Abstract
Epstein-Barr virus (EBV) is associated with several human malignancies where it expresses limited subsets of latent proteins. Of the latent proteins, latent membrane protein 1 (LMP1) is a potent transforming protein that constitutively induces multiple cell signaling pathways and contributes to EBV-associated oncogenesis. Regulation of LMP1 expression has been extensively described during the type III latency of EBV. Nevertheless, in the majority of EBV-associated tumors, the virus is commonly found to display a type II latency program in which it is still unknown which viral or cellular protein is really involved in maintaining LMP1 expression. Here, we demonstrate that LMP1 activates its own promoter pLMP1 through the JNK signaling pathway emerging from the TES2 domain. Our results also reveal that this activation is tightly controlled by LMP1, since pLMP1 is inhibited by LMP1-activated NF-κB signaling pathway. By using our physiological models of EBV-infected cells displaying type II latency as well as lymphoblastoid cell lines expressing a type III latency, we also demonstrate that this balanced autoregulation of LMP1 is shared by both latency programs. Finally, we show that this autoactivation is the most important mechanism to maintain LMP1 expression during the type II latency program of EBV.
Epstein-Barr virus (EBV) is a widespread herpesvirus, found in more than 90% of healthy adults and persisting, in the vast majority of individuals, as a lifelong and asymptomatic infection. EBV classically infects B cells, causing sometimes a benign disease (infectious mononucleosis) but also malignant disorders, such as B-lymphoproliferative diseases, in patients with severe immunodeficiency. In immunocompetent hosts, EBV is also associated with other malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, peripheral or nasal NK/T-cell lymphomas, and gastric, breast, and hepatocellular adenocarcinomas (46, 50). In all EBV-associated tumors, the virus displays mainly a latency program of infection with a restricted pattern of gene expression, which can be classified in three types. Type I latency, during which only the EBV-encoded nuclear antigen 1 (EBNA1) is expressed, is found in Burkitt's lymphoma. Type II latency is characterized by coexpression of EBNA1 and latent membrane proteins LMP1, LMP2A, and LMP2B and is found in nasopharyngeal carcinoma, Hodgkin's disease, NK/T-cell lymphomas, and AIDS-related non-Hodgkin's lymphomas. Type III latency with expression of the five EBNAs and three latent membrane proteins is restricted to B lymphomas of immunodeficient patients (50).
Cellular models allowing investigation of the roles of LMP1 during type II and type III latencies have been developed. In vitro, EBV can infect and immortalize resting B cells to yield permanent growth of lymphoblastoid cell lines (LCLs) displaying a full latency III program (27). We previously showed that EBV can also infect and transform T cells and monocytes. We have described and extensively characterized two cell lines (TE1 and NC5) in which EBV was found to express a type II latency (20, 39, 40). Moreover, as already shown by others studying LCLs (27), we have demonstrated by two different approaches (i.e., antisense oligonucleotides and our original dominant-negative mutant, LMP1-CT) that LMP1 is essential for proliferation and survival of both of our EBV-transformed models of type II latency (2, 39). Owing to this essential role in EBV-dependent oncogenesis and since it can transform rodent fibroblasts (61) and sensitizes transgenic mice to lymphomas (31), LMP1 is considered the main EBV oncogene.
LMP1 is a 63-kDa plasma membrane protein with six transmembrane segments, which mimics a constitutively activated cell surface receptor of the tumor necrosis factor superfamily (29). Two signaling domains mediating its signaling properties have been identified in the cytoplasmic C-terminal region of LMP1. These domains named TES1 (for transforming effector site 1) or TES2 contain critical residues responsible for the binding of adapters and thus, for inducing several specific signal transduction pathways (24, 25, 28, 37). The nuclear factor κB (NF-κB) and c-Jun N-terminal kinase (JNK) signaling pathways are the most important, since their activation results in the overexpression of most LMP1 target genes (37). These genes include those encoding for antiapoptotic proteins (Bfl1, Mcl1, TRAF1, etc.) (10, 12), various cell surface markers (CD83, CD44, etc.) (13, 62), cellular receptors (CD40, epidermal growth factor receptor, etc.), and intercellular adhesion molecules (ICAM1, LFA1, etc.) (10, 62).
Since LMP1 is the main EBV oncogene, regulation of its expression in EBV-associated cancer cells is an important issue. Transcription of the LMP1 gene can be initiated from two promoters in the viral genome. pLMP1 (also named ED-L1) is the proximal promoter, while TR-L1 is the distal one located in the terminal repeats (7, 23, 51). Both viral and cellular factors regulate the activities of the LMP1 promoters during EBV latency. LMP1 regulation driven by EBNA2 is the best described, but it occurs only in latency III-expressing cells such as LCLs. EBNA2-dependent transactivation involves only pLMP1 (63) and needs cellular transcription factors, such as RBP-Jκ (60), PU.1 (26), and ATF2/c-Jun (53). This transactivation is further enhanced by two other latent proteins, EBNA-LP and EBNA3C (45, 67). Alternatively, pLMP1 can be activated in an EBNA2-independent manner by several cellular proteins, including interferon regulatory factor 7 (IRF7) (42), STAT3 (7), Notch1 (22), Sp1 (34), USF, ATF1, and CREB1 (53). pLMP1 activity can also be inhibited by a Max-Mad1 and histone deacetylase (HDAC) complex (54). By contrast with pLMP1, TR-L1 activation does not require EBNA2 but could be due to action of cellular Sp1/Sp3 (57) or STAT3 (7) transcription factors. Importantly, some of the regulation mechanisms are cell type specific, as exemplified by the lack of pLMP1 transactivation by EBNA2 in epithelial cells (17, 63).
Surprisingly, among all these regulation mechanisms of LMP1 expression, only the EBNA2-dependent one has been shown to be absolutely essential for LMP1 expression. Thus, since EBNA2 is expressed only during the type III latency program of EBV (50), the driving process leading to expression of the LMP1 oncogene in type II latency remains elusive. However, it is important to understand this type of latency program, since it is commonly found in most EBV-associated malignancies. In regard to its strong transactivation abilities mediated by several signaling pathways, we decided to investigate whether LMP1 itself can induce its own expression and what roles the JNK and NF-κB signaling pathways might have in this scenario.
MATERIALS AND METHODS
Cell lines, culture conditions, and drug treatments.
HEK 293 (a human embryonic kidney cell line) was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, penicillin, and streptomycin. PRI (a lymphoblastoid cell line) (41) and TE1 (a monocytic cell line) (39) are both transformed by EBV and were propagated in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM glutamine, 1 mM sodium pyruvate, 1% nonessential amino acids, penicillin, streptomycin, and gentamicin. All cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Where indicated, cells were treated with sulfasalazine (5-[4-(2-pyridylsulfamoyl)phenylazo]salicylic acid) (Sigma) or SP600125 (1,9-pyrazoloanthrone) (Calbiochem) for 16 h. They were both reconstituted in dimethyl sulfoxide (DMSO) (Sigma).
Stable cell clones were cultured in the presence of hygromycin (Euromedex) at 150 μg/ml and 20% fetal calf serum. Twenty million PRI cells were electroporated with 20 μg of pRT1-IκBm or pRT1-LMP1-CT at 250 V and 960 μF (5, 33, 41). Following the manufacturer's instructions (Amaxa Biosystems), one million TE1 cells were nucleofected with 1 μg of pRT1-LMP1-CT in Amaxa's solution V with the V-01 setting of an Amaxa Nucleofector (Amaxa Biosystems). After 4 days, hygromycin was added at 50 μg/ml. Hygromycin concentration was progressively increased up to 150 μg/ml during the first 2 weeks of selection. After 4 weeks of culture in the presence of hygromycin, induction was performed with doxycycline: cells were washed once in RPMI 1640 medium and resuspended in standard medium without hygromycin, and doxycycline (Sigma) was then added at 2 μg/ml.
