Abstract
Sulfolobus turreted icosahedral virus (STIV) was the first icosahedral virus characterized from an archaeal host. It infects Sulfolobus species that thrive in the acidic hot springs (pH 2.9 to 3.9 and 72 to 92°C) of Yellowstone National Park. The overall capsid architecture and the structure of its major capsid protein are very similar to those of the bacteriophage PRD1 and eukaryotic viruses Paramecium bursaria Chlorella virus 1 and adenovirus, suggesting a viral lineage that predates the three domains of life. The 17,663-base-pair, circular, double-stranded DNA genome contains 36 potential open reading frames, whose sequences generally show little similarity to other genes in the sequence databases. However, functional and evolutionary information may be suggested by a protein's three-dimensional structure. To this end, we have undertaken structural studies of the STIV proteome. Here we report our work on A197, the product of an STIV open reading frame. The structure of A197 reveals a GT-A fold that is common to many members of the glycosyltransferase superfamily. A197 possesses a canonical DXD motif and a putative catalytic base that are hallmarks of this family of enzymes, strongly suggesting a glycosyltransferase activity for A197. Potential roles for the putative glycosyltransferase activity of A197 and their evolutionary implications are discussed.
The last few years have seen a dramatic increase in the number of viruses known to infect hyperthermophilic organisms (42, 52). Many of these viruses infect archaeal hosts belonging to the Crenarchaea phyla and exhibit unprecedented diversity with respect to both morphology and genomic sequence, necessitating the creation of new viral families to reflect their unique characteristics (42, 52). Compared to the study of viruses infecting the domains Eukarya and Bacteria, the study of archaeal viruses is still in its infancy. Little is known regarding archaeal virus life cycles, virus-host relationships, genetics, or biochemistry. This is particularly true for viruses of the Crenarchaea, and further study of these unique viruses is clearly needed. Importantly, while such studies will lead to a greater understanding of the viruses themselves, they are also expected to provide genetic, biochemical, and evolutionary insight into their crenarchaeal hosts and the requirements for life in the harsh environments in which these organisms often thrive.
Sulfolobus turreted icosahedral virus (STIV) was the first hyperthermophilic virus described with icosahedral capsid architecture. STIV infects Sulfolobus species resident in the acidic hot springs (pH 2.9 to 3.9 and 72 to 92°C) of Yellowstone National Park (54). It encapsidates a circular, double-stranded DNA genome of 17,663 bp containing 36 open reading frames, whose predicted products generally exhibit a lack of similarity to sequences in the publicly available databases, thus precluding functional assignment. The major capsid protein (B345), however, has been isolated from purified virus by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and identified by mass spectrometry (54). Purified virus has also allowed an electron cryomicroscopy reconstruction of STIV revealing a pseudo-T=31 icosahedral capsid with large turret-like projections extending from the vertices and an apparent internal membrane (54). The capsid architecture of STIV has generated much interest because it is common to viruses from all three domains of life, suggesting that this viral lineage shares a common ancestor that predates the fundamental evolutionary events that gave rise to the Archaea, Bacteria, and Eukarya (6, 29, 54). The recent crystal structure of the STIV major capsid protein (29) shows stunning structural homology to the major capsid proteins of bacteriophage PRD1 (7) and the eukaryotic viruses Paramecium bursaria Chlorella virus 1 (PBCV-1) (41) and adenovirus (56), further supporting this evolutionary lineage (29).
It is highly unlikely that most of the STIV proteome consists of unique protein folds serving novel functions. Rather, like the major capsid protein, the general lack of sequence similarity in the STIV open reading frames is probably due to the unique environment in which these viruses live and to the evolutionary distance between them and their bacterial and eukaryotic homologues. With respect to protein evolution, it is well established that similarity between related protein structures will persist longer than similarities in their corresponding amino acid sequences. While the structure of the STIV major capsid protein is an excellent example of this, there are many other examples in which protein structure has suggested functional and evolutionary relationships (6, 7, 11, 29, 31, 32, 39, 55, 61, 62). To this end, we have initiated crystallographic studies of the STIV proteome. Like similar studies of Sulfolobus spindle-shaped virus 1 (31, 32), structural analysis of the STIV gene products is expected to make specific suggestions regarding the functions and roles that these proteins play in the STIV life cycle.
Here we report on the structure of A197, one of the first STIV proteins to lend itself to crystallographic study. A197 is a 197-residue protein whose sequence provides little insight into its function. For example, standard BLAST (3) and CD searches (36) with default settings fail to identify proteins with similar sequence or structure. We have determined the crystal structure of STIV A197 and identify it as a glycosyltransferase. To our knowledge, this represents the first structure of a viral or archaeal glycosyltransferase displaying the GT-A fold. Potential roles for A197 in the STIV life cycle and their evolutionary implications are discussed.
MATERIALS AND METHODS
Cloning.
The A197 open reading frame was amplified by nested PCR directly from viral particles purified as previously described (54). The PCR primers added attB sites to facilitate ligase-free cloning using the Gateway system (Invitrogen), a Shine-Dalgarno sequence to facilitate efficient translation, and a C-terminal six-His tag to facilitate protein purification via nickel affinity chromatography. The internal forward and reverse primers were 5′-TTCGAAGGAGATAGAACCATGAGAACGCTTTTTTTTATAC-3′ and 5′-GTGATGGTGATGGTGATGCTTTATTCTAGCTACGTGAT-3′, respectively, while the external forward and reverse primers were 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGTGATGGTGATGGTGATG-3′, respectively. The sequence of the resulting entry clone was determined using ABI BigDye terminator cycle sequencing and is mutation free. The six-His-tagged A197 construct was then inserted into destination vector pDEST14 (Invitrogen), yielding the expression vector pEXP14-A197, for protein expression in Escherichia coli.
Expression, purification, and characterization of A197.
Typically, pEXP14-A197 was transformed into E. coli BL21-CodonPlus(DE3)-RIL (Stratagene), and a single colony was used to inoculate 5 ml of LB medium and grown overnight, with subsequent serial expansion to a 10-liter fermentor (New Brunswick; BIOFLO 2000) batch culture, all at 37°C. Medium for batch fermentation was as recommended by the manufacturer (53). All media contained 100 μg/ml ampicillin. Fermentor batch cultures were supplemented with filtered air at a flow rate of 10 liters/minute and stirred at 300 to 500 rpm. Cells were grown to an optical density at 600 nm of 0.8 to 1.5, and protein expression was then induced by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After an additional 4 to 6 hours of growth, cells were harvested by centrifugation (IEC PR-7000 M) at 6,000 × g for 20 min and pellets were stored at −80°C until needed.
For purification, cell pellets were thawed and resuspended in lysis buffer (10 mM Tris-HCl, pH 8.0, 400 mM NaCl) at 10 ml/g of wet cell pellet mass. Phenylmethylsulfonyl fluoride (0.1 mM) was added to the cell suspension, and cells were lysed by passage through a microfluidizer (Microfluidics Corporation, Newton, MA) or a Power laboratory press (American Instrument Co., Inc., Silver Spring, MD), depending on the volume of the cell suspension. The cell lysate was incubated at 65°C for 20 min to denature many of the contaminating E. coli proteins and clarified by centrifugation (Beckman; Avanti J-30I) at 22,000 × g for 20 min. The resulting cleared lysate was applied to a column containing a 3- to 5-ml bed volume of HIS-Select nickel affinity gel (Sigma-Aldrich), washed with at least 10 column volumes of wash buffer (lysis buffer plus 5 mM imidazole), and eluted with elution buffer (10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 200 mM imidazole). Fractions containing A197 were then pooled and loaded onto a Superdex 75 gel filtration column (Amersham Biosciences) for further purification and buffer exchange into a minimal buffer (10 mM Tris-HCl, pH 8.0, 50 mM NaCl) for crystallization. Peak fractions were pooled and concentrated using a 10-kDa molecular mass cutoff (Amicon ultracentrifugal filter devices; Millipore) to 10.5 mg/ml. Protein concentrations were determined by Bradford assay (9) using “protein assay reagent” (Bio-Rad) and bovine serum albumin as a standard. Protein yield was typically 2 to 5 mg/g of cell pellet. Molecular weight and purity were assessed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Dynamic light scattering (DLS) measurements were made on a Brookhaven Instruments ZetaPals (phase analysis light scattering) particle size/ζ potential analyzer. DLS was measured at 90o using a 661-nm diode laser, and the correlation functions were fitted using a constrained least-squares analysis as described previously (2).
For expression and purification of selenomethionine-incorporated A197, pEXP14-A197 was transformed into E. coli B834(DE3)pLysS (Novagen), a methionine auxotroph. Methionine auxotrophy was confirmed for a single colony, which was then used to inoculate 5 ml of medium, essentially as described previously (60), but supplemented with 1 ng/ml biotin, 50 ng/ml l-(+)-selenomethionine, and 100 μg/ml ampicillin, followed by serial expansion to a 10-liter batch culture (New Brunswick; BIOFLO 2000) in the same medium. After growth to optical density at 600 nm of 0.6 to 0.8, protein expression was induced and the protein purified as described above for native protein.
Crystallization and data collection.
Purified A197 was crystallized by hanging drop vapor diffusion at 22°C. Drops were assembled with 2 μl of A197 (10.5 mg/ml in 10 mM Tris-HCl, pH 8.0, 50 mM NaCl) mixed with 2 μl of well solution (0.1 M MES [morpholineethanesulfonic acid], pH 6.5, 1.2 to 1.6 M ammonium sulfate, 10% 1,4-dioxane). Selenomethionine-incorporated A197 crystals were grown under identical conditions. Single rod-shaped or wedge-shaped crystals were isolated, and cryoprotectant was introduced with a quick soak (10 to 60 seconds) in well solution supplemented with 22.5 to 25% glucose. Crystals were then flash frozen in liquid nitrogen.
Multiwavelength anomalous diffraction was used to solve the structure. Data were collected to 2.28-Å resolution at the Se-K edge (peak, remote, and inflection wavelengths) on beamline 11-1 at the Stanford Synchrotron Radiation Laboratory (SSRL) (Table 1). An additional single-wavelength data set to 1.86-Å resolution was collected on beamline 1-5 at SSRL and used for refinement (Table 1). Data were integrated, scaled, and reduced in space group I222 (Table 1) using the HKL2000 software package (43).
TABLE 1.
Data collectionc
| Parameter | Resulta for data set:
|
|||
|---|---|---|---|---|
| Se-Edge | Se-Peak | Se-Remote | Refinement | |
| Wavelength (Å) | 0.97966 | 0.97935 | 0.94642 | 0.97845 |
| Cell constants (a, b, c; Å; α = β = γ = 90°) | 66.54, 70.96, 81.77 | 66.54, 70.96, 81.77 | 66.54, 70.96, 81.77 | 66.42, 70.56, 81.27 |
| Resolution range (Å) | 30-2.28 (2.36-2.28) | 30-2.28 (2.36-2.28) | 30-2.28 (2.36-2.28) | 29.1-1.86 (1.93-1.86) |
| Unique reflections | 8,389 (893) | 8,403 (896) | 8,346 (890) | 15,646 (1,575) |
| Avg redundancy | 3.5 (3.6) | 3.5 (3.6) | 3.5 (3.6) | 8.6 (8.5) |
| I/σ | 22.9 (11.2) | 23.2 (11.7) | 23.4 (11.9) | 18.2 (4.1) |
| Completeness (%) | 92.3 (99.9) | 92.3 (99.9) | 91.9 (99.9) | 95.5 (99.1) |
| Rsymb (%) | 5.0 (8.7) | 5.3 (8.6) | 4.7 (8.2) | 5.1 (17.5) |
Numbers in parenthesis refer to the highest-resolution shell.
Rsym = 100 · ΣhΣiIi(h) − <I(h)>/ΣhI(h), where Ii(h) is the ith measurement of reflection h and <I(h)> is the average value of the reflection intensity.
Data were integrated, scaled, and reduced in space group I222 using the HKL2000 software package (43).
In order to identify manganese binding sites, crystals of A197 were soaked in mother liquor supplemented with 10 mM MnSO4 · H2O for 4 h. Crystals were flash frozen in liquid nitrogen as described above, and data to 3.0-Å resolution were collected in house using a Rigaku RUH3R X-ray generator producing Cu Kα X-rays and a MAR345 image plate detector. The data were integrated and scaled using HKL2000 (43).
Structure determination and refinement.
SOLVE (59) was used to determine the positions of the selenium substructure and to calculate initial phases. Four Se sites were identified, corresponding to one A197 molecule per asymmetric unit. The SOLVE output was then used in RESOLVE (57, 58) for density modification and initial model building. The resulting electron density map was of good quality, allowing the best parts of various models output by RESOLVE to be manually assembled into a composite model using O (27), followed by refinement with REFMAC5 (5, 40). The model was then further refined against the 1.86-Å data set. The refinement included the use of TLS (translation/libration/screw) parameters in which the model was broken into 12 TLS groups (1, 1 to 27; 2, 28 to 41; 3, 42 to 52; 4, 53 to 67; 5, 68 to 81; 6, 82 to 91; 7, 92 to 107; 8, 108 to 150; 9, 151 to 166; 10, 167 to 176; 11, 177 to 189; 12, 190 to 198) as suggested by the TLS motion determination home (http://skuld.bmsc.washington.edu/∼tlsmd/) (44-46). Iterative rounds of model building and refinement with O and REFMAC5 resulted in a final model with an Rcryst of 17.1% and an Rfree of 20.4% (Table 2). The final model has good stereochemistry, with all residues in allowed regions of the Ramachandran plot (33) (Table 2). Amino acids 139 to 147 were not modeled due to the lack of interpretable electron density, presumably due to disorder in this region of the protein. The protein quaternary structure server (http://pqs.ebi.ac.uk/) was used to calculate the surface area at the putative dimer interface. Structural comparisons were performed using the DALI (http://www.ebi.ac.uk/dali) (26) and VAST (http://www.ncbi.nlm.nih.gov/Structure/VAST/) (22) servers. Structural figures were generated with PYMOL (http://www.pymol.org) (17).
TABLE 2.
Model refinement
| Parameter | Value |
|---|---|
| Rcrysta (%) | 17.1 |
| Rfreea (%) | 20.4 |
| Real space CCb (%) | 95.5 |
| Mean B value (overall; Å2) | 18.3 |
| Coordinate error (based on maximum likelihood; Å) | 0.082 |
| RMSD from ideality: | |
| Bonds (Å) | 0.015 |
| Angles (°) | 1.545 |
| Ramachandran plotc: | |
| Most favored (%) | 90.6 |
| Additional allowed (%) | 9.4 |
| PDB accession code | 2C0N |
Rcryst = Σ∥Fo−Fc∥/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes used in refinement. Rfree is calculated as Rcryst, but using the “test” set of structure factor amplitudes that were withheld from refinement (4.9%).
Correlation coefficient (CC) is agreement between the model and 2mFo − DFc density map.
Calculated using PROCHECK (33).
Coordinates.
Atomic coordinates and structure factors for A197 are on deposit in the Protein Data Bank (www.pdb.org) under accession code 2C0N.
RESULTS
The A197 construct used in this study codes for the 197 amino acids of the native protein plus an additional C-terminal six-His tag, for a total of 203 residues with a calculated mass of 24,090 Da. A197 elutes from the Superdex 75 size exclusion column as a single peak with an apparent molecular mass of approximately 20 kDa. In contrast, dynamic light scattering indicates a monodisperse solution with an average particle diameter of approximately 9.5 nm, suggesting that A197 is present as a higher-order oligomer in solution.
A197 crystallizes in space group I222 with one copy of the A197 polypeptide in the asymmetric unit. The structure was initially solved at a resolution of 2.28 Å by multiwavelength anomalous diffraction using selenomethionyl-incorporated protein. The final model was refined against data collected on a second crystal to a resolution of 1.86 Å. Details on data collection and model refinement are presented in Tables 1 and 2.
Structure of A197.
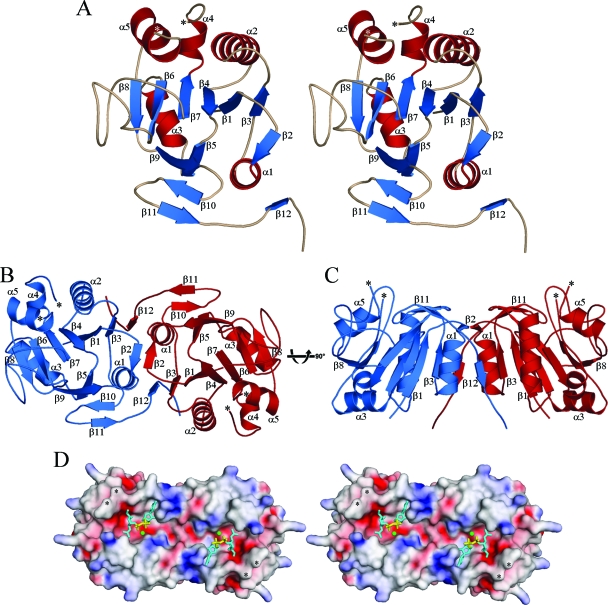
The structure of the A197 monomer reveals a six-stranded, predominantly parallel, α/β/α sandwich that is flanked by a four-stranded antiparallel β-sheet with an extended C terminus. This C-terminal tail shows significant interaction with a neighboring molecule in the crystal (Fig. 1). Residues 139 to 147, which connect helices α4 and α5, are disordered in the crystal structure and thus are not included in the model. The topology of the central β-sheet is 3, 1, 4, 7, 6, 8, with strand 7 running in the antiparallel direction, while the flanking four-stranded antiparallel β-sheet is composed successively of β-strands 5, 9, 10, and 11.
FIG. 1.
(A) Stereo image of the A197 monomer. The ribbon diagram depicts secondary structural elements of A197 (β-strands are blue, α-helices are red, and loops are wheat), which are labeled in ascending order from the N terminus to the C terminus. *, N- and C-terminal ends of the disordered region within the α4-α5 loop (residues 139 to 147). (B) Ribbon diagram of the A197 homodimer looking into the putative active site. One monomer is blue, and the second is red. The orientation of the blue monomer is rotated slightly counterclockwise with respect to that depicted in Fig. 1A. Secondary structural elements and the termini of the disordered region within the α4-α5 loops are labeled as in panel A. (C) Relative to panel B, the A197 homodimer has been rotated 90° about the depicted axis. The putative active site now runs along the top of the figure. (D) Stereo image of the electrostatic surface of the A197 homodimer. The orientation is identical to that in panel B, serving to highlight the putative active site. *, termini of the disordered region within the α4-α5 loop. The putative donor substrate-binding sites are marked with stick representations of the superpositioned donor substrate of GnT I. The electrostatic potential was mapped to the surface of A197 with SPOCK (13), using a probe radius of 1.4 Å, a temperature of 353°K, an ionic strength of 0.15 M, and protein and solvent dielectric constants of 4 and 80, respectively. The color ramp of the surface is from −15 kT/e (red, acidic) to 15 kT/e (blue, basic). All images were prepared and rendered using PyMOL (17).
In concert with a neighboring molecule of A197, β-strands 2, 3, and 12 are involved in the formation of intermolecular β-sheets (Fig. 1B and C). β-strand 2 interacts across a crystallographic twofold axis, hydrogen bonding to an equivalent β-strand in the neighboring molecule to form a two-stranded antiparallel β-sheet. β-strand 12 interacts with the same neighboring molecule, hydrogen bonding to β-strand 3 in an antiparallel fashion. This results in extension of the central β-sheet of the α/β/α sandwich to seven strands, showing a topology of 12, 3, 1, 4, 7, 6, 8, with strands 12 and 7 running antiparallel to the remaining strands. These interactions suggest that the dimer seen in the crystal is related to the higher-order oligomer indicated by dynamic light scattering. This putative dimer exhibits overall dimensions of approximately 80 Å by 45 Å by 40 Å. Relative to known dimer interfaces, this putative dimer interface is extensive, with occlusion of 1,950 Å2 of solvent-accessible surface area per monomer and a buried surface that is rich in hydrophobic residues.
The top edge of the central β-sheet, along with β-strand 2, creates the “floor” of a large pocket that is lined by elements of helix α2, helix α5, and the β4-β5 loop on one side and by the N-terminal half of the long β6-β7 loop, β-strand 5, β-strand 9, and much of the β9-β10 loop on the other. Formation of the crystallographic dimer results in an extended cleft that spans the dimer interface (Fig. 1D). The largest pocket on a protein surface is usually involved in ligand recognition and, in the case of enzymes, will mark the active site (34). Thus, this pocket is likely to play an important role in the activity of A197.
Structural homology to glycosyltransferases.
A DALI search (26) for structurally similar proteins identifies members of the glycosyltransferase superfamily as the closest structural homologues. Glycosyltransferases catalyze the transfer of a sugar moiety from an activated donor sugar onto saccharide and nonsaccharide acceptors (15). Members of the glycosyltransferase superfamily exhibit extreme diversity in their amino acid sequences, reflecting the large number of different donor and acceptor molecules that are utilized by this class of enzymes. Sequence similarity and biochemical characterization have been used to group the glycosyltransferase superfamily into 85 distinct families (as of May 2006) designated GT1 to GT85 (14, 15). However, this number is certain to grow as more of these genes are identified and their products characterized. Characteristics of the various families along with current membership are available from the carbohydrate-active enzyme (CAZy) database (http://afmb.cnrs-mrs.fr/CAZY/).
In contrast to the diverse sequence composition of these enzymes, structural studies have identified only two predominant folds, designated GT-A and GT-B, though a third fold related to the GT-A fold has been found in sialyltransferase CstII from Campylobacter jejuni (10, 12, 16). Recent structural work in a number of laboratories has allowed the GT-A and GT-B folds to be assigned to many of the GT families (10). Importantly, our DALI search with A197 identified glycosyltransferases that exhibit the GT-A fold. Interestingly, three of the four most similar matches are to eukaryotic glycosyltransferases. These show highly significant Z scores (≥9) and root mean square deviations (RMSDs) near 3.0 Å for superposition of structurally equivalent Cα atoms, despite less than 15% sequence identity over equivalent residues.
The structure of A197 differs from the canonical GT-A fold in a few minor ways. First, the canonical fold shows a seven-stranded central β-sheet, whereas there are only six strands in A197. The β-strand running along the outside edge of the central β-sheet and the preceding α-helix are not present. In A197, however, dimer formation supplies a replacement in the form of β-strand 12, though it runs in the opposite direction (antiparallel). A second minor difference is the addition of β-strands 10 and 11 to extend the size of the flanking antiparallel β-sheet, though extended versions of this flanking β-sheet have been seen before. For example, the nucleotide-diphospho-sugar transferase from Bacillus subtilis, SpsA (11a) also shows a four-stranded antiparallel β-sheet, though the connectivity differs from that in A197. Apart from the addition to this flanking β-sheet in A197, there is little decoration of its GT-A fold. In contrast, most enzymes with glycosyltransferase activity are significantly larger than A197 and often contain additional domains. Thus, this putative viral glycosyltransferase represents a minimal form of this large, extremely diverse, and highly important class of enzymes.
A197 shows greatest structural similarity to the catalytic domain of rabbit N-acetylglucosaminyltransferase I (GnT I) (62), an inverting glycosyltransferase in CAZy family GT13 (Fig. 2A and B). DALI identifies 149 structurally equivalent residues, with 9% sequence identity, whose Cα atoms superpose with an RMSD of 2.9 Å. A refined superposition of the two proteins with LSQMAN (30) identifies a highly conserved structural core consisting of 129 equivalent Cα atoms with 12.4% identity and an RMSD of 1.78 Å.
FIG. 2.
Ribbon diagrams of A197 and the equivalent domains of two structural neighbors. The relative orientations are the same in all panels and are identical to that in Fig. 1A. (A) Residues 1 to 197 of the A197 monomer. (B) Residues 106 to 317 of rabbit GnT I (PDB identifier [ID] 1FOA) (62). Secondary structural elements that are shared with A197 are labeled accordingly. The donor substrate (UDP-GlcNAc) is depicted with sticks, and the coordinated divalent manganese cation (Mn2+) is shown as a pink sphere in the active site. The C-terminal end of the structure is marked with dots to indicate the continuation of the polypeptide leading to an additional domain that is not present in A197. (C) Residues 75 to 310 of human GlcAT-I (PDB ID 1FGG) (48). Labels are consistent with those in panels A and B. UDP in the donor-binding site and Galβ1-3Gal in the acceptor-binding site are shown as sticks. The coordinated manganese cation is shown as a pink sphere. (D) Superposition of A197 (blue), GnT I (orange), and GlcAT-I (yellow) with detail of key GT-A glycosyltransferase active-site features. Residues constituting the DXD motif (Asp61, Glu62, and Asp63) and catalytic base (Asp151) of A197 are shown in sticks, as are the equivalent residues in GnT I (Glu211, Asp212, Asp213, and Asp291, respectively) and GlcAT-I (Asp194, Asp195, Asp196, and Glu281, respectively). For clarity, only the ribbon diagram of A197 is shown. The green mesh depicts difference electron density (contoured at 6σ) for the manganese binding site; it is found adjacent to the DXD motif and coincides nicely with the expected metal binding site. In GnT I and GlcAT-I, the DXD motif is involved in coordination of the diphosphate moiety of the donor substrate through the intervening Mn2+ ion, while the catalytic base is appropriately positioned for proton extraction from the hydroxyl moiety of the acceptor sugar substrate. Superpositions were prepared using LSQKAB (28). All images were prepared and rendered using PyMOL (17).
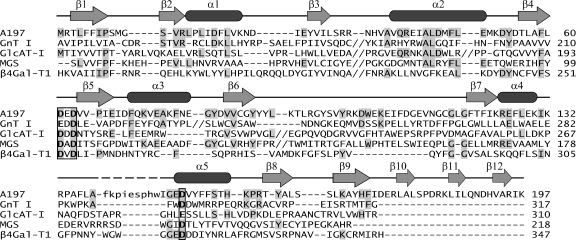
While the sequences of the many glycosyltransferase families are highly divergent, those that adopt the GT-A fold generally possess a DXD motif. The DXD typically serves to coordinate a divalent cation, usually Mn2+, which in turn interacts with the donor substrate through the diphosphate group (10). This motif is invariantly found in a short loop or β-turn that connects an internal strand of the central β-sheet to the first β-strand in the flanking β-sheet (10). Such is the case for A197, where the DXD motif is situated in the short loop that connects strands β4 and β5 and is composed of residues Asp61, Glu62, and Asp63. Superposition of A197 upon its closest structural neighbors (Fig. 2D) and the resulting structure-based sequence alignment (Fig. 3) show that this motif aligns very well, suggesting that the DXD motif in A197 will likewise interact with a Mn2+ ion. Accordingly, we see very strong difference density at this position in a low-resolution difference map (3.0 Å) for crystals that were soaked with Mn2+ (Fig. 2D).
FIG. 3.
Structure-based sequence alignment of A197 (PDB identifier [ID] 2C0N) with GT-A domains of close structural neighbors. GnT I, residues 106 to 317 of rabbit N-acetylglucosaminyltransferase I (PDB ID 1FOA; family GT13; inverting mechanism) (62); GlcAT-I, residues 75 to 310 of human β1,3-glucuronyltransferase I (PDB ID 1FGG; family GT43; inverting mechanism) (48); MGS, residues 2 to 99 of Rhodobacter marinus mannosylglycerate synthase (PDB ID 2BO4; family GT78; retaining mechanism) (19); β4Gal-T1, residues 180 to 347 of bovine β1,4-galactosyltransferase T1 (PDB ID 1FGX; family GT7; inverting mechanism) (21). The secondary structural elements of A197 are mapped above the alignment (arrows denote β-strands, and rounded rectangles denote α-helices). Long stretches of sequence that are not structurally equivalent to A197 have been removed from the alignment for conciseness and are marked by double shills. The missing residues of the A197 α4-α5 loop are in lowercase in the alignment and are marked with dashes in the secondary structure map. Identical residues are highlighted in gray, and the DXD motif and catalytic base are in boldface and are boxed. The structure-based sequence alignment was created with the 3DCoffee@igs web server (http://www.igs.cnrs-mrs.fr/Tcoffee/) (51), using Msap_pair to compute the library and slight manual adjustment around the gaps with consideration of the pairwise structural alignments created by LSQMAN (30).
In addition to the DXD motif, glycosyltransferase enzymes exhibiting the GT-A fold commonly possess a catalytic base, Glu or Asp, that is thought to deprotonate a hydroxyl group on the acceptor substrate, thus assisting nucleophilic attack on the donor sugar (48, 62). The structural superposition (Fig. 2D) and resulting structure-based sequence alignment (Fig. 3) identify Asp151 at the N terminus of helix α5 as the putative catalytic base in A197. Thus, active-site features that are hallmarks of glycosyltransferase activity, the DXD motif and a putative catalytic base, are preserved within the proper structural context of the GT-A fold, strongly suggesting that A197 is indeed a glycosyltransferase.
Donor substrate binding site.
The structure of GnT I (62) was solved as a complex with its donor substrate, UDP-N-acetylglucosamine, and Mn2+ (Fig. 2B). Thus, superposition of GnT I and the donor substrate upon A197 suggests probable binding sites for an activated sugar substrate and a metal cofactor in the active site of A197. The donor substrate is predicted to sit within the extended active-site pocket that runs across the surface of A197, with the uridine moiety situated over the C-terminal ends of β1 and β3, adjacent to the N-terminal end of α2 (Fig. 1D). The diphosphate moiety is predicted to span the C terminus of β4 and the N terminus of β7, with the sugar moiety situated over the N terminus of β7, adjacent to helices α2 and α5. The superposed Mn2+ ion is positioned between the β4-β5 loop and the phosphodiester bond of the UDP, a position that corresponds nicely with the major peak seen in the low-resolution difference maps from Mn2+-soaked crystals of A197 (Fig. 1D and 2D).
Interestingly, there is a loop adjacent to the active site in GnT I that remains disordered until the donor substrate is bound. Closure of the loop over the donor substrate not only serves to hold it in place but also serves an important role in the binding of the acceptor substrate, providing a structural explanation for the ordered substrate binding that is seen in GnT I (62). Similarly, nine residues of the loop connecting helices α4 and α5 of A197 are disordered in the absence of substrate. However, relative to GnT I, this disordered loop is situated on the opposite side of the substrate-binding pocket. Nevertheless, its position would allow it to fulfill a similar role in closing over the substrate, possibly serving to complete the formation of an acceptor-binding site.
Acceptor substrate binding site.
The DALI search also identified human β1,3-glucuronyltransferase I (GlcAT-I) (47, 48), an inverting glycosyltransferase in CAZy family GT43, as a close structural homologue (3.1-Å RMSD and 13% identity over 148 structurally equivalent residues) (Fig. 2C). Once again, a refined superposition of GlcAT-I onto A197 with LSQMAN identifies the same highly conserved structural core (120 residues, 15% sequence identity, 1.83-Å RMSD). Interestingly, while the structure of the C-terminal extension in A197 differs significantly from that seen in GlcAT-I, both proteins utilize the C-terminal tail in the formation of an extensive dimer interface. However, unlike A197, the active-site clefts of each monomer do not merge across the dimer interface.
Importantly, the GlcAT-I structure was solved in complex with UDP, Mn2+, and an acceptor substrate analog, Galβ1-3Galβ1-4Xyl (48). The superposition of GlcAT-I on A197 suggests that the acceptor substrate for A197 will bind adjacent to strand β6, in a groove formed predominantly by the long intervening region between β6 and β7. The acceptor substrate will then be positioned adjacent to the N terminus of α5, an optimal arrangement relative to the catalytic base and the expected position of the donor substrate. A second structure of GlcAT-I has been solved with UDP-glucuronic acid, the full donor substrate, in the active site (47). Not surprisingly, the glucuronic acid moiety binds in a manner similar to that seen for the N-acetylglucosamine in GnT I, further indicating that the relative positions of the donor and acceptor substrates in A197 are reliably indicated by superposition of its closest structural homologues.
DISCUSSION
Like the majority of the STIV genome, the predicted amino acid sequence of A197 provides little insight into its role in the viral life cycle. Thus, we initiated crystallographic studies of A197 with the specific goal of identifying structural homologues that might assist in making a functional assignment. The resulting structure does indeed identify close homologues; it is remarkably similar to several glycosyltransferases exhibiting the GT-A fold.
Due to the large number of potential substrates, limited sequence similarity and three-dimensional structural information are generally insufficient to allow reliable prediction of substrates for a given glycosyltransferase. In the absence of biochemical characterization with the known substrates, it is also difficult to predict whether the enzyme will catalyze sugar transfer with inversion or retention of stereochemistry at the anomeric carbon. For example, all of the structural homologues aligned in Fig. 3 utilize different substrates, and three of the four are inverting enzymes (GnT I, GlcAT-I, and β4Gal-T1), while mannosylglycerate synthase (MGS) is retaining. Yet the GT-A fold, the DXD motif, and the catalytic base are common to all. MGS illustrates the difficulty in assigning specific properties to a glycosyltransferase in the absence of biochemical characterization. Sequence-based assignment had placed it in inverting family GT2; however, subsequent characterization showed that it utilizes a retaining mechanism; hence MGS was reclassified as the founding member of family GT78 (19). Thus, in the case of A197, the identity of donor and acceptor substrates and elucidation of mechanistic details await biochemical and genetic studies.
A197 is one of the smallest glycosyltransferases that we are aware of, composed of only the core catalytic GT-A fold and lacking additional functional domains. Thus, its structure may define the minimal components necessary for glycosyltransferase activity. The presence of minimal, “stripped-down” structural motifs is common in viruses (37), where the need for genetic efficiency within the viral genome is one common explanation for their occurrence. A197 has yet to be assigned to a GT family in the CAZy database. However, the lack of sequence similarity to other known glycosyltransferases suggests that A197 represents yet another sequence-based glycosyltransferase family.
The CAZy database includes a number of viral glycosyltransferases, including annotated glycosyltransferase genes from other crenarchaeal viruses AFV1 (8), AFV2 (24), ARV1 (63), SIFV (4), SIRV1 (49, 50), SIRV2 (49), and STSV1 (65). These crenarchaeal viral glycosyltransferases have been placed in family GT4, a family that utilizes the retaining mechanism of sugar transfer and lacks structural representation (15). We note that GT4 also includes a glycosyltransferase from PBCV-1 (23, 37), a eukaryotic virus that is believed to be evolutionarily related to STIV. Thus, glycosyltransferases are not uncommon to viral genomes and are frequently found in viruses infecting the Crenarchaea.
Viruses commonly decorate their proteins with sugars as a means of regulating interactions with their hosts. While viruses can accomplish this task by utilizing the host's glycosylation machinery, it is apparent that many viruses also encode their own proteins for specialized glycosylation needs (37). Virally encoded glycosyltransferases are known to serve a variety of functions and target a wide variety of acceptor substrates, including DNA, proteins, and small molecules. For example, some lytic bacteriophages glycosylate their DNA to protect it from host restriction enzymes while others alter the glycosylation of cell surface antigens to induce serotype conversion of the host bacteria during lysogeny or, in the case of lytic phage, to induce receptor conversion, preventing retention of progeny on the host cell debris (37).
In eukaryotic systems, many baculoviruses utilize virally encoded glycosyltransferases to glycosylate host ecdysteroids. This modification alters the biological activity of the hormone, suppressing development of the infected host. Glycosyltransferase-encoding genes are also common in the phycodnavirus family. This includes PBCV-1, where these virally encoded enzymes have been implicated in glycosylation of its major capsid protein, Vp54. While eukaryotic viral proteins are typically glycosylated by host-encoded enzymes located in the endoplasmic reticulum (ER) and Golgi apparatus, Vp54 is apparently glycosylated in an ER and Golgi apparatus-independent manner by virally encoded glycosyltransferases that are expressed in the cytosol. Thus, it has been suggested that glycosylation of Vp54 reflects an ancestral pathway that existed prior to ER and Golgi apparatus formation (23, 37).
This is particularly interesting in light of the putative evolutionary relationship between PBCV-1 and STIV. The similarities between these viruses are not limited to their overall capsid architecture and the structures of their major capsid proteins. In this issue Maaty et al. show that, like Vp54 in PBCV-1, the STIV major capsid protein (B345), is also glycosylated (35). While A197 might serve any number of functional roles, the similarities to PBCV-1 with respect to capsid architecture, fold of the major capsid protein, glycosylation of the major capsid protein, and the occurrence of a virally encoded glycosyltransferase suggest that A197 participates in glycosylation of the major capsid protein in STIV and is likely involved in particle maturation.
Maaty et al. also observe that the STIV major capsid protein gives a horizontal spot train on two-dimensional gels, indicating a series of increasingly acidic species compared to the bacterially expressed protein, which is not glycosylated (35). This suggests that glycosylation introduces negative charge through addition of acidic sugar moieties such as galacturonic acid, glucuronic acid, and iduronic acid (18). Alternatively, the spot train might represent sulfation of neutral saccharides (18).
In addition to the obvious structural and functional roles for protein glycosylation, increased glycosylation has been shown to correlate with environmental extremes of temperature, pH, and salt concentration (1, 18, 25, 38, 64, 66). Thus, it has been suggested that increased protein glycosylation may stabilize extracellular proteins in these harsh environments (1, 18, 25, 38, 64, 66). In this light, it is not surprising that many crenarchaeal viruses apparently encode glycosyltransferases.
Glycosyltransferases are an enormous and extremely diverse class of enzymes that are thought to have evolved from a common ancestor, particularly those of the GT-A superfamily (20). As glycosyltransferases are ubiquitous to the three domains of life, they probably predate the evolutionary events that established these domains. Likewise, there is accumulating evidence that STIV is part of a viral lineage that predates the split of the three domains. Identification of a glycosyltransferase in STIV extends the observed similarities among double-stranded DNA viruses inhabiting the three domains of life beyond those of capsid architecture and the conserved fold of the major capsid protein. The last virus common to the STIV/PBCV-1 lineage is likely to have carried a glycosyltransferase gene in its genome. With the development of immunity, however, descendant viruses infecting multicellular eukaryotic organisms would have found it advantageous to lose this feature, relying instead on enzymes supplied by their hosts.
In summary, the structure of A197 provides substantial insight into its function and its relevance to the STIV life cycle. It is yet another demonstration that structure can reveal functional and evolutionary relationships.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-0236344 and MCB-0132156) and the National Aeronautics and Space Administration (NAG5-8807). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The Macromolecular Diffraction Laboratory at Montana State University was supported, in part, by a grant from the Murdock Foundation.
REFERENCES
- 1.Albers, S.-V., S. M. Koning, W. N. Konings, and A. J. M. Driessen. 2004. Insights into ABC transport in Archaea. J. Bioenerg. Biomembr. 36:5-15. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M., D. Willits, M. Young, and T. Douglas. 2003. Constrained synthesis of cobalt oxide nanomaterials in the 12-subunit protein cage from Listeria innocua. Inorg. Chem. 42:6300-6305. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, H. P., W. Zillig, U. Ziese, I. Holz, M. Crosby, T. Utterback, J. F. Weidmann, J. K. Kristjanson, H. P. Klenk, K. E. Nelson, and C. M. Fraser. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267:252-266. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, S. 1994. The CCP4 suite—programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 6.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 7.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 8.Bettstetter, M., X. Peng, R. A. Garrett, and D. Prangishvili. 2003. AFV1, a novel virus infecting hyperthermophilic Archaea of the genus Acidianus. Virology 315:68-79. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Breton, C., L. Snajdrova, C. Jeanneau, J. Koca, and A. Imberty. 2006. Structures and mechanisms of glycosyltransferases. Glycobiology 16:29R-37R. [DOI] [PubMed] [Google Scholar]
- 11.Buehner, M., G. C. Ford, D. Moras, K. W. Olsen, and M. G. Rossmann. 1973. D-glyceraldehyde-3-phosphate dehydrogenase: three-dimensional structure and evolutionary significance. Proc. Natl. Acad. Sci. USA 70:3052-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Charnock, S. J., and G. J. Davies. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38:6380-6385. [DOI] [PubMed] [Google Scholar]
- 12.Chiu, C. P. C., A. G. Watts, L. L. Lairson, M. Gilbert, D. Lim, W. W. Wakarchuk, S. G. Withers, and N. C. J. Strynadka. 2004. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 11:163-170. [DOI] [PubMed] [Google Scholar]
- 13.Christopher, J. A. 1998. SPOCK: the structural properties observation and calculation kit. http://quorum.tamu.edu/.
- 14.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 15.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes. http://afmb.cnrs-mrs.fr/CAZY/.
- 16.Davies, G. J., T. M. Gloster, and B. Henrissat. 2005. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr. Opin. Struct. Biol. 15:637-645. [DOI] [PubMed] [Google Scholar]
- 17.DeLano, W. L. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- 18.Eichler, J., and M. W. W. Adams. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69:393-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint, J., E. Taylor, M. Yang, D. N. Bolam, L. E. Tailford, C. Martinez-Fleites, E. J. Dodson, B. G. Davis, H. J. Gilbert, and G. J. Davies. 2005. Structural dissection and high-throughput screening of mannosylglycerate synthase. Nat. Struct. Mol. Biol. 12:608-614. [DOI] [PubMed] [Google Scholar]
- 20.Franco, O. L., and D. J. Rigden. 2003. Fold recognition analysis of glycosyltransferase families: further members of structural superfamilies. Glycobiology 13:707-712. [DOI] [PubMed] [Google Scholar]
- 21.Gastinel, L., C. Cambillau, and Y. Bourne. 1999. Crystal structures of the bovine β4 galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 18:3546-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibrat, J.-F., T. Madej, and S. H. Bryant. 1996. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 6:377-385. [DOI] [PubMed] [Google Scholar]
- 23.Graves, M. V., C. T. Bernadt, R. Cerny, and J. L. Van Etten. 2001. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 285:332-345. [DOI] [PubMed] [Google Scholar]
- 24.Haring, M., G. Vestergaard, K. Brugger, R. Rachel, R. A. Garrett, and D. Prangishvili. 2005. Structure and genome organization of AFV2, a novel archaeal lipothrixvirus with unusual terminal and core structures. J. Bacteriol. 187:3855-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hettmann, T., C. L. Schmidt, S. Anemuller, U. Zahringer, H. Moll, A. Petersen, and G. Schafer. 1998. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius. A novel highly glycosylated, membrane-bound b-type hemoprotein. J. Biol. Chem. 273:12032-12040. [DOI] [PubMed] [Google Scholar]
- 26.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 27.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch, W. 1976. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. Sect. A 32:922-923. [Google Scholar]
- 29.Khayat, R., L. Tang, E. T. Larson, C. M. Lawrence, M. Young, and J. E. Johnson. 2005. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. USA 102:18944-18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleywegt, G. 1996. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr Sect. D Biol. Crystallogr. 52:842-857. [DOI] [PubMed] [Google Scholar]
- 31.Kraft, P., D. Kummel, A. Oeckinghaus, G. H. Gauss, B. Wiedenheft, M. Young, and C. M. Lawrence. 2004. Structure of D-63 from sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function. J. Virol. 78:7438-7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft, P., A. Oeckinghaus, D. Kummel, G. H. Gauss, J. Gilmore, B. Wiedenheft, M. Young, and C. M. Lawrence. 2004. Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein. J. Virol. 78:11544-11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 34.Liang, J., H. Edelsbrunner, and C. Woodward. 1998. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 7:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maaty, W. S. A., A. C. Ortmann, M. Dlakić, K. Schulstad, J. Hilmer, L. Liepold, B. Weidenheft, T. Douglas, M. Young, and B. Bothner. 2006. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J. Virol. 80:7625-7635. [DOI] [PMC free article] [PubMed]
- 36.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markine-Goriaynoff, N., L. Gillet, J. L. Van Etten, H. Korres, N. Verma, and A. Vanderplasschen. 2004. Glycosyltransferases encoded by viruses. J. Gen. Virol. 85:2741-2754. [DOI] [PubMed] [Google Scholar]
- 38.Mengele, R., and M. Sumper. 1992. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 267:8182-8185. [PubMed] [Google Scholar]
- 39.Moult, J., and E. Melamud. 2000. From fold to function. Curr. Opin. Struct. Biol. 10:384-389. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallgr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 41.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortmann, A. C., B. Wiedenheft, T. Douglas, and M. Young. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat. Rev. Microbiol., in press. [DOI] [PubMed]
- 43.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 44.Painter, J., and E. A. Merritt. 2005. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr. Sect. D Biol. Crystallgr. 61:465-471. [DOI] [PubMed] [Google Scholar]
- 45.Painter, J., and E. A. Merritt. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. Sect. D Biol. Crystallgr. 62:439-450. [DOI] [PubMed] [Google Scholar]
- 46.Painter, J., and E. A. Merritt. 2006. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39:109-111. [Google Scholar]
- 47.Pedersen, L. C., T. A. Darden, and M. Negishi. 2002. Crystal structure of β1,3-glucuronyltransferase I in complex with active donor substrate UDP-GlcUA. J. Biol. Chem. 277:21869-21873. [DOI] [PubMed] [Google Scholar]
- 48.Pedersen, L. C., K. Tsuchida, H. Kitagawa, K. Sugahara, T. A. Darden, and M. Negishi. 2000. Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I. J. Biol. Chem. 275:34580-34585. [DOI] [PubMed] [Google Scholar]
- 49.Peng, X., H. Blum, Q. She, S. Mallok, K. Brugger, R. A. Garrett, W. Zillig, and D. Prangishvili. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226-234. [DOI] [PubMed] [Google Scholar]
- 50.Peng, X., A. Kessler, H. Phan, R. A. Garrett, and D. Prangishvili. 2004. Multiple variants of the archaeal DNA rudivirus SIRV1 in a single host and a novel mechanism of genomic variation. Mol. Microbiol. 54:366-375. [DOI] [PubMed] [Google Scholar]
- 51.Poirot, O., K. Suhre, C. Abergel, E. O'Toole, and C. Notredame. 2004. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucleic Acids Res. 32:W37-W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prangishvili, D., and R. A. Garrett. 2005. Viruses of hyperthermophilic crenarchaea. Trends Microbiol. 13:535-542. [DOI] [PubMed] [Google Scholar]
- 53.Research and Development Laboratory. 1996. Fundamentals of fermentation: techniques for benchtop fermentors, part I. New Brunswick Scientific, Edison, N.J.
- 54.Rice, G., L. Tang, K. Stedman, F. Roberto, J. Spuhler, E. Gillitzer, J. E. Johnson, T. Douglas, and M. Young. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. USA 101:7716-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossmann, M. G., A. D. B. Malcolm, and W. F. Bodmer. 1981. Evolution of glycolytic enzymes. Philos. Trans. R. Soc. London Biol. Sci. 293:191-203. [DOI] [PubMed] [Google Scholar]
- 56.Rux, J. J., P. R. Kuser, and R. M. Burnett. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77:9553-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terwilliger, T. C. 2002. Automated main-chain model-building by template-matching and iterative fragment extension. Acta Crystallogr. Sect. D Biol. Crystallgr. 59:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terwilliger, T. C. 2000. Maximum likelihood density modification. Acta Crystallogr. Sect. D Biol. Crystallgr. 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D Biol. Crystallgr. 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thoma, R., G. Obmolova, D. A. Lang, M. Schwander, P. Jenö, R. Sterner, and M. Wilmanns. 1999. Efficient expression, purification and crystallisation of two hyperthermostable enzymes of histidine biosynthesis. FEBS Lett. 454:1-6. [DOI] [PubMed] [Google Scholar]
- 61.Unligil, U. M., and J. M. Rini. 2000. Glycosyltransferase structure and mechanism. Curr. Opin. Struct. Biol. 10:510-517. [DOI] [PubMed] [Google Scholar]
- 62.Unligil, U. M., S. Zhou, S. Yuwaraj, M. Sarkar, H. Schachter, and J. M. Rini. 2000. X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily. EMBO J. 19:5269-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vestergaard, G., M. Haring, X. Peng, R. Rachel, R. A. Garrett, and D. Prangishvili. 2005. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336:83-92. [DOI] [PubMed] [Google Scholar]
- 64.Wang, C., M. Eufemi, C. Turano, and A. Giartosio. 1996. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry 35:7299-7307. [DOI] [PubMed] [Google Scholar]
- 65.Xiang, X., L. Chen, X. Huang, Y. Luo, Q. She, and L. Huang. 2005. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J. Virol. 79:8677-8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zahringer, U., H. Moll, T. Hettmann, Y. A. Knirel, and G. Schafer. 2000. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur. J. Biochem. 267:4144-4149. [DOI] [PubMed] [Google Scholar]