Abstract
The cell wall of the unicellular green alga Chlamydomonas reinhardtii consists predominantly of Hyp-rich glycoproteins, which also occur in the extracellular matrix of multicellular green algae and higher plants. In addition to the Hyp-rich polypeptides, the insoluble glycoprotein framework of the Chlamydomonas cell wall contains minor amounts of 14-3-3 proteins, as revealed by immunochemical studies and mass spectroscopic analysis of tryptic peptides. Polypeptides immunologically related to the 14-3-3 proteins also were found in the culture medium of Chlamydomonas. The levels of two of these 14-3-3–related polypeptides were decreased in the culture medium of the wall-deficient mutant cw-15. These findings indicate that 14-3-3 proteins are involved in the cross-linking of Hyp-rich glycoproteins in the Chlamydomonas cell wall.
INTRODUCTION
Cell walls of higher plants consist not only of cellulose and other polysaccharides but also contain insoluble, Hyp-rich glycoproteins and their soluble precursors, the extensins (Kieliszewski and Lamport, 1994; Knox, 1995; Lamport, 2001). Hyp-rich glycoproteins are the predominant constituents of the extracellular matrix of volvocine algae, which range from unicellular Chlamydomonas reinhardtii to multicellular organisms in the genus Volvox (Woessner and Goodenough, 1994). The multilayered cell wall of the unicellular green alga Chlamydomonas consists of an insoluble, Hyp-rich glycoprotein framework and several chaotrope-soluble, Hyp-containing glycoproteins (Roberts, 1974; Monk et al., 1983; Goodenough and Heuser, 1985; Imam et al., 1985; Roberts et al., 1985; Voigt, 1988; Voigt et al., 1991). The insoluble wall fraction can be isolated from intact cells by successive extractions with different detergent-containing buffers (Voigt, 1984; Vogeler et al., 1990). After chemical deglycosylation with hydrofluoric acid (HF)/pyridine, the “sac-like” morphology of the highly purified insoluble wall component was destroyed and at least part of its polypeptide constituents became soluble in SDS-containing buffers (Vogeler et al., 1990). Isodityrosine has been detected in hydrolysates of Chlamydomonas cell walls (Waffenschmidt et al., 1993), indicating that peroxidase-catalyzed cross-linking of cell wall glycoproteins via Tyr side chains, which has been observed for the extensins of higher plants (Fry, 1982; Epstein and Lamport, 1984; Biggs and Fry, 1990), also occurs in Chlamydomonas. Furthermore, it has been reported that a transglutaminase also catalyzes the cross-linking of cell wall proteins in Chlamydomonas (Waffenschmidt et al., 1999).
The size of the Chlamydomonas cell is limited by the size of the insoluble glycoprotein framework of the wall, which completely surrounds the protoplast as a sac-like structure (Voigt, 1984; Imam et al., 1985). Therefore, enlargement of this insoluble glycoprotein framework is a prerequisite for cell growth. As revealed by pulse-labeling and pulse-chase experiments with 3H-Pro and 35S-Met, turnover of the insoluble framework of the Chlamydomonas cell wall takes place during the cell enlargement period accompanied by a release of large polypeptide fragments into the culture medium (Voigt, 1985, 1986). The resulting “holes” in the glycoprotein framework are repaired by the simultaneous incorporation of soluble precursors. The pulse-labeling and pulse-chase experiments also indicated that these soluble precursors of the insoluble framework of the Chlamydomonas cell wall are constituents of the salt-soluble wall fraction (Voigt, 1986). This conclusion was corroborated by the finding that an antibody raised against the deglycosylation products of the highly purified insoluble cell wall component reacted specifically with a 150-kD glycoprotein, a major constituent of the chaotrope-soluble wall fraction of Chlamydomonas (Voigt et al., 1996).
Screening of a Chlamydomonas cDNA expression library with the antibody against the deglycosylation products of the purified insoluble cell wall component of Chlamydomonas (Voigt et al., 1996) resulted in the isolation of four cDNA clones that encode a Chlamydomonas 14-3-3 protein (Liebich and Voigt, 1995). Therefore, we have investigated the cross-reactivity of the anti–cell wall antibody with this 14-3-3 protein. Peptide scan analysis (Frank, 1992) using 83 pentadecapeptides derived from the open reading frame (ORF) of the previously cloned Chlamydomonas 14-3-3 cDNA (Liebich and Voigt, 1995) revealed that this particular cell wall antibody recognized several epitopes distributed over almost the entire amino acid sequence of the 14-3-3 protein. The conclusion that the insoluble component of the Chlamydomonas cell wall contains a polypeptide that is related immunologically to the 14-3-3 proteins was further corroborated by immunochemical studies using antibodies raised against individual polypeptides released from the highly purified insoluble fraction of the Chlamydomonas cell wall during chemical deglycosylation and by mass spectroscopic analyses of tryptic peptides of the insoluble wall fraction. Here, we report that 14-3-3 proteins also occur extracellularly. Furthermore, our data provide evidence that 14-3-3 proteins are involved in the formation of the insoluble glycoprotein framework of the cell wall.
RESULTS
Characterization of Cell Wall Antibodies
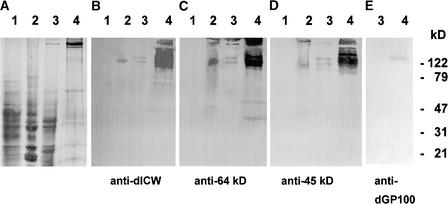
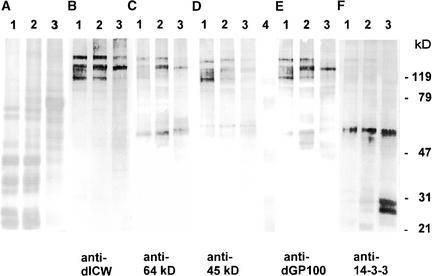
As reported previously, defined polypeptides are released from the highly purified, insoluble cell wall fraction of Chlamydomonas by treatment with anhydrous HF/pyridine (Vogeler et al., 1990). Polyclonal antibodies were raised against the mixture of these deglycosylated cell wall polypeptides (anti-dICW) and against two prominent constituents of this fraction with apparent molecular masses of 64 and 45 kD (anti-dICW-64kDa and anti-dICW-45kDa). The specificity of these antibodies was tested by protein gel blot analysis of cytosol, microsomes, and LiCl extracts of intact cells. LiCl extracts of intact wild-type cells contain soluble precursors of the insoluble cell wall fraction (Voigt et al., 1991, 1996). The cell wall antibodies did not cross-react with constituents of the cytosol (Figures 1A to 1D, lanes 1) but recognized high-molecular-mass components (>120 kD) present in both the microsomes and the LiCl extracts (Figures 1A to 1D, lanes 2 and 3). The amounts of the high-molecular-mass polypeptides recognized by these antibodies were considerably higher in the LiCl extracts of wild-type cells (Figures 1B to 1D, lanes 4) than in the LiCl extracts of wall-deficient cw-15 cells (Figures 1B to 1D, lanes 3), indicating that these polypeptides are constituents of the chaotrope-soluble cell wall layers and presumably precursors of the insoluble glycoprotein framework of the Chlamydomonas cell wall.
Figure 1.
SDS-PAGE and Protein Gel Blot Analyses of Chlamydomonas Cytosol, Microsomes, and LiCl Extracts of Intact cw-15 and Wild-Type Cells.
Samples corresponding to 30 μg of protein were fractionated by SDS-PAGE on slab gels containing 12.5% (w/v) acrylamide. After electrophoresis, the gels were either stained for protein with Coomassie Brilliant Blue (A) or blotted onto polyvinylidene difluoride (PVDF) membranes ([B] to [E]). The protein gel blots were probed with anti-dICW (B), anti-dICW-64kDa (C), anti-dICW-45kDa (D), or anti-dGP100 (E). Lanes 1, cytosol; lanes 2, microsomal fraction; lanes 3, LiCl extract of intact cw-15 cells; lanes 4, LiCl extract of intact wild-type cells.
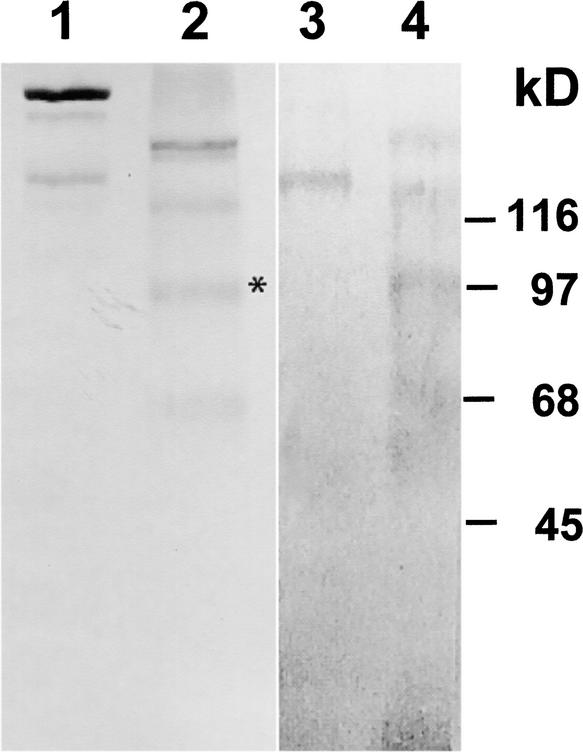
These high-molecular-mass polypeptides did not cross-react with antibodies raised against a recombinant 14-3-3 protein (anti-β-Gal/14-3-3; data not shown). A polyclonal antibody (anti-dGP100) raised against the 100-kD component of the chemically deglycosylated, LiCl-soluble cell wall fraction (Figure 2, lane 2, asterisk) exclusively recognized a 150-kD component present in the LiCl extracts of intact wild-type cells (Figure 1E, lane 4) but absent in the LiCl extracts of the wall-deficient mutant cw-15 (Figure 1E, lane 3). However, when protein gel blots containing the deglycosylation products of the purified LiCl-soluble, high-molecular-mass cell wall glycoproteins (Figure 2, lane 2) were probed with this anti-dGP100 antibody, all of the polypeptides showed cross-reactivity (Figure 2, lane 4). The same results also were obtained when these protein gel blots were probed with anti-dICW, anti-dICW-65kDa, and anti-dICW-45kDa (data not shown). Cross-reactivity of anti-dGP100 with the chemically deglycosylated precursors of the insoluble cell wall fraction apparently is attributable to the high Hyp content of these cell wall polypeptides (Voigt et al., 1991), especially given the fact that this cross-reactivity was not observed in the case of the glycosylated cell wall precursors (Figure 2, lane 3). Anti-dICW-65kDa and anti-dICW-45kDa exclusively recognized LiCl-soluble precursors of the insoluble cell wall fraction (Figures 1C and 1D, lanes 4) whose apparent molecular masses were substantially higher than those of the corresponding cell wall antigens (Table 1). The deglycosylation products of these LiCl-soluble cell wall glycoproteins (Figure 2, lane 2) also were considerably larger than the 64- and 45-kD polypeptides released from the insoluble cell wall fraction. Therefore, these 64- and 45-kD components must be derived from the larger precursors by proteolytic processing.
Figure 2.
SDS-PAGE and Protein Gel Blot Analyses of Purified, LiCl-Soluble High-Molecular-Mass Cell Wall Glycoproteins and Their Deglycosylation Products.
Untreated and chemically deglycosylated cell wall glycoproteins (30 μg of protein) were separated by SDS-PAGE on gel slabs containing 7% (w/v) acrylamide. After electrophoresis, the gels were either stained for protein with Coomassie Brilliant Blue (lanes 1 and 2) or blotted onto PVDF membranes (lanes 3 and 4). The protein gel blots were probed with anti-dGP100 (lanes 3 and 4). Lanes 1 and 3, untreated cell wall glycoproteins; lanes 2 and 4, cell wall glycoproteins treated for 3 h with trimethylsilyl trifluoromethanesulfonate at room temperature. The deglycosylated polypeptide that was used as an antigen to increase the anti-dGP100 antibody is marked with an asterisk.
Table 1.
Polyclonal Antibodies Used in This Study
| Antibody | Antigen | Reference |
|---|---|---|
| Anti-β-Gal/14-3-3a | Recombinant (E. coli) β-Gal:(Chlamydomonas) 14-3-3 fusion protein | Voigt et al. (2001) |
| Anti-dGP100 | 100-kD deglycosylation product of the 150-kD chaotrope-soluble cell wall glycoprotein of Chlamydomonas | Voigt et al. (1996) |
| Anti-dICW | Mixture of polypeptides released from the purified insoluble glycoprotein framework of the Chlamydomonas cell wall by chemical deglycosylation with anhydrous HF/pyridine |
Voigt et al. (1996) |
| Anti-dICW-64kDa | 64-kD polypeptide purified from the mixture of polypeptides released from the purified insoluble glycopro- tein framework of the Chlamydomonas cell wall by chemical deglycosylation with anhydrous HF/pyridine | This report |
| Anti-dICW-45kDa | 45-kD polypeptide purified from the mixture of polypeptides released from the purified insoluble glycopro- tein framework of the Chlamydomonas cell wall by chemical deglycosylation with anhydrous HF/pyridine | This report |
Depleted from anti-(E. coli) β-galactosidase (β-Gal) Igs by preincubation with PVDF membrane–bound (E. coli) β-Gal.
As reported previously, partial proteolysis of the insoluble cell wall fraction is a prerequisite for cell enlargement (Voigt, 1985, 1986; Voigt et al., 1996) and is accompanied by a release of cell wall macromolecules into the culture medium (Voigt, 1985, 1986). The size of the polypeptide fragments retained in the insoluble cell wall layer obviously is not determined exclusively by the distribution of cleavage sites because the N-terminal amino acid sequences of the 64- and 45-kD constituents were found to be rather heterogenous (Table 2). Mixtures of amino acid residues were cleaved off during each round of Edman degradation, in contrast to their LiCl-soluble precursors (Table 2). These findings indicate that the 64- and 45-kD constituents of the chemically deglycosylated insoluble cell wall fraction do not represent single molecular entities but are mixtures composed of polypeptide fragments derived by proteolytic processing of various precursors (Figure 2, lane 2).
Table 2.
N-Terminal Amino Acid Sequences of Polypeptide Constituents of the Insoluble Cell Wall Fraction of Chlamydomonas and Its LiCl-Soluble Precursors
| N-Terminal Amino Acid Sequence
|
||||||
|---|---|---|---|---|---|---|
| Cell Wall Fraction | Apparent Molecular Mass | 1 | 2 | 3 | 4 | 5 |
| Insoluble | 64 kD | AGDSTILM | GNALR | VLGIEQA | DVNQV | PNESG |
| Insoluble | 45 kD | GSAMLVI | QNAGV | DSTEL | STQILD | GENR |
| LiCl soluble | 200 kD | S | X | X | X | X |
| LiCl soluble | 135 kD | ?a | ||||
| LiCl soluble | 100 kD | I | N | I | P | N |
The N terminus is blocked.
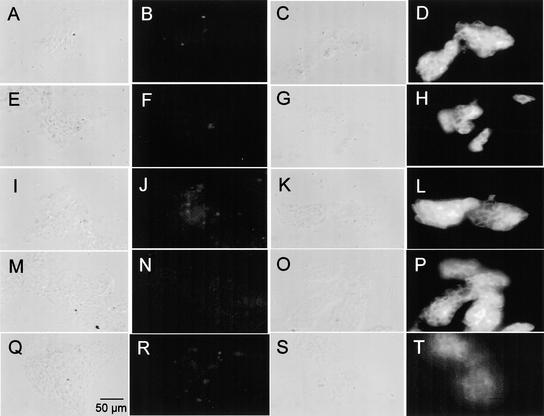
The insoluble glycoprotein framework of the Chlamydomonas cell wall contains polypeptide constituents that are related immunologically to the 14-3-3 proteins. The highly purified insoluble cell wall component revealed immunofluorescence not only with anti-dICW, anti-dICW-64kDa, and anti-dICW-45kDa (Figures 3D, 3H, and 3L) but also with the antibodies against the 100-kD deglycosylation product of the 150-kD chaotrope-soluble cell wall glycoprotein (anti-dGP100; Figure 3P) and, to a lesser extend, with anti-β-Gal/14-3-3 (Figure 3T). No immunofluorescence was observed with any of the corresponding preimmune sera under the same conditions (Figures 3B, 3F, 3J, 3N, and 3R). Immunofluorescence studies with formaldehyde-fixed wild-type cells revealed that the different cell wall antibodies, but not the antibody raised against the recombinant 14-3-3 protein (anti-β-Gal/14-3-3), reacted with the cell surface of Chlamydomonas wild-type cells (data not shown).
Figure 3.
Immunofluorescence Analysis of the Purified, Insoluble Cell Wall Component of Chlamydomonas.
(A), (C), (E), (G), (I), (K), (M), (O), (Q), and (S) Phase-contrast micrographs.
(B), (D), (F), (H), (J), (L), (N), (P), (R), and (T) Fluorescence micrographs.
(A) and (B) Anti-dICW preimmune serum.
(C) and (D) Anti-dICW Igs.
(E) and (F) Anti-dICW-64kDa preimmune serum.
(G) and (H) Anti-dICW-64kDa Igs.
(I) and (J) Anti-dICW-45kDa preimmune serum.
(K) and (L) Anti-dICW-45kDa Igs.
(M) and (N) Anti-dGP100 preimmune serum.
(O) and (P) Anti-dGP100 Igs.
(Q) and (R) Anti-β-Gal/14-3-3 preimmune serum.
(S) and (T) Antibodies against the recombinant Chlamydomonas 14-3-3 protein (anti-β-Gal/14-3-3).
Bar in (Q) = 50 μm for all panels.
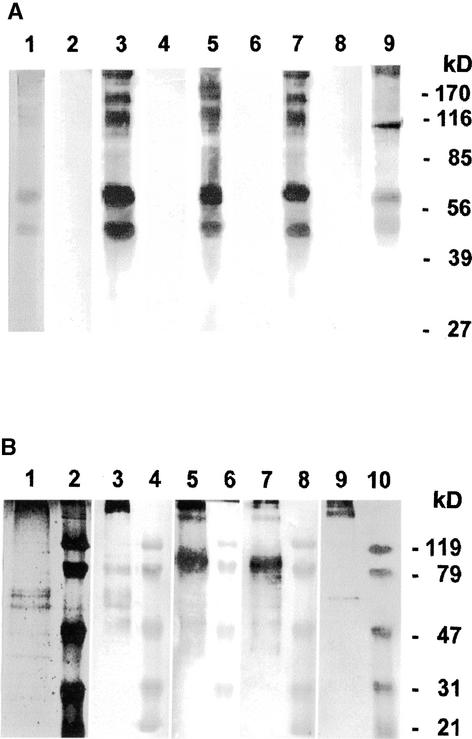
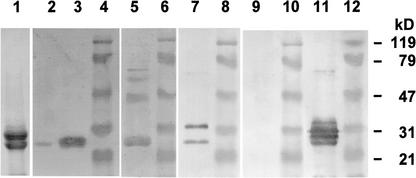
Distinct polypeptides are released from the purified insoluble glycoprotein framework of the Chlamydomonas cell wall by chemical deglycosylation with anhydrous HF/pyridine (Voigt et al., 1996). When these deglycosylation products were subjected to protein gel blot analyses, all of the solubilized polypeptides (Figure 4A, lane 1) were recognized by the cell wall antisera anti-dICW, anti-dICW-64kDa, and anti-dICW-45kDa (Figure 4A, lanes 3, 5, and 7). The prominent 64- and 45-kD components (Figure 4A, lane 1) cross-reacted with the antibody raised against the recombinant Chlamydomonas 14-3-3 protein (Figure 4A, lane 9). Additionally, the latter antibody reacted strongly with a 107-kD component (Figure 4A, lane 9) that was considerably less reactive toward the different cell wall antibodies (Figure 4A, lanes 3, 5, and 7). None of the polypeptides released from the insoluble wall fraction by treatment with HF/pyridine (Figure 4A, lane 1) reacted with any of the various preimmune sera (Figure 4A, lanes 2, 4, 6, and 8). As reported recently, Chlamydomonas cells contain four 14-3-3 isoforms whose subunits have apparent molecular masses of 32, 30, 27, and 24 kD (Voigt et al., 2001). In this range, no polypeptides were detected in the deglycosylation products of the insoluble Chlamydomonas cell wall fraction by anti-β-Gal/14-3-3 Ig (Figure 4A, lane 9). The cross-reacting 107-, 64-, and 45-kD components are intrinsic constituents of the insoluble framework of the Chlamydomonas cell wall.
Figure 4.
SDS-PAGE and Protein Gel Blot Analyses of Polypeptides Released from the Purified Insoluble Cell Wall Fraction of Chlamydomonas by Treatment with Anhydrous HF/Pyridine and Trimethylsilyl Trifluoromethanesulfonate in the Cold.
The freeze-dried, highly purified insoluble wall fraction was treated with anhydrous HF/pyridine or trimethylsilyl trifluoromethanesulfonate as described in Methods. Twenty micrograms (lane 1) or 2 μg (other lanes) of solubilized polypeptides was fractionated by SDS-PAGE on slab gels containing 12.5% (w/v) acrylamide. After electrophoresis, the gels were either stained for protein with Coomassie Brilliant Blue or blotted onto PVDF membranes.
(A) Polypeptides released by treatment with anhydrous HF/pyridine. Lane 1, gel stained for protein; lanes 2 and 3, protein gel blots probed with anti-dICW (lane 3) and the corresponding preimmune serum (lane 2); lanes 4 and 5, protein gel blots probed with anti-dICW-64kDa (lane 5) and the corresponding preimmune serum (lane 4); lanes 6 and 7, protein gel blots probed with anti-dICW-45kDa (lane 7) and the corresponding preimmune serum (lane 6); lanes 8 and 9, protein gel blots probed with antibodies against the recombinant 14-3-3 protein (lane 9) and the corresponding preimmune serum (lane 8).
(B) Polypeptides released by treatment with trimethylsilyl trifluoromethanesulfonate. Lane 1, gel stained for protein; lanes 2, 4, 6, 8, and 10, prestained protein molecular mass markers (apparent molecular masses are given at right); lane 3, protein gel blot probed with anti-dICW; lane 5, protein gel blot probed with anti-dICW-64kDa; lane 7, protein gel blot probed with anti-dICW-45kDa; lane 9, protein gel blot probed with antibodies against the recombinant 14-3-3 protein.
This conclusion was further corroborated by analyses of polypeptides released from the insoluble glycoprotein framework of the Chlamydomonas cell wall by incomplete deglycosylation with trimethylsilyl trifluoromethanesulfonate under mild conditions. The pattern of polypeptides released under these conditions (Figure 4B, lane 1) differed considerably from that observed after treatment with HF/pyridine (Figure 4A, lane 1). The predominant portion of the released polypeptides was observed in the stacking gel (data not shown). Furthermore, prominent polypeptides with apparent molecular masses of 38, 47, 59, 64, 67, 74, 79, 100, and >300 kD were detected by SDS-PAGE analysis (Figure 4B, lane 1). Most of these components were recognized by the cell wall antibodies anti-dICW, anti-dICW-64kDa, and anti-dICW-45kDa (Figure 4B, lanes 3, 5, and 7). The relative intensities of these bands, however, were rather different (Figure 4B, lanes 1, 3, 5, and 7). The antibodies against the recombinant Chlamydomonas 14-3-3 protein reacted strongly with a 63-kD component (Figure 4B, lane 9) that cross-reacted weakly with the different cell wall antibodies (Figure 4B, lanes 3, 5, and 7). In addition to the 63-kD component, anti-β-Gal/14-3-3 recognized polypeptides with apparent molecular masses of >300 kD (Figure 4B, lane 9). These findings indicate that the polypeptides that are related immunologically to the 14-3-3 proteins are cross-linked to the insoluble glycoprotein framework of the Chlamydomonas cell wall.
Reactivities of Different Cell Wall Antibodies toward Epitopes of a Chlamydomonas 14-3-3 Protein
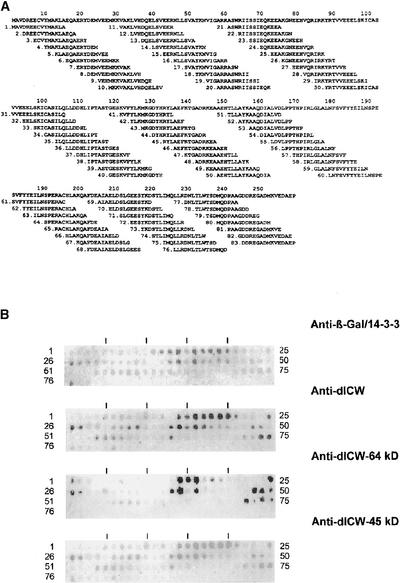
Screening of a Chlamydomonas cDNA expression library with anti-dICW (Voigt et al., 1996) resulted in the isolation of four cDNA clones that encode a Chlamydomonas 14-3-3 protein (Liebich and Voigt, 1995). By contrast, a polyclonal antibody raised against the corresponding recombinant 14-3-3 protein (anti-β-Gal/14-3-3) was shown to react specifically with the insoluble cell wall fraction of Chlamydomonas, as revealed by immunofluorescence studies (Figure 3T), indicating that the glycoprotein framework of the Chlamydomonas cell wall contains at least one polypeptide that is related immunologically to this particular 14-3-3 protein. Therefore, we comparatively analyzed the reactivities of anti-β-Gal/14-3-3 and anti-dICW, anti-dICW-64kDa, and anti-dICW-45kDa toward epitopes of this particular Chlamydomonas 14-3-3 protein by peptide-scan analyses (Figure 5).
Figure 5.
Comparative Epitope Analyses of the Polyclonal Antibody Raised against the Recombinant 14-3-3 Protein and Different Polypeptides of the Insoluble Cell Wall Fraction.
(A) Eighty-three overlapping pentadecapeptides derived from the ORF of the previously cloned Chlamydomonas 14-3-3 cDNA and representing the entire amino acid sequence were synthesized by spot synthesis using cellulose paper as a solid support (Frank, 1992).
(B) After treatment with blocking solution, the cellulose-bound scan peptides were incubated with the different antibodies. After extensive washing, bound IgGs were detected by incubation with alkaline phosphatase–coupled anti-rabbit IgG and subsequent visualization of the indirectly bound alkaline phosphatase as described in Methods.
To this end, 83 overlapping pentadecapeptides (Figure 5A) were generated by spot synthesis on a cellulose membrane (Frank, 1992) that, together, represented the entire amino acid sequence of the ORF derived from the previously cloned Chlamydomonas 14-3-3 cDNA (Liebich and Voigt, 1995). The anti-β-Gal/14-3-3 recognized several pentadecapeptide sequences (Figure 5B) distributed over almost the entire 29.5-kD ORF of the cloned Chlamydomonas 14-3-3 cDNA (Figure 5A). A rather similar pattern was observed for the anti-dICW antibody used to screen the cDNA expression library, which resulted in the isolation of 14-3-3 clones. The relative reactivities of both antisera toward these peptides, however, were rather different (Figure 5B). Peptides 7, 10, 31 to 34, 54 to 57, 63, 74, 75, and 81 reacted more strongly with anti-dICW (Figure 5B), whereas peptides 11 to 13, 28, 29, and 76 to 78 revealed weaker signals with this particular antiserum than with anti-β-Gal/14-3-3 (Figure 5B). The anti-dICW-64kDa antiserum recognized peptides 14 to 16, 25, 26, 38, 39, 41, 48 to 50, 72, 74, and 75 (Figure 3B). The anti-dICW-45kDa serum (Figure 5B), on the other hand, cross-reacted with several pentadecapeptides derived from the ORF of the previously cloned Chlamydomonas 14-3-3 cDNA (Liebich and Voigt, 1995), which also were recognized by anti-dICW (Figure 4B) and by anti-β-Gal/14-3-3 (Figure 4B). These data clearly show that the insoluble cell wall fraction of Chlamydomonas and its 64- and 45-kD deglycosylation products contain polypeptides that are related immunologically to the 14-3-3 proteins.
Differential Reactivities of the Various Cell Wall Antibodies toward the 14-3-3 Isoforms of Chlamydomonas
When the mixture of 14-3-3 proteins partially purified from Chlamydomonas cells was analyzed by SDS-PAGE (Figure 6, lane 1), two predominant polypeptides with apparent molecular masses of 30 and 27.5 kD were observed. Protein gel blots of the same 14-3-3 preparation probed with the anti-β-Gal/14-3-3 (Figure 6, lane 11) showed the presence of two additional 14-3-3 isoforms with apparent molecular masses of 32 and 25 kD and a faint band at 63 kD that might be a residual 14-3-3 dimer. None of these polypeptides reacted with anti-dGP100 raised against the 100-kD deglycosylation product of a 150-kD glycoprotein (Figure 6, lane 9), which was shown to be a precursor of the insoluble fraction of the Chlamydomonas cell wall (Voigt et al., 1996). Anti-dICW exclusively recognized the 27.5-kD 14-3-3 isoform (Figure 6, lanes 2 and 3). Anti-dICW-45kDa reacted preferentially with the 32-kD 14-3-3 isoform but also recognized the 27.5-kD isoform (Figure 6, lane 7). Anti-dICW-64kDa recognized the 27.5-kD isoform but not the other 14-3-3 isoforms and cross-reacted strongly with contaminating polypeptides with apparent molecular masses of 58 and 45 kD (Figure 6, lane 5) that were not recognized by the other antibodies (Figure 6, lanes 2, 3, 7, 9, and 11). However, considerably increased incubation times in the presence of 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium chloride were required to visualize these bands compared with the other antibodies, indicating that the reactivity of this particular antibody was rather low.
Figure 6.
SDS-PAGE and Comparative Protein Gel Blot Analyses of the Chlamydomonas 14-3-3 Proteins with Polyclonal Antibodies Raised against Different Polypeptides of the Insoluble Fraction of the Chlamydomonas Cell Wall.
Twenty (lane 1), 0.2 (lane 2), or 2 μg (other lanes) of purified 14-3-3 proteins were fractionated by SDS-PAGE on slab gels containing 12.5% (w/v) acrylamide. After electrophoresis, the gels were either stained for protein with Coomassie Brilliant Blue (lane 1) or blotted onto PVDF membranes. Lanes 4, 6, 8, 10, and 12, prestained protein molecular mass markers (apparent molecular masses are given at right); lanes 2 and 3, protein gel blots probed with anti-dICW; lane 5, protein gel blot probed with anti-dICW-64kDa; lane 7, protein gel blot probed with anti-dICW-45kDa; lane 9, protein gel blot probed with anti-dGP100; lane 11, protein gel blot probed with antibodies against the recombinant 14-3-3 protein.
Precursors of the Insoluble Cell Wall Fraction
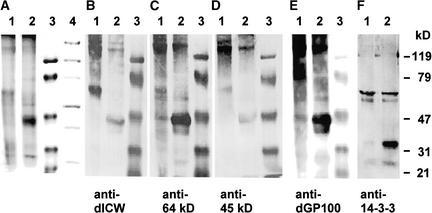
Our findings that the insoluble glycoprotein framework of the Chlamydomonas cell wall contains polypeptides that are related immunologically to the 14-3-3 proteins (Figures 3 to 6) prompted us to search for their soluble precursors. Cell wall precursors, like other secreted polypeptides, must pass the endoplasmic reticulum (ER) and the dictyosomes. Therefore, the crude microsomes were fractionated by sucrose gradient centrifugation to obtain the ER, the dictyosomes, and the plasma membrane, which were detected by marker enzyme activities. Again, the corresponding polypeptides (Figure 7, lanes 1 to 3) were subjected to comparative protein gel blot analyses using different cell wall antibodies (Figures 7B to 7E) and the anti-β-Gal/14-3-3 antibody (Figure 7F). The different cell wall antibodies (anti-dICW, anti-dICW-64kDa, anti-dICW-45kDa, and anti-dGP100) reacted preferentially with high-molecular-mass components with apparent molecular masses of >120 kD (Figures 7B to 7E). The finding that the same patterns of cross-reacting high-molecular-mass polypeptides were obtained with all of these antibodies (Figures 7B to 7E) was astonishing because the corresponding antigens were rather different (Table 1). A 150-kD component was predominant in the ER (Figures 7B to 7E, lanes 3) as in the crude microsomal fraction (Figures 1B to 1D, lanes 2). Increased amounts of other high-molecular-mass polypeptides were detected in the dictyosomes (Figures 7B to 7E, lanes 2) and in the plasma membrane fraction (Figures 7B to 7E, lanes 1), which presumably are generated by post-translational modification in the dictyosomes.
Figure 7.
Comparative SDS-PAGE and Protein Gel Blot Analyses of Chlamydomonas Membranes.
Samples corresponding to 30 μg of protein were fractionated by SDS-PAGE on slab gels containing 12.5% (w/v) acrylamide. After electrophoresis, the gels were either stained for protein with Coomassie Brilliant Blue (A) or blotted onto PVDF membranes ([B] to [F]). The protein gel blots were probed with anti-dICW (B), anti-dICW-64kDa (C), anti-dICW-45kDa (D), anti-dGP100 (E), and antibodies against the recombinant Chlamydomonas 14-3-3 protein (F). Lanes 1, plasma membrane; lanes 2, dictyosomes; lanes 3, ER.
Almost the same patterns of high-molecular-mass polypeptides were detected by the various cell wall antibodies in the dictyosomes (Figures 7B to 7E, lanes 2), in the plasma membrane fraction (Figures 7B to 7E, lanes 1), and in the LiCl extracts of intact cells (Figures 7B to 7D, lanes 3 and 4). None of these high-molecular-mass polypeptides cross-reacted with the anti-β-Gal/14-3-3 antibody, which recognized only the known 14-3-3 isoforms (Figure 7F, lanes 1 to 3). As reported recently (Voigt et al., 2001), the 27.5-kD component was predominant in the ER fraction, which also contained the 30-kD isoform and, to a lesser extent, the 24-kD isoform (Figure 7F, lane 3). Different patterns of 14-3-3 isoforms were detected in the plasma membrane (Figure 7F, lane 1) and dictyosome fractions (Figure 7F, lane 2), as reported recently (Voigt et al., 2001). An additional 64-kD component, however, was detected in all of these fractions (Figure 7F, lanes 1 to 3), which might be a residual 14-3-3 dimer.
The findings described above clearly show that the predominant precursors of the insoluble glycoprotein framework of the Chlamydomonas cell wall are high-molecular-mass polypeptides (glycoproteins) that are modified post-translationally in the dictyosomes and are not related immunologically to the 14-3-3 proteins. Therefore, the constituents of the insoluble glycoprotein framework of the Chlamydomonas cell wall that are immunologically related to the 14-3-3 proteins must be minor components of this cell wall fraction, whose soluble precursors presumably are intrinsic 14-3-3 isoforms associated with the microsomal membranes.
Characterization of 14-3-3–Related Proteins by Analysis of Tryptic Fragments of the Insoluble Cell Wall Fraction of Chlamydomonas
The data described above indicate that 14-3-3–related polypeptides are cross-linked to the insoluble cell wall fraction of Chlamydomonas. Therefore, we used mass spectrometry to analyze tryptic fragments of the highly purified, insoluble cell wall fraction for the presence of 14-3-3–related peptides. No tryptic fragments of the previously described Chlamydomonas 14-3-3 protein (Voigt et al., 2001) were detected in this peptide fraction by matrix-assisted laser-desorption ionization time-of-flight analysis. However, some of the peptide fragments obtained revealed amino acid sequences that are similar to sequence motifs of the known 30-kD 14-3-3 isoform (Table 3). The peptides VAVLANEQELSVEER and NLLSVSYK match positions 31 to 45 and 46 to 53, respectively, of the amino acid sequence of the 30-kD isoform (Table 3). The scan peptides 11 to 16 (Figure 5A) contain eight or more of these amino acid residues. All of these scan peptides reacted with anti-β-Gal/14-3-3, but only the scan peptides 14 to 16 were recognized by the different cell wall antibodies (Figure 5B). This difference apparently is attributable to the substitution of the sequence VHD (positions 35 to 37 of the 30-kD isoform) by ANE (Table 3), indicating that the 30-kD 14-3-3 isoform is not contained in the cell wall.
Table 3.
Analysis of 14-3-3–Related Peptides Released by Trypsin from the Insoluble Cell Wall Fraction of Chlamydomonas
| Peptides of the Insoluble Cell Wall Fraction | Corresponding Peptide Fragments of the 30-kD 14-3-3 Isoform |
Position in the Amino Acid Sequence of the 30-kD 14-3-3 Isoform |
|---|---|---|
| VAVLANEQELSVEER | VAKLVHDQELSVEER | 31 to 45 |
| NLLSVSYK | NLLSVAYK | 46 to 53 |
| YLVPSASTTEAAVFYLK | HLIPTASTGESKVFYLK | 110 to 126 |
| AFDEAISDLDSLGEDSYK | AFDEAIAELDSLGEESYK | 201 to 218 |
| DNLTLWTSEM | DNLTLWTSDM | 299 to 238 |
| VVLYSAFAAR | Not identified | – |
| MITAIGLVK | Not identified | – |
| EINPNR | Not identified | – |
| EQFDTFGDTAAIR | Not identified | – |
| ESILNALLSK | Not identified | – |
| AGVDPSADPAVVR | Not identified | – |
| MLFTPALAFSSTILR | Not identified | – |
| ETNSAFYPAYTMLGVK | Not identified | – |
| EVSYGLYLSGIAGVFLR | Not identified | – |
| EVILPNSDK | Not identified | – |
After digestion of the purified insoluble cell wall fraction, peptides were examined by matrix-assisted laser-desorption ionization time-of-flight analysis and electrospray ionization tandem mass spectrometry. Sequences are shown in one-letter code. The 30-kD 14-3-3 isoform is the same as that shown in Figure 5. The underlined residues are different from the previously cloned 30-kD 14-3-3 isoform.
The peptide YLVPSASTTEAAVFYLK matches positions 110 to 126 of the 30-kD isoform (Table 3). This sequence was contained largely in the scan peptides 37 to 41 (Figure 5A). All of the antibodies reacted strongly with the scan peptides 38, 39, and 41 (particularly with peptide 39) and only weakly with peptide 40 (Figure 5B). The peptide AFDEAISDLDSLGEDSY matches positions 201 to 218 of the 30-kD isoform (Table 3). This amino acid sequence was more or less contained (approximately six amino acid residues) in the scan peptides 65 to 71 (Figure 5A). Only scan peptide 68 reacted with anti-β-Gal/14-3-3, anti-dICW, and anti-dICW-45kDa (Figure 5B). The peptide DNLTLWTSEM matches positions 229 to 238 of the 30-kD isoform (Table 3). More than five of these amino acid residues were contained in the scan peptides 74 to 78 (Figure 5A), which were recognized by all of the antibodies (Figure 5B). All of these peptides match a single 14-3-3 isoform that is being sequenced at present (J. Voigt, S. Stevanovic, M. Schirle, M. Fausel, J. Maier, K.-H. Adam, and O. Marquardt, unpublished results). The other peptides do not match any protein sequence present in the databases. Presumably, they are fragments of the Hyp-rich cell wall glycoproteins. Some of these peptides match partial amino acid sequences of LiCl-soluble cell wall glycoproteins (J. Voigt and M. Kiess, unpublished results).
Analyses of Macromolecules Accumulated in the Culture Medium
As reported previously, the culture medium of Chlamydomonas wild-type strains contains fragments of the insoluble glycoprotein framework of the Chlamydomonas cell wall that are released both during the cell enlargement period (Liebich and Voigt, 1995; Voigt et al., 1996) and during the liberation of zoospores by degradation of the mother cell wall (Schlösser, 1966; Mihara and Hase, 1975; Waffenschmidt et al., 1988). The culture medium of wall-deficient strains such as cw-15, however, contains constituents of the chaotrope-soluble wall layers and components normally localized in the periplasm (Loppes and Deltour, 1975; Voigt, 1986; Voigt et al., 1997). Accordingly, the patterns of polypeptides accumulated in the culture medium of the wall-deficient strain cw-15 (Figure 8A, lane 1) and the Chlamydomonas wild-type strain (Figure 8A, lane 2) were rather different. Apart from the high-molecular-mass components (>120 kD), the culture medium of the wild-type strain contained a predominant polypeptide of 45 kD (Figure 8A, lane 2) that was absent from the culture medium of the wall-deficient strain (Figure 8A, lane 1) and was recognized by all of the antibodies raised against the different deglycosylated cell wall polypeptides (Figures 8B to 8E, lanes 2) but did not cross-react with the antibodies raised against the recombinant Chlamydomonas 14-3-3 protein (Figure 8F, lane 2).
Figure 8.
Comparative SDS-PAGE and Protein Gel Blot Analyses of Macromolecules Accumulated in the Culture Medium of the Chlamydomonas Wild-Type Strain and the Cell Wall–Deficient Mutant Strain cw-15.
Macromolecules (corresponding to 20 μg of protein) accumulated in the culture medium were fractionated by SDS-PAGE on slab gels containing 12.5% (w/v) acrylamide. After electrophoresis, the gels were either stained for protein with Coomassie Brilliant Blue (A) or blotted onto PVDF membranes ([B] to [F]) and probed with anti-dICW (B), anti-dICW-64kDa (C), anti-dICW-45kDa (D), anti-dGP100 (E), and antibodies against the recombinant Chlamydomonas 14-3-3 protein (F). Lanes 1, macromolecules accumulated in the culture medium of the wall-deficient mutant strain cw-15; lanes 2, macromolecules accumulated in the culture medium of wild-type Chlamydomonas; lanes 3 and 4, protein molecular mass markers (apparent molecular masses are given at right).
On the other hand, the culture medium of the wall-deficient strain cw-15 contained greater amounts of polypeptides with apparent molecular masses of 65 and 150 kD (Figure 8A, lane 1) compared with the wild type (Figure 8A, lane 2), which also were recognized by all of the antibodies raised against the different deglycosylated cell wall polypeptides (Figures 8B to 8E, lanes 1) but did not cross-react with the anti-β-Gal/14-3-3 serum (Figure 8F, lane 1). The anti-β-Gal/14-3-3 antibody recognized four polypeptides accumulated in the culture medium of both the wall-deficient strain cw-15 (Figure 8F, lane 1) and the wild type (Figure 8F, lane 2) with apparent molecular masses of 25, 33, 59, and 64 kD. The relative amounts of the 25-kD component and especially the 33-kD component were decreased considerably in the culture medium of the wall-deficient strain cw-15 (Figure 8F, lane 1) compared with the wild type (Figure 8F, lane 2).
DISCUSSION
The cell wall of Chlamydomonas consists of an insoluble Hyp-rich glycoprotein framework and several chaotrope-soluble glycoproteins (Roberts, 1974; Monk et al., 1983; Goodenough and Heuser, 1985; Imam et al., 1985; Roberts et al., 1985). In addition to the Hyp-rich glycoproteins, 14-3-3 proteins are minor constituents of the insoluble cell wall fraction, as revealed by immunochemical studies (Figures 3 to 6) and mass spectroscopic analysis of tryptic peptides (Table 3).
The 14-3-3 proteins were discovered during a systematic study of brain-specific proteins (Moore and Perez, 1967). The name 14-3-3 originates from their migration pattern on DEAE-cellulose chromatography and subsequent electrophoresis of brain proteins (Moore and Perez, 1967). In the meantime, they have been found in all eukaryotic organisms studied to date (Aitken et al., 1992; Wang and Shakes, 1996; Ferl et al., 2002). Mammalian cells and higher plants contain up to 15 different 14-3-3 genes (Wang and Shakes, 1996; Aitken, 2002; Ferl et al., 2002). In the yeast Saccharomyces cerevisiae and Schizosaccharomyces pombe, which contain only two 14-3-3 genes, inactivation of both genes is lethal (Ford et al., 1994; van Heusden et al., 1995). In animal cells and higher plants, most studies have concentrated on the functions of 14-3-3 proteins in the cytosol, including their effects on metabolic enzymes (Huber et al., 2002; Comparot et al., 2003) and signal transduction (Aitken, 1996, 2002; Roberts et al., 2002; Sehnke et al., 2002).
In higher plants, members of this protein family have been shown to be constituents of the G-box binding complex involved in transcriptional control (De Vetten et al., 1992; Lu et al., 1992; Bihn et al., 1997). Previously, 14-3-3 proteins were detected in the nuclei of human cells (Todd et al., 1998). Some brain isoforms were found to be associated with the plasma membrane and synaptic vesicles (Jones et al., 1995). A role in vesicular trafficking has been observed for the budding yeast (S. cerevisiae) 14-3-3 homologs (Gelperin et al., 1995). The four 14-3-3 isoforms of Chlamydomonas interact differentially with the ER, dictyosomes, and plasma membrane (Voigt et al., 2001). Plant plasma membrane pump H+-ATPases are activated by a complex of the fungal toxin fusicoccin with 14-3-3 (Oecking et al., 1994; Baunsgaard et al., 1998; Fullone et al., 1998). Furthermore, 14-3-3 proteins occur in chloroplasts and plant mitochondria, where they regulate ATP synthase activities (Bunney et al., 2001; Aducci et al., 2002; Jarvis and Soll, 2002; Soll, 2002). Although 14-3-3 proteins have been found in several organelles, their presence in the extracellular matrix has not been described.
The 14-3-3 proteins are highly helical and exist as homodimers and heterodimers (Aitken, 1996, 2002). Each subunit contains an amphipathic groove that is formed by hydrophobic amino acid residues of the C-terminal domain and basic residues of the N-terminal domain and acts as a binding site for phosphorylated proteins containing the motif R(S)XSpXP (where Sp represents phosphoserine) or specific unphosphorylated peptides (Muslin et al., 1996; Ichimura et al., 1997; Zhang et al., 1997; Petosa et al., 1998). Therefore, 14-3-3 proteins are assumed to be adapter proteins that may function in an analogous manner to SH2 (Src Homology 2) domains (Aitken, 1996, 2002; Muslin et al., 1996) or act as chaperones (Jackson-Constan et al., 2001; Jeanclos et al., 2001; Jarvis and Soll, 2002; Soll, 2002; Yaffe, 2002).
Our observation that 14-3-3 proteins are accumulated in the culture medium of Chlamydomonas and that the levels of two 14-3-3 components were decreased in the culture medium of the wall-deficient mutant cw-15 compared with the wild type (Figure 8) indicate that these 14-3-3 components are involved in the generation of the insoluble glycoprotein framework of the Chlamydomonas cell wall. We assume that they affect the cross-linking of soluble glycoprotein precursors by acting as adapters or chaperones.
As shown by Ender et al. (2002), two main Hyp-rich components (pherophorins DZ1 and DZ2) of the extracellular matrix of Volvox carteri are able to self-assemble and cross-link in an apparently autocatalytic reaction. Cross-linking of pherophorin DZ1 occurs via carbohydrate side chains by the formation of Ara-5′-phospho-5′-Ara phosphodiester bonds (Ender et al., 2002), as described previously for the extracellular matrix glycoproteins SSG 185 and pherophorin S (Holst et al., 1989). To date, such a cross-linking mechanism has not been described for the cell wall glycoproteins of Chlamydomonas, although polypeptides were found to be released from the highly purified insoluble glycoprotein framework of this volvocine green alga during chemical deglycosylation by anhydrous HF/pyridine (Vogeler et al., 1990; Voigt et al., 1996; this report). Molecular genetic evidence indicates that V. carteri evolved from a unicellular ancestor similar to Chlamydomonas during the past 50 million years (Rausch et al., 1989). Two mechanisms for the cross-linking of cell wall glycoproteins have been described for Chlamydomonas: isodityrosine formation by peroxidase-catalyzed cross-linking of Tyr residues (Waffenschmidt et al., 1993) and cross-linking by transglutaminase (Waffenschmidt et al., 1999). Simultaneous binding of two soluble cell wall glycoproteins to one 14-3-3 molecule could facilitate their cross-linking by peroxidase or transglutaminase or an autocatalytic reaction. Therefore, it is possible that 14-3-3 proteins have a general function in the formation of the extracellular matrix of unicellular and multicellular organisms.
However, this model does not explain why 14-3-3 proteins are cross-linked to the insoluble glycoprotein framework of the Chlamydomonas cell wall. If they act as chaperones, they should be liberated after cross-linking of the soluble glycoprotein precursors. Indeed, the 14-3-3 components, whose level in the culture medium is diminished in the case of the wall-deficient mutant compared with the wild type (Figure 8), have apparent molecular masses of 25 and 33 kD, which correspond to those of free 14-3-3 subunits, whereas the 14-3-3 polypeptides released from the insoluble wall fraction by treatment with HF/pyridine are considerably larger (45 and 64 kD; Figure 4A). Therefore, we assume that only a small portion of the 14-3-3 proteins that occur in the cell wall are linked covalently to the insoluble cell wall fraction by peroxidase-catalyzed isodityrosine formation and/or transglutaminase activity (Waffenschmidt et al., 1993, 1999), whereas the predominant portion remains soluble and is accumulated subsequently in the culture medium.
The portion of the 14-3-3 proteins cross-linked to the insoluble cell wall fraction obviously has the same fate as the unsolubilized Hyp-rich glycoproteins. The prominent 64- and 45-kD polypeptides that are released from the insoluble cell wall fraction during treatment with anhydrous HF/pyridine cross-react with antibodies against the recombinant Chlamydomonas 14-3-3 protein (anti-β-Gal/14-3-3; Figure 4A, lanes 1 and 9). On the other hand, antibodies raised against these two polypeptides (anti-dICW-64kDa and anti-dICW-45kDa) cross-react with 14-3-3 proteins (Figures 5 and 6) but preferentially recognize high-molecular-mass glycoproteins that are the precursors of the insoluble glycoprotein framework of the Chlamydomonas cell wall (Voigt et al., 1996) and their deglycosylation products (Figures 1, 2, and 7). Chemical deglycosylation of these chaotrope-soluble precursors of the insoluble cell wall fraction resulted in a mixture of polypeptides with apparent molecular masses of 200, 135, 100, and 72 kD (Figure 2, lane 2), which are larger than the prominent 64- and 45-kD polypeptides released from the insoluble cell wall fraction during chemical deglycosylation with anhydrous HF/pyridine. Therefore, after cross-linking of these precursors to the insoluble glycoprotein framework of the wall, the size of their polypeptide backbones must be reduced by proteolytic processes to reveal the 64- and 45-kD fragments.
Partial proteolysis of the insoluble cell wall fraction is a prerequisite for cell enlargement (Voigt, 1985, 1986; Voigt et al., 1996) and is accompanied by a release of macromolecules into the culture medium (Voigt, 1985, 1986). The size of the polypeptide fragments retained in the insoluble cell wall layer apparently is not determined only by the distribution of putative cleavage sites but also by the carbohydrate chains and the associated chaotrope-soluble glycoproteins. As shown by Edman degradation, the 65- and 45-kD constituents of the insoluble cell wall fraction are not homogeneous but consist of polypeptides with rather different N-terminal amino acid sequences (Table 2). Therefore, the fragmentation pattern is largely dependent on the overall structure of the cell wall.
When peptide mixtures generated by tryptic digestion of the purified insoluble wall fraction were analyzed by mass spectrometry, several fragments were found whose amino acid sequences were similar, but not identical, to the amino acid sequence of the previously described Chlamydomonas 14-3-3 protein (Liebich and Voigt, 1995). These findings demonstrate that this particular 30-kD 14-3-3 isoform is not a constituent of the insoluble cell wall fraction but that other 14-3-3 isoforms are cross-linked to this wall component, presumably those that are accumulated in the culture medium (Figure 8).
METHODS
Strains and Growth Conditions
The Chlamydomonas reinhardtii wild-type strain 137C (mating type +) was obtained from R.P. Levine (Harvard University, Cambridge, MA). The cell wall–deficient mutant cw-15 derived from the wild-type strain 137C (mating type +) by Davies and Plaskitt (1971) was supplied by the Sammlung von Algenkulturen (University of Göttingen, Germany). Cells were grown at 21°C and 20,000 lux in a high-salt medium supplemented with 0.2% (w/v) sodium acetate, as described previously (Voigt and Münzner, 1987). Cell densities were determined by duplicate hemocytometer counting.
Subcellular Fractions
Cells were harvested by centrifugation at 6000g for 10 min at 4°C. The cells were washed and resuspended to a final cell density of 0.5 to 1 × 109 cells/mL in ice-cold homogenization buffer consisting of 25 mM Tris-HCl, pH 7.0, 2 mM DTT, and 0.1 mM EDTA. After addition of the protease inhibitors phenylmethylsulfonyl fluoride (final concentration of 0.1 mM) and chymostatin (final concentration of 5 μg/mL), the cells were disrupted using a tight-fitting Potter-Elvehjem homogenizer. The homogenate was centrifuged at 20,000g for 20 min to remove cellular debris, chloroplasts, and mitochondria. The 20,000g supernatant then was centrifuged for 4 h at 100,000g in the Beckman ultracentrifuge rotor Ti60 to obtain the 100,000g pellet (i.e., the microsomal fraction) and the supernatant (i.e., the cytosol).
Sucrose Density Gradient Centrifugation of Microsomal Membranes
Microsomal membranes were fractionated by sucrose density gradient centrifugation, and the different membrane fractions were identified by measuring the activities of marker enzymes, as described previously (Voigt et al., 2001).
Isolation of 14-3-3 Proteins
The 14-3-3 proteins were isolated from the cytosol of Chlamydomonas as described by Voigt et al. (2001).
Extraction of Salt-Soluble Cell Wall Components
Salt-soluble cell wall components were extracted with aqueous LiCl (3 M) from intact cells, as described previously (Voigt, 1986). Cells were harvested by centrifugation at 6000g for 10 min and resuspended in fresh culture medium to a final cell density of 2 × 108 cells/mL. After addition of the same volume of aqueous LiCl (6 M), the suspensions were incubated at 0°C for 3 h and subsequently centrifuged at 20,000g for 30 min. The supernatants were stored at −20°C until use.
Isolation of the Insoluble Cell Wall Component
The insoluble cell wall component of Chlamydomonas was isolated by successive extractions of intact cells with different detergent-containing buffers, treated with α-amylase, reextracted with detergent-containing buffers, and finally washed with distilled water, as described previously (Voigt, 1984; Vogeler et al., 1990).
Chemical Deglycosylation
Anhydrous Hydrofluoric Acid (HF) /Pyridine
The purified insoluble wall component was freeze-dried and subjected to chemical deglycosylation by treatment with anhydrous HF/pyridine (Vogeler et al., 1990). After 3 h at 0°C, the reaction was quenched by the addition of an equal volume of ice-cold water and 8 volumes of cold acetone. After 1 h at 0°C, the precipitates were collected by centrifugation at 20,000g for 30 min, washed twice with aqueous acetone (80% [v/v]), dried, and dissolved overnight in a small volume of urea-SDS buffer (Voigt, 1986). Individual cell wall polypeptides were isolated by preparative SDS-PAGE according to Laemmli (1970) on gels containing 10% (w/v) acrylamide.
Trimethylsilyl Trifluoromethanesulfonate
The reagent was prepared by mixing 2 g of trimethylsilyl trifluoromethanesulfonate with 6.5 mL of anhydrous trifluoroacetic acid and 0.5 mL of thioanisole, cooled to 0°C, and added to the freeze-dried insoluble wall fraction (1 mL of reagent/mg of cell wall dry mass). After 3 h at 0 or 30°C, the reaction was quenched by the addition of 1 mL of distilled water and 8 mL of ice-cold acetone (100%). The precipitate was collected by centrifugation at 20,000g for 30 min at 4°C. The supernatant was discarded, and the precipitate was washed twice with 80% (v/v) aqueous acetone and subsequently vacuum-evaporated until completely dry.
Determination of Protein
Quantitation of proteins was performed by the method of Bradford (1976) using BSA as a standard.
Preparation of Antibodies
Polyclonal antibodies were raised in rabbits against the 64- and 45-kD polypeptides released from the insoluble wall fraction by chemical deglycosylation with HF/pyridine and isolated by preparative SDS-PAGE according to Laemmli (1970). After staining with Coomassie Brilliant Blue G 250 and destaining, the corresponding bands were cut from the gels. The gel pieces were destained completely with isopropanol:acetic acid:water (25:10:65), treated extensively with distilled water, homogenized, suspended in 2 volumes of 20 mM sodium phosphate, pH 7.5, containing 0.15 M NaCl and 0.1% (w/v) SDS, and incubated overnight at 37°C. These suspensions were used as antigens.
Preparation of the antibodies against the unfractionated deglycosylation products of the insoluble cell wall fraction (anti-dICW Ig), against the 100-kD deglycosylation product of the chaotrope-soluble 150-kD cell wall glycoprotein (anti-dGP100 Ig), and against the β-galactosidase:14-3-3 fusion protein (anti-β-Gal/14-3-3 Ig) has been described previously (Voigt et al., 1996, 2001). All antibodies used in this study and their corresponding antigens are listed in Table 1.
SDS-PAGE and Protein Gel Blot Analyses
Macromolecules present in the LiCl extracts and in the culture medium were precipitated by the addition of trichloroacetic acid (final concentration of 10% [w/v]) in the cold, and the precipitates were collected by centrifugation. The pellets were washed twice with distilled water and redissolved in a small volume of urea-SDS buffer (Voigt, 1986). After the addition of sample buffer (Laemmli, 1970) containing bromphenol blue as a tracking dye, the polypeptides were separated by SDS-PAGE according to Laemmli (1970) on gel slabs (83 mm × 65 mm × 0.15 mm) containing 10 to 12.5% (w/v) acrylamide. After electrophoresis, the gels were either stained with Coomassie Brilliant Blue G 250 or blotted electrophoretically onto polyvinylidene difluoride membranes (Porablot; Macherey-Nagel, Düren, Germany) as described by Towbin et al. (1979). After pretreatment of the blots with 3% (w/v) BSA in PBS for 3 h at 37°C or overnight at 7°C, the protein gel blots were probed with polyclonal antibodies raised in rabbits against the different cell wall polypeptides and against the recombinant Chlamydomonas 14-3-3 protein depleted from anti–Escherichia coli β-Gal Igs diluted 1:300 with PBS containing 1% (w/v) BSA. After 1 h at 37°C, the blots were washed four times with PBS for 30 min at room temperature followed by treatment for 1 h at 37°C with alkaline phosphatase–coupled goat anti-rabbit IgG antibodies (Amersham-Buchler, Braunschweig, Germany) diluted 1:800 with PBS containing 1% (w/v) BSA. After incubation, the blots were washed at least four times with PBS for 30 min at room temperature. Finally, the indirectly bound alkaline phosphatase was detected by its enzymatic activity using 5-bromo-4-chloro-3-indolyl phosphate as a substrate in the presence of nitroblue tetrazolium chloride (Hawkes et al., 1982).
Epitope Analysis
A total of 83 overlapping pentadecapeptides representing the entire amino acid sequence of the Chlamydomonas 14-3-3 protein derived from the open reading frame of the previously cloned Chlamydomonas 14-3-3 cDNA (Liebich and Voigt, 1995) were generated by spot synthesis using a cellulose paper sheet as a solid support (Frank, 1992). These overlapping pentadecapeptides were used to determine the epitope specificities of the different antibodies, as described previously (Frank, 1992; Voigt et al., 2001). After incubation with the antibody and extensive washing, bound rabbit IgGs were measured by incubation with alkaline phosphatase–coupled goat anti-rabbit IgG (Amersham-Buchler) and subsequent detection of the indirectly bound alkaline phosphatase via its enzyme activity using 5-bromo-4-chloro-3-indolyl phosphate as a substrate in the presence of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. After documentation of the results, the membrane was stripped as described by Frank (1992).
Immunofluorescence
Aliquots of the purified insoluble cell wall material were incubated with 3% (w/v) BSA in PBS overnight at 4°C and then treated for 16 to 20 h at 4°C with either preimmune serum or antibodies diluted 1:200 with 1% (w/v) BSA in PBS. After washing with PBS (four times), the samples were incubated for 4 h at room temperature with fluorescein isothiocyanate–conjugated goat anti-rabbit IgG (Sigma, Deisenhofen, Germany) diluted 1:200 with 1% (w/v) BSA in PBS. Finally, the samples were washed four times with PBS and examined using Olympus T2 epifluorescence optics with a filter set for fluorescein isothiocyanate (BH-2; Olympus, Tokyo, Japan).
Determination of N-Terminal Amino Acid Sequences
Cell wall polypeptides were deglycosylated chemically by treatment with trimethylsilyl trifluoromethanesulfonate at room temperature for 3 h. The deglycosylated polypeptides then were purified by SDS-PAGE according to Laemmli (1970) and blotted electrophoretically onto polyvinylidene difluoride membranes (Porablot; Macherey-Nagel) as described by Towbin et al. (1979). After staining with Coomassie Brilliant Blue G 250, the protein bands were cut from the membrane and destained completely with methanol. N-terminal amino acid sequences were determined by automated Edman degradation using a gas-phase sequencer (model 470A; Applied Biosystems, Weiterstadt, Germany).
Mass Spectrometry
Proteolysis with trypsin of the purified insoluble cell wall fraction was performed essentially as described for in-gel digestion of protein bands cut from SDS-PAGE gels (Shevchenko et al., 1996). The cell wall material was digested for 3 h with porcine trypsin (sequencing grade, modified; Promega, Madison, WI) at a concentration of 67 ng/μL in 25 mM ammonium bicarbonate, pH 8.1, at 37°C. Before peptide mass mapping and sequencing of tryptic fragments by tandem mass spectrometry, peptide mixtures were extracted from the insoluble cell wall material using 1% formic acid followed by two changes of 50% acetonitrile. The combined extracts either were subjected to HPLC separation on a SMART system (Pharmacia, Freiburg, Germany), and individual fractions were analyzed by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (G2025A; Hewlett-Packard, Waldbronn, Germany) and microsequencing (Procise 494A; Applied Biosystems), or they were vacuum-dried until only 1 to 2 μL was left and desalted using ZipTips according to the manufacturer's instructions (Millipore, Bedford, MA).
Matrix-assisted laser-desorption ionization time-of-flight analysis from the matrix α-cyano-4-hydroxycinnamic acid/nitrocellulose prepared on the target using the fast evaporation method (Arnott et al., 1998) was performed on a Bruker Reflex III mass spectrometer (Bruker Daltonik, Bremen, Germany) equipped with a N2 337-nm laser and gridless pulsed-ion extraction. Sequence verifications of some fragments were performed by nanoelectrospray tandem mass spectrometry on a Q-Tof I mass spectrometer (Micromass, Manchester, UK) equipped with a nanoflow electrospray ionization source. Gold-coated glass capillary nanoflow needles were obtained from Protana (type Medium NanoES spray capillaries; Odense, Denmark). Database searches (NCBInr, nonredundant protein database) were performed using MASCOT software from Matrix Science (London, UK) (Perkins et al., 1999).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The SWISSPROT database accession number for Chlamydomonas 14-3-3 cDNA is P52908.
Acknowledgments
The authors thank M. Kiess (Gesellschaft für Biotechnologische Forschung) for the determination of N-terminal amino acid sequences and S. Stevanovic and M. Schirle (Institute for Cell Biology, University of Tübingen) for their generous support during the analysis of the peptide fragments by mass spectrometry. This work was supported by Grants VO 327/3 and VO 327/7 from the Deutsche Forschungsgemeinschaft.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010611.
References
- Aducci, P., Camoni, L., Marra, M., and Visconti, S. (2002). From cytosol to organelles: 14-3-3 proteins as multifunctional regulators of plant cell. IUBMB Life 53, 49–55. [DOI] [PubMed] [Google Scholar]
- Aitken, A. (1996). 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 6, 342–347. [DOI] [PubMed] [Google Scholar]
- Aitken, A. (2002). Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation: Chromosome location of mammalian isoforms and variants. Plant Mol. Biol. 50, 993–1010. [DOI] [PubMed] [Google Scholar]
- Aitken, A., Collinge, D.B., van Heusden, B.P.H., Isobe, T., Roseboom, P.H., Rosenfeld, G., and Soll, J. (1992). 14-3-3 proteins: A highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 17, 498–501. [DOI] [PubMed] [Google Scholar]
- Arnott, D., O'Connell, K.L., King, K.L., and Stults, J.T. (1998). An integrated approach to proteome analysis: Identification of proteins associated with cardiac hypertrophy. Anal. Biochem. 258, 1–18. [DOI] [PubMed] [Google Scholar]
- Baunsgaard, L., Fuglsang, A.T., Jahn, T., Korthout, H.A., de Boer, A.H., and Palmgren, M.G. (1998). The 14-3-3 proteins associate with the plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 13, 661–671. [DOI] [PubMed] [Google Scholar]
- Biggs, K.J., and Fry, S.C. (1990). Solubilization of covalently bound extensin from Capsicum cell walls. Plant Physiol. 92, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihn, E.A., Paul, A.L., Wang, S.W., Erdos, G.W., and Ferl, R.J. (1997). Localization 14-3-3 proteins in the nuclei of Arabidopsis and maize. Plant J. 12, 1439–1445. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bunney, T.D., van Walraven, H.S., and de Boer, A.H. (2001). 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc. Natl. Acad. Sci. USA 98, 4249–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot, S., Lingiah, G., and Martin, T. (2003). Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J. Exp. Bot. 54, 595–604. [DOI] [PubMed] [Google Scholar]
- Davies, D.R., and Plaskitt, A. (1971). Genetical and structural analyses of cell wall formation in Chlamydomonas reinhardtii. Genet. Res. 17, 33–43. [Google Scholar]
- De Vetten, N.C., Lu, G., and Ferl, R.J. (1992). A maize protein associated with the G-box binding complex has homology to brain regulatory proteins. Plant Cell 4, 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender, F., Godl, K., Wenzl, S., and Sumper, M. (2002). Evidence for autocatalytic cross-linking of hydroxyproline-rich glycoproteins during extracellular matrix assembly in Volvox. Plant Cell 14, 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, L., and Lamport, D.T.A. (1984). An intramolecular linkage involving isodityrosine in extensin. Phytochemistry 23, 1241–1246. [Google Scholar]
- Ferl, R.J., Manak, M.S., and Reyes, M.F. (2002). The 14-3-3s. Genome Biol. 3, Reviews 3010. [DOI] [PMC free article] [PubMed]
- Ford, J.C., Al-Khodairy, F., Fotou, E., Sheldrick, K.S., Griffith, D.J.F., and Carr, A.M. (1994). 14-3-3 protein homologues required for the DNA damage checkpoint in fission yeast. Science 265, 533–535. [DOI] [PubMed] [Google Scholar]
- Frank, R. (1992). Spot synthesis: An easy technique for the positionally addressable, parallel chemical synthesis on membrane support. Tetrahedron 48, 9217–9232. [Google Scholar]
- Fry, S.C. (1982). Isodityrosine, a new cross linking amino acid from plant cell wall glycoproteins. Biochem. J. 204, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullone, M.R., Visconti, S., Marra, M., Fogliani, V., and Aducci, P. (1998). Fusicoccin effect on the in vitro interaction between plant 14-3-3 proteins and plasma membrane H+-ATPase. J. Biol. Chem. 273, 7698–7702. [DOI] [PubMed] [Google Scholar]
- Gelperin, D., Weigle, J., Nelson, K., Roseboom, P., Irie, K., Matsumoto, K., and Lemmon, S. (1995). 14-3-3 proteins: Potential roles in vesicular transport and Ras signalling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92, 11539–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1985). The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze, deep-etch technique. J. Cell Biol. 101, 1550–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes, R., Nidag, E., and Gordon, J. (1982). A dot-immunobinding assay for monoclonal and other antibodies. Anal. Biochem. 119, 142–147. [DOI] [PubMed] [Google Scholar]
- Holst, O., Christoffel, V., Fründ, R., Moll, H., and Sumper, M. (1989). A phosphodiester bridge between two arabinose residues as a structural element of an extracellular matrix glycoprotein of Volvox carteri. Eur. J. Biochem. 181, 345–350. [DOI] [PubMed] [Google Scholar]
- Huber, S.C., MacKintosh, C., and Kaiser, W.M. (2002). Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol. Biol. 50, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Ichimura, T., Ito, M., Itagaki, V., Takahashi, M., Horigome, T., Omata, S., Ohno, S., and Isobe, