Abstract
Vaccinia virus does not grow in Chinese hamster ovary (CHO-K1) cells in the absence of a viral host range factor, cowpox protein CP77. In this study, CP77 was fused to the C terminus of green fluorescence protein (GFP-CP77) and a series of nested deletion mutants of GFP-CP77 was constructed for insertion into a vaccinia virus host range mutant, VV-hr, and expressed from a viral early promoter. Deletion mapping analyses demonstrated that the N-terminal 352 amino acids of CP77 were sufficient to support vaccinia virus growth in CHO-K1 cells, whereas the C-terminal residues 353 to 668 were dispensable. In yeast two-hybrid analyses, CP77 bound to a cellular protein, HMG20A, and GST pulldown analyses showed that residues 1 to 234 of CP77 were sufficient for this interaction. After VV-hr virus infection of CHO-K1 cells, HMG20A was translocated from the nucleus to viral factories and bound to the viral genome via the HMG box region. In control VV-hr-infected CHO-K1 cells, binding of HMG20A to the viral genome persisted from 2 to 8 h postinfection (h p.i.); in contrast, when CP77 was expressed, the association of HMG20A with viral genome was transient, with little HMG20A remaining bound at 8 h p.i. This indicates that dissociation of HMG20A from viral factories correlates well with CP77 host range activity in CHO-K1 cells. Finally, in cells expressing a CP77 deletion protein (amino acids 277 to 668) or a ΔANK5 mutant that did not support vaccinia virus growth and did not contain the HMG20A binding site, HMG20A remained bound to viral DNA, demonstrating that the binding of CP77 to HMG20A is essential for its host range function. In summary, our data revealed that a novel cellular protein, HMG20A, the dissociation of which from viral DNA is regulated by CP77, providing the first cellular target regulated by viral host range CP77 protein.
Poxviruses fail to grow in certain host cells, a phenomenon known as host restriction (13, 15, 20, 22, 28, 38, 66, 73). In these restrictive cells, poxvirus enters the cell efficiently but fails to complete its life cycle as a result of cell-type-specific blockages (see reference 39 and references therein). A family of poxviral host range (hr) genes has been identified, and their expression supports virus growth in restrictive cells. Vaccinia virus, the prototype of the poxvirus family, has two hr genes, the K1L and C7L genes, which support virus growth in RK13 and HeLa cells, respectively (23, 48, 65). Growth of vaccinia virus in Chinese hamster ovary (CHO-K1) cells requires an hr gene, the CP77 (or V025) gene, from cowpox virus (33, 48, 53, 57, 63). The modified vaccinia virus Ankara strain requires E3L protein to grow in primary chicken embryo fibroblasts and HeLa cells (27, 35). Other poxviruses, such as myxoma virus, have several hr genes (M-T2, M-T4, M-T5, and M11L genes) that support virus growth in different rabbit cell types (4, 17, 18, 43, 71, 72). The SPI-1 gene in rabbitpox and its orthologue in vaccinia virus supports virus growth in PK15 and A549 cells, respectively (1, 36, 59).
The mechanism by which each viral hr protein overcomes host restriction could be unique to each protein and its cellular context. Vaccinia E3L protein suppresses both the 2′-5′-oligoadenylate synthetase/RNase L pathway and PKR in various human cell lines (5, 27, 35). Myxoma M-T5 regulates cell cycle progression at the G0/G1 checkpoint to protect infected cells from host antiviral responses induced by cell cycle arrest (31). In addition, M-T5 regulates cellular Akt signaling and is an important tropism factor for myxoma virus infection in certain human cancer cells (72).
Although the CP77 gene was initially recognized as the hr gene for vaccinia virus growth in CHO-K1 cells, it can replace the K1L and C7L genes in supporting vaccinia virus growth in RK13 and HeLa cells (15, 29, 48, 54). In contrast, C7L and K1L genes do not support vaccinia virus growth in CHO-K1 cells, suggesting a unique function of the CP77 gene in this cell type (45, 48, 63). Without CP77, the growth of vaccinia virus in CHO-K1 and HeLa cells was blocked at translation of viral intermediate RNA and, in RK13 cells, at translation of viral early RNA (29, 53, 54). It has been suggested that the CP77 gene suppresses apoptosis in restrictive cells; however, suppression of apoptosis by adenoviral E1B, bcl-2, or caspase inhibitors does not rescue virus growth in these restrictive cells (11, 19, 29, 30, 52, 54). Despite its obvious importance in antagonizing host restriction, the molecular basis of the hr activity of the CP77 gene remains unclear. Moreover, CP77 gene orthologues are present in other poxvirus genomes (http://www.poxvirus.org). Monkeypox virus encodes a homologous protein of 660 amino acids (aa) in length with 91% sequence identity. Variola virus contains a shorter homologous protein of 452 residues with 87% sequence identity. Although vaccinia virus is very close to cowpoxvirus, the CP77 gene orthologue in vaccinia virus genome either is deleted (in strain Copenhagen) or becomes fragmented due to nonsense mutations (in strain WR). The role of CP77 gene orthologues in monkeypox or variola virus life cycle is not known.
This study analyzed the structure of CP77 to determine the domains that are important for its host range activity. A cellular protein that binds to CP77 was also identified and its involvement in vaccinia virus host restriction in CHO cells investigated.
MATERIALS AND METHODS
Cell cultures, viruses, yeast strains, and reagents.
BHK-21 cells were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS). CHO-K1 cells were cultured in F-12 medium supplemented with 5% FBS. 293T cells were cultured in Dulbecco's modified Eagle's medium with 10% FBS. Isolation of the vaccinia virus host range mutant VV-hr has been described previously (13). VV-hr has an 18-kb deletion of the viral genome which includes two genes, designated K1L and C7L genes, that are known to be essential for VV multiplication in various human cell lines (23, 45). Stocks of VV-hr and recombinant viruses containing green fluorescent protein (GFP), wild-type GFP-CP77, and GFP-CP77 deletion constructs were grown in BHK-21 cells. Antibodies (Abs) against GFP were purchased from Clontech, Inc. The preparation of the rabbit anti-G8R antiserum was described previously (29). Horseradish peroxidase-conjugated goat anti-mouse Ab was obtained from Piers Inc., and alkaline phosphatase-conjugated goat anti-rabbit Ab was obtained from Calbiochem Inc. Yeast strains L40 (MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS::(lexAop) HIS3-4 URA3::(lexAop) lacZ-8 GAL4) and AMR70 (MATα trp1 leu2 his3 URA3::lexA-lacZ) were used for transformation of the LexA-DNA binding domain and Gal4 activation domain fusion proteins (70). The yeast strains were maintained at 30°C on yeast extract-peptone-dextrose plates with 40 μg/ml adenine.
Construction of CP77 deletion mutant plasmids.
The CP77 open reading frame (ORF) was amplified by PCR from the cowpox virus genome and cloned into a yeast shuttle vector, pGBT9 (Clontech, Inc.). The resulting plasmid, pGBT9-CP77, was digested with EcoRI and BamHI, and the CP77 ORF insert was cloned into pcDNA3 to obtain pcDNA3-CP77, which was digested with BamHI, and the CP77 ORF was purified and subsequently cloned into pEFGP-C1 to obtain pEGFP-C1-CP77. CP77 deletion mutants were generated by digestion of pEGFP-C1-CP77 with restriction enzymes as follows. Plasmid CP77(79-668) was produced by digestion of EGFP-CP77 with HindIII and AccI, filling in with Klenow fragment, and religation; CP77(176-668) by digestion with BglII and partial MluI treatment, filling in with Klenow fragment, and religation; CP77(278-668) by digestion with XhoI and religation; CP77(353-668) by digestion with EcoRV and XhoI, filling in with Klenow fragment, and religation; CP77(441-668) by digestion with HindIII and ScaI and filling in with Klenow fragment; CP77(1-440) by digestion with ScaI and XbaI, filling in with Escherichia coli DNA polymerase I in the presence of dTTP/dCTP, and religation; and CP77(1-504) by digestion with XbaI and religation. All plasmid constructs were confirmed by sequencing.
The coding region of each of the aforementioned CP77 deletion proteins in pEGFP-C1 was excised out with NheI and BamHI [for CP77(79-668)], NheI and MluI [for CP77(176-668), CP77(278-668), CP77(353-668), and CP77(441-668)], NheI and DraI [for CP77(1-440)], or NheI and XbaI [for CP77(1-504]. Each of the coding sequences was cloned into a SmaI-digested pSC11-360 vector, and the CP77 deletion genes were expressed from a previously described synthetic early promoter (29). As a control, the coding region of GFP, obtained by digesting pEGFP-C1 with NheI and MluI, was cloned into SmaI-digested pSC11-360.
To generate the constructs CP77(1-352) and CP77(79-352), in vitro mutagenesis was performed using a QuikChange XL site-directed mutagenesis kit (Stratagene) as described below. For EGFP-CP77(1-352), the primers 5′ATAACATTCAGCGATATCGATTGATCATAATCAGCCATACCA 3′ and 5′TGGTATGGCTGATTATGATCAATCGATATCGCTGAATGTTAT 3′ were used with plasmid pSC11-360/EGFP-CP77 [CP77 (1-440)] as the template for the PCR (95°C for 50 s, 55°C for 50 s, and 68°C for 22 min) for 18 cycles of amplification. For EGFP-CP77(79-352), the primers 5′-ACATTCAGCGATATCGATTAATAGATCTCTCTAGTGGAATAC-3′ and 5′-GTATTCCACTAGAGAGATCTATTAATCGATATCGCTGAATGT-3′ were used with plasmid pSC11-360/EGFP-CP77(79-668) as the template in the PCR (95°C for 1 min, 95°C for 50 s, 55°C for 50 s, and 68°C for 22 min) for 18 cycles. The PCR products were treated with DpnI (New England BioLab) for 1 h and transformed into E. coli. Plasmids were isolated from the colonies and sequenced. To generate the ΔANK5 construct, in vitro mutagenesis was performed using a QuikChange XL site-directed mutagenesis kit (Stratagene), as described in the user's manual, to remove the internal aa 191 to 231.
Generation and purification of recombinant vaccinia viruses expressing CP77 deletion proteins.
BHK-21 cells (4 × 105) were seeded overnight and infected with VV-hr at a multiplicity of infection (MOI) of 0.1 PFU per cell at 37°C for 1 h. The cells were then transfected with 2 μg of plasmid in 10 μl of Lipofectamine (Invitrogen, Inc.) for 3 h and then cultured in culture medium and harvested at 4 days postinfection (p.i.). Cell lysates, prepared by three cycles of freeze-thawing in phosphate-buffered saline (PBS) containing 0.05% bovine serum albumin and 10 mM MgCl2, were serially diluted for isolation of individual plaques on BHK-21 cells under 1% top agarose with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (150 μg/ml) staining for lacZ expression. Blue plaques were isolated and the recombinant viruses were purified three times or more until 100% purity was achieved.
One-step virus growth and virus titer determination analyses.
CHO-K1 cells (6.5 × 105) in 60-mm-diameter culture dishes were infected for 1 h at 37°C with VV-hr-GFP or VV-hr-GFP-CP77 at an MOI of 5 PFU per cell and harvested 0, 2, 4, 6, 12, 16, 24, and 48 h p.i. Alternatively, CHO-K1 cells were infected for 1 h at 37°C with each of the CP77 deletion viruses at an MOI of 5 PFU per cell and harvested at 0 and 24 h p.i. Virus titers on BHK-21 cells were determined as described previously (29).
Immunoblot analysis.
BHK-21 (8 × 105 cells) or CHO-K1 (7 × 105 cells) in 60-mm-diameter culture dishes were infected with CP77 mutant viruses at an MOI of 5 PFU per cell; then, the cells were harvested at 2 and 6 h p.i. and lysed with sodium dodecyl sulfate (SDS) sample buffer, and the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to a nitrocellulose membrane at 250 mA for 3 h according to the manufacturer's protocol (Bio-Rad). After incubation with 0.2% I-block (Applied Biosystems Inc.) in PBS containing 0.1% Tween 20 (PBST) for 1 h, the membrane was incubated with primary antibody (a 1:4,000 dilution for anti-GFP Ab or a 1:1,000 dilution for anti-G8R Ab), washed five times for 5 min with PBST, and incubated with horseradish peroxidase-conjugated goat anti-mouse Ab (1:2,000) or AP-conjugated goat anti-rabbit Ab (1:1,000). After five washes with PBST, the blots were developed using the ECL system (Amersham Inc.) or CDP-Star (Applied Biosystems Inc.) as described by the manufacturer.
Yeast two-hybrid screening for cellular proteins that interact with CP77.
Yeast two-hybrid screening was performed as described previously (32, 70). The full-length CP77 ORF was fused to LexA-DNA binding domain in the vector pBTM116 and used as a bait for yeast two-hybrid screening analyses with a human HeLa cDNA library (Clontech, Inc.) as described previously (70). Interactions between CP77 and cellular target proteins encoded by the cDNA library activated HIS3 and lacZ reporter genes. HIS3 confers upon yeast the ability to grow on histidine-free plates supplemented with 5 mM 3-amino-1,2,4-triazole, whereas lacZ produces β-galactosidase was detected colorimetrically by filter assays. Several cDNA candidates including HMG20A were isolated from a total of 1.5 × 106 yeast colonies. The specificity of HMG20A binding to CP77 bait was further tested with control vectors containing other cDNA baits, such as lamin, transforming acidic coiled-coil protein 3 (TACC3), and the PH domain of Etk (ETK-PH) (32), and no interactions between HMG20A and the control baits mentioned above were detected.
GST pulldown analyses.
GST, GST-CP77, GST-CP77(1-352), GST-CP77(1-234), GST-CP77(1-175), and GST-CP77(1-131) were overexpressed in E. coli BL21(DE3) and purified by glutathione Sepharose 4B (Amersham Bioscience) beads as described previously (24). For the pulldown assays, 10 μg of each purified GST fusion protein was bound to glutathione-agarose beads and incubated with lysates of HeLa cells that had been transfected with pcDNA3.1/HMG20A-His-V5, infected with vTF7-3 and harvested at 8 h p.i. After being mixed for 2 h at 4°C in pulldown buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM EDTA, 0.5% NP-40, 2 μg/ml of aprotinin, 1 μg/ml of leupeptin, 0.7 μg/ml of pepstatin A, and 1 mM phenylmethylsulfonyl fluoride), the samples were washed five times in the same buffer and then boiled for 5 min in SDS sample buffer. The proteins were then resolved by SDS-PAGE and subjected to immunoblot analysis using anti-V5 Ab (1:5,000).
Immunofluorescence microscopy.
To monitor protein expression in virus-infected cells, CHO-K1 cells (1 × 105) were seeded on coverslips in 12-well plates. The next day, the cells were transfected with plasmid pDsRed, pHMG20A-DsRed, or pΔHMG-box-DsRed (see below), incubated overnight, and then infected for 1 h with recombinant VV-hr-GFP or VV-hr-GFP-CP77 at an MOI of 10 PFU per cell. At different times p.i., the cells were fixed for 25 min at room temperature in 4% paraformaldehyde, washed in PBS, permeabilized for 5 min at room temperature in 0.2% Triton X-100, and stained for 5 min with DAPI (4′,6′-diamidino-2-phenylindole) (0.5 μg/ml). In experiments in which cells needed to be treated with rifampin to block virion assembly, it was added at a final concentration of 100 μg/ml into the culture medium after virus infection and remained in the medium until cell harvesting at 8 h p.i. Cell images were collected by confocal laser scanning microscopy (Carl Zeiss, Gottingen, Germany), using a 100 × objective lens. For quantification, roughly 150 cells were individually counted in each experiment to determine the number of cells containing HMG20A at viral factories (HMG20A+ cells) or not containing HMG20A at viral factories, and the percentage of HMG20A+ cells was determined accordingly.
Construction of HMG20A and deletion plasmids expressing HMG20A-DsRed fusion proteins.
The pDsRed2 vector (Clontech, Inc.) was digested with AgeI, filled in with Klenow fragment, and digested with NotI, and then the DNA fragment containing the DsRed2 ORF was isolated by using agarose gel purification. Human HMG20A cDNA (IRAKp961D0315) was purchased from the Ressourcenzentrum f Genomforschung resource center, and the coding region was amplified by PCR using the primers 5′-ATGGAAAACTTGATGACTAGCTCC-3′ and 5′-ACGATCGAGTCTGTTCACAACTTC-3′ and cloned into the vector pcDNA3.1/V5-His (Invitrogen), resulting in pcDNA3.1/HMG20A-V5-His, which was subsequently sequenced. This plasmid was digested with EcoRV and NotI and the DsRed2 ORF DNA fragment (purified as described above) inserted to obtain plasmid pHMG20A-DsRed. The pΔHMG-box-DsRed plasmid was generated from pHMG20A-DsRed, using an in vitro mutagenesis kit (Stratagene, Inc.), and the primers 5′-CCTCTTCGAGACAGCAATGCACAGAAAACAGAGGCCTACAAG-3′ and 5′-CTTGTAGGCCTCTGTTTTCTGTGCATTGCTGTCTCGAAGAGG-3′ to remove internal sequences encoding amino acids 103 to 170.
RESULTS
Construction and expression of nested deletion mutants of the GFP-CP77 fusion protein.
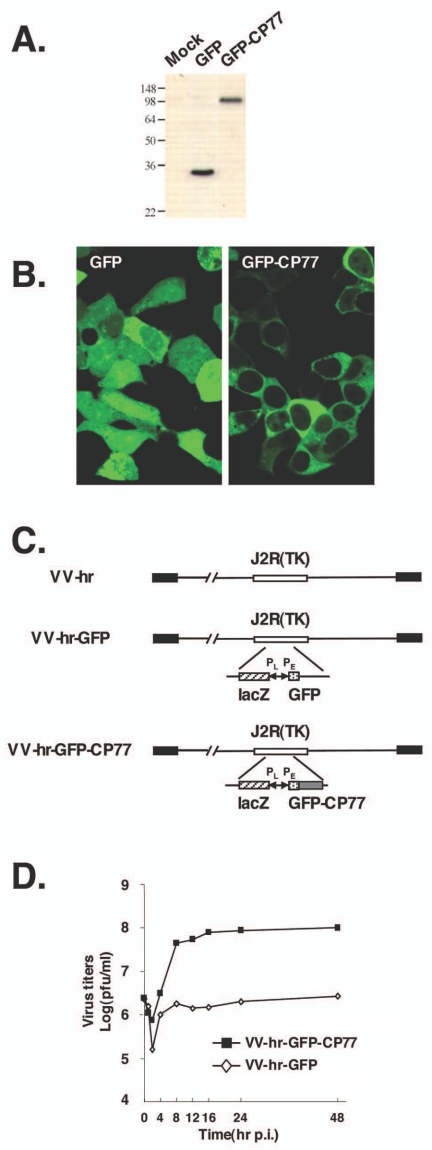
CP77 was fused to the C terminus of GFP, resulting in the detection of a 110-kDa fusion protein on immunoblots (Fig. 1A) (57, 63). Immunofluorescence studies of transfected 293T cells showed that more GFP-CP77 accumulated in the cytoplasm than in the nucleus, whereas GFP was evenly distributed in the two compartments (Fig. 1B). To ensure that the GFP-CP77 fusion protein retained its host range activity, we inserted GFP or GFP-CP77 ORF into the tk (J2R) locus of the previously described vaccinia virus host range mutant, VV-hr (Fig. 1C) (13). Both GFP and GFP-CP77 in VV-hr were expressed from a viral early promoter, and the resulting recombinant VV-hr-GFP and VV-hr-GFP-CP77 viruses were isolated. Analysis of one-step growth curves revealed that VV-hr-GFP-CP77 virus established a productive infection in CHO-K1 cells, resulting in virus titers 53 times higher than those of the control VV-hr-GFP virus at 24 h p.i. (Fig. 1D). The virus yield of VV-hr-GFP-CP77 was comparable to that produced by VV-36hr, a recombinant VV-hr virus expressing the wild-type CP77 (data not shown) (14, 29). These data showed that the GFP-CP77 fusion protein retained its biological activity of overcoming restriction of VV-hr virus in CHO-K1 cells.
FIG. 1.
Construction and expression of GFP-CP77. (A) Immunoblot analysis of GFP and GFP-CP77, expressed from the cytomegalovirus promoter, in transfected 293T cells with anti-GFP Ab (1:4,000) (B) Confocal microscopy of GFP and GFP-CP77 in transfected 293T cells. (C) Construction of recombinant VV-hr expressing GFP or GFP-CP77. VV-hr is a vaccinia virus mutant that contains an 18-kb deletion at the left end of the viral genome (13). VV-hr-GFP and VV-hr-GFP-CP77 were generated by inserting an expression cassette containing GFP or GFP-CP77 ORF driven by an early promoter (PE) and the lacZ gene driven by a viral late promoter (PL) into the tk (J2R) locus, as described previously (29). (D) One-step growth curve analysis of recombinant VV-hr viruses in CHO-K1 cells. CHO-K1 cells were infected with VV-hr-GFP or VV-hr-GFP-CP77 at an MOI of 5 PFU per cell, and cell lysates were harvested at 0, 2, 4, 8, 12, 16, 24, or 48 h p.i. for virus titer determination on BHK-21 cells.
N-terminal residues 1 to 352 of CP77 are sufficient for the host range activity in CHO-K1 cells.
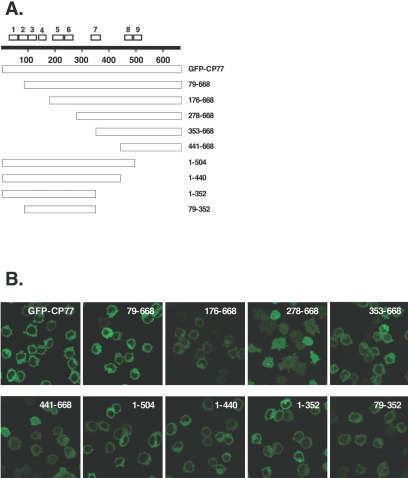
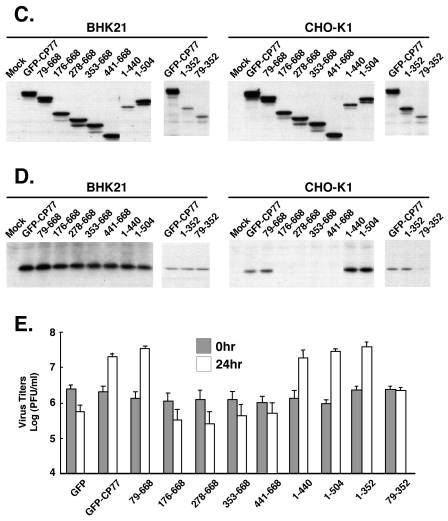
To determine which structural domain of CP77 was important for its host range activity, a series of GFP-CP77 N-terminal and C-terminal deletion constructs was generated (Fig. 2A), and these constructs were inserted into the tk locus of VV-hr for expression from a viral early promoter, as described above for full-length GFP-CP77. Recombinant viruses expressing the CP77 deletion proteins were isolated from BHK-21 cells permissive for VV-hr, as described in Materials and Methods (29). As expected, all the recombinant viruses grew well in BHK-21 cells (data not shown). When CHO-K1 cells were infected with recombinant VV-hr viruses expressing GFP-CP77 deletion proteins, all the CP77 deletion proteins were predominantly expressed in the cytoplasm, except for CP77(278-668), which was expressed in both the nucleus and cytoplasm (Fig. 2B). When CHO-K1 cells were infected with these recombinant viruses, with the exception of CP77(1-440) and CP77(79-352), which were expressed at lower levels, comparable expression of CP77 deletion proteins was seen at 2 h p.i. (Fig. 2C). No major difference of proteins expression was detected between CHO-K1 and BHK-21 cells. Since host restriction in CHO-K1 cells is reported to be at the level of the translation of viral intermediate genes (53), detection of viral intermediate proteins in infected CHO-K1 cells serves as a diagnostic marker for hr activity. When infected BHK-21 and CHO-K1 cells were harvested at 6 h p.i. for immunoblot analyses, as expected, G8R was expressed efficiently in permissive BHK-21 cells in all cases (Fig. 2D, left panel), while in CHO-K1 cells, it was expressed in cells expressing full-length CP77, CP77(79-668), CP77(1-440), CP77(1-504), or CP77(1-352) but not in those expressing CP77(176-668), CP77(278-668), CP77(353-668), CP77(441-668), or CP77(79-352) (Fig. 2D, right panel). Since CP77(1-440) was expressed at a low level and still retained activity, the loss of hr activity in the other CP77 deletion proteins is not due to differences in protein stability. The only exception is CP77(79-352), which was also expressed at a low level, suggesting that its loss of hr activity may be due to protein instability. We therefore concluded that residues 1 to 352 of CP77 were sufficient for supporting VV-hr growth in CHO-K1 cells and that the C-terminal half (residues 353 to 668) was not needed. Virus progeny production in CHO-K1 cells infected with the CP77 deletion mutant viruses, measured by virus titers at 24 h p.i., supported this conclusion (Fig. 2E).
FIG. 2.
GFP-CP77 deletion mutant viruses in infected CHO-K1 cells. (A) CP77 deletion constructs used. The white boxes at the top represent the nine ankyrin repeats in CP77. (B) Immunofluorescence analysis of CP77 deletion proteins. CHO-K1 cells were infected with each of the recombinant viruses expressing CP77 deletion proteins at an MOI of 5 PFU per cell, fixed with paraformaldehyde at 2 h p.i., and photographed using confocal microscopy. (C) Immunoblots of CP77 deletion proteins in infected BHK-21 and CHO-K1 cells. Cells were infected with the CP77 deletion mutant viruses shown in panel A and harvested at 2 h p.i. for immunoblot analysis using anti-GFP Ab (1:4,000). (D) Immunoblots of viral G8R protein in infected BHK-21 and CHO-K1 cells. Cells were infected with the CP77 deletion mutant viruses shown in panel A and harvested at 6 h p.i. for immunoblot analysis using anti-G8R Ab (1:1,000). (E) Growth of CP77 deletion viruses in CHO-K1 cells. CHO-K1 cells were infected as described for panel B and harvested at 0 or 24 h p.i., and virus titers in cell lysates were determined by plaque assays on BHK-21 cells.
CP77 binds via N-terminal residues 1 to 234 to a novel cellular protein, HMG20A, in vitro.
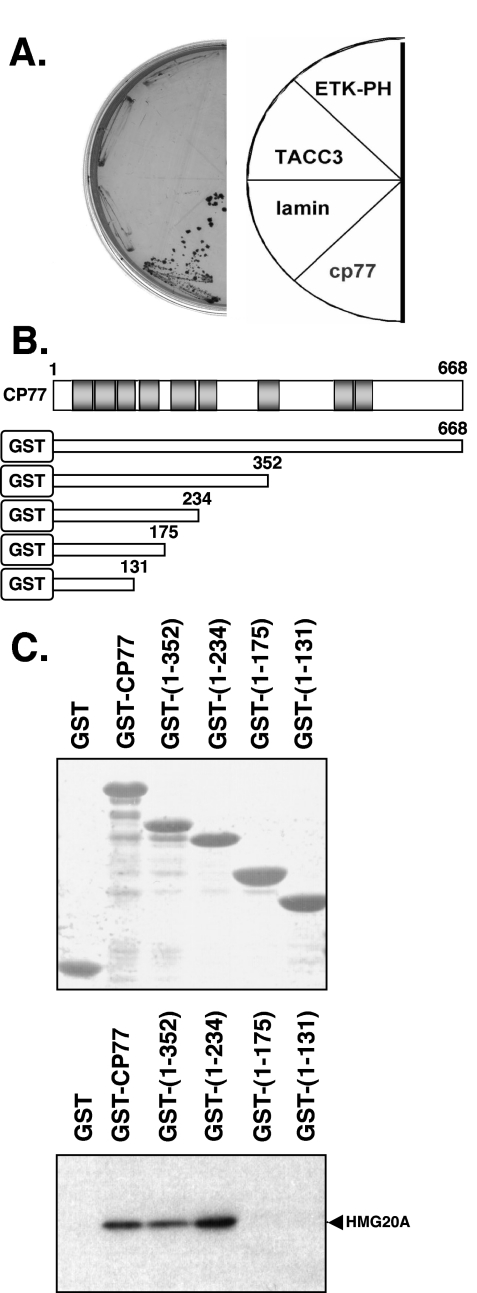
To examine the molecular mechanism of CP77 function, we performed yeast two-hybrid screening, using full-length CP77 as a bait, as described in Materials and Methods, and identified a cellular protein, HMG20A, which bound to CP77. This singly isolated yeast clone contained a full-length cDNA encoding HMG20A (64). HMG20A interacted with CP77 specifically and not with other control bait proteins, such as lamin, TACC3, and ETK-PH (Fig. 3A) (32).
FIG. 3.
The N-terminal region from aa 1 to 234 of CP77 binds to HMG20A in vitro. (A) Yeast two-hybrid analyses identified that HMG20A specifically interacts with CP77 bait and not with baits such as lamin, TACC3, and ETK-PH. (B) The diagram shows CP77 protein with the nine ankyrin repeats (gray boxes). The GST-CP77 deletion constructs used are shown below; the numbers represent the C-terminal amino acids in each constructs. (C). GST pulldown with purified recombinant GST-CP77 deletion proteins. Each purified proteins were separated on SDS-PAGE, stained with Coomassie blue, and photographed. For the pulldown assays, 10 μg of each purified recombinant protein bound to glutathione-agarose beads was incubated with virus-infected cell lysates overexpressing V5-tagged HMG20A. The beads were then washed, and the pulled-down HMG20A protein was analyzed in immunoblot with anti-V5 Ab (1:5,000).
HMG20A, which belongs to the HMG box family, is a protein of 347 residues (64). To confirm that HMG20A bound to CP77, we expressed and purified GST and GST-CP77 from bacteria (Fig. 3B). In addition, GST was fused with different N-terminal fragments of CP77 (residues 1 to 352, 1 to 234, 1 to 175, and 1 to 131) to determine the minimal region of CP77 for binding to HMG20A. These proteins were purified and used to perform GST pulldown analyses with V5-tagged HMG20A ectopically expressed in virus-infected cells (Fig. 3C). The results demonstrated that HMG20A bound to GST-CP77 but not to GST. Besides, constructs CP77(1-352) and CP77(1-234) pulled down HMG20A, whereas constructs CP77(1-175) and CP77(1-131) did not. The results demonstrated that the CP77(176-233) residues, containing the fifth ankyrin repeat (ANK5), were necessary and that N-terminal aa 1 to 234 were sufficient to bind HMG20A in vitro.
HMG20A is a nuclear protein and is translocated to viral factories, where it colocalizes with GFP-CP77 in infected CHO-K1 cells.
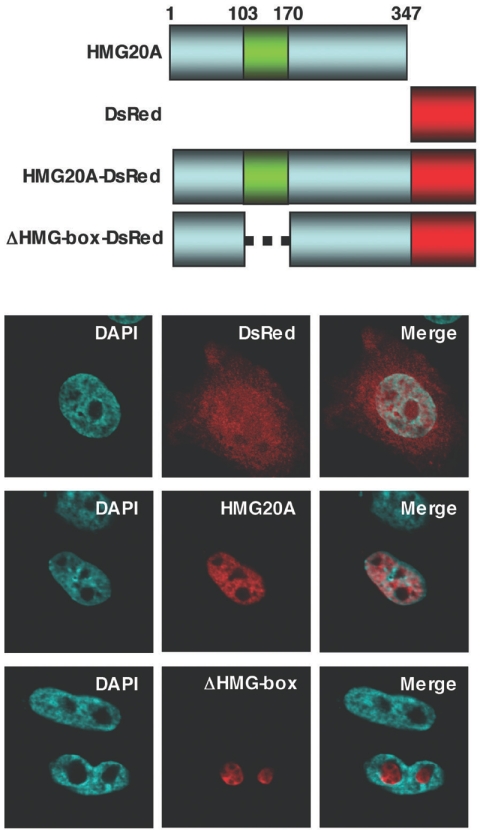
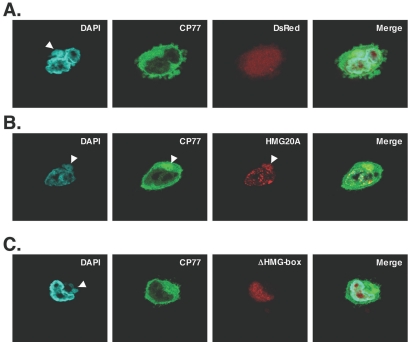
We then generated a construct coding for a fluorescent fusion protein consisting of full-length HMG20A fused to the N terminus of DsRed (Fig. 4). We also constructed a HMG20A mutant, ΔHMG box, in which the DNA binding domain, the HMG box, was deleted by the removal of residues 103 to 170 (55). When the plasmids were transfected into CHO-K1 cells, the control DsRed protein was spread throughout the cell, HMG20A protein was localized exclusively in the nucleus, and the ΔHMG box mutant protein was localized in the nucleolus. These transfected CHO-K1 cells were then infected for 1 h with VV-hr-GFP-CP77 at an MOI of 10 PFU per cell and fixed at 4 h p.i. for immunofluorescence analysis (Fig. 5). Control DsRed protein spread throughout the cell, with no specific location in cells (Fig. 5A). In contrast, a small fraction of HMG20A was translocated into the cytoplasm after virus infection and colocalized with GFP-CP77 (Fig. 5B) at the viral factories. This shows that the translocated HMG20A binds to viral genome within the viral factories, since the ΔHMG box mutant, lacking the DNA binding domain, was not localized to viral factories (Fig. 5C).
FIG. 4.
HMG20A is a nuclear protein. A schematic representation of the HMG20A constructs is shown at the top of the figure. The green box represents the HMG box region that binds to DNA. CHO-K1 cells were transfected with pDsRed, pHMG20A-DsRed, or pΔHMG-box-DsRed plasmids that express DsRed fusion protein from a cytomegalovirus promoter and cell images collected at 24 h posttransfection by confocal laser scanning microscopy (Carl Zeiss, Gottingen, Germany) using a 100 × objective lens with excitation/emission wavelengths of 563 nm/582 nm for DsRed, 359 nm/461 nm for DAPI, and 484 nm/510 nm for enhanced GFP.
FIG. 5.
Colocalization of CP77 with HMG20A at viral factories in infected CHO-K1 cells. CHO-K1 cells on coverslips were transfected with pDsRed (A), pHMG20A-DsRed (B), and pΔHMG-box-DsRed (C) plasmids and then infected with recombinant VV-hr-GFP-CP77 at an MOI of 10 PFU per cell. At 4 h p.i., the cells were fixed, permeabilized, and stained with DAPI (0.5 mg/ml) for 5 min. Cell images were collected with confocal laser scanning microscopy (Carl Zeiss, Gottingen, Germany) using a 100 × objective lens. The white arrows show viral factories where CP77 colocalized with HMG20A.
CP77 regulates the dissociation of HMG20A from viral factories in infected CHO-K1 cells.
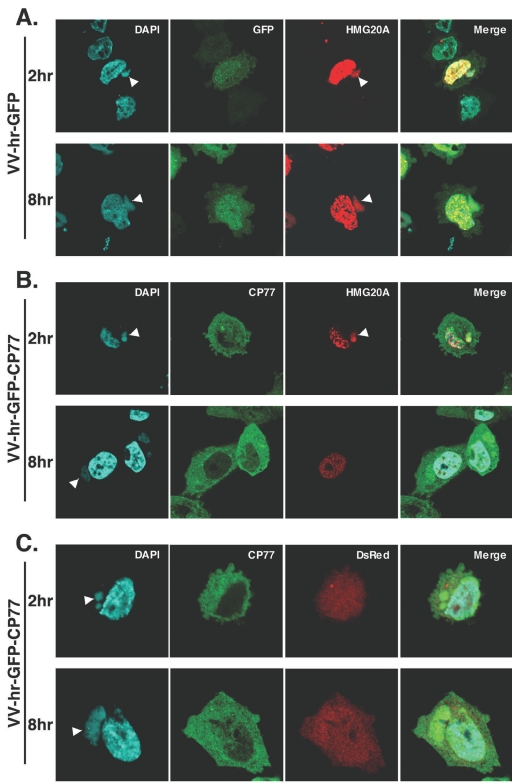
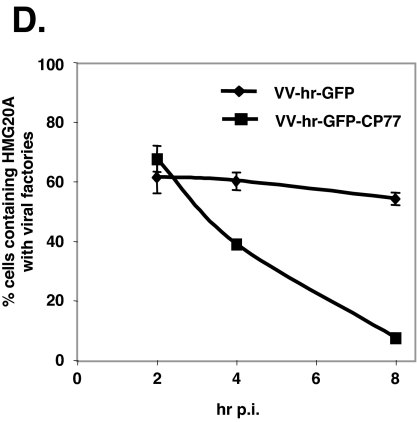
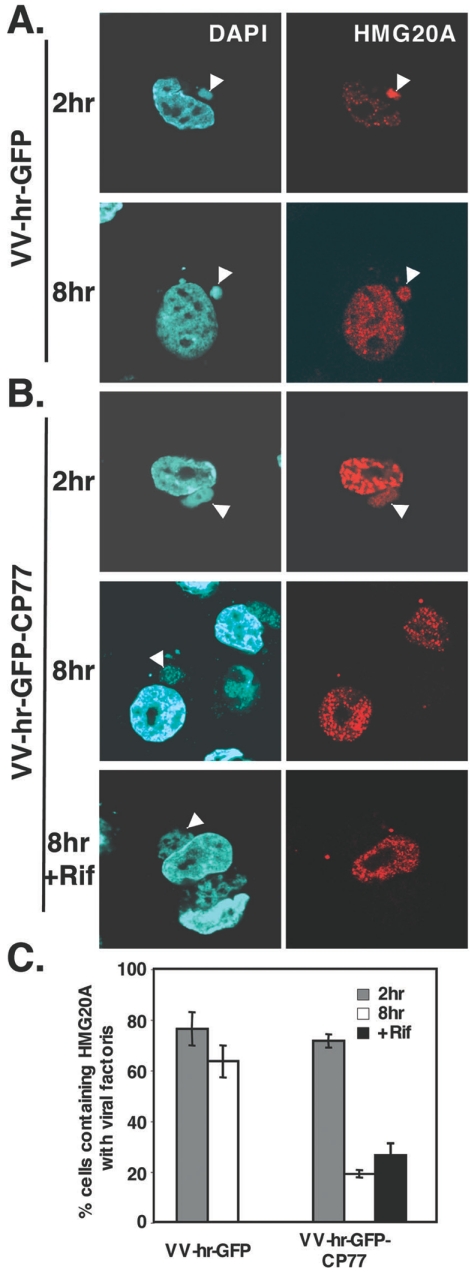
To understand how HMG20A is involved in the virus life cycle and how it is kinetically regulated by CP77, CHO-K1 cells were transfected with control DsRed or HMG20A, infected with VV-hr-GFP or VV-hr-GFP-CP77, and fixed at 2 and 8 h p.i. To our surprise, translocation of HMG20A to cytoplasmic viral factories was seen at 2 h p.i. in cells infected with either VV-hr-GFP (Fig. 6A) or VV-hr-GFP-CP77 (Fig. 6B), indicating that nuclear export of HMG20A after virus infection does not require CP77. Consistent with the data in Fig. 5, HMG20A was specifically colocalized with GFP-CP77 (Fig. 6B) but not with GFP (Fig. 6A) at the viral factories. As expected, control DsRed did not localize in viral factories (Fig. 6C). At 8 h p.i., the association of HMG20A with viral factories persisted in cells infected with VV-hr-GFP (Fig. 6A) but not in cells infected with VV-hr-GFP-CP77 (Fig. 6B), suggesting that CP77 causes dissociation of HMG20A from viral factories. Figure 6D shows the numbers of infected cells containing HMG20A-positive viral factories at 2, 4, and 8 h p.i. The data showed that 61.4%, 60%, and 54.1% of VV-hr-GFP-infected cells contained HMG20A-associated viral factories at 2, 4, and 8 h p.i., respectively, while in VV-hr-GFP-CP77-infected cells, the corresponding values were 68%, 39%, and 7.5%. The data therefore revealed two important points about the involvement of HMG20A in vaccinia virus infections. Firstly, CP77-independent translocation of nuclear HMG20A to cytoplasmic viral factories occurred early after vaccinia virus infection, and secondly, CP77-dependent dissociation of HMG20A from viral factories was seen at 4 to 8 h p.i, concomitant with viral intermediate gene expression in the cells.
FIG. 6.
HMG20A dissociates from viral factories in CHO-K1 cells expressing CP77. (A to C) CHO-K1 cells were transfected with HMG20A-DsRed (A and B) or DsRed (C), infected with recombinant VV-hr-GFP (A) or VV-hr-GFP-CP77 (B and C) at an MOI of 10 PFU per cell, and then collected at 2 and 8 h p.i., fixed, permeabilized, and stained with DAPI (0.5 mg/ml). Cell images were collected by confocal laser scanning microscopy (Carl Zeiss, Gottingen, Germany) using a 100 × objective lens. The white arrowheads show the locations of viral factories in the cells. (D) Quantification of cells containing HMG20A associated with viral factories. For each experiment performed at 2, 4, and 8 h p.i., ∼150 cells were individually counted to obtain the numbers of cells containing HMG20A at viral factories and cells not containing HMG20A at viral factories, and the percentage of cells with HMG20A at viral factories was calculated.
To demonstrate that CP77-dependent dissociation of HMG20A did not occur as a consequence of virus growth, we monitored HMG20A dissociation from viral factories when virion assembly is inhibited by rifampin (Fig. 7). Rifampin is a drug that prevents formation of the crescent-shaped viral membranes and subsequent steps in virus morphogenesis without a significant effect on viral gene expression and DNA or protein synthesis (41, 42, 61). CHO-K1 cells were infected with VV-hr-GFP-CP77 and subsequently treated with rifampin at a dose of 100 μg/ml. Control infections with VV-hr-GFP (Fig. 7A) or VV-hr-GFP-CP77 (Fig. 7B) on cells that were not treated with rifampin were performed in parallel. As expected, in rifampin-treated cells, viral protein synthesis remained active, while the VV-hr-GFP-CP77 titer was effectively reduced 800-fold by the drug (data not shown). More importantly, dissociation of HMG20A from viral factories at 8 h p.i was not inhibited by rifampin in cells infected with VV-hr-GFP-CP77 (Fig. 7B), and the quantified results, as shown in Fig. 7C, also supported the same conclusion. The findings thus strongly supported the concept that CP77 regulates HMG20A dissociation in a direct way.
FIG. 7.
HMG20A dissociates from viral factories in CHO-K1 cells does not require virion assembly. CHO-K1 cells were transfected with HMG20A-DsRed, infected with recombinant VV-hr-GFP (A) or VV-hr-GFP-CP77 (B) at an MOI of 10 PFU per cell, and then collected at 2 and 8 h p.i., fixed, permeabilized, and stained with DAPI (0.5 mg/ml). When rifampin was used, it was added to the infected cells after virus infection at a final concentration of 100 μg/ml and remained in medium until cell harvesting. Cell images were collected by confocal laser scanning microscopy (Carl Zeiss, Gottingen, Germany) using a 100 × objective lens. The white arrowheads show the locations of viral factories in the cells. (C) Quantification of cells containing HMG20A associated with viral factories. For each experiment performed at 2 and 8 h p.i., ∼150 cells were individually counted to obtain the numbers of cells containing HMG20A at viral factories and cells not containing HMG20A at viral factories, and the percentage of cells with HMG20A at viral factories was calculated.
CP77 binding to HMG20A is essential for HMG20A dissociation from viral factories in infected CHO-K1 cells.
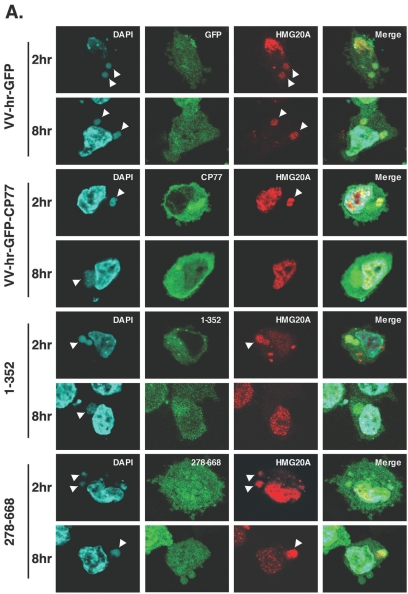
To demonstrate that CP77 binding to HMG20A is required for dissociation of HMG20A from viral factories, we employed two additional recombinant viruses, VV-hr-CP77(1-352), a CP77 mutant virus that contains the HMG20A binding region and has host range function, and VV-hr-CP77(278-668), which does not. CHO-K1 cells were transfected with HMG20A, infected for 1 h with VV-hr-GFP, VV-hr-GFP-CP77, VV-hr-GFP-CP77(1-352), or VV-hr-GFP-CP77(278-668) at an MOI of 10 PFU per cell, and fixed at 2 and 8 h p.i. for immunofluorescence analysis. As shown in Fig. 8A, translocation of nuclear HMG20A to cytoplasmic viral factories occurred at 2 h p.i in all four sets of infected cells, confirming that nuclear export of HMG20A after virus infection does not require CP77. At 8 h p.i., dissociation of HMG20A from viral factories occurred in cells expressing full-length GFP-CP77 or GFP-CP77(1-352), both of which were shown to bind to HMG20A in vitro. In contrast, HMG20A remained bound to viral factories in cells expressing control GFP or GFP-CP77(278-668), which lacks the HMG20A binding site at residues 1 to 234. The quantified immunofluorescence data are shown in Fig. 8B. Together, the results showed that CP77 binding to HMG20A is required for dissociation of HMG20A from viral factories.
FIG. 8.
Binding of CP77 to HMG20A is required for HMG20A dissociation from viral factories. (A) CHO-K1 cells were transfected with plasmid expressing HMG20A-DsRed, then infected with VV-hr-GFP, VV-hr-GFP-CP77, VV-hr-GFP-CP77(1-352), or VV-hr-GFP-CP77(278-668) as described above, and fixed at 2 and 8 h p.i. for confocal immunofluorescence analyses. The white arrowheads point to viral factories. (B) Quantification of cells containing HMG20A associated with viral factories. For each experiment, ∼150 cells were individually counted to obtain the numbers of cells containing HMG20A at viral factories or not containing HMG20A at viral factories, and the percentage of cells showing HMG20A at viral factories was calculated.
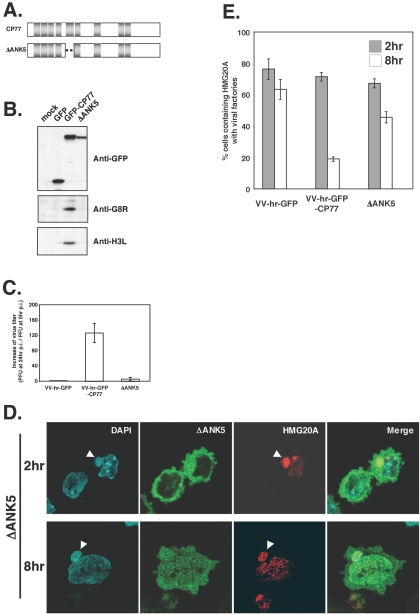
Although GFP-CP77(278-668) provided good evidence that HMG20A binding is important for CP77 activity, one might still argue that the conclusion is flawed due to the fact that CP77(79-668) grew well in CHO-K1 cells and that CP77(278-668) had residues 235 to 277 removed in addition to aa 1 to 234. Thus, we constructed an additional CP77 mutant virus, a ΔANK5 virus, that had the ANK5 domain from aa 191 to 231 deleted, within a region shown to be necessary for CP77 binding to HMG20A (aa 175 to 234) in GST pulldown analyses (Fig. 3). Expression of ΔANK5 protein was detected in the infected CHO-K1 cells; however, viral intermediate (G8R) and late (H3L) proteins were not detected (Fig. 9B), suggesting that the CP77 ΔANK5 protein has lost the hr activity. Consistently, in one-step growth analyses, the ΔANK5 virus titer increased a minimal 6.5-fold at 24 h p.i. when the full-length GFP-CP77 grew 127-fold, showing a decrease of 20 times in virus yield (Fig. 9C). Most importantly, in cells expressing ΔANK5 protein, HMG20A was translocated to viral factories at 2 h p.i. and remained bound to viral factories at 8 h p.i. (Fig. 9D). As shown in Fig. 9E, in cells infected with ΔANK5 mutant, most (∼70%) of the HMG20A that already translocated to viral factories at 2 h p.i. remained bound to viral DNA at 8 h p.i., whereas in cells expressing full-length GFP-CP77 most (∼75%) of HMG20A that bound to viral DNA at 2 h p.i. became dissociated at 8 h p.i. (Fig. 9E), demonstrating that CP77 hr activity requires CP77 binding to HMG20A to dissociate HMG20A from viral factories.
FIG. 9.
The ANK5 domain is required for CP77 h activity and for HMG20A dissociation from viral factories. (A) Schematic representation of the CP77 ΔANK5 mutant construct. The dotted line represents the deletion of aa 191 to 231. The gray boxes represent ankyrin repeats. (B) Immunoblot analyses of viral proteins in cells infected with ΔANK5 mutant virus. CHO-K1 cells were mock infected or infected with VV-hr-GFP, VV-hr-GFP-CP77, or ΔANK5 as described in the text and harvested at 2 and 6 h p.i. for immunoblotting using anti-GFP Ab (1:4,000), anti-G8R Ab (1:1,000), or anti-H3L Ab (1:2,000), respectively. (C) The CP77 ΔANK5 mutant has lost the hr activity. CHO-K1 cells were mock infected or infected with VV-hr-GFP, VV-hr-GFP-CP77, or ΔANK5 virus as described in the legend to Fig. 2E; cells were harvested at 0 and 24 h p.i., and virus titers in lysates were determined on BHK-21 cells. (D) The ΔANK5 mutant protein did not dissociate HMG20A from viral factories at 8 h p.i. CHO-K1 cells were transfected with a plasmid expressing HMG20A-DsRed, then infected with ΔANK5 mutant virus as described above, and fixed at 2 and 8 h p.i. for confocal immunofluorescence analyses. The white arrowheads point to viral factories. (E) Quantification of cells containing HMG20A associated with viral factories. Cells were individually counted as described in the text to obtain the numbers of cells containing HMG20A at viral factories or not containing HMG20A at viral factories, and the percentage of cells showing HMG20A at viral factories was calculated.
DISCUSSION
This study showed that the N-terminal region 1 to 352 of CP77 was sufficient for rescuing vaccinia virus growth in CHO-K1 cells. We also demonstrated that CP77 binds to a cellular protein, HMG20A, as shown by yeast two-hybrid analysis, GST pulldown analyses in vitro, and immunofluorescence studies of virus-infected cells. Since HMG20A is a nuclear protein, our original hypothesis was that CP77 binds to HMG20A and promotes nuclear exit of the latter. However, our data argue against this. First, in cells infected with VV-hr-GFP, translocation of nuclear HMG20A to viral factories in the cytoplasm readily occurred at an early stage, suggesting that virus infection triggered nuclear export of HMG20A. In addition, expression of CP77 resulted in no further increase in cytoplasmic accumulation of HMG20A. Thus, it appears that the early translocation of HMG20A from the nucleus to viral factories is not regulated by CP77. Instead, our results revealed that the subsequent dissociation of HMG20A from viral factories at 4 to 8 h p.i. requires CP77.
Several pieces of evidence supported the idea that HMG20A plays an important role in CP77 host range activity. First, full-length CP77 dissociates HMG20A from viral factories in the infected cells. Dissociation of HMG20A does not require virion morphogenesis, implying a direct regulation. Second, the minimal active CP77 construct [CP77(1-352)] also retains the HMG20A binding site and the ability to dissociate HMG20A from factories, supporting the idea that HMG20A binding and dissociation correlate well with CP77 function. Third, two CP77 mutant proteins, CP77(278-668) and the ΔANK5 protein, lacking the HMG20A binding site, were unable to dissociate HMG20A from viral DNA and had little hr activity, demonstrating that CP77 hr activity requires HMG20A binding to allow HMG20A to be dissociated from viral factories. Together, our data showed that HMG20A dissociation from viral DNA was temporally regulated through its interaction with CP77 in CHO-K1 cells. We previously tried to coimmunoprecipitate HMG20A with CP77 in infected cells but without success (data not shown). It could be that only a small fraction of nuclear HMG20A is translocated to viral factories and that the interaction of CP77 with HMG20A is transient, making coimmunoprecipitation difficult.
HMG20A belongs to a family of proteins containing the HMG box domain, a motif consisting of approximately 70 amino acids and forming three α-helices that bind to the minor groove of DNA in a non-sequence-specific manner (10, 67, 68). HMG proteins are chromosome remodeling proteins that recognize distorted DNA structures, such as cruciforms or chemical-induced adducts (6, 50, 51, 69). They can also induce DNA bending by binding to the minor groove in DNA (47). HMG box-containing proteins are therefore considered important in chromosome remodeling during DNA replication, recombination, or repair (8). In addition, certain HMG box-containing proteins can affect gene transcription by interacting with transcription factors at the local sites (7, 10). In Saccharomyces cerevisiae, the closest match to HMG20A is NHP6A, a nonhistone chromatin-binding protein that is involved in potentiating transcription (47). In mammalian cells, HMG20B, which shares 40% identity with HMG20A, was identified as breast cancer 2 (BRCA2)-associated factor 35 (BRAF35) (37, 64). BRAF35 is a component of a large BRCA2-containing protein complex and is transiently localized on mitotic chromosomes during the prophase-to-metaphase transition, suggesting a role in the early phase of chromosome condensation prior to segregation (37). Some BRAF35 is present in a non-BRCA2- containing six-subunit BRAF-histone-deacetylase complex, which mediates the repression of neuron-specific genes, suggesting that BRAF35 may regulate gene expression through remodeling of chromatin structure (25). Finally, HMG20A was recently shown to recruit histone methytransferase MLL to the promoter of neuronal-specific genes, resulting in increased histone methylation and, consequently, transcriptional activation (74). Although these results uncovered opposite roles for the chromosomal remodeling proteins HMG20A and HMG20B in neuronal gene transcriptional regulation (74), we found no evidence for HMG20B binding to CP77 in our yeast two-hybrid screening.
What could be the role of HMG20A in the vaccinia virus life cycle in cells? One possibility is that HMG20A may serve as an adaptor to recruit CP77 to viral factories. Known as a chromatin remodeling protein, HMG20A may bind to postreplicative DNA intermediates and recruit CP77 to viral factories. Vaccinia virus DNA replication requires two distinct processes, the synthetic and processing phases (3, 12, 41). The initial synthetic phase generates multiple-branched DNA intermediates with hairpins at the concatemer junction (12, 16, 41, 44). With Holliday junctions and crossover branches, these postreplicative concatemeric DNA intermediates could be stabilized by binding to HMG20A in infected cells. In the absence of CP77 expression, the processing of viral DNA is blocked and HMG20A remains bound to the DNA concatemers. In the presence of CP77, the interaction of HMG20A and CP77 at viral factories allows CP77 to interact with other factors to rescue virus growth, leading to concatemer resolution into unit-length viral genome and, thus, HMG20A dissociation from viral DNA (16, 41). A second possibility is that, since HMG20A has been shown to regulate gene transcription in neuronal cells (74), it may affect viral intermediate gene transcription such that CP77-dependent dissociation of HMG20A affects virus translation in CHO-K1 cells. It is conceivable that the binding of HMG20A to the viral genome may alter DNA architecture and facilitate the assembly of protein complexes that control intermediate gene transcription. On the other hand, viral intermediate translation occurred only in cells infected with CP77, which triggers dissociation of HMG20A, implying that HMG20A may negatively regulate the viral intermediate transcription-translation switch. Clearly, other hypotheses are possible, and more study is needed in the future. Although there is no precedent for cellular protein affecting vaccinia viral genome architecture, two HMG box-containing proteins have been reported to be important for virus growth (21, 62). A cellular HMG box-containing protein, T160, is involved in the replication of murine cytomegalovirus, as suppression of T160 expression impairs murine cytomegalovirus replication (21). Another cellular HMG box-containing protein, HMGB1, enhances the replication and transcription activator-mediated viral gene expression of gamma-2 herpesvirus (62). Besides HMG20A protein, several host nuclear factors associated with vaccinia viral factories in infected cells have been identified (9, 46, 60). Whether HMG20A influences the binding of these cellular factors to the viral genome is not known.
Host restriction is also described for other viruses, such as human immunodeficiency virus (HIV). Growth of HIV is restricted by a cellular APOBEC3G protein (26, 49), which belongs to a family of cellular deaminases that introduce catastrophic mutations into the HIV genome (34, 75). To counteract that lethal effect, the HIV Vif protein binds to APOBEC3G protein to promote its degradation (40, 56). Cells permissive of HIV expressed minimal or no APOBEC3G mRNA whereas restrictive cells expressed abundant levels of APOBEC 3G mRNA (58). It will be interesting to determine whether expression of HMG20A transcripts is more abundant in the restrictive cells. Alternatively, it is also possible that other mechanisms, such as intracellular translocation of HMG20A, could be regulated differently in permissive cells than in restrictive cells.
CP77 contains nine ankyrin repeats which are known to function as protein interaction domains. A myxoma host range protein, the M-T5 protein, contains seven ankyrin-repeat domains and was shown to bind to cellular E3 ligase complex member cullin-1 to regulate cell cycle (31) and to bind to Akt protein to regulate host kinase signaling (72). Although HMG20A was first identified in cellular protein binding to CP77, there might be other cellular factors regulated by CP77. It is worth noting that the growth of CP77 ΔANK5 mutant virus in CHO-K1 cells was severely compromised; however, a small increase (sixfold) in virus titer at 24 h p.i. was observed, implying that CP77 ΔANK5 may retain modest hr activity through modulating other cellular factors. More experiments to identify these cellular factors in the future are awaited.
Acknowledgments
This work was supported by grants from the Academia Sinica and the National Science Council (NSC93-2320-B-001-031) of the Republic of China.
REFERENCES
- 1.Ali, A. N., P. C. Turner, M. A. Brooks, and R. W. Moyer. 1994. The SP-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology 202:305-314. [DOI] [PubMed] [Google Scholar]
- 2.Bair, C.-H., C.-S. Chung, I. A. Vasilevskaya, and W. Chang. 1996. Isolation and characterization of a Chinese hamster ovary mutant cell line with altered sensitivity to vaccinia virus killing. J. Virol. 70:4655-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baroudy, B. M., S. Venkatesan, and B. Moss. 1983. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp. Quant. Biol. 47:723-729. [DOI] [PubMed] [Google Scholar]
- 4. Barry, M., S. Hnatiuk, K. Mossman, S. F. Lee, L. Boshkov, and G. McFadden. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239:360-377. [DOI] [PubMed] [Google Scholar]
- 5.Beattie, E., K. L. Denzler, J. Tartaglia, M. E. Perkus, E. Paoletti, and B. L. Jacobs. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi, M. E., M. Beltrame, and G. Paonessa. 1989. Specific recognition of cruciform DNA by nuclear protein HMG1. Science 243:1056-1059. [DOI] [PubMed] [Google Scholar]
- 7.Bonaldi, T., G. Langst, R. Strohner, P. B. Becker, and M. E. Bianchi. 2002. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 21:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branzei, D., and M. Foiani. 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17:568-575. [DOI] [PubMed] [Google Scholar]
- 9.Broyles, S. S., X. Liu, M. Zhu, and M. Kremer. 1999. Transcription factor YY1 is a vaccinia virus late promoter activator. J. Biol. Chem. 274:35662-35667. [DOI] [PubMed] [Google Scholar]
- 10.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, C. S., I. A. Vasilevskaya, S. C. Wang, C. H. Bair, and W. Chang. 1997. Apoptosis and host restriction of vaccinia virus in RK13 cells. Virus Res. 52:121-132. [DOI] [PubMed] [Google Scholar]
- 12.DeLange, A. M. 1989. Identification of temperature-sensitive mutants of vaccinia virus that are defective in conversion of concatemeric replicative intermediates to the mature linear DNA genome. J. Virol. 63:2437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drillien, R., F. Koehren, and A. Kirn. 1981. Host range deletion mutant of vaccinia virus defective in human cells. Virology 111:488-499. [DOI] [PubMed] [Google Scholar]
- 14.Drillien, R., D. Spehner, and A. Kirn. 1982. Complementation and genetic linkage between vaccinia virus temperature-sensitive mutants. Virology 119:372-381. [DOI] [PubMed] [Google Scholar]
- 15.Drillien, R., D. Spehner, and A. Kirn. 1978. Host range restriction of vaccinia virus in Chinese hamster ovary cells: relationship to shutoff of protein synthesis. J. Virol. 28:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckert, D., O. Williams, C. A. Meseda, and M. Merchlinsky. 2005. Vaccinia virus nicking-joining enzyme is encoded by K4L (VACWR035). J. Virol. 79:15084-15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, H., M. Barry, S. F. Lee, X. Sun, K. Graham, J. Stone, R. C. Bleackley, and G. McFadden. 2000. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J. Exp. Med. 191:1487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, H., M. Barry, X. Sun, S. F. Lee, C. Frantz, L. G. Berthiaume, G. McFadden, and R. C. Bleackley. 2002. The myxoma poxvirus protein, M11L, prevents apoptosis by direct interaction with the mitochondrial permeability transition pore. J. Exp. Med. 196:1127-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, H., and G. McFadden. 2002. Poxviruses and apoptosis: a time to die. Curr. Opin. Microbiol. 5:395-402. [DOI] [PubMed] [Google Scholar]
- 20.Fenner, F., and J. F. Sambrook. 1966. Conditional lethal mutants of rabbitpox virus. II. Mutants (p) that fail to multiply in PK-2a cells. Virology 28:600-609. [DOI] [PubMed] [Google Scholar]
- 21.Gariglio, M., P. Foresta, C. Sacchi, M. Lembo, L. Hertel, and S. Landolfo. 1997. Suppression of high mobility group protein T160 expression impairs mouse cytomegalovirus replication. J. Gen. Virol. 78:665-670. [DOI] [PubMed] [Google Scholar]
- 22.Gemmell, A., and F. Fenner. 1960. Genetic studies with mammalian poxviruses III White (u) mutants of rabbitpox virus. Virology 11:219-235. [DOI] [PubMed] [Google Scholar]
- 23.Gillard, S., D. Spehner, R. Drillien, and A. Kirn. 1986. Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc. Natl. Acad. Sci. USA 83:5573-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 25.Hakimi, M. A., D. A. Bochar, J. Chenoweth, W. S. Lane, G. Mandel, and R. Shiekhattar. 2002. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA 99:7420-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 27.Hornemann, S., O. Harlin, C. Staib, S. Kisling, V. Erfle, B. Kaspers, G. Hacker, and G. Sutter. 2003. Replication of modified vaccinia virus Ankara in primary chicken embryo fibroblasts requires expression of the interferon resistance gene E3L. J. Virol. 77:8394-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hruby, D. E., D. L. Lynn, R. C. Condit, and J. R. Kates. 1980. Cellular differences in the molecular mechanisms of vaccinia virus host range restriction. J. Gen. Virol. 47:485-488. [DOI] [PubMed] [Google Scholar]
- 29.Hsiao, J. C., C. S. Chung, R. Drillien, and W. Chang. 2004. The cowpox virus host range gene, CP77, affects phosphorylation of eIF2 alpha and vaccinia viral translation in apoptotic HeLa cells. Virology 329:199-212. [DOI] [PubMed] [Google Scholar]
- 30.Ink, B. S., C. S. Gilbert, and G. I. Evan. 1995. Delay of vaccinia virus-induced apoptosis in nonpermissive Chinese hamster ovary cells by the cowpox virus CHOhr and adenovirus E1B 19K genes. J. Virol. 69:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston, J. B., G. Wang, J. W. Barrett, S. H. Nazarian, K. Colwill, M. Moran, and G. McFadden. 2005. Myxoma virus M-T5 protects infected cells from the stress of cell cycle arrest through its interaction with host cell cullin-1. J. Virol. 79:10750-10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jui, H. Y., R. J. Tseng, X. Wen, H. I. Fang, L. M. Huang, K. Y. Chen, H. J. Kung, D. K. Ann, and H. M. Shih. 2000. Protein-tyrosine phosphatase D1, a potential regulator and effector for Tec family kinases. J. Biol. Chem. 275:41124-41132. [DOI] [PubMed] [Google Scholar]
- 33.Kotwal, G., and B. Moss. 1988. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virol. 167:524-537. [PubMed] [Google Scholar]
- 34.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig, H., Y. Suezer, Z. Waibler, U. Kalinke, B. S. Schnierle, and G. Sutter. 2006. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J. Gen. Virol. 87:1145-1155. [DOI] [PubMed] [Google Scholar]
- 36.Macen, J., A. Takahashi, K. B. Moon, R. Nathaniel, P. C. Turner, and R. W. Moyer. 1998. Activation of caspases in pig kidney cells infected with wild-type and CrmA/SPI-2 mutants of cowpox and rabbitpox viruses. J. Virol. 72:3524-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marmorstein, L. Y., A. V. Kinev, G. K. Chan, D. A. Bochar, H. Beniya, J. A. Epstein, T. J. Yen, and R. Shiekhattar. 2001. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell 104:247-257. [DOI] [PubMed] [Google Scholar]
- 38.McClain, M. E. 1965. The host range and plaque morphology of rabbitpox virus (RPu+) and its u mutants on chick fibroblasts PK-2a and L929 cells. Aust. J. Exp. Biol. Med. Sci. 43:31-44. [DOI] [PubMed] [Google Scholar]
- 39.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 41.Merchlinsky, M., and B. Moss. 1989. Resolution of vaccinia virus DNA concatemer junctions requires late-gene expression. J. Virol. 63:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moss, B., E. N. Rosenblum, E. Katz, and P. M. Grimley. 1969. Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature 224:1280-1284. [DOI] [PubMed] [Google Scholar]
- 43.Mossman, K., S. F. Lee, M. Barry, L. Boshkov, and G. McFadden. 1996. Disruption of M-T5, a novel myxoma virus gene member of poxvirus host range superfamily, results in dramatic attenuation of myxomatosis in infected European rabbits. J. Virol. 70:4394-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moyer, R. W., and R. L. Graves. 1981. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell 27:391-401. [DOI] [PubMed] [Google Scholar]
- 45.Oguiura, N., D. Spehner, and R. Drillien. 1993. Detection of a protein encoded by the vaccinia virus C7L open reading frame and study of its effect on virus multiplication in different cell lines. J. Gen. Virol. 74:1409-1413. [DOI] [PubMed] [Google Scholar]
- 46.Oh, J., and S. S. Broyles. 2005. Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J. Virol. 79:12852-12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paull, T. T., M. J. Haykinson, and R. C. Johnson. 1993. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 7:1521-1534. [DOI] [PubMed] [Google Scholar]
- 48.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 49.Pham, P., R. Bransteitter, and M. F. Goodman. 2005. Reward versus risk: DNA cytidine deaminases triggering immunity and disease. Biochemistry 44:2703-2715. [DOI] [PubMed] [Google Scholar]
- 50.Pil, P. M., and S. J. Lippard. 1992. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science 256:234-237. [DOI] [PubMed] [Google Scholar]
- 51.Pöhler, J. R., D. G. Norman, J. Bramham, M. E. Bianchi, and D. M. Lilley. 1998. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J. 17:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramsey-Ewing, A., and B. Moss. 1998. Apoptosis induced by a postbinding step of vaccinia virus entry into Chinese hamster ovary cells. Virology 242:138-149. [DOI] [PubMed] [Google Scholar]
- 53.Ramsey-Ewing, A., and B. Moss. 1995. Restriction of vaccinia virus replication in CHO cells occurs at the stage of viral intermediate protein synthesis. Virology 206:984-993. [DOI] [PubMed] [Google Scholar]
- 54.Ramsey-Ewing, A. L., and B. Moss. 1996. Complementation of a vaccinia virus host-range K1L gene deletion by the nonhomologous CP77 gene. Virology 222:75-86. [DOI] [PubMed] [Google Scholar]
- 55.Read, C. M., P. D. Cary, C. Crane-Robinson, P. C. Driscoll, and D. G. Norman. 1993. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 21:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10:291-297. [DOI] [PubMed] [Google Scholar]
- 57.Shchelkunov, S. N., P. F. Safronov, A. V. Totmenin, N. A. Petrov, O. I. Ryazankina, V. V. Gutorov, and G. J. Kotwal. 1998. The genomic sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology 243:432-460. [DOI] [PubMed] [Google Scholar]
- 58.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 59.Shisler, J. L., S. N. Isaacs, and B. Moss. 1999. Vaccinia virus serpin-1 deletion mutant exhibits a host range defect characterized by low levels of intermediate and late mRNAs. Virology 262:298-311. [DOI] [PubMed] [Google Scholar]
- 60.Slezak, K., M. Michalik, A. Kowalczyk, and H. Rokita. 2004. YY1 is recruited to the cytoplasm of vaccinia virus-infected human macrophages by the Crm1 system. Virus Res. 102:177-184. [DOI] [PubMed] [Google Scholar]
- 61.Sodeik, B., G. Griffiths, M. Ericsson, B. Moss, and R. W. Doms. 1994. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J. Virol. 68:1103-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song, M. J., S. Hwang, W. Wong, J. Round, D. Martinez-Guzman, Y. Turpaz, J. Liang, B. Wong, R. C. Johnson, M. Carey, and R. Sun. 2004. The DNA architectural protein HMGB1 facilitates RTA-mediated viral gene expression in gamma-2 herpesviruses. J. Virol. 78:12940-12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spehner, D., S. Gillard, R. Drillien, and A. Kirn. 1988. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J. Virol. 62:1297-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sumoy, L., L. Carim, M. Escarceller, M. Nadal, M. Gratacos, M. A. Pujana, X. Estivill, and B. Peral. 2000. HMG20A and HMG20B map to human chromosomes 15q24 and 19p13.3 and constitute a distinct class of HMG-box genes with ubiquitous expression. Cytogenet. Cell Genet. 88:62-67. [DOI] [PubMed] [Google Scholar]
- 65.Sutter, G., A. Ramsey-Ewing, R. Rosales, and B. Moss. 1994. Stable expression of the vaccinia virus K1L gene in rabbit cells complemetns the host range defect of a vaccinia virus mutant. J. Virol. 68:4109-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tagaya, I., T. Kitamura, and Y. Sano. 1961. A new mutant of dermovaccinia virus. Nature 192:381-382. [DOI] [PubMed] [Google Scholar]
- 67.Thomas, J. O. 2001. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 29:395-401. [DOI] [PubMed] [Google Scholar]
- 68.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 69.Travers, A. 2000. Recognition of distorted DNA structures by HMG domains. Curr. Opin. Struct. Biol. 10:102-109. [DOI] [PubMed] [Google Scholar]
- 70.Vojtek, A. B., and S. M. Hollenberg. 1995. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 255:331-342. [DOI] [PubMed] [Google Scholar]
- 71.Wang, G., J. W. Barrett, S. H. Nazarian, H. Everett, X. Gao, C. Bleackley, K. Colwill, M. F. Moran, and G. McFadden. 2004. Myxoma virus M11L prevents apoptosis through constitutive interaction with Bak. J. Virol. 78:7097-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, G., J. W. Barrett, M. Stanford, S. J. Werden, J. B. Johnston, X. Gao, M. Sun, J. Q. Cheng, and G. McFadden. 2006. Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc. Natl. Acad. Sci. USA 103:4640-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wyatt, L. S., M. W. Carroll, C. P. Czerny, M. Merchlinsky, J. R. Sisler, and B. Moss. 1998. Marker rescue of the host range restriction defects of modified vaccinia virus Ankara. Virology 251:334-342. [DOI] [PubMed] [Google Scholar]
- 74.Wynder, C., M. A. Hakimi, J. A. Epstein, A. Shilatifard, and R. Shiekhattar. 2005. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat. Cell Biol. 7:1113-1117. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]