of the VZV IE62 protein with an N-terminal GST tag. pGEX-IE62 (1-238), pGEX-IE62 (1-248), pGEX-IE62 (1-258), pGEX-IE62 (1-268), pGEX-IE62 (1-278), and pGEX-IE62 (1-288) were generated by inserting each of the respective IE62 coding sequences between the BamHI (EcoRI for 1-238 only) and SalI sites in the pGEX-4T-3 vector. pGEX-IE62 (1-299d10) and pGEX-IE62 (1-299d20) were derived from pGEX-IE62 (1-299) by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) to delete 20 amino acids (238 to 258) in the IE62 gene. Generation of pGEX-IE62 (1-43), pGEX-IE62 (1-226), pGEX-IE62 (1-299), and pGEX-IE62 (1-406) was previously described by Spengler et al. (46). The pGST-IE62AD, pGST-USF1AD, and pGST-VP16AD expression plasmids were constructed using the pGEX-4T-3 plasmid. Sequences coding for the N-terminal 107 amino acids of the IE62 protein were inserted between the BamHI and SalI sites. Sequences encoding the USF1 activation domain (aa 1 to 155) and the VP16 activation domain (aa 410 to 490) were inserted between the BamHI and XhoI sites.
Purification of recombinant proteins.
Escherichia coli DH5α transformed with pHF2 expressing His-USF1/ΔN or pQE-USF1 expressing His-USF1 was grown in 250-ml LB broth cultures and induced with 0.1 or 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), respectively. Three hours postinduction, cells were pelleted and resuspended in 10 ml lysis buffer (20 mM HEPES, pH 7.9, 500 mM NaCl, and 10 mM imidazole). The His-tagged proteins were purified using a HisTrap kit following the manufacturer's instructions (Amersham Biosciences, Uppsala, Sweden). Protein eluates were then loaded on a PD-10 desalting column (Amersham Biosciences, Uppsala, Sweden) and eluted with buffer D (20 mM HEPES, pH 7.9, 100 mM KCl, 20% glycerol, and 0.2 mM EDTA). The VZV IE62 protein was expressed in recombinant baculovirus and purified from infected cell cultures as previously described (46). GST-IE62 (1-299) was expressed and purified via glutathione-agarose chromatography as previously described (46). Purified proteins were stored at −80°C prior to use.
Transient-transfection and reporter gene assays.
Transient-transfection assays were performed as previously described (55) in a human melanoma cell line (MeWo) that supports VZV replication. Transfections were performed in triplicate using 12-well plates. Briefly, 2 × 105 cells were seeded in each well 24 h before transfection. One microgram of reporter vector and 0.02 μg of IE62-expressing plasmid or 0.5 μg of USF fusion protein-expressing plasmid were transfected in each assay with Lipofectamine reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Various amounts of the empty cloning vector, pcDNA, were transfected along with the pCMV62 and pGU plasmids to equalize the amount of both total DNA and CMV promoter in each set of transfections. The empty pQE-tri vector was used in the same way in transfections with the USF-expressing plasmids. The pCMV · SPORT · βGal vector (Gibco, Carlsbad, CA) was used as an internal control reporter for transfections with the pRFL reporter vector. The pEF1α-RL vector expressing Renilla luciferase under the control of the EF1α promoter was used as the internal control reporter for transfections with the Luc reporter vectors. Cells were collected 48 h posttransfection and were lysed in lysis buffer (50 mM HEPES, pH 7.4, 250 mM NaCl, 1% NP-40, 1 mM EDTA). Dual luciferase activities were normalized to the beta-galactosidase activities. Firefly luciferase activities were normalized to the Renilla luciferase activities. Transfection experiments were repeated at least three times, and each set of transfection conditions in a given experiment was used in triplicate. The data from representative experiments are presented as the means of triplicate transfections. Error bars indicate standard deviations. Statistical significance was determined by a one-way analysis of variance test followed by Tukey's post hoc test.
EMSA.
A double-stranded DNA oligomer (IDT, Coralville, IA) 22 bp in length was used in the electrophoretic mobility shift assays (EMSAs). The wild-type USF probe contained the wild-type USF binding motif (underlined) with the flanking sequence derived from the ORF28/29 regulatory element: GTGTAATCACGTGATTTGTTG. The duplex oligomer was end labeled with [γ-32P]ATP using T4 kinase (Invitrogen, Carlsbad, CA). His-USF1/ΔN was purified as described above. Recombinant human Sp1 (rhSp1) was obtained from Promega (Madison, WI).
One hundred femtomoles of the labeled probes (1 × 105 cpm) was incubated with 20 ng of purified protein in a 20-μl reaction mixture containing 40 mM HEPES, pH 7.9, 100 mM NaCl, 10 mM MgCl2, 200 μg/ml bovine serum albumin (BSA), 12% glycerol, 0.05% NP-40, 1 mM dithiothreitol, and 1 μg poly(dI-dC). Anti-USF1 and anti-Sp1 antibodies were obtained from Santa Cruz Biologicals (Santa Cruz, CA). The samples were analyzed by electrophoresis on a 5% polyacrylamide (37.5:1 acrylamide/bisacrylamide) gel followed by autoradiography.
Magnetic bead recruitment assays.
Nuclear extracts of uninfected and infected MeWo cells were prepared as previously described (55). Biotinylated oligomers containing the relevant promoter sequences were generated by PCR using 5′-end-biotinylated sense-strand primer (5′-biotinylated promoter) or antisense-strand primer (3′-biotinylated promoter). The 5′-biotinylated ORF28/29 promoter oligomer was the 221-bp intergenic region that is inserted in the pRFL/WT vector. The 3′-biotinylated 132-bp USF-TATA promoter was derived from the pUSFTALuc vector by PCR using a GL-1 primer and 5′-biotinylated GL-2 primer. The 3′-biotinylated USFm-TATA promoter was PCR amplified from the pUSFmTALuc vector using the same set of primers as that for the biotinylated USF-TATA promoter.
Magnetic bead recruitment assays were performed as previously described (3, 21). Briefly, 10 pmol of 5′- or 3′-biotinylated promoter was conjugated to 50 μl Dynabeads M-280-streptavidin (Dynal, Oslo, Norway). The promoter-coupled beads were blocked with 100 μl blocking buffer (20 mM HEPES, pH 7.9, 100 mM KCl, 8 mM MgCl2, 10 μM ZnSO4, 0.1 mM EDTA, 0.01% Triton X-100, and 50 mg/ml BSA) and then incubated with 250 μg nuclear protein extract derived from MeWo cells in 50 μl binding buffer (20 mM HEPES, pH 7.9, 100 mM KCl, 8 mM MgCl2, 10 μM ZnSO4, 0.1 mM EDTA, 0.01% Triton X-100) supplemented with 5 μg/ml heparin and 2 μg poly(dI-dC) at room temperature for 30 min. The beads were subjected to three washes with 400 μl binding buffer, and bound proteins were eluted in 50 μl 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and analyzed by SDS-PAGE followed by immunoblotting. The presence of the USF1 and IE62-USF1 fusion proteins was determined using the anti-USF1 (C-20) antibody (Santa Cruz). The polyclonal anti-IE62 antibody was previously described (46). Monoclonal anti-TBP antibody was obtained from Neoclone (Madison, WI).
His-tag protein affinity pull-down assays.
Full-length and truncated His-tagged USF1 fusion proteins were expressed in E. coli BL21, and the lysates were used in protein affinity pull-down assays in conjunction with purified IE62 and GST-IE62 (1-299) as previously described (37). Bound proteins were eluted with 50 μl 2× SDS-PAGE sample buffer by being boiled for 5 min and then analyzed by SDS-PAGE and immunoblotting.
GST-tag protein affinity pull-down assays.
Following induction with IPTG crude lysates of E. coli expressing GST and GST-IE62 fusions were prepared and clarified as previously described (30). Two-hundred-microliter aliquots of the bacterial lysates were added to 50 μl glutathione-Sepharose beads and incubated for 1 h at 4°C. In the capture assay with purified recombinant protein, 100 ng His-USF1/ΔN with 1 mg/ml BSA in 200 μl buffer D (20 mM HEPES, pH 7.9, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) was added to the protein-coupled beads. Following repeated washings with 0.1% Triton X-100 in buffer D, bound protein was eluted with 50 μl 2× SDS-PAGE sample buffer by being boiled for 5 min and then analyzed by SDS-PAGE and immunoblotting. In assays with nuclear extracts, 50 μl extract (15 μg/μl protein) in 250 μl buffer D was added to the protein-coupled beads. Incubation was performed for 3 h at 4°C and followed by the washing and elution steps described above.
Coimmunoprecipitation assays.
MeWo cells were grown in 12-well plates and transfected with 0.02 μg pcDNA, 0.02 μg pCMV62, or 0.01 μg pCMV62d20 per well. Cells were lysed 48 h posttransfection with the addition of 200 μl lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40). Lysates from three wells transfected with the same plasmid were pooled, and coimmunoprecipitations were performed with monoclonal anti-IE62 antibody coupled to protein G-Sepharose 4 Fast Flow beads (GE Healthcare Bio-Sciences, Uppsala, Sweden) as previously described (30). Bound proteins were eluted by being boiled in 2× SDS-PAGE sample buffer, separated by SDS-PAGE, and analyzed by immunoblotting.
RESULTS
USF can synergize with IE62 to activate model minimal promoters.
The cellular transcription factor USF has previously been shown to mediate IE62 activation of native VZV promoters (23, 30). USF binding sites have been predicted to be present in a number of VZV genes, and the physiological significance of USF-mediated IE62 activation in VZV replication has been well documented (37, 39). To explore the functional interaction of the VZV IE62 protein and USF in terms of promoter activation, model firefly luciferase reporter vectors (Fig. 1A) were generated containing either a consensus binding site for USF fused 25 bp upstream of the TATA element (TATAAAA) derived from the ADMLP or the TATA element alone. The IE62 protein has been previously shown to be capable of activating expression from reporter plasmids containing only this consensus TATA element (32). Thus, these model promoters should represent the minimal cis-acting elements required for IE62 activation in the presence and absence of USF, respectively.
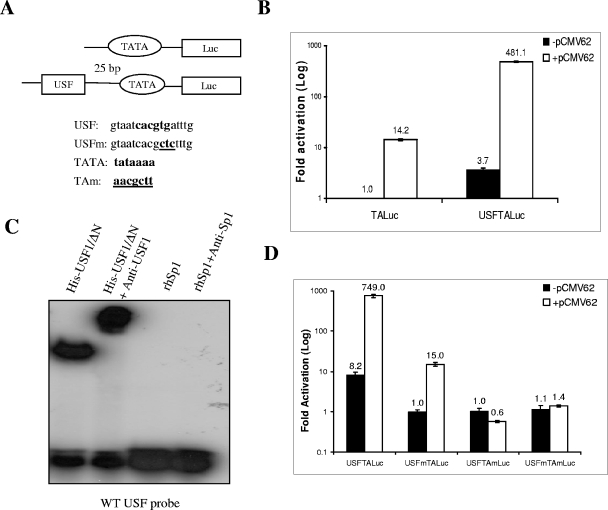
FIG. 1.
Synergistic IE62-USF activation of a model minimal promoter. (A) Schematic of the model luciferase reporter vector, pUSFTAluc, indicating the relative positions and sequences of the USF site and TATA element. Consensus binding motifs are shown in boldface. Mutations are underlined. (B) Results of luciferase assays. One microgram of each reporter plasmid including the basic pTALuc plasmid, which lacks the USF binding site, was cotransfected with or without 0.02 μg of the IE62-expressing plasmid, pCMV62, into MeWo cells. The luciferase activity expressed from the pTALuc reporter in the absence of IE62 was normalized to 1. The promoter activities resulting from the presence of USF, IE62, or both are reported as induction (n-fold) of the luciferase activity over the pTALuc level. The open and solid bars represent promoter activity in the absence and presence of IE62, respectively. (C) Results of EMSAs confirming that recombinant USFΔN binds to the consensus binding site inserted into pUSFTALuc and that the complex is supershifted by anti-USF1 antibody. Recombinant Sp1 and anti-Sp1 antibody were used as negative controls. (D) Control transfection assays showing the requirement of the TATA element for both USF and IE62 activation. These results were normalized to the activity observed with the pUSFTALuc reporter in the absence of IE62. Luciferase assay data in panels B and D represent the averages of triplicate transfections. The average values are shown above each bar, and the error bars represent standard deviations.
In the first series of experiments, we wished to determine if this cellular factor could synergize with IE62 in the context of the ADMLP TATA element and to determine the extent of the individual contributions of IE62 and USF to activation of the promoter in each other's absence. The minimal reporter vector pTALuc, containing only the TATA element, and the chimeric reporter vectors were cotransfected with an IE62-expressing plasmid, pCMV62, into MeWo cells, a melanoma cell line that supports productive VZV infection. The results of the transient-transfection assays are shown in Fig. 1B. The luciferase activity obtained with the pTALuc reporter in the absence of IE62, representing the basal level of core promoter activity, was normalized to 1. IE62-regulated promoter activity and the activities of the promoter containing the USF binding site in both the absence and presence of IE62 are reported as induction (n-fold) of luciferase activities in reference to this basal activity. Addition of the USF site to the minimal promoter containing only the TATA element resulted in an increase of 3.7-fold in the absence of IE62. In comparison IE62 activated the minimal pTALuc reporter by 14.2-fold. Thus, a maximum of 17- to 18-fold activation would be expected if the contributions of USF and IE62 were additive. In marked contrast to this prediction, the presence of IE62 activated the model promoter by 500-fold, indicating synergistic activation some 28- to 29-fold greater than the additive contributions.
The binding of USF was examined by EMSA using a 22-bp duplex DNA oligomer containing the USF site present in the reporter vector. As shown in Fig. 1C, a USF binding complex was observed with the probe in the presence of a purified recombinant USF protein containing aa 105 to 310 of USF1, which encompass a portion of the activation domain and the complete USR and bHLH-Zip domains of USF1. Similar results have previously been reported in studies using MeWo cell nuclear extracts, which also showed that USF binding was virtually abolished in the context of a probe containing the USFm mutation (23, 37). Thus, the binding of USF correlated with the increased activity of the promoter in the absence of IE62.
A second set of experiments examined the dependence of USF and IE62 activation on the presence of a functional TATA element within the promoter. Three additional reporter vectors were constructed using the pUSFTA-Luc sequence. One (pUSFmTALuc) contained a mutant USF binding site, the second (pUSFTAmLuc) contained a mutant TATA element, and the third (pUSFmTAmLuc) contained both mutant USF and TATA elements (Fig. 1A). As shown in Fig. 1D (where the data were normalized to the activity observed with the USFmTALuc reporter in the absence of IE62), a functional TATA element was necessary for both basal and synergistic activation by USF and IE62. Thus, TBP binding to a specific site within the promoter appears to be a requirement for USF- and IE62-mediated activation.
Very similar levels of activation (14.2-fold versus 15-fold) in the presence of IE62 were observed with the TALuc and the USFmTALuc reporters, respectively, indicating that the mutation of the USF binding site completely ablated USF activity. Further, while the basal levels of the USFTALuc expression in the two experiments differ by approximately a factor of 2, the relative increase seen in the presence of IE62 is of the same order of magnitude: 130-fold in Fig. 1B and 91-fold in Fig. 1D. These data indicate that the effect of the presence of IE62 is superimposed on the basal level of transactivation in its absence.
IE62/USF synergy at the VZV ORF28/29 regulatory element.
A consensus USF binding site has been shown to be essential for IE62-mediated activation of the two divergent, overlapping unidirectional promoters contained within the VZV ORF28/29 intergenic regulatory element (Fig. 2A), which controls expression of the viral DNA polymerase (ORF28) and major single-stranded DNA binding protein (ORF29) (25, 55). Both of these promoters contain atypical TATA elements that are also required for IE62 activation. An Sp1 binding site located within the unique portion of the ORF28 promoter plays a secondary role in the context of the complete regulatory element to further augment IE62-mediated activation of both genes, based on work with bidirectional reporter plasmids (55). In the next series of experiments, we wished to determine the extent of synergy between USF and IE62 at this complex native viral regulatory element.
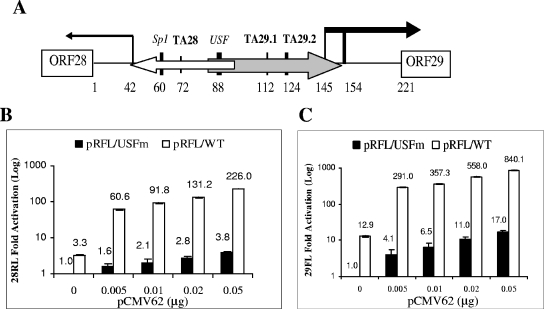
FIG. 2.
Synergistic IE62-USF activation of the VZV ORF28/29 regulatory element. (A) Schematic of the VZV ORF28/29 regulatory element showing authenticated transcription factor binding sites and TATA elements. The locations of the two overlapping minimal promoters are shown as open (ORF28) and gray (ORF29) arrows, respectively. The difference in thickness reflects their levels of transcription efficiency in the presence of IE62. The vertical lines capped by an arrow indicate the positions of transcription start sites. The difference in thickness of the two bold vertical lines over the ORF29 gene transcription start sites indicates that one is preferentially utilized (55). (B) Results of transfection experiments showing expression of Renilla luciferase (ORF28 position) activity from the wild-type (open bars) and mutant USFm (solid bars) dual luciferase reporter plasmids in the presence of increasing amounts of the pCMV62 expression plasmid. The level of Renilla luciferase activity observed with the pRFL/USFm reporter in the absence of IE62 was normalized to 1. (C) Results of transfection experiments showing expression of firefly luciferase (ORF29 position) activity from the wild-type (open bars) and USFm (solid bars) dual luciferase reporter plasmids in the presence of increasing amounts of the pCMV62 expression plasmid. The level of firefly luciferase activity observed with the pRFL/USFm reporter in the absence of IE62 was normalized to 1. Luciferase assay data in panels B and C represent the averages of triplicate transfections. The average values are shown above each bar, and the error bars represent standard deviations.
To answer this question, two previously described (55) dual luciferase bidirectional reporter vectors were utilized. These were pRFL/WT, with Renilla luciferase in the position of the ORF28 gene and firefly luciferase in the position of the ORF29 gene, and pRFL/USFm, which contains a mutated USF site. The basal activities of the two reporter genes in the pRFL/USFm plasmid in the absence of IE62 were normalized to 1. The activities of both pRFL/USFm and the wild-type pRFL/WT reporters in the presence of increasing amounts of pCMV62 were determined relative to this value. The results are presented in Fig. 2B, showing the activity in the direction of ORF28, and Fig. 2C, showing that in the direction of ORF29. Comparison of the activities of the relative levels of Renilla luciferase (ORF28 activity) with the wild-type and mutant reporter vectors shows that the presence of the USF site contributed a 3.3-fold increase in activation. Transfection of 0.02 and 0.05 μg of the IE62 expression plasmid resulted in expression levels 2.8- and 3.8-fold, respectively, higher than those observed with pRFL/USFm alone. Thus, the maximum additive levels expected based on the individual contributions would be 6.1 and 7.1, respectively. In contrast, the levels of expression observed with the wild-type promoter and IE62 under the same experimental conditions were 131- and 226-fold, respectively. These values represent activation 20- to 30-fold higher than the additive values and show a remarkable congruence with the levels of synergy observed with the simple model promoter.
Similar synergy was observed with activation of the firefly luciferase reporter representing the ORF29 gene. In this case, the activity from the wild-type regulatory element was 12.9-fold higher than that seen with the mutant in the absence of IE62, suggesting that either the sequence or the placement of the two TATA elements involved in the expression of the ORF29 gene is slightly more efficient than that of the ORF28 TATA element. Transfection of 0.02 and 0.05 μg of the IE62 expression plasmid resulted in expression levels 11.0- and 17.0-fold higher than those observed with pRFL/USFm alone. Once again, in contrast to predicted additive values (23.9- and 29.9-fold), the levels of expression observed with the wild-type promoter and IE62 under the same experimental conditions were 558- and 840-fold higher, respectively, values some 23 and 28 times higher than predicted. Thus, equivalent levels of synergistic activation mediated by the presence of IE62 and USF were observed on both model and native promoters.
The activation domain of USF1 is necessary and sufficient to mediate synergy with IE62.
In order to shed light on the mechanism of USF synergism with IE62, the region of USF1 that is required for synergy with IE62 was investigated. USF1 was chosen out of the two USF isoforms, since previous work showed that USF1 was the predominant form of USF bound to the ORF28/29 regulatory element (23, 24). The initial approach was to examine the effect of exogenously expressed full-length USF1 and USF1 fragments (Fig. 3A) containing the C-terminal DNA binding and dimerization domains with various extensions towards the N-terminal activation domain on endogenous USF-mediated IE62 activation. All of these ectopically expressed USF1 proteins would be predicted to compete with endogenous USF for binding to the target promoter, and truncations that remove the region of USF essential for IE62 activation would result in a dominant-negative effect in reporter assays.
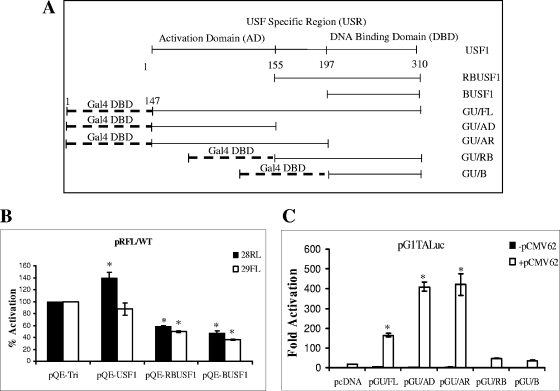
FIG. 3.
Identification of the region of USF1 that is involved in mediating IE62 activation. (A) Schematic depiction of the USF1 fragments expressed via pQE-tri plasmid and the Gal4-USF fusion proteins with the DNA binding domain (DBD) of Gal4 fused with different fragments of the USF1 protein. (B) Examination of the effect of ectopic expression of the full-length and truncated USF1 proteins on IE62 activation of the VZV ORF28/29 regulatory element. One microgram pRFL/WT reporter vector and 0.02 μg pCMV62 plasmid were cotransfected with 0.5 μg each of the USF1-expressing plasmids and the control vector pQE-tri. The solid bars represent the promoter activities in the direction of ORF28, and the open bars represent that in the direction of ORF29. The endogenous USF1-mediated IE62 activation of the individual luciferase reporter genes in the presence of the empty pQE-tri plasmid was normalized to 100%. (C) Analysis of the Gal4-USF1 fusion proteins in mediation of IE62 activation of the pG1TALuc vector. One microgram pG1TALuc reporter vector and 0.02 μg pCMV62 plasmid were cotransfected with 0.5 μg each of the Gal4-USF1 fusion protein-expressing plasmids and the control vector pcDNA as indicated in the figure. Open and closed bars represent activity in the presence and absence of IE62, respectively. Luciferase assay data in panels B and C represent the averages of triplicate transfections. The error bars represent standard deviations. *, P < 0.05.
The pQE-USF1, pQE-RBUSF1, and pQE-BUSF1 plasmids (Table 1) were cotransfected with the pCMV62 plasmid and the pRFL/WT reporter into MeWo cells. As shown in Fig. 3B, overexpression of full-length USF1 did not significantly alter IE62 activation of the reporter in the ORF29 position whereas expression of the Renilla luciferase reporter in the ORF28 position was increased by 1.4-fold. While this difference was statistically significant, it is quite small relative to other differences in activity reported here. Similar slight increases (1.5-fold) with a reporter construct containing the ORF28 promoter driving luciferase expression were reported by Qyang et al. (36) upon ectopic expression of USF1. This modest increase in expression suggests that the ectopically expressed full-length USF1, probably by increased overall concentration, compensates for the lower efficiency of IE62-driven expression from the ORF28 side versus the ORF29 side of the bidirectional regulatory element seen with endogenous levels of USF.
In contrast, cotransfection of both pQE-RBUSF1 and pQE-BUSF1 reduced the level of IE62 stimulation, suggesting that the activation domain of USF1, which is absent in RBUSF1 and BUSF1, plays a significant role in mediating IE62 activation. To further identify the domain or domains of USF1 that functionally interact(s) with IE62 and to eliminate interference from endogenous USF in the interaction of IE62 with the truncated USF1 constructs, Gal4-USF fusion constructs (GU) expressing the Gal4 DNA binding domain (aa 1 to 147) fused to various domains of USF1 were created (Table 1). As diagrammed in Fig. 3A, pGU/FL contained full-length USF, pGU/AD contained only the N-terminal activation domain, pGU/AR contained the activation domain and the USR, pGU/RB contained the USR and DNA binding domains, and pGU/B contained only the DNA binding domain. A chimeric reporter plasmid (pG1TALuc) containing a single copy of the 17-nucleotide consensus Gal4 binding site was constructed to allow evaluation of the activities of the Gal4-USF1 fusion proteins. The Gal4 binding site was inserted 25 bp upstream of the TATA box, mimicking the position of the USF binding site in the minimal model promoters.
The results of cotransfections with the reporter plasmid and the individual Gal4-USF1 effector plasmids are presented in Fig. 3C. IE62 alone activated expression of the reporter 17-fold over basal expression, in good agreement with the level of activation observed with the TALuc and USFmTALuc reporters. A 10-fold increase in activation above that seen with IE62 alone was observed when both IE62 and the GU/FL fusion protein were present. The GU/AD fusion, containing only the activation domain of USF1, mediated IE62 activation to a higher level (24-fold above IE62 alone) than did the full-length USF1. The GU/AR fusion supported a similar level of activation. In contrast, the presence of the GU/RB and GU/B fusions containing only the bHLH-Zip domain of USF1 resulted in levels of activation which were only two- to threefold above those with IE62 alone. Thus, the activation domain of USF1 appears to be both necessary and sufficient to mediate significant IE62 activation.
Mapping of the regions of IE62 and USF responsible for direct physical interaction.
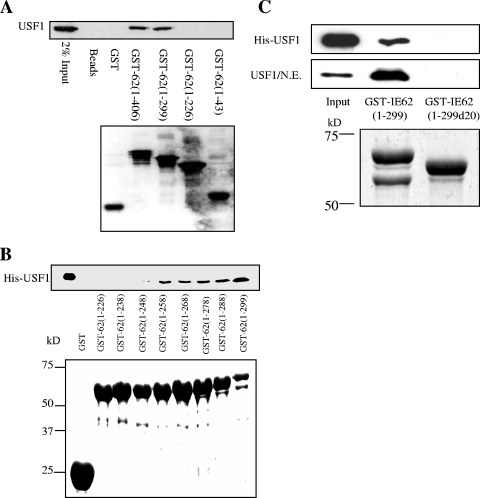
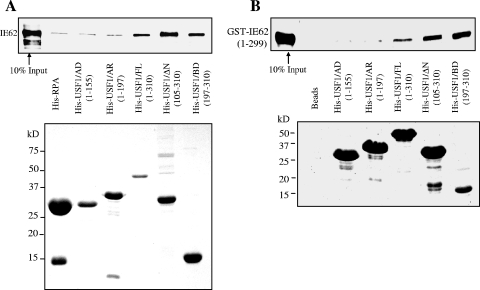
A direct physical interaction has been demonstrated to be able to occur between USF and IE62, and such an interaction has been suggested to be part of the mechanism of joint USF-IE62 activation of promoters (23-25, 37). We next wished to determine if a direct physical interaction is involved in synergistic USF-IE62 activation. Previous work from this laboratory has shown that both USF and Sp1 are capable of binding to IE62 in the absence of other viral proteins (30, 37). Amino acids 226 to 299 of IE62 were shown to be necessary for the interaction with Sp1 in protein pull-down experiments. The same set of GST-IE62 fusion proteins containing N-terminal fragments of IE62 utilized in that study, GST-IE62 (1-43), GST-IE62 (1-226), GST-IE62 (1-299), and GST-IE62 (1-406), was used in initial screens utilizing MeWo cell nuclear extracts and showed that USF1 also binds to this region of IE62 (Fig. 4A).
FIG. 4.
GST pull-down analysis of the region of IE62 that interacts with USF1. (A) Preliminary mapping of the region of IE62 that interacts with USF. Truncated GST-tagged IE62 proteins were coupled to glutathione Sepharose beads and incubated with nuclear extracts of MeWo cells. GST alone was used as a control. The binding of USF to the GST-IE62 fusions was examined by Western blotting (upper panel). The lower panel shows a Western blot assay using anti-GST antibody documenting the levels of the GST and GST-IE62 fusion proteins eluted from the beads. (B) Fine mapping of the USF binding region of IE62 using a series of progressive 10-amino-acid C-terminal deletions of GST-IE62 (1-299). The upper panel shows an immunoblot analysis of the level of purified HIS-USF1 binding. The lower panel is a Coomassie blue stain showing the levels of the GST-IE62 fusions eluted from the glutathione beads. (C) Binding of recombinant full-length USF1 and USF1 present in MeWo cell nuclear extracts to GST-IE62 (1-299) and GST-IE62 (1-299d20). The top two panels are an immunoblot analysis of bound USF1. The bottom panel is a Coomassie blue stain showing the levels of the two fusion proteins eluted from the glutathione beads.
To fine-map the region of IE62 required for interaction with USF1, a second set of GST-IE62 fusions was constructed in which 10 amino acids were successively removed from the carboxy terminus of GST-IE62 (1-299), ultimately yielding GST-IE62 (1-239). These GST fusion proteins along with GST-IE62 (1-226) were then tested for USF1 binding in protein pull-down assays. Purified recombinant His-USF1 bound equivalently to all of the GST-IE62 fusions through GST-IE62 (1-258). Markedly reduced binding was observed with GST-IE62 (1-248), and no binding was observed with GST-IE62 (1-238) and shorter fusions, suggesting that amino acids 238 to 258 of IE62 are critical for USF1 binding (Fig. 4B). Similar results were obtained with purified His-USF1/ΔN, an N-terminal truncation lacking the first 104 amino acids of USF1, and USF1 present in MeWo cell nuclear extracts (data not shown). The essential role of this region of IE62 in binding USF1 was confirmed by deletion mutagenesis, in which amino acids 238 to 258 were deleted from the GST-IE62 (1-299) construct. The ability of the GST-IE62 fusion protein carrying this deletion to interact with USF1 was examined in GST pull-down assays using purified full-length His-tagged USF1 and MeWo cell nuclear extracts. As shown in Fig. 4C, the 20-amino-acid deletion completely ablated the interaction of USF1 with IE62 in the context of both purified proteins and nuclear extracts. The data obtained with bacterially expressed IE62 fragments and bacterially expressed USF1 further indicate that no other viral or eukaryotic cellular factors are required for this interaction.
In order to map the region(s) of USF1 required for or involved in binding of IE62, protein affinity pull-down assays were performed using His-tagged, bacterially expressed full-length USF1, His-tagged fragments of USF1, and purified, recombinant IE62. The USF1 fragments included His-USF/AD (aa 1 to 155), which contained the entire N-terminal activation domain; His-USF/AR (aa 1 to 197), containing the activation domain and USR; His-USF/ΔN (aa 105 to 310); and His-USF1/BD (aa 197 to 301), which contains only the bHLH-Zip domain at the C terminus of USF1 (Table 1). His-tagged RPA14, a component of the heterotrimeric human replication protein A (RPA) complex, was coexpressed in E. coli with the RPA32 subunit and served as a negative control (37). As shown in Fig. 5, neither His-USF1/AD nor -AR was able to pull down IE62 above levels observed in controls. However, USF1/FL, -ΔN, and -BD all clearly interacted with IE62 at levels above background.
FIG. 5.
His-tagged protein affinity pull-down analysis of the region of USF1 that interacts with IE62. (A) E. coli-expressed full-length and truncated His-USF1 proteins were coupled to Ni-nitrilotriacetic acid magnetic agarose beads and incubated with recombinant IE62 protein. His-RPA14 was coexpressed with RPA32 in E. coli and used as a negative control. The binding of IE62 was examined by immunoblotting (upper panel). Coomassie blue staining shows the levels of the bound His-tagged proteins present on the beads (lower panel). (B) His-USF protein pull-down assays using purified bacterially expressed GST-IE62 (1-299) fusion protein. The binding of GST-IE62 (1-299) was examined by immunoblotting with anti-GST antibody (upper panel). Coomassie blue staining shows the levels of the bound His-tagged proteins present on the beads (lower panel).
To exclude the possibility that insect cell proteins that potentially copurify with the full-length IE62 interfered with or altered the interactions between IE62 and the USF1 fragments, a second series of His-USF1 affinity assays was performed using purified GST-IE62 (1-299). The results are presented in Fig. 5B and are essentially identical to those obtained with the full-length baculovirus-expressed IE62, indicating that no contaminating proteins from either source of IE62 influence the results. These data also indicate that no additional sequences beyond amino acids 1 to 299 of IE62 are involved in the interaction in vitro. The bHLH-Zip domain of USF1 is, therefore, the minimal domain required for interaction with IE62. This domain maps outside of the region that functionally interacts with IE62, suggesting that USF1 and IE62 can cooperate in a mechanism that does not require direct physical interaction.
Direct physical interaction is dispensable for synergistic promoter activation by IE62 and USF.
Although the domain required for physical interaction between USF1 and IE62 does not correspond to the functionally required domain, the possibility of a physical interaction augmenting the functional interplay between USF1 and IE62 in situ remained. Moreover, the functional mapping data were obtained using the artificial Gal4-USF fusions and a model promoter, leaving open the question of whether a direct physical interaction between intact IE62 and USF is required for synergistic activation of native viral promoters. We therefore investigated these questions in the context of the ORF28/29 regulatory element.
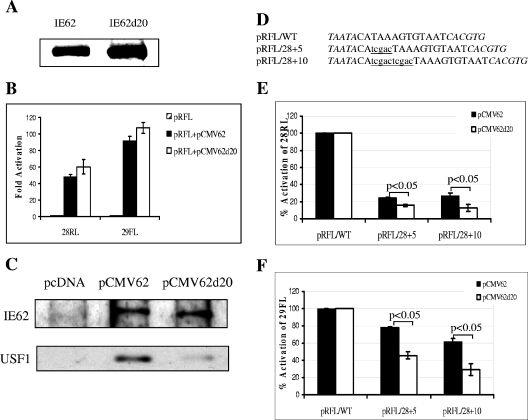
An effector plasmid, pCMV62d20, was generated which expressed IE62 carrying the aa 238 to 258 deletion. As shown in Fig. 6A, the mutant IE62, IE62d20, was readily expressed in MeWo cells. Expression levels of IE62d20 were consistently somewhat higher than those for wild-type IE62 using equivalent amounts of the two expression plasmids. The transactivating activity of the IE62 deletion mutant was then assessed in transient-transfection assays using the pRFL/WT reporter. IE62d20 was found to be as efficient as wild-type IE62 in activation of the promoter mediated through the native USF binding site (Fig. 6B). The somewhat higher level of activation seen with the mutant protein is most likely due to the above-mentioned greater levels of IE62d20 expression. Experiments with the model USFTALuc reporter also showed no loss in the transactivation activity of IE62d20 compared to IE62 (data not shown).
FIG. 6.
Transactivation activity of IE62d20 mediated by USF. (A) Immunoblot analysis using polyclonal anti-IE62 antibody to detect expression of IE62 and IE62d20 in MeWo cells. Sixteen micrograms of pCMV62 and pCMV62d20 plasmids was transfected into MeWo cells in 100-mm petri dishes. Forty-eight hours posttransfection, cell extracts of the transfected MeWo cells were isolated and resolved by SDS-PAGE. (B) Results of transient-transfection assays. One microgram of pRFL/WT and pRFL/Sp1sub reporter vector was cotransfected with or without 0.02 μg pCMV62 and pCMV62d20 into MeWo cells. Striped bars represent the luciferase activities derived from the reporter vector alone, which were normalized to 1. Solid bars represent the wild-type IE62-mediated activities, and the open bars represent the IE62d20-mediated activities, both of which are reported as induction (n-fold) of the luciferase activity over the basal level. Data represent the averages of triplicate transfections. The error bars indicate standard deviations. (C) Results of coimmunoprecipitation experiments using anti-IE62 monoclonal antibody and extracts derived from cells transfected with pcDNA, pCMV62, and pCMV62d20. Detection of the levels of wild-type and mutant IE62 and USF1 was performed by immunoblotting using rabbit polyclonal antibodies against the respective proteins. (D) Positions and sequences of the 5- and 10-bp insertions between the ORF28 TATA element and the USF site within the ORF28/29 regulatory element. The TATA element and USF site are shown in italics. The insertions are shown in lowercase and underlined. The designations of the resulting dual luciferase reporter vectors are listed at the left. (E) Results of transfection assays showing the effects of the insertions on transactivation by IE62 (solid bars) and IE62d20 (open bars) in the context of the Renilla luciferase (ORF28) reporter. (F) Results of transfection assays showing the effects of the insertions on transactivation by IE62 (solid bars) and IE62d20 (open bars) in the context of the firefly luciferase (ORF29) reporter. Data in panels E and F represent the averages of triplicate transfections. The error bars indicate standard deviations. Statistical significance of the differences observed with IE62 and IE62d20 was determined by one-way analysis of variance followed by Tukey's post hoc test.
These results raised the possibility that IE62d20 could interact with USF1 to the same extent as wild-type IE62 upon expression of the two IE62 molecules in eukaryotic cells. Such an interaction might occur via a second domain, separate from that identified in the in vitro experiments, that became available following posttranslational modification of IE62. In order to explore this possibility, MeWo cells were transfected with the pCMV62 and pCMV62d20 plasmids and the expressed IE62 molecules were immunoprecipitated with monoclonal anti-IE62 antibody. The precipitates were then analyzed for the presence of IE62 or IE62d20 and USF1 by immunoblotting. The results are presented in Fig. 6C and show that while equivalent amounts of IE62 and IE62d20 were precipitated, very low levels of USF1 were coprecipitated with IE62d20 compared to wild-type IE62. Thus, the deletion of amino acids 238 to 258 disrupted the direct interaction of IE62 with USF1 in situ as well as in vitro.
The position of the USF site relative to the TATA element has been shown to be important for IE62-mediated synergy, with the optimal distance between the two elements determined by Meier et al. (23) being 24 bp. We reasoned, therefore, that the ability of IE62 to participate in synergistic activation of promoters containing USF sites at distances greater than the optimal distance could involve direct physical interaction in order to maintain a specific geometry of factors within the preinitiation complex. In order to test this hypothesis, two additional reporter vectors were generated in which 5 and 10 bp were inserted between the USF binding site and the TATA element for ORF28 in the pRFL/WT plasmid. The ability of wild-type IE62 and IE62d20 to activate these regulatory elements was then tested, and the results are shown in Fig. 6E and 6F.
The 5- and 10-bp insertions affected the ability of both wild-type IE62 and IE62d20 to transactivate the Renilla reporter gene and also affected the ability of both proteins to transactivate the firefly luciferase present in the ORF29 position, although to a lesser extent. This latter finding suggests a coordinate regulation of expression based on the overall architecture of this regulatory element. There was a greater (approximately twofold) loss of activation of both reporter genes with IE62d20 compared to wild-type IE62 upon alteration of the promoter architecture via the 5- and 10-bp insertions, and these differences were determined to be statistically significant. Thus, a direct physical interaction appears to be dispensable for synergistic activation mediated by IE62 in combination with the cellular transcription factor USF in the context of the promoters used in this study. However, this interaction does contribute to the efficiency of activation based on the relative positions of the cis-acting elements present.
IE62 and USF increase TBP/TFIID binding to promoters.
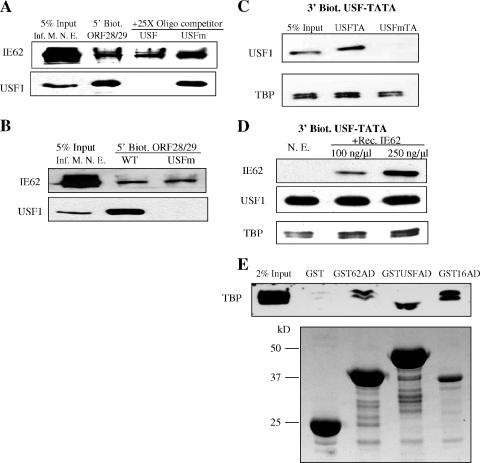
A consensus IE62 binding site (ATCGT) is rarely present in native viral promoters (38, 39) and is not present in either the native (23, 55) or model promoters used in this study. Thus, recruitment of IE62 by cellular trans-acting factors is intuitively an obvious potential component of its mechanism for promoter activation. While a direct interaction between IE62 and USF appears to be dispensable for synergistic activation based on the data presented above, it is possible that the presence of USF at the promoter could influence the interaction of other factors with IE62, thus indirectly increasing IE62 recruitment. In order to examine the influence of USF on recruitment of IE62 to promoters, a DNA fragment containing the ORF28/29 regulatory element was incubated with nuclear extracts derived from VZV-infected MeWo cells in magnetic bead recruitment assays. Nonbiotinylated oligomers containing the wild-type consensus USF binding site or the mutated site were added to the incubation, and the binding of IE62 and USF1 to the promoter in the presence and absence of competitor was examined. The results, presented in Fig. 7A, showed that IE62 bound to the ORF28/29 regulatory element to similar levels irrespective of whether or not USF was bound.
FIG. 7.
Analysis of the binding of IE62, USF, and TBP to promoters. (A) The effect of USF binding on IE62 recruitment to the ORF28/29 regulatory element. The 210-bp 5′-biotinylated ORF28/29 regulatory element (5′ Biot. ORF28/29) was conjugated to magnetic beads and incubated with nuclear extracts derived from VZV-infected MeWo cells (Inf. M. N. E.). Oligomers (22 bp) containing the wild-type or mutant USF binding site (USF or USFm, respectively) were used as competitors in the incubation. The presence or absence of IE62 and USF1 in eluates was determined by immunoblotting. (B) Immunoblot analysis of binding of IE62 present in infected cell nuclear extracts to the ORF28/29 regulatory element containing either the wild-type or mutant USF binding site (5′ Biot. ORF28/29 WT and USFm, respectively). (C) USF1 and TBP binding to the model USF-TATA promoter. Bead-immobilized 132-bp 3′-biotinylated USF-TATA promoter sequences containing the mutant or wild-type USF binding site (USFm or USF, respectively) were incubated with 250 μg nuclear extracts of uninfected MeWo cells. The levels of USF1 and TBP were determined by immunoblotting. (D) Effect of IE62 on TBP binding to the wild-type model promoter. Bead-immobilized 3′-biotinylated USF-TATA promoter was incubated with nuclear extracts of uninfected MeWo cells (N. E.) with or without preincubation with purified recombinant IE62 (Rec. IE62) present in increasing amounts. The presence of IE62, USF1, and TBP stably associated with the promoter was determined by immunoblotting following elution. (E) Interaction of the IE62, USF1, and VP16 ADs present as GST fusions in protein pull-down assays. The activation domain fusions were expressed in E. coli using the pGST-IE62AD, pGST-USF1AD, and pGST-VP16AD plasmids. The upper panel is an immunoblot showing the levels of TBP/TFIID detected in eluates from glutathione beads. The lower panel is a Coomassie blue-stained gel showing the levels of the fusion proteins and GST which coeluted from the beads. The difference in the position of the TBP band between the GSTUSFAD lane and the other lanes is due to distortion resulting from the high level of the recombinant GSTUSFAD fusion, which migrates with a mobility very similar to that of TBP.
Although USF binding to the wild-type ORF28/29 regulatory element showed no effect on IE62 binding, it remained a formal possibility that the loss of synergistic IE62 activation of promoters carrying the USFm mutation could occur through ablation or reduction of IE62 promoter binding due to the presence of the mutated site in a mechanism independent of USF binding. In order to investigate this question, magnetic bead recruitment assays were performed with the ORF28/29 regulatory element containing either the wild-type or mutant USF binding site using infected cell extracts. As shown in Fig. 7B, the USF binding site mutation eliminated USF binding but no significant effect on IE62 binding was observed. These results indicate that IE62 and USF appear to have no effect on each other's binding in the context of promoter elements shown to be synergistically activated by their combined action.
The TATA element was required for activation by IE62 and USF in the first series of experiments presented in this work, and both proteins have been suggested to stabilize TBP/TFIID binding in previous studies (30, 41). Therefore, TBP/TFIID binding to promoters in the presence and absence of USF and IE62 was also examined. To exclude the possibility that other transcription factors that bind to the ORF28/29 regulatory element may influence or offset the effects of USF and IE62 in this regard, TBP binding was examined by magnetic bead recruitment assays using promoter elements containing only wild-type or mutant USF binding sites upstream of the ADMLP TATA element. Densitometric analysis of the immunoblot data shown in Fig. 7C indicates that TBP binding to the USFm-TATA promoter was reduced by threefold compared to binding to the USF-TATA promoters in assays with nuclear extracts derived from uninfected MeWo cells. To examine the effect of the presence of IE62 on the levels of TBP bound in the presence of USF, increasing amounts of purified recombinant IE62 were preincubated with nuclear extracts derived from uninfected MeWo cells prior to the addition of the biotinylated promoter. As shown in Fig. 7D, increased IE62 binding correlated with increased TBP binding to the promoter (approximately 2.5-fold under these conditions) with USF binding being unaffected. These experiments were repeated twice with similar results regarding decreases and increases (two- to threefold) in the levels of TBP bound, respectively.
Since the activation domain of USF1 was sufficient to mediate synergy with IE62, we wished to determine if it was also capable of interaction with TBP/TFIID in protein pull-down experiments. While USF has been suggested to be involved in stabilization of TBP at promoters based on DNase footprinting (41), no information was available on the region of the protein involved. IE62 has also been shown to interact with TBP. While the site of this interaction (aa 273 to 734) mapped to a region other than the IE62 acidic activation domain (32), these data were obtained using in vitro-translated fragments of IE62 and purified TBP, which rarely exists in mammalian cells separate from the TATA-associated factors (TAFs) that, along with TBP, make up the general transcription factor TFIID (11, 28). Thus, the possibility remained that the IE62 activation domain might interact with a component of a larger complex containing TBP. The VP16 activation domain has been shown to interact with TBP/TFIID (13, 48) and acted as a positive control. The results are presented in Fig. 7E and show that the activation domain of USF1 was capable of capturing TBP or a complex containing TBP in these pull-down experiments. This represents the first demonstration of this finding for a specific region of USF. The IE62 activation domain was also capable of capturing TBP in these pull-down assays, suggesting that it may interact with one of the TAFs or another component of a larger transcription complex containing or stably interacting with TBP/TFIID.
DISCUSSION
The VZV IE62 protein is known to be a potent and promiscuous transcriptional activator. While this function of IE62 has been well established for over a decade, little information is available concerning the molecular details of the mechanism or mechanisms involved. One of the most intriguing aspects of the IE62 activation mechanism lies in the fact that while IE62 can activate promoters containing only a TATA element, much higher levels of activation have been observed with promoters containing binding sites for cellular transcription factors which, like IE62, contain potent activation domains (5, 14, 23, 30, 32). In this study, functional and physical interactions between IE62 and the ubiquitous cellular transcription factor USF were examined in the context of model minimal promoters and a complex native VZV regulatory element in order to assess the level of synergy involved and to probe the molecular details of the observed activation.
The first series of experiments assessed the level of functional interaction between IE62 and USF on model promoters containing consensus USF and TATA binding motifs. The individual contributions of IE62 and USF toward activation of these promoters were similar to levels seen for each in other studies (16, 32). However, when both factors were present, the observed activation was 23- to 30-fold greater than the simple additive level of their individual contributions. This represents the first determination of the level of synergy between the VZV IE62 protein and a cellular transcription factor. Extension of this analysis to a complex native regulatory element, the ORF28/29 regulatory element, which contains two unidirectional, partially overlapping promoters for the viral DNA polymerase and major DNA binding protein, showed that the level of synergy observed was the same as that for the model promoter. This suggests that the functional interaction between IE62 and USF is the same in this case despite the presence of atypical TATA elements and an Sp1 consensus binding site.
A straightforward possibility regarding the mechanism of IE62 synergy would involve recruitment of IE62 and its activation domain to the promoter via an interaction with USF. Such a model would be in keeping with prior data showing a direct physical interaction between the two proteins (30, 37). The increase in activation observed could thus potentially be due to the correct positioning of the IE62 activation domain for contact with the cellular transcription apparatus with or without the need for the activation domain of the cellular factor. The competition experiments using full-length and truncated forms of USF1 expressed ectopically, however, clearly indicate that the USF1 activation domain is involved in the activation observed in the presence of IE62. This was confirmed in the experiments using Gal4 fusions of USF1, which showed that the activation domain was necessary and sufficient for mediation of synergy with IE62. Thus, both activation domains are required based on these results regarding USF1 and previous analyses of IE62 (4, 34). The increased synergy observed with the Gal4 fusions containing only the activation domain and the activation domain plus the USR suggests that the activation domain of full-length USF1 in the GU/FL construct may be partially masked in the fusion protein. Alternatively steric inhibition of the interaction between the USF AD in the full-length fusion and IE62 could exist, resulting in lower activity of this protein than of the GU/AD construct. These results are in contrast with, but not necessarily in opposition to, the results of Qyang et al. (36), which suggested that the USR of USF2 rather than the activation domain was required for IE62-mediated activation based on similar competition experiments. This previous study was performed in Saos-2 cells, in which USF is expressed but inactive in the absence of IE62 due to the lack of a cellular coactivator. Thus, there may be several mechanisms by which IE62 can synergize with USF in activation of promoters depending on the presence or absence of the native USF cofactor, and possibly whether the synergy is mediated via USF1 or USF2.
The experiments using the USF1-Gal4 fusions indicated that the activation domain of USF1 is required for synergy but did not rule out the possibility that the two proteins also need to be in direct physical contact. However, the mapping of the regions of IE62 and USF1 required for their direct physical interaction and subsequent experiments assessing the ability of the IE62d20 mutant to activate the promoters within the wild-type ORF28/29 regulatory element indicate that this is not the case. These results are therefore in contrast to the mechanism of action of the adenovirus E1A protein which, unlike IE62, does not appear to require either specific or nonspecific DNA binding for transcriptional activation (57) but where the need for a direct physical interaction with the DNA binding domains of cellular transcription factors was demonstrated (17, 56).
The lack of a requirement for direct physical interaction between synergistic partners documented here shows some similarity to recent findings concerning the cellular factors Ets-1 and Pit-1. No effect on the synergistic activity of these proteins at the rat prolactin promoter was observed when Ets-1 proteins carrying mutations on their Pit-1 interaction surface, which significantly reduced binding of the two factors in vitro, were used in in situ assays (6). Ets-1 and Pit-1, however, both require specific DNA binding sites at the promoter. Likewise, human immunodeficiency virus TAT, which is currently not believed to require a direct interaction with the numerous cellular factors with which it can synergize, is tethered to the promoter via interaction with the nascent mRNA (19, 29). Thus, the requirement for DNA binding but lack of a need for a specific binding site within the promoter or a direct interaction with a required cellular transcription factor exhibited by IE62 appears to present a new mechanistic variation on the theme of synergy between transactivating proteins.
This does not, however, totally preclude the possibility that a direct physical interaction between IE62 and USF may be important for activation of specific promoters. The data obtained in this work showing a decrease in the efficiency of IE62 activation in the case of the IE62d20 protein compared with wild-type IE62 upon alteration of the structure of the ORF28/29 regulatory element clearly indicate that this could be the case. Thus, promoter architecture and the presence or absence of tissue-specific factors involved in VZV gene expression could result in the requirement for such an interaction. It is interesting, in this regard, that while numerous mutations within the IE62 gene have been identified in the live attenuated VZV vaccine (1, 8, 9, 18, 35), none of these mutations occur within the region of IE62 that contains the interaction sites for USF mapped in this work and for Sp1 mapped by Peng et al. (30).
The influence of IE62 and USF in the context of their binding to promoter elements and the influence of their presence on TBP/TFIID binding were also examined. USF and IE62 did not appear to influence each other's presence at either model or native promoter elements under any of the experimental conditions used in this study. Increased TBP/TFIID binding was observed in the presence of a consensus USF site within a model minimal promoter element compared to one containing a mutant site incapable of binding USF. Addition of recombinant IE62 showed a further increase in TBP/TFIID bound in the presence of USF. These results suggest that IE62 and USF stabilization of TBP/TFIID binding is part of the mechanism of synergistic activation.
Both the USF1 and IE62 activation domains were capable of capturing TBP/TFIID in GST pull-down assays. These results represent the first such information concerning the ability of a specific portion of either USF1 or USF2 to interact directly or indirectly with TBP/TFIID. Similarly novel are the findings for the activation domain of IE62, the target or targets of which have not previously been identified. These results differ from those of Perera (32), who showed that purified recombinant TBP bound to a fragment of IE62 encompassing amino acids 273 to 734. This suggests that the activation domain of IE62 may interact either with one of the TAFs associated with TBP or with another component of the general transcription apparatus that interacts stably with TFIID under the experimental conditions utilized in the pull-down experiments.
Previous work from several laboratories concerning IE62-mediated activation has demonstrated a requirement for both the N-terminal acidic activation domain and DNA binding activity of IE62 (4, 34, 38) and the requirement for the presence of a TATA element in the promoter (32). These findings, in conjunction with the data gathered in this study, allow the proposal of the following model for a mechanism of USF-IE62 activation. In this model, IE62 is not recruited to the promoter via interaction with the USF bound to the promoter. Instead, it recognizes and stabilizes TBP/TFIID bound to the promoter. The nonspecific DNA binding activity of IE62 may be required to enable the protein to track along the DNA until it encounters the bound TBP/TFIID. IE62 could further stabilize the complex through its nonspecific DNA binding activity or by interaction with one or more TBP-associated factors within TFIID. USF is also involved in interaction with and stabilization of TBP/TFIID. It is possible that this binding and stabilization could occur alternatively with that of IE62 and thus there would be no influence of IE62 and USF on the other's interaction with the promoter.
While the presence of both IE62 and USF increased TBP/TFIID binding to promoter elements in the magnetic bead recruitment assays, these increases were relatively modest compared to the level of synergy observed. USF is known to exist and bind as a dimer (10, 12), and native IE62 is also believed to be dimeric by analogy with herpes simplex virus ICP4 and based on the ability of its DNA binding region to dimerize (43, 51). Thus, interaction of IE62 and USF with TBP/TFIID could occur through one activation domain, allowing the second activation domain to be in the correct geometric position to allow cooperative interplay with elements of the cellular transcription apparatus, resulting in stabilization of the preinitiation complex. The activation domains of IE62 and USF could both physically and functionally interact with a coactivator protein such as the human Mediator complex, which is known to be required for the activities of Sp1 (40), the VP16 activation domain (26, 27, 54), and the adenovirus E1A protein (3). Conversely, IE62 could interact with other transcriptional components including the general transcription factors or one or more subunits of the polymerase II holoenzyme. These possibilities remain to be explored.
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases, AI18449 (W.T.R. and J.H.).
REFERENCES
- 1.Argaw, T., J. I. Cohen, M. Klutch, K. Lekstrom, T. Yoshikawa, Y. Asano, and P. R. Krause. 2000. Nucleotide sequences that distinguish Oka vaccine from parental Oka and other varicella-zoster virus isolates. J. Infect. Dis. 181:1153-1157. [DOI] [PubMed] [Google Scholar]
- 2.Betz, J. L., and S. G. Wydoski. 1993. Functional interaction of varicella-zoster virus gene 62 protein with the DNA sequence bound by herpes simplex virus ICP4 protein. Virology 195:793-797. [DOI] [PubMed] [Google Scholar]
- 3.Cantin, G. T., J. L. Stevens, and A. J. Berk. 2003. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. USA 100:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, J. I., D. Heffel, and K. Seidel. 1993. The transcriptional activation domain of varicella-zoster virus open reading frame 62 protein is not conserved with its herpes simplex virus homolog. J. Virol. 67:4246-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1998. Varicella-zoster virus gene 21: transcriptional start site and promoter region. J. Virol. 72:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval, D. L., A. Jean, and A. Gutierrez-Hartmann. 2003. Ras signaling and transcriptional synergy at a flexible Ets-1/Pit-1 composite DNA element is defined by the assembly of selective activation domains. J. Biol. Chem. 278:39684-39696. [DOI] [PubMed] [Google Scholar]
- 7.Giniger, E., S. M. Varnum, and M. Ptashne. 1985. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40:767-774. [DOI] [PubMed] [Google Scholar]
- 8.Gomi, Y., T. Imagawa, M. Takahashi, and K. Yamanishi. 2000. Oka varicella vaccine is distinguishable from its parental virus in DNA sequence of open reading frame 62 and its transactivation activity. J. Med. Virol. 61:497-503. [DOI] [PubMed] [Google Scholar]
- 9.Gomi, Y., H. Sunamichi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregor, P. D., M. Sawadogo, and R. G. Roeder. 1990. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 4:1730-1740. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann, A., T. Oelgeschlager, and R. G. Roeder. 1997. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc. Natl. Acad. Sci. USA 94:8928-8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hough, P. V., I. A. Mastrangelo, J. S. Wall, J. F. Hainfeld, M. Sawadogo, and R. G. Roeder. 1987. The gene-specific initiation factor USF (upstream stimulatory factor) bound at the adenovirus type 2 major late promoter: mass and three-dimensional structure. Proc. Natl. Acad. Sci. USA 84:4826-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingles, C. J., M. Shales, W. D. Cress, S. J. Triezenberg, and J. Greenblatt. 1991. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351:588-590. [DOI] [PubMed] [Google Scholar]
- 14.Ito, H., M. H. Sommer, L. Zerboni, H. He, D. Boucaud, J. Hay, W. Ruyechan, and A. M. Arvin. 2003. Promoter sequences of varicella-zoster virus glycoprotein I targeted by cellular transactivating factors Sp1 and USF determine virulence in skin and T cells in SCIDhu mice in vivo. J. Virol. 77:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschbaum, B. J., P. Pognonec, and R. G. Roeder. 1992. Definition of the transcriptional activation domain of recombinant 43-kilodalton USF. Mol. Cell. Biol. 12:5094-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, S.-S., S.-H. Kwon, J.-S. Sung, M.-Y. Han, and Y.-M. Park. 2003. Cloning and characterization of the rat Hsf2 promoter: a critical role of proximal E-box element and USF protein in Hsf2 regulation in different compartments of the brain. Biochim. Biophys. Acta 1625:52-63. [DOI] [PubMed] [Google Scholar]
- 17.Liu, F., and M. R. Green. 1994. Promoter targeting by adenovirus E1A through interaction with different cellular DNA-binding domains. Nature 368:520-525. [DOI] [PubMed] [Google Scholar]
- 18.Loparev, V. N., T. Argaw, P. R. Krause, M. Takayama, and D. S. Schmid. 2002. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J. Clin. Microbiol. 38:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loregian, A., K. Bortolozzo, S. Boso, B. Sapino, M. Betti, M. A. Biasolo, A. Caputo, and G. Palu. 2003. The Sp1 transcription factor does not directly interact with the HIV-1 Tat protein. J. Cell. Physiol. 196:251-257. [DOI] [PubMed] [Google Scholar]
- 20.Luo, X., and M. Sawadogo. 1996. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol. Cell. Biol. 16:1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch, J. M., T. K. Kenyon, C. Grose, J. Hay, and W. T. Ruyechan. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 302:71-82. [DOI] [PubMed] [Google Scholar]
- 22.McKee, T. A., and C. M. Preston. 1991. Identification of two protein binding sites within the varicella-zoster virus major immediate early gene promoter. Virus Res. 20:59-69. [DOI] [PubMed] [Google Scholar]
- 23.Meier, J. L., X. Luo, M. Sawadogo, and S. E. Straus. 1994. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol. Cell. Biol. 14:6896-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier, J. L., and S. E. Straus. 1995. Interactions between varicella-zoster virus IE62 and cellular transcription factor USF in the coordinate activation of genes 28 and 29. Neurology 45:S30-S32. [DOI] [PubMed] [Google Scholar]
- 25.Meier, J. L., and S. E. Straus. 1993. Varicella-zoster virus DNA polymerase and major DNA-binding protein genes have overlapping divergent promoters. J. Virol. 67:7573-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittler, G., T. Stuhler, L. Santolin, T. Uhlmann, E. Kremmer, F. Lottspeich, L. Berti, and M. Meisterernst. 2003. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 22:6494-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers, L. C., C. M. Gustafsson, K. C. Hayashibara, P. O. Brown, and R. D. Kornberg. 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakajima, N., M. Horikoshi, and R. G. Roeder. 1988. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol. Cell. Biol. 8:4028-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagtakhan, A. S., and S. E. Tong-Starksen. 1997. Interactions between Tat of HIV-2 and transcription factor Sp1. Virology 238:221-230. [DOI] [PubMed] [Google Scholar]
- 30.Peng, H., H. He, J. Hay, and W. T. Ruyechan. 2003. Interaction between the varicella zoster virus IE62 major transactivator and cellular transcription factor Sp1. J. Biol. Chem. 278:38068-38075. [DOI] [PubMed] [Google Scholar]
- 31.Perera, L. P., J. D. Mosca, M. Zadeghi-Zadeh, W. T. Ruyechan, and J. Hay. 1992. The varicella zoster virus immediate early protein IE62 can positively regulate its cognate promoter. Virology 191:346-354. [DOI] [PubMed] [Google Scholar]
- 32.Perera, L. P. 2000. The TATA motif specifies the differential activation of minimal promoters by varicella zoster virus immediate-early regulatory protein IE62. J. Biol. Chem. 275:487-496. [DOI] [PubMed] [Google Scholar]
- 33.Perera, L. P., J. D. Mosca, W. T. Ruyechan, and J. Hay. 1992. Regulation of varicella-zoster virus gene expression in human T lymphocytes. J. Virol. 66:5298-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perera, L. P., J. D. Mosca, W. T. Ruyechan, G. S. Hayward, S. E. Straus, and J. Hay. 1993. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J. Virol. 67:4474-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinlivan, M. L., A. A. Gershon, S. P. Steinberg, and J. Breuer. 2004. Rashes occurring after immunization with a mixture of viruses in the Oka vaccine are derived from a single clone of virus. J. Infect. Dis. 190:793-796. [DOI] [PubMed] [Google Scholar]
- 36.Qyang, Y., X. Luo, T. Lu, P. M. Ismail, D. Krylov, C. Vinson, and M. Sawadogo. 1999. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol. Cell. Biol. 19:1508-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahaus, M., N. Desloges, M. Yang, W. T. Ruyechan, and M. H. Wolff. 2003. Transcription factor USF, expressed during the entire phase of varicella-zoster virus infection, interacts physically with the major viral transactivator IE62 and plays a significant role in virus replication. J. Gen. Virol. 84:2957-2967. [DOI] [PubMed] [Google Scholar]
- 38.Ruyechan, W. T. 2004. Mechanism(s) of activation of varicella zoster virus promoters by the VZV IE62 protein. Recent Res. Dev. Virol. 6:145-172. [Google Scholar]
- 39.Ruyechan, W. T., H. Peng, M. Yang, and J. Hay. 2003. Cellular factors and IE62 activation of VZV promoters. J. Med. Virol. 70(Suppl. 1):S90-S94. [DOI] [PubMed] [Google Scholar]
- 40.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 41.Sawadogo, M. 1988. Multiple forms of the human gene-specific transcription factor USF. II. DNA binding properties and transcriptional activity of the purified HeLa USF. J. Biol. Chem. 263:11994-12001. [PubMed] [Google Scholar]
- 42.Sawadogo, M., M. W. Van Dyke, P. D. Gregor, and R. G. Roeder. 1988. Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J. Biol. Chem. 263:11985-11993. [PubMed] [Google Scholar]
- 43.Shepard, A. A., and N. A. DeLuca. 1991. Activities of heterodimers composed of DNA-binding- and transactivation-deficient subunits of the herpes simplex virus regulatory protein ICP4. J. Virol. 65:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirito, M., Q. Lin, T. Maity, and M. Sawadogo. 1994. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sirito, M., S. Walker, Q. Lin, M. T. Kozlowski, W. H. Klein, and M. Sawadogo. 1992. Members of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 2:231-240. [PMC free article] [PubMed] [Google Scholar]
- 46.Spengler, M. L., W. T. Ruyechan, and J. Hay. 2000. Physical interaction between two varicella zoster virus gene regulatory proteins, IE4 and IE62. Virology 272:375-381. [DOI] [PubMed] [Google Scholar]
- 47.Straus, S. E., J. Owens, W. T. Ruyechan, H. E. Takiff, T. A. Casey, G. Vande Woude, and J. Hay. 1982. Molecular cloning and physical mapping of varicella-zoster virus DNA. Proc. Natl. Acad. Sci. USA 79:993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stringer, K. F., C. J. Ingles, and J. Greenblatt. 1990. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature 354:783-786. [DOI] [PubMed] [Google Scholar]
- 49.Tyler, J. K., K. E. Allen, and R. D. Everett. 1994. Mutation of a single lysine residue severely impairs the DNA recognition and regulatory functions of the VZV gene 62 transactivator protein. Nucleic Acids Res. 22:270-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyler, J. K., and R. D. Everett. 1993. The DNA binding domain of the varicella-zoster virus gene 62 protein interacts with multiple sequences which are similar to the binding site of the related protein of herpes simplex virus type 1. Nucleic Acids Res. 21:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyler, J. K., and R. D. Everett. 1994. The DNA binding domains of the varicella-zoster virus gene 62 and herpes simplex virus type 1 ICP4 transactivator proteins heterodimerize and bind to DNA. Nucleic Acids Res. 22:711-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Workman, J. L., R. G. Roeder, and R. E. Kingston. 1990. An upstream transcription factor, USF (MLTF), facilitates the formation of preinitiation complexes during in vitro chromatin assembly. EMBO J. 9:1299-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, C. L., and K. W. Wilcox. 1991. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J. Virol. 65:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, F., R. DeBeaumont, S. Zhou, and A. M. Naar. 2004. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 101:2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, M., J. Hay, and W. T. Ruyechan. 2004. The DNA element controlling expression of the varicella-zoster virus open reading frame 28 and 29 genes consists of two divergent unidirectional promoters which have a common USF site. J. Virol. 78:10939-10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zu, Y. L., T. Maekawa, S. Matsuda, and S. Ishii. 1991. Complete putative metal finger and leucine zipper structures of CRE-BP1 are required for E1A-induced trans-activation. J. Biol. Chem. 266:24134-24139. [PubMed] [Google Scholar]
- 57.Zu, Y. L., Y. Takamatsu, M. J. Zhao, T. Maekawa, H. Handa, and S. Ishii. 1992. Transcriptional regulation by a point mutant of adenovirus-2 E1A product lacking DNA binding activity. J. Biol. Chem. 267:20181-20187. [PubMed] [Google Scholar]