Plasmids and antibodies.
pSVHA, pSVHA-LMP1, pSVHA-LMP1-TM, and LMP1-CT constructs were all previously described (2). pSVHA-LMP1-Tes1mut and pSVHA-LMP1-Tes2mut mutants were generated by site-directed mutagenesis; codons 204 to 208 in TES1 were mutated from PXQXT to AXAXA, and the codons 384 to 386 YYD in TES2 were deleted. All constructs were sequenced to confirm that they contained the desired mutation. The JNKAPF and MKK7-JNKfus (36) expression vectors were a generous gift of R. J. Davis. The IκBm and p65 expression plasmids were a generous gift of J. Hiscott. The reporter plasmids used in this study carry the firefly luciferase under the control of different regulating sequences. The pLMP-luc construct was obtained by inserting the pLMP1 sequence into the enhancer- and promoter-less vector pGL2-basic (Promega). pLMP1 is defined as nucleotides 169479 to 169841 of the EBV B95-8 sequence, which corresponds to positions −326 to +37 relative to the transcription initiation site. The κB luciferase reporter construct has five NF-κB-responsive elements in tandem and was from Stratagene. The gal4-luc and Gal4-Jun vectors were a generous gift of B. Derijard and M. Ptashne. The normalizing vector pRLnull has no promoter sequences to drive expression of the Renilla luciferase gene and was from Promega. The pRT1-IκBm and pRT1-LMP1-CT vectors carry a doxycycline-inducible version of IκBm or LMP1-CT, respectively (5, 33, 41). Plasmids were purified by use of Nucleobond-EF kits (Macherey-Nagel) according to the manufacturer's instructions.
The anti-LMP1 monoclonal antibody was obtained from the S12 hybridoma culture supernatant. Anti-EBNA2 (PE2) monoclonal antibody was purchased from Dako, anti-JNK1 (sc-474), anti-p65 (sc-109-G), anti-IκBα (sc-1643), anti-TRAF1 (sc-875), anti-ICAM1 (sc-8439), and anti-β-actin (sc-8432) antibodies were obtained from Santa Cruz, and anti-hemagglutinin (anti-HA) antibody (MMS-101P) was purchased from Covance Research Products.
Transfections and luciferase assays.
For transient-expression experiments in epithelial cells, cells were seeded in 12-well plates and were cotransfected using polyethylenimine reagent (ExGen 500; Euromedex) in OptiMEM (Gibco) the following day. Routinely, 80-ng amounts of reporter constructs were cotransfected with 10 ng of the normalizing vector (pRLnull) and the indicated amounts of the effector plasmids. Cells were incubated with reagents for 5 h at 37°C, and then the medium was replaced by fresh culture medium with serum for 48 h.
To measure the reporter activity, cells were harvested 48 h after transfections, with passive lysis buffer (Promega) after washing with 1× phosphate-buffered saline (PBS). Luciferase assay was performed using a Dual Luciferase assay system (Promega) according to the manufacturer's instructions, and firefly and Renilla luciferase activities were measured with a Lumat LB 9507 (Berthold). In all experiments, the ratio of firefly luciferase activity to Renilla luciferase activity was calculated for each condition, and the changes in induction when empty vectors were used is shown. Three replicate samples were used in each experiment, and standard deviations (SD) are shown. Each experiment was repeated at least twice with independent plasmid preparations to assess reproducibility, and representative results are shown.
TE1 cells were transiently transfected by nucleofection in buffer V (Amaxa Biosystems) with an Amaxa Nucleofector (Amaxa Biosystems) according to the manufacturer's instructions. Five million cells were resuspended in 100 μl of buffer V, 2 μg of plasmid was added, and the cells were pulsed with the Nucleofector set at V-01. After the cells were transfected, they were immediately transferred in 2 ml of fresh culture medium at 37°C in a 12-well plate for 40 h.
Western blot analysis.
For control Western blots from transactivation experiments, protein concentrations of lysates from luciferase assays were determined by a bicinchoninic acid assay (Pierce). One microgram of protein was used for detection of LMP1. For all the other Western blots, viable cells were counted by using the trypan blue method. After two washes in cold PBS, total protein extracts were obtained by direct lysis of equal amounts of viable cells in 3× Laemmli buffer boiled at 95°C for 5 min. Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then electrotransferred to Immobilon-P membranes (Millipore). Membranes were blocked with 0.2% casein in PBS containing 0.1% Tween before incubation with specific antibodies. The specific primary antibodies were applied at dilutions of 1:4 for S12 hybridoma culture supernatant, 1:500 for anti-EBNA2, anti-JNK1, anti-p65, anti-IκBα, anti-TRAF1, anti-ICAM1, and anti-β-actin, and 1:2,500 for anti-HA. Appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch) were applied at a dilution of 1:10,000. Specific signals were detected by enhanced chemiluminescence following the manufacturer's protocol (ECL kit; Amersham Bioscience).
RNA extraction, reverse transcription, and real-time PCR quantification.
Following the manufacturer's instructions, total RNA was collected using TRIzol reagent (Invitrogen) and cDNA was made using a SuperScript II reverse transcriptase (RT) kit (Invitrogen). The specific primers for LMP1 used were sense (5′-AGCCCTCCTTGTCCTCTATTCCTT-3′) and antisense (5′-ACCAAGTCGCCAGAGAATCTCCAA-3′). For housekeeping genes, the primers were as follows: β-actin sense (5′-GGGTCAGAAGGATTCCTATG-3′) and antisense (5′-GGTCTCAAACATGATCTGGG-3′), glyceraldehyde-3-phosphate dehydrogenase sense (5′-CCATCAATGACCCCTTCATTG-3′) and antisense (5′-CTTGACGGTGCCATGGAATT-3′), and hypoxanthine phosphoribosyltransferase sense (5′-CCCTGGCGTCGTGATTAG-3′) and antisense (5′-ATGGCCTCCCATCTCCTT-3′). The cDNA was amplified using the FastStart DNA Master SYBR green I kit (Roche) according to the manufacturer's instructions. Fifty cycles (1 cycle consisting of 5 s at 95°C, 5 s at 64°C, and 13 s at 72°C) were performed in a LightCycler (Roche) thermocycler. In each case, LMP1 and the three housekeeping genes were detected simultaneously and under the same conditions. A melting curve analysis was performed to verify the specificity of the products and the cycle threshold (CT) value was obtained for each gene in each condition. The values for the relative quantification of LMP1 mRNA levels are calculated by the ΔΔCT method. In each experimental condition, the mean value of the CT values for the three housekeeping genes is calculated and then subtracted from the CT value of the LMP1 gene. The ΔCT value for the control condition (DMSO or no doxycycline) is subtracted from the ΔCT value of the corresponding treated condition (ΔΔCT). The relative LMP1 mRNA levels in the different conditions are then calculated as follows: 2−ΔΔCT (38). Data are shown with the value of the control condition arbitrarily set at 100.
RESULTS
LMP1 activates its own promoter through its TES2 domain.
To determine whether LMP1 activates its own expression, we performed transient-transfection assays in HEK 293 human epithelial cells, using the reporter plasmid pLMP-luc, carrying the luciferase coding sequence under the control of the proximal LMP1 promoter (pLMP1; −326 to +37), and a mammalian expression vector for LMP1 (pSVHA-LMP1).
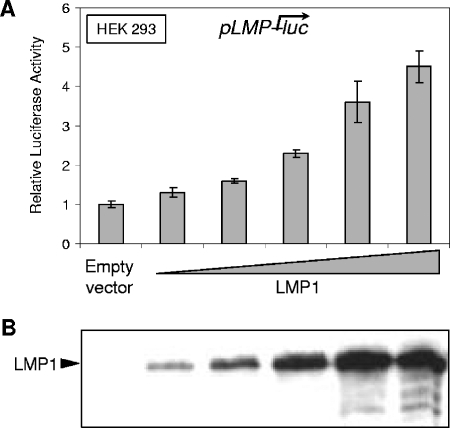
HEK 293 cells were cotransfected with increasing amounts of expression vector together with the pLMP-luc reporter. In cells transfected with LMP1, the LMP1 promoter activity was increased in a dose-dependent manner and reached up to more than fourfold compared with that produced by the empty vector alone (Fig. 1A). Western blot analysis performed with an anti-LMP1 antibody (S12) confirmed an appropriate dose-dependent expression of exogenous LMP1 in this experiment (Fig. 1B). Similar results were obtained with other cell lines (data not shown), indicating that LMP1 transactivates its own promoter in a non-cell-specific manner.
FIG. 1.
LMP1 upregulates its own promoter in a dose-dependent manner. (A) Dose-dependent transactivation of the LMP1 promoter by LMP1. HEK 293 cells were cotransfected with 80 ng of the firefly luciferase-based pLMP-luc reporter plasmid in conjunction with increasing doses (0 to 0.8 μg) of LMP1 expression vector. For each point, the amounts of DNA were completed with the corresponding empty vector (pSVHA). At 48 h posttransfection, we measured firefly luciferase activities, which were then normalized for transfection efficiency (on the basis of Renilla luciferase activity measured from cotransfected pRLnull reporter, which was included in all transfections). The promoter activity was expressed as activation (n-fold) over the corresponding control empty vector. Representative results are shown as means ± SD values (error bars) of an experiment with three replicate samples. (B) Dose-dependent expression of LMP1. Equal amounts (1 μg) of protein extracts from the cells described above were analyzed by Western blotting with the S12 antibody.
To map the region of LMP1 protein that mediates activation of LMP1 promoter activity, LMP1 mutants were generated and cotransfected together with the luciferase reporter construct pLMP-luc in HEK 293 cells. The LMP1 mutants used in this study were LMP1-TM in which all the cytoplasmic carboxy-terminal region is deleted and LMP1-Tes1mut and LMP1-Tes2mut that carry mutations in critical residues in either TES1 (the PXQXT motif transformed in AXAXA) or TES2 (the last three amino acids were deleted), respectively. All these LMP1 constructs are N-terminally tagged with an HA epitope and are schematically depicted in Fig. 2A. They were separately transfected into HEK 293 cells and were found to be expressed at their expected size by Western blot analysis performed with an anti-HA antibody (Fig. 2B).
FIG. 2.
Mutation in TES2 abrogates the upregulating effect of LMP1 on its own promoter. (A) Schematic representation of the mutants used. Wild-type LMP1 is composed of a short N-terminal cytoplasmic region fused to the HA peptide, six transmembrane domains, and a long C-terminal cytoplasmic region responsible of signaling. Domains involved in signal transduction (transforming effector sites TES1 and TES2) are depicted by light gray boxes. LMP1-TM is deleted of the C-terminal cytoplasmic region of LMP1. For LMP1-Tes1mut and LMP1-Tes2mut, point mutations generated in either TES1 or TES2 are indicated. (B) Expressed levels of wild-type LMP1 and its mutants. HEK 293 cells were transiently transfected with 0.4 μg of plasmid DNA for each construct. Equal amounts (1 μg) of protein extracts from the cells indicated were analyzed by Western blotting with anti-HA antibody. The filled arrowhead indicates the wild-type and point-mutated versions of LMP1, while the open arrowhead points to LMP1-TM. The filled star designates a LMP1 cleavage product. (C) Comparison of the effects of LMP1 mutants on the LMP1 promoter. HEK 293 cells were cotransfected with 80 ng of pLMP-luc and 0.4 μg of expression plasmids encoding LMP1 or its mutant versions as indicated. Cells were harvested and analyzed for luciferase activity as described above. Representative results are shown as means ± SD values (error bars) of an experiment with three replicate samples.
As expected from a mutant lacking the entire signaling region, LMP1-TM was completely inactive in inducing LMP1 promoter activity. More interestingly, LMP1-Tes2mut was also severely impaired in its transactivating properties of the LMP1 promoter. By contrast, LMP1-Tes1mut displayed transactivating properties similar to that obtained with wild-type LMP1 (Fig. 2C). These results indicate that LMP1 induces its own expression through at least one signaling pathway emerging from its TES2 domain.
LMP1 activates its own promoter via the JNK signaling pathway.
Various reports demonstrated that JNK is the sole signaling pathway emerging specifically from TES2 in HEK 293 cells (15, 28). We thus investigated whether this pathway is implicated in pLMP1 transactivation by LMP1. For this purpose, we transiently cotransfected in HEK 293 cell-specific dominant-negative or constitutively activated forms of the c-Jun N-terminal kinase together with pSVHA-LMP1 and pLMP-luc.
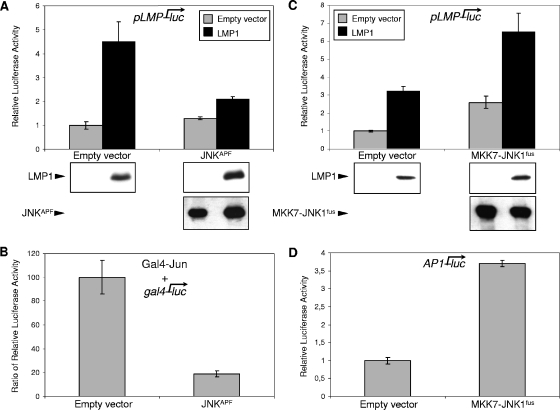
JNK is a mitogen-activated protein kinase (MAPK) activated by dual phosphorylation within the activation loop on the motif Thr-Pro-Tyr by MAPK-kinases (MAPKK or MKK) (9). To inhibit JNK, we decided to use a nonactivable form of the kinase, called JNKAPF, in which both key amino acids of the activation loop, threonine and tyrosine, were replaced by alanine and phenylalanine, respectively. As shown in Fig. 3B, overexpression of this dominant-negative form of JNK severely impaired the induction of a JNK-dependent reporter construct (Gal4-Jun/gal4-luc) by LMP1. The cotransfection assay with the pLMP-luc reporter construct showed that this JNK inhibition also prevented the upregulation of pLMP1 by LMP1 (Fig. 3A). For a control, we showed by Western blotting that cotransfection of exogenous LMP1 and JNKAPF led to only a slight increase in their levels of expression, showing that this inhibition was not due to a nonspecific decreased expression of LMP1 induced by JNKAPF (Fig. 3A).
FIG. 3.
The JNK signaling pathway induced by LMP1 is essential for pLMP1 upregulation. (A) Inhibition of LMP1 autoactivation by overexpression of JNKAPF. HEK 293 cells were cotransfected with 80 ng of pLMP-luc and 0.24 μg of expression plasmid encoding JNKAPF or its corresponding empty vector together with (black bars) or without (gray bars) 0.24 μg of pSVHA-LMP1. (B) Overexpression of JNKAPF effectively inhibits the LMP1-induced JNK signaling. c-Jun-dependent transcription activity was monitored by using a Gal4-Jun fusion protein and a gal4-luc reporter. HEK 293 cells were cotransfected with 80 ng of a construct encoding Gal4-Jun, 80 ng of a reporter plasmid, gal4-luc, containing Gal4-responsive elements, and 0.24 μg of pSVHA-LMP1 or the corresponding empty vector with or without 0.24 μg of expression plasmid encoding JNKAPF. Data shown are the ratios of luciferase activity measured in the presence of LMP1 to luciferase activity observed in the absence of LMP1. (C) Artificial activation of JNK induces pLMP1 activity and enhances LMP1 autoactivation. HEK 293 cells were cotransfected with 80 ng of pLMP-luc and 0.24 μg of expression plasmid encoding the fusion protein MKK7-JNKfus or its corresponding empty vector together with (black bars) or without (gray bars) 0.24 μg of pSVHA-LMP1. (D) Expression of MKK7-JNKfus activates the JNK signaling pathway. HEK 293 cells were cotransfected with 80 ng of a reporter plasmid AP1-luc, containing AP1-responsive elements, with or without 0.4 μg of expression plasmid encoding MKK7-JNKfus. In each case, cells were harvested and analyzed for luciferase activity and expression of LMP1 and JNKAPF or MKK7-JNKfus (under panels A and C) as described above. Representative results are shown as means ± SD values (error bars) of an experiment with three replicate samples.
Overexpression of JNK is not sufficient per se to specifically induce the JNK signaling pathway. For this purpose, Lei et al. have fused MKK7 (one of the JNK-activating upstream kinases) with JNK, resulting in constitutively activated JNK (36). As shown in a transient-transfection assay in HEK 293 cells (Fig. 3D), this fusion protein, called MKK7-JNKfus, properly activated AP1-luc, a reporter construct responding to the JNK signaling pathway. Then, to confirm the pLMP1 inducibility by JNK, we transiently cotransfected pLMP-luc with MKK7-JNKfus in the presence or absence of LMP1. The enforced activation of the JNK signaling pathway by means of the MKK7-JNK fusion protein, either alone or together with LMP1, upregulated the pLMP1 activity (Fig. 3C). Since no cross talk between MKK7-JNK fusion protein and endogenous JNK was found (36), the rather additive effects of LMP1 and MKK7-JNK fusion protein on pLMP1 are not surprising. We also verified by Western blot analysis that LMP1 expression was not affected by JNK activation, whereas LMP1 coexpression only slightly downregulated MKK7-JNK expression (Fig. 3C).
LMP1 inhibits its promoter through the NF-κB signaling pathway.
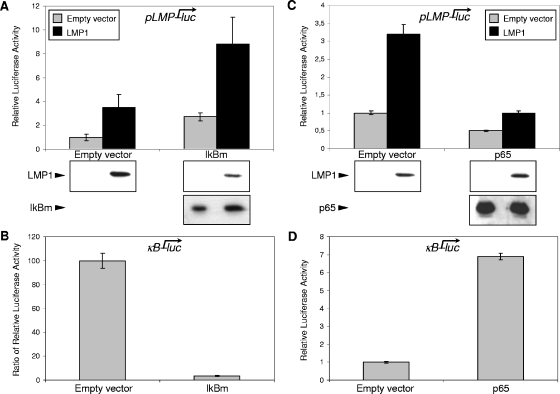
We next wanted to investigate whether the NF-κB signaling pathway is also implicated in the regulation of the pLMP1 promoter by LMP1. In noninduced conditions, NF-κB exists in a cytoplasmic complex with an inhibitor protein, IκBα (59). The activation of NF-κB mainly occurs via IκB kinase (IKK)-mediated phosphorylation of IκBα at serines 32 and 36 (56, 59). This phosphorylation targets IκBα for ubiquitination and proteasome-mediated degradation, thereby releasing NF-κB to enter the nucleus and activate several genes (59). We chose to specifically inhibit NF-κB complexes by transfection of a constitutively active IκBα mutated on serines 32 and 36 (IκBm). For a control, we monitored NF-κB activation by LMP1 and the inhibitory abilities of IκBm. For this purpose, we used a NF-κB-dependent reporter construct (κB-luc). As shown in Fig. 4B, LMP1-induced κB transcriptional activity was efficiently inhibited when cells were cotransfected with the IκBm construct. Then, we transiently cotransfected pLMP-luc with IκBm in the presence or absence of LMP1. In the presence of LMP1, the upregulated pLMP1 activity was remarkably enhanced (more than threefold) by the cotransfected IκBm (Fig. 4A). This enhancement of pLMP1 activation was not due to an increased LMP1 expression in transfected HEK 293 cells, since LMP1 expression was slightly reduced when cotransfected with IκBm. Conversely, IκBm expression was slightly enhanced by LMP1 coexpression (Fig. 4A, Western blots). pLMP1 activity was also enhanced (more than twofold) by expression of IκBm in the absence of LMP1 protein (Fig. 4A, gray bars). The result indicated that a molecule in the NF-κB signaling pathway might play a role in regulating the basal pLMP1 activity.
FIG. 4.
The NF-κB signaling pathway inhibits the LMP1 promoter. (A) Increased pLMP1 activity and enhanced LMP1 autoactivation by IκBm overexpression. HEK 293 cells were cotransfected with 80 ng of pLMP-luc and 0.24 μg of expression plasmid encoding IκBm or its corresponding empty vector together with (black bars) or without (gray bars) 0.24 μg of pSVHA-LMP1. (B) Overexpression of IκBm abolishes the LMP1-induced NF-κB signaling pathway. HEK 293 cells were cotransfected with 80 ng of a reporter construct κB-luc, containing NF-κB-responsive elements, and 0.24 μg of pSVHA-LMP1 or its corresponding empty vector with or without 0.24 μg of expression plasmid encoding IκBm. Data shown are the ratios of luciferase activity measured in the presence of LMP1 to luciferase activity observed in the absence of LMP1. (C) NF-κB p65 overexpression inhibits pLMP1 and impairs LMP1 autoactivation. HEK 293 cells were cotransfected with 80 ng of pLMP-luc and 0.24 μg of expression plasmid encoding p65 or its corresponding empty vector together with (black bars) or without (gray bars) 0.24 μg of pSVHA-LMP1. (D) Activation of NF-κB signaling by overexpression of p65. HEK 293 cells were cotransfected with 80 ng of a reporter plasmid κB-luc with or without 0.24 μg of expression plasmid encoding p65. In each case, cells were harvested and analyzed for luciferase activity and expression of LMP1 and IκBm or p65 (blots at the bottom of panels A and C) as described above. Representative results are shown as means ± SD values (error bars) of an experiment with three replicate samples.
We next investigated the effect of enforced NF-κB activation on pLMP1 activity. To artificially activate NF-κB, we transiently overexpressed p65, the main transactivator within the Rel-NF-κB family, in HEK 293 cells. As expected, transfection of a construct coding for p65 markedly increased activity of the κB-luc reporter (Fig. 4D). By contrast, we observed a ca. 50% decrease in the pLMP1 basal activity when p65 was overexpressed (Fig. 4C, gray bars). This inhibitory effect on the LMP1 promoter was further confirmed in LMP1-induced conditions. Indeed, when coexpressed with p65, LMP1 protein was impaired in pLMP1 transactivation (Fig. 4C). As assessed by Western blotting, neither LMP1 nor p65 expression yields were affected by cotransfection (Fig. 4C). These results indicate that inhibition was not due to a side effect on expression of the exogenous LMP1 protein.
Opposite effects of JNK and NF-κB signaling pathways on endogenous LMP1 expression in EBV-infected cells displaying type II or type III latency.
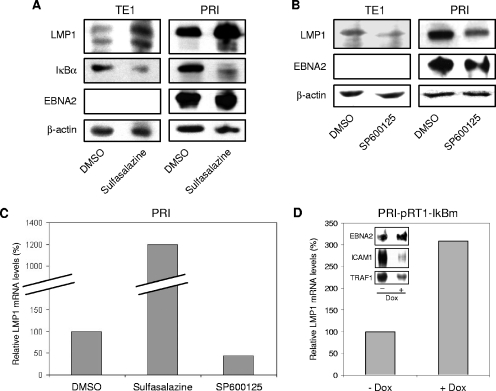
The experiments described above clearly show a positive and negative influence of JNK and NF-κB transduction pathways, respectively, on the LMP1 promoter activity. To determine whether the expression of LMP1 from the endogenous EBV genome was affected by these pathways, we used the LCL PRI (type III latency) (41) and the TE1 cell line (type II latency) that we previously characterized (39). Both cell lines have been shown to be dependent on LMP1 for their survival and proliferation (27, 39). These cell lines were treated with pharmacological inhibitors hitting either the NF-κB (sulfasalazine) or the JNK (SP600125) pathway. Sulfasalazine and SP600125 inhibit their target pathway by blocking kinase activity of IKKs and JNK, respectively (4, 64). Expression of LMP1 in cells was assessed by immunoblotting. Whatever the cell line tested, LMP1 protein levels were increased when sulfasalazine was used (or BAY11-7082, another NF-κB inhibitor; data not shown) and decreased when SP600125 was used (Fig. 5A and B, top row). The same membranes were also probed with an anti-β-actin antibody to demonstrate equal loading (Fig. 5A and B, bottom row). Another control was to ensure that these compounds were efficient in inhibiting their corresponding pathways in the cell lines tested. We showed that the NF-κB target IκBα and the JNK target c-Jun were properly downregulated by treatment with inhibitors of their respective pathways (Fig. 5A, second row, and data not shown). In addition, since the viral EBNA2 protein is the best characterized transactivator of the LMP1 promoter, we monitored its expression by Western blot analysis. Figure 5A and B show that neither sulfasalazine nor SP600125 significantly changed the EBNA2 expression in PRI cells. In TE1 cells, EBNA2 was not expressed under normal or treated conditions (Fig. 5A and B). Thus, an increase in LMP1 expression after treatment of these cells with sulfasalazine was not a consequence of EBNA2 reexpression.
FIG. 5.
Opposite roles of the JNK and NF-κB pathways on endogenous LMP1 expression at the mRNA and protein levels. Inhibition of (A) NF-κB activity upregulates, whereas inhibition of (B) JNK activity downregulates the endogenous LMP1 protein levels in PRI and TE1 cell lines. The cells were treated with vehicle (DMSO), sulfasalazine (1 mM and 5 mM for TE1 and PRI cells, respectively), or SP600125 (20 μM) for 16 h. Equal amounts of cells were harvested and directly lysed in 3× Laemmli buffer. Protein lysates were analyzed by Western blotting for the expression of LMP1, IκBα, and EBNA2. Equal loading of proteins in each lane was confirmed by probing the membrane with anti-β-actin antibody. (C) Sulfasalazine increases, whereas SP600125 decreases the LMP1 mRNA levels in PRI cells. PRI cells were treated with either vehicle (DMSO) or sulfasalazine (5 mM) or SP600125 (20 μM) for 16 h. Cells were then harvested, and mRNAs were extracted and subjected to real-time RT-PCR as described in Materials and Methods. LMP1 mRNA levels were normalized to the mRNA levels of three housekeeping genes (β-actin, glyceraldehyde-3-phosphate dehydrogenase, and hypoxanthine phosphoribosyltransferase), and the normalized levels in the untreated condition were arbitrarily assigned a value of 100. (D) The constitutively active form of IκBα inhibits NF-κB target genes and increases the levels of LMP1 transcripts in PRI cells. Where indicated, the PRI-pRT1-IκBm stable cell line were treated with doxycycline (2 μg/ml) (+ Dox) to induce the expression of IκBm. After induction for 24 h, the relative LMP1 mRNA levels were quantified by real-time RT-PCR as described above. After induction for 48 h, protein lysates were collected as described above and analyzed by Western blotting for the expression of EBNA2, ICAM1, and TRAF1.
The transactivating effects of LMP1 on its own promoter shown in transient-expression assays suggested that modulation of LMP1 protein expression could be a consequence of transcriptional regulation. To ascertain this point, we analyzed LMP1 mRNA levels in treated PRI cells by using real-time quantitative RT-PCR. Figure 5C shows that, in agreement with their action on protein levels, the JNK inhibitor hampered LMP1 mRNA accumulation (ca. 50%), whereas the NF-κB inhibitor increased it (up to 12-fold). We decided to confirm the effect of sulfasalazine on LMP1 mRNA with another approach to inhibit NF-κB in these cells. PRI-pRT1-IκBm is a previously characterized PRI cell line carrying a doxycycline-inducible version of the NF-κB inhibitory molecule IκBm (41). When we induced IκBm by doxycycline in this cell line, we clearly detected an increase of LMP1 proteins (data not shown) and transcripts correlating with a strong decrease in expression of two NF-κB target genes (ICAM1 and TRAF1) and no change in EBNA2 expression (Fig. 5D). This confirms that NF-κB-induced repression of LMP1 occurred at the mRNA level.
Altogether, these results show that, whatever the status of EBNA2 expression in a latently EBV-infected cell, the NF-κB and JNK signaling pathways can decrease or increase, respectively, the endogenous expression of LMP1 mRNA and proteins.
Endogenous LMP1 expression is mainly due to LMP1 signaling in EBV-infected cells expressing a type II latency.
Although LMP1 is supposed to be the major inducer of NF-κB and JNK pathways in EBV-infected cells, contribution of some LMP1-independent signals to this induction cannot be excluded. Moreover, LMP1 can activate other signaling pathways in addition to NF-κB and JNK. In a previous study, we have demonstrated that a construct containing the cytoplasmic C-terminal tail of LMP1 fused to green fluorescent protein (LMP1-CT) could selectively impair the LMP1-induced signaling pathways by hampering the binding of critical adapters to LMP1 (2). Then, to determine the influence of pathways specifically induced by LMP1 on its own promoter regulation, we inhibited the LMP1-dependent signaling by expressing this dominant-negative version of LMP1 in EBV-infected cell lines and measured LMP1 expression.
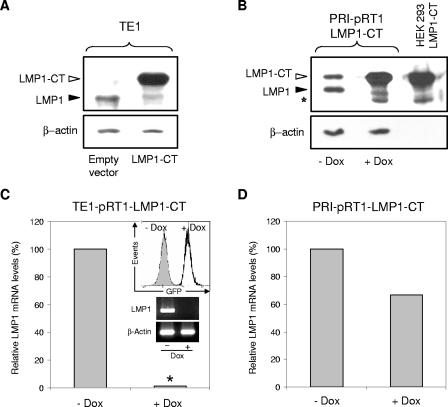
We transiently transfected LMP1-CT or the corresponding empty vector in TE1 cells. LMP1 expression, monitored by immunoblotting, was greatly diminished by LMP1-CT in these type II latency-expressing cells (Fig. 6A), confirming the autoactivation loop already shown through transactivation assays in HEK 293 cells. We also developed stably transfected PRI cells with an episomal expression vector (pRT1-LMP1-CT) allowing a doxycycline-dependent expression of LMP1-CT (5, 33). In these PRI-pRT1-LMP1-CT cells, induction of LMP1-CT also led to a significant decrease of LMP1 expression (Fig. 6B). However, this decrease seems weaker than that obtained by LMP1-CT induction in TE1 cells (compare Fig. 6A and B). To measure LMP1 mRNA levels by real-time quantitative RT-PCR in TE1 cells, we established stably transfected TE1 cells with the episomal expression vector pRT1-LMP1-CT. In these TE1-pRT1-LMP1-CT cells, doxycycline highly induces expression of LMP1-CT (Fig. 6C, graph in insert). Thus, we verified that the reduced expression of endogenous LMP1 protein in TE1 and PRI cells was mainly due to a LMP1-CT-induced decrease in the LMP1 mRNA pool (Fig. 6C and D). This decrease was almost total in TE1 cells, since the level of LMP1 mRNA was below the detection threshold of the real-time quantitative RT-PCR. A gel analysis of a semiquantitative RT-PCR was performed to confirm and visualize this loss of LMP1 expression in TE1 cells (Fig. 6C, gels in insert). For an important control of our approach, we verified that all the LMP1 mRNA in TE1 cells were actually synthesized from the proximal LMP1 promoter (pLMP1) studied here. Indeed, quantitative RT-PCR experiments performed with primers specific for distal TR-L1 transcription failed to amplified any transcript (data not shown). Note that the weaker decrease observed in LMP1 protein levels in PRI-pRT1-LMP1-CT cells is also found at the mRNA levels (compare Fig. 6C and D). In agreement with these Western blot and RT-PCR results (Fig. 6), we also obtained a similar difference in LMP1 expression reduction between TE1 and PRI cells when we measured LMP1 expression at the cell membrane by flow cytometry analysis in LMP1-CT-expressing cells (data not shown).
FIG. 6.
The dominant-negative form of LMP1, LMP1-CT, downregulates the mRNA and protein levels of LMP1 in EBV-infected cells. (A) Transient overexpression of LMP1-CT results in a decrease in LMP1 protein levels in TE1 cells. TE1 cells were transiently transfected by nucleofection (see Materials and Methods) with 2 μg of the LMP1-CT expression vector or the corresponding empty vector. Forty hours after nucleofection, protein lysates were collected as described above and analyzed by Western blotting with the S12 antibody. The open arrowhead points to full-length LMP1-CT, while the filled arrowhead indicates the endogenous wild-type LMP1. (B) Conditional LMP1-CT overexpression results in a decrease in LMP1 protein levels in PRI cells. Where indicated, the PRI-pRT1-LMP1-CT stable cell line was treated with doxycycline (2 μg/ml) (+ Dox) to induce the expression of LMP1-CT. After induction for 48 h, protein lysates were collected as described above and analyzed by Western blotting with the S12 antibody. The open arrowhead points to full-length LMP1-CT, while the filled arrowhead indicates endogenous wild-type LMP1. The open star designates a LMP1-CT cleavage product. In the rightmost lane, 1 μg of protein lysate from HEK 293 cells transfected only with the LMP1-CT expression vector was loaded to clearly localize the full-length and cleaved products of LMP1-CT. (C) Induction of LMP1-CT expression results in a disappearance of LMP1 mRNA in TE1 cells. The TE1-pRT1-LMP1-CT stable cell line was treated as described above. After induction for 48 h, LMP1-CT expression was measured by green fluorescent protein (GFP) fluorescence detection with a flow cytometer (inset), and the relative LMP1 mRNA levels were quantified by real-time RT-PCR as described above. The asterisk indicates undetectable levels of amplification. Total RNAs were also subjected to semiquantitative RT-PCR followed by electrophoresis on ethidium bromide-stained 1.5% agarose gel (insets). (D) Induction of LMP1-CT expression results in a decrease in LMP1 mRNA levels in PRI cells. The PRI-pRT1-LMP1-CT stable cell line was treated as described above. After induction for 48 h, the relative LMP1 mRNA levels were quantified by real-time RT-PCR as described above.
Thus, whatever the type of latency displayed, our data favor the existence of a positive autoregulation of LMP1 in EBV-infected cells expressing this viral protein. Moreover, this mechanism seems to be of major incidence for the expression of LMP1 during the type II latency program of EBV.
DISCUSSION
In this work, we addressed the question of LMP1 expression during the type II latency program of EBV. We demonstrated that LMP1, through its TES2 domain, can activate its own promoter (pLMP1) by inducing the JNK signaling pathway. Moreover, we also showed that this JNK-dependent autoactivation of LMP1 is tightly regulated, since LMP1 activation of NF-κB simultaneously exerts an inhibition of pLMP1 activity. We demonstrated that these opposite effects of LMP1-induced JNK and NF-κB signaling pathways on pLMP1 activity are effective in both latency programs in which EBV expresses LMP1 (i.e., type II and III), stressing the physiological relevance of this autoregulation. Overall, our results in different cell lines show that this balance is always in favor of a positive autoregulatory loop of LMP1. Moreover, we show that this loop is the major inducer of LMP1 expression in EBV-infected cells displaying a type II latency.
Although several pLMP1 sites (Sp1 and CRE) and transcription factors activating LMP1 expression (Sp1, c-Jun, ATF2, ATF1, and CREB1) are potential targets of the JNK signaling pathway (1, 9, 21, 35), the influence of this pathway on LMP1 expression had never been tested. It is particularly striking for pLMP1, since its CRE site has been shown to be very important for an EBNA2-independent induction of LMP1 expression (53). Here, we showed that the JNK pathway was actually implicated in the activation of LMP1 expression, and more particularly, when LMP1 itself induced this signaling pathway. Furthermore, this self-activation of LMP1 was shown essential for its own expression when EBNA2 was not expressed (i.e., in type II latency cells).
We also demonstrated that LMP1 can inhibit its own expression in a NF-κB-dependent manner. Indeed, whereas most of the NF-κB target genes are reported to be regulated positively by this transcription factor family, a growing number of NF-κB-repressed genes and their various repression processes have begun to be described (6, 18, 59). In the present study, we found that inhibition of NF-κB supported better LMP1 expression. Furthermore, this was confirmed by LMP1 promoter repression induced by p65 overexpression. The ability of p65 to inhibit target genes has also been noted in several recent publications. For example, direct binding of NF-κB to DNA has been implicated in tumor necrosis factor alpha-mediated inhibition of human papillomavirus type 16 long control region (passive inhibition through squelching of coactivators) (18) and in UV-C- and daunorubicin-induced repression of antiapoptotic genes (active repression by recruitment of corepressors) (6). Other reports suggest that NF-κB can inhibit target gene expression independently of DNA binding. First, NF-κB can sequester coactivators under its active form (free in nucleus) (58) and/or sequester corepressors under its inactive form (retained by IκB in cytoplasm) (16). Second, NF-κB p65 can recruit repression complexes though indirect binding to target promoters (3). In either case, the function of another transcription factor that normally regulates the target gene promoter is attenuated, resulting in a reduction in target gene expression.
In the case of LMP1 target gene regulation, few examples of NF-κB-mediated repression, corresponding to some of the mechanisms described above, have already been reported. Recently, Grimm et al. demonstrated that LMP1 inhibits Bax through a direct binding of p50/p65 heterodimer on the promoter (19). Alternatively, another NF-κB-repressed target gene of LMP1, CD99, lacks a NF-κB consensus sequence in its promoter, suggesting an indirect NF-κB-mediated recruitment of corepressors or a squelching effect of coactivators (34). Lack of a NF-κB binding element in the LMP1 promoter suggests that DNA binding of p65 is not required for the p65-mediated inhibition of pLMP1. Furthermore, our preliminary data with an overexpressed CBP coactivator support that squelching effects would not be implicated (data not shown). Finally, the ability of trichostatin A (inhibitor of histone deacetylases, a group of corepressors) to upregulate LMP1 in many cases suggests that recruitment of corepressors, such as HDACs, may be involved (44, 48). Altogether, these observations seem to support an indirect NF-κB-mediated recruitment of HDACs to pLMP1.
Accumulating data indicate that NF-κB activation antagonizes the JNK signaling cascade via induction of target genes (GADD45β, XIAP, thioredoxin, Mn-superoxide dismutase, and FHC) whose products inhibit different activating events upstream of JNK (11, 47). This cross talk could be an explanation for the opposite effect of JNK and NF-κB pathways in LMP1 autoregulation. However, control experiments performed in HEK 293 cells showed no modification in the activity of one pathway (data not shown), while we successfully inhibited or activated the other pathway (Fig. 3B and D and Fig. 4B and D). Therefore, this negative cross talk between NF-κB and JNK signaling pathways does not seem effective in the case of an induction by LMP1, as it was already shown for interleukin 1 (IL-1) (55). Consequently, even though we cannot totally rule out the additional presence of this cross talk in physiological target cells of EBV, both activation by JNK and repression by NF-κB of the LMP1 promoter would be most likely independent. Thus, further investigation is needed to precisely delineate the cis- and trans-acting factors involved in the balanced mechanisms of repression and activation that we described here.
During its type III latency program, EBV expresses all the nuclear antigens (EBNAs) and particularly EBNA2 and EBNA-LP (50). These two factors form a very potent activator/coactivator complex to induce expression of LMP1 from its proximal promoter (45, 67). In this context, it is worthy to note that the strong variation measured in LMP1 expression at protein and mRNA levels in PRI cells after inhibition of either the NF-κB or JNK signal points out the importance of the regulation induced by each of these pathways. As exemplified by the pharmacological inhibition, the NF-κB or JNK signaling pathway individually contributes to LMP1 expression during both type II and type III latencies. On the other hand, as suggested by inhibition of the whole LMP1 signaling by LMP1-CT, combined effects of NF-κB and JNK signaling seem to have an predominant role in induction of LMP1 expression only during type II latency. Indeed, following the specific inhibition of LMP1 signal, LMP1 expression dropped by only 33% in cells with type III latency, whereas it was completely shut off in those displaying type II latency (Fig. 6C and D). This is certainly due to the strong pLMP1 activation driven by EBNA2. Subsequently, LMP1 autoactivation represents only a minor part of the LMP1 expression process in type III latency. This is supported by a similar cell-specific phenomenon (independent of latency) observed concerning the induction of another LMP1 target gene, CD40. Contrary to T lymphocytes, B lymphocytes express CD40 constitutively at high levels (49). Therefore, when LMP1 is expressed in B cells, it can overinduce only rather slightly the expression of this gene. It is actually what we measured in the B-cell line PRI when we specifically inhibited the LMP1 signal. CD40 expression dropped only by 9%, whereas this decrease reached 43% in the NC5 T-cell line (data not shown). Consequently, the minor effect of LMP1 on its own expression measured during type III latency, along with the lower levels of LMP1 detected in cells expressing type II latency, seem to reveal a difference between EBNA2- and LMP1-dependent mechanisms concerning their relative strength of pLMP1 transactivation.
Since the LMP1 gene is located immediately upstream of the LMP2B gene and is transcribed in the direction opposite that of LMP2B, the region between these two genes, where pLMP1 is located, serves as a bidirectional promoter for both LMP1 and LMP2B (32). Therefore, we wondered whether LMP2B could also be regulated by LMP1. With an RT-PCR approach, we noted that only the PRI cell line expressed LMP2B and that there was no difference in its expression when LMP1-CT was induced or not in the PRI cells with the pRT1-LMP1-CT vector. Such a dissociation in the regulation of LMP1 and LMP2B expression was already shown in the case of an upregulation of LMP1 by IL-10 (30).
Recently, two other groups have found several lines of evidence suggesting that LMP1 could regulate its own promoter by using mechanisms different from that described here. The first proposed mechanism was a positive autoregulatory loop implicating the IL-6 cytokine (8). In their epithelial cellular models, Chen et al. have actually demonstrated that LMP1 induced cellular secretion of IL-6 which, in turn, activated STAT3 transcription factor. Furthermore, they showed that overexpression of a constitutively active STAT3 in an epithelial model of EBV type II latency enhanced the expression of LMP1, mainly by transactivating the distal LMP1 promoter located within the terminal repeats (TR-L1). Another study suggests a more direct regulatory circuit centered on the cellular transcription factor IRF7 (42). Ning et al. have clearly shown that IRF7 transactivated the LMP1 promoter without involvement of any viral factor (especially EBNA2) in B lymphocytes. Previously, Zhang et al. had already shown that LMP1 itself could induce both the transcription and phosphorylation of IRF7 (65, 66). Furthermore, Ning et al. recently showed that a dominant-negative form of IRF7 decreased LMP1 expression in recombinant EBV-infected MDA-MB-231 cell lines expressing a type II latency profile (43). Consequently, they have suggested the presence of a regulatory circuit where LMP1 could induce its own expression via IRF7 induction.
Even if our results cannot fully exclude them, these two mechanisms do not seem to be present and effective in our cellular models. First, it was shown that both IL-6 and IRF7 are positively regulated by LMP1 in a NF-κB-dependent manner (14, 66), whereas we found here that the NF-κB signaling pathway rather inhibited expression of LMP1. We previously showed that TE1 cells do not secrete detectable amounts of IL-6 in the absence of activation (39). Furthermore, quantitative RT-PCR experiments performed with TR-L1-specific primers in these cells failed to detect any transcript expressed from this IL-6-sensitive promoter (data not shown). This strongly suggests that LMP1 is mainly expressed from its proximal promoter (pLMP1) in TE1 cells and does not favor a role for this cytokine in the expression of LMP1 in this model of EBV-infected cells displaying type II latency.
Before this study, the only known mechanism responsible for LMP1 expression in latently EBV-infected cells was dependent and centered on EBNA2. EBNA2 is a strong pLMP1 transactivator expressed only during type III latency (50, 63). Here, we clearly demonstrated that EBV has developed at least one other way to maintain the expression of LMP1, its major oncogene. This alternative mechanism is the JNK-dependent autoactivation of LMP1 itself. We show that this mechanism is functional in both type II and III latencies. However, it allows activation of LMP1 expression only when the main activating process of LMP1 expression (i.e., pLMP1 transactivation by EBNA2) cannot be set up. Therefore, our study is the first demonstration of a mechanism sustaining LMP1 expression during the type II latency program, which is commonly found in most EBV-associated malignancies. Interestingly, it was reported that EBV strains with an intact wild-type cyclic AMP response element (CRE) in pLMP1 were significantly more frequent in individuals with Hodgkin's disease than in both infectious mononucleosis and asymptomatic EBV carriers (52). Moreover, as already mentioned, this site binds several transcription factors, which can be activated by the JNK signaling pathway, and has been shown to be very important for an EBNA2-independent pLMP1 activation (53). These results together with ours could suggest a pathogenic role of LMP1 autoactivation in malignant diseases where EBV expresses a type II latency program.
Acknowledgments
We thank R. J. Davis (Howard Hughes Medical Institute, Worcester, MA) for the generous gift of the JNKAPF and MKK7-JNKfus expression vectors, J. Hiscott (Lady Davis Institute, McGill University, Montréal, Québec, Canada) for providing the IκBm and p65 expression plasmids, and B. Derijard (Université de Nice-Sophia Antipolis, Nice, France) and M. Ptashne (Memorial Sloan-Kettering Cancer Center, New York, NY) for the generous gift of the gal4-luc and Gal4-Jun vectors. We thank Catherine Leroy and Brigitte Quatannens for skillful technical assistance and Sylvie Reveneau, Irène Joab, and Alain Sergeant for helpful discussions.
This study was supported by grants from the Association de Recherche contre le Cancer and the Ligue Nationale contre le Cancer, Comité du Pas-de-Calais. G.G. was supported by grants from the Ministère délégué à l'Enseignement Supérieur et à la Recherche and the Association de Recherche contre le Cancer, and C.L.C. was supported by the Ligue contre le Cancer, Comité de la Corrèze.
REFERENCES
- 1.Adamson, A. L., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser, J. Swenson, and S. Kenney. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adriaenssens, E., A. Mougel, G. Goormachtigh, E. Loing, V. Fafeur, C. Auriault, and J. Coll. 2004. A novel dominant-negative mutant form of Epstein-Barr virus latent membrane protein-1 (LMP1) selectively and differentially impairs LMP1 and TNF signaling pathways. Oncogene 23:2681-2693. [DOI] [PubMed] [Google Scholar]
- 3.Aguilera, C., R. Hoya-Arias, G. Haegeman, L. Espinosa, and A. Bigas. 2004. Recruitment of IκBα to the hes1 promoter is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 101:16537-16542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornkamm, G. W., C. Berens, C. Kuklik-Roos, J. M. Bechet, G. Laux, J. Bachl, M. Korndoerfer, M. Schlee, M. Hölzel, A. Malamoussi, R. D. Chapman, F. Nimmerjahn, J. Mautner, W. Hillen, H. Bujard, and J. Feuillard. 7 September 2005, posting date. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33:e137. [Online.] doi: 10.1093/nar/gni137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, K. J., S. Rocha, and N. D. Perkins. 2004. Active repression of anti-apoptotic gene expression by RelA(p65) NF-κB. Mol. Cell 13:853-865. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., J. M. Lee, Y. Zong, M. Borowitz, M. H. Ng, R. F. Ambinder, and S. D. Hayward. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 75:2929-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H., L. Hutt-Fletcher, L. Cao, and S. D. Hayward. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J. Virol. 77:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 10.Devergne, O., E. D. Cahir McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djavaheri-Mergny, M., D. Javelaud, J. Wietzerbin, and F. Besancon. 2004. NF-κB activation prevents apoptotic oxidative stress via an increase of both thioredoxin and MnSOD levels in TNFα-treated Ewing sarcoma cells. FEBS Lett. 578:111-115. [DOI] [PubMed] [Google Scholar]
- 12.D'Souza, B. N., L. C. Edelstein, P. M. Pegman, S. M. Smith, S. T. Loughran, A. Clarke, A. Mehl, M. Rowe, C. Gelinas, and D. Walls. 2004. Nuclear factor κB-dependent activation of the antiapoptotic bfl-1 gene by the Epstein-Barr virus latent membrane protein 1 and activated CD40 receptor. J. Virol. 78:1800-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudziak, D., A. Kieser, U. Dirmeier, F. Nimmerjahn, S. Berchtold, A. Steinkasserer, G. Marschall, W. Hammerschmidt, G. Laux, and G. W. Bornkamm. 2003. Latent membrane protein 1 of Epstein-Barr virus induces CD83 by the NF-κB signaling pathway. J. Virol. 77:8290-8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eliopoulos, A. G., M. Stack, C. W. Dawson, K. M. Kaye, L. Hodgkin, S. Sihota, M. Rowe, and L. S. Young. 1997. Epstein-Barr virus-encoded LMP1 and CD40 mediate IL-6 production in epithelial cells via an NF-κB pathway involving tumor necrosis factor receptor-associated factors. Oncogene 14:2899-2916. [DOI] [PubMed] [Google Scholar]
- 15.Eliopoulos, A. G., E. R. Waites, S. M. Blake, C. Davies, P. Murray, and L. S. Young. 2003. TRAF1 is a critical regulator of JNK signaling by the TRAF-binding domain of the Epstein-Barr virus-encoded latent infection membrane protein 1 but not CD40. J. Virol. 77:1316-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinosa, L., J. Inglés-Esteve, A. Robert-Moreno, and A. Bigas. 2003. IκBα and p65 regulate the cytoplasmic shuttling of nuclear corepressors: crosstalk between Notch and NFκB pathways. Mol. Biol. Cell 14:491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fåhraeus, R., A. Jansson, A. Sjoblom, T. Nilsson, G. Klein, and L. Rymo. 1993. Cell phenotype-dependent control of Epstein-Barr virus latent membrane protein 1 gene regulatory sequences. Virology 195:71-80. [DOI] [PubMed] [Google Scholar]
- 18.Fontaine, V., E. van der Meijden, J. de Graaf, J. ter Schegget, and L. Struyk. 2000. A functional NF-κB binding site in the human papillomavirus type 16 long control region. Virology 272:40-49. [DOI] [PubMed] [Google Scholar]
- 19.Grimm, T., S. Schneider, E. Naschberger, J. Huber, E. Guenzi, A. Kieser, P. Reitmeir, T. F. Schulz, C. A. Morris, and M. Sturzl. 2005. EBV latent membrane protein-1 protects B cells from apoptosis by inhibition of BAX. Blood 105:3263-3269. [DOI] [PubMed] [Google Scholar]
- 20.Groux, H., F. Cottrez, C. Montpellier, B. Quatannens, J. Coll, D. Stehelin, and C. Auriault. 1997. Isolation and characterization of transformed human T-cell lines infected by Epstein-Barr virus. Blood 89:4521-4530. [PubMed] [Google Scholar]
- 21.Higuchi, H., A. Grambihler, A. Canbay, S. F. Bronk, and G. J. Gores. 2004. Bile acids up-regulate death receptor 5/TRAIL-receptor 2 expression via a c-Jun N-terminal kinase-dependent pathway involving Sp1. J. Biol. Chem. 279:51-60. [DOI] [PubMed] [Google Scholar]
- 22.Hofelmayr, H., L. J. Strobl, C. Stein, G. Laux, G. Marschall, G. W. Bornkamm, and U. Zimber-Strobl. 1999. Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J. Virol. 73:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson, G. S., P. J. Farrell, and B. G. Barrell. 1985. Two related but differentially expressed potential membrane proteins encoded by the EcoRI Dhet region of Epstein-Barr virus B95-8. J. Virol. 53:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi, K. M., K. M. Kaye, and E. D. Kieff. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 94:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU. 1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 19:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieser, A., E. Kilger, O. Gires, M. Ueffing, W. Kolch, and W. Hammerschmidt. 1997. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 16:6478-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilger, E., A. Kieser, M. Baumann, and W. Hammerschmidt. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 17:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kis, L. L., M. Takahara, N. Nagy, G. Klein, and E. Klein. 2006. IL-10 can induce the expression of EBV-encoded latent membrane protein-1 (LMP-1) in the absence of EBNA-2 in B-lymphocytes, in Burkitt lymphoma-, and in NK-lymphoma derived cell lines. Blood 107:2928-2935. [DOI] [PubMed] [Google Scholar]
- 31.Kulwichit, W., R. H. Edwards, E. M. Davenport, J. F. Baskar, V. Godfrey, and N. Raab-Traub. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. USA 95:11963-11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laux, G., F. Dugrillon, C. Eckert, B. Adam, U. Zimber-Strobl, and G. W. Bornkamm. 1994. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J. Virol. 68:6947-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Clorennec, C., I. Youlyouz-Marfak, E. Adriaenssens, J. Coll, G. W. Bornkamm, and J. Feuillard. 2006. EBV latency III immortalization program sensitizes B-cells to induction of CD95 mediated apoptosis via LMP1: role of NF-κB, STAT1 and p53. Blood 107:2070-2078. [DOI] [PubMed] [Google Scholar]
- 34.Lee, I., M. K. Kim, E. Y. Choi, A. Mehl, K. C. Jung, M. C. Gil, M. Rowe, and S. H. Park. 2001. CD99 expression is positively regulated by Sp1 and is negatively regulated by Epstein-Barr virus latent membrane protein 1 through nuclear factor-κB. Blood 97:3596-3604. [DOI] [PubMed] [Google Scholar]
- 35.Lee, R. J., C. Albanese, R. J. Stenger, G. Watanabe, G. Inghirami, G. K. Haines III, M. Webster, W. J. Muller, J. S. Brugge, R. J. Davis, and R. G. Pestell. 1999. pp60v-src induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60v-src signaling in breast cancer cells. J. Biol. Chem. 274:7341-7350. [DOI] [PubMed] [Google Scholar]
- 36.Lei, K., A. Nimnual, W. X. Zong, N. J. Kennedy, R. A. Flavell, C. B. Thompson, D. Bar-Sagi, and R. J. Davis. 2002. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH2-terminal kinase. Mol. Cell. Biol. 22:4929-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, H. P., and Y. S. Chang. 2003. Epstein-Barr virus latent membrane protein 1: structure and functions. J. Biomed. Sci. 10:490-504. [DOI] [PubMed] [Google Scholar]
- 38.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 39.Masy, E., E. Adriaenssens, C. Montpellier, P. Crépieux, A. Mougel, B. Quatannens, G. Goormachtigh, N. Faumont, F. Meggetto, C. Auriault, H. Groux, and J. Coll. 2002. Human monocytic cell lines transformed in vitro by Epstein-Barr virus display a type II latency and LMP-1-dependent proliferation. J. Virol. 76:6460-6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montpellier, C., P. Crépieux, B. Quatannens, B. Delobel, M. F. Croquette, D. Stehelin, C. Auriault, H. Groux, and J. Coll. 1997. Homologous T and B cells immortalized in vitro by the Epstein-Barr virus exhibit differential genetic and functional features. Int. J. Oncol. 11:87-96. [DOI] [PubMed] [Google Scholar]
- 41.Najjar, I., F. Baran-Marszak, C. Le Clorennec, C. Laguillier, O. Schischmanoff, I. Youlyouz-Marfak, M. Schlee, G. W. Bornkamm, M. Raphael, J. Feuillard, and R. Fagard. 2005. Latent membrane protein 1 regulates STAT1 through NF-κB-dependent interferon secretion in Epstein-Barr virus-immortalized B cells. J. Virol. 79:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning, S., A. M. Hahn, L. E. Huye, and J. S. Pagano. 2003. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. J. Virol. 77:9359-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ning, S., L. E. Huye, and J. S. Pagano. 2005. Interferon regulatory factor 5 represses expression of the Epstein-Barr virus oncoprotein LMP1: braking of the IRF7/LMP1 regulatory circuit. J. Virol. 79:11671-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishikawa, J., L. L. Kis, A. Liu, X. Zhang, M. Takahara, K. Bandobashi, C. Kiss, N. Nagy, K. Okita, G. Klein, and E. Klein. 2004. Upregulation of LMP1 expression by histone deacetylase inhibitors in an EBV carrying NPC cell line. Virus Genes 28:121-128. [DOI] [PubMed] [Google Scholar]
- 45.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagano, J. S., M. Blaser, M. A. Buendia, B. Damania, K. Khalili, N. Raab-Traub, and B. Roizman. 2004. Infectious agents and cancer: criteria for a causal relation. Semin. Cancer Biol. 14:453-471. [DOI] [PubMed] [Google Scholar]
- 47.Papa, S., F. Zazzeroni, C. G. Pham, C. Bubici, and G. Franzoso. 2004. Linking JNK signaling to NF-κB: a key to survival. J. Cell Sci. 117:5197-5208. [DOI] [PubMed] [Google Scholar]
- 48.Park, J. H., and D. V. Faller. 2002. Epstein-Barr virus latent membrane protein-1 induction by histone deacetylase inhibitors mediates induction of intercellular adhesion molecule-1 expression and homotypic aggregation. Virology 303:345-363. [DOI] [PubMed] [Google Scholar]
- 49.Quezada, S. A., L. Z. Jarvinen, E. F. Lind, and R. J. Noelle. 2004. CD40/CD154 interactions at the interface of tolerance and immunity. Annu. Rev. Immunol. 22:307-328. [DOI] [PubMed] [Google Scholar]
- 50.Rickinson, A., and E. Kieff. 2001. Epstein-Barr virus, p. 2573-2627. In D. Knipe, P. Howley, D. Griffin, R. Lamb, M. Martin, B. Roizman, and S. Straus (ed.), Virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 51.Sadler, R. H., and N. Raab-Traub. 1995. The Epstein-Barr virus 3.5-kilobase latent membrane protein 1 mRNA initiates from a TATA-less promoter within the first terminal repeat. J. Virol. 69:4577-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandvej, K., B. S. Andresen, X. G. Zhou, N. Gregersen, and S. Hamilton-Dutoit. 2000. Analysis of the Epstein-Barr virus (EBV) latent membrane protein 1 (LMP-1) gene and promoter in Hodgkin's disease isolates: selection against EBV variants with mutations in the LMP-1 promoter ATF-1/CREB-1 binding site. Mol. Pathol. 53:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sjöblom, A., W. Yang, L. Palmqvist, A. Jansson, and L. Rymo. 1998. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J. Virol. 72:1365-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sjöblom-Hallen, A., W. Yang, A. Jansson, and L. Rymo. 1999. Silencing of the Epstein-Barr virus latent membrane protein 1 gene by the Max-Mad1-mSin3A modulator of chromatin structure. J. Virol. 73:2983-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, G., Y. Minemoto, B. Dibling, N. H. Purcell, Z. Li, M. Karin, and A. Lin. 2001. Inhibition of JNK activation through NF-κB target genes. Nature 414:313-317. [DOI] [PubMed] [Google Scholar]
- 56.Traenckner, E. B., H. L. Pahl, T. Henkel, K. N. Schmidt, S. Wilk, and P. A. Baeuerle. 1995. Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 14:2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai, C. N., C. M. Lee, C. K. Chien, S. C. Kuo, and Y. S. Chang. 1999. Additive effect of Sp1 and Sp3 in regulation of the ED-L1E promoter of the EBV LMP 1 gene in human epithelial cells. Virology 261:288-294. [DOI] [PubMed] [Google Scholar]
- 58.Vasudevan, K. M., S. Gurumurthy, and V. M. Rangnekar. 2004. Suppression of PTEN expression by NF-κB prevents apoptosis. Mol. Cell. Biol. 24:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viatour, P., M. P. Merville, V. Bours, and A. Chariot. 2005. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 30:43-52. [DOI] [PubMed] [Google Scholar]
- 60.Waltzer, L., F. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 62.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, F., S. F. Tsang, M. G. Kurilla, J. I. Cohen, and E. Kieff. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber, C. K., S. Liptay, T. Wirth, G. Adler, and R. M. Schmid. 2000. Suppression of NF-κB activity by sulfasalazine is mediated by direct inhibition of IκB kinases α and β. Gastroenterology 119:1209-1218. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, L., and J. S. Pagano. 2001. Interferon regulatory factor 7 mediates activation of Tap-2 by Epstein-Barr virus latent membrane protein 1. J. Virol. 75:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, L., L. Wu, K. Hong, and J. S. Pagano. 2001. Intracellular signaling molecules activated by Epstein-Barr virus for induction of interferon regulatory factor 7. J. Virol. 75:12393-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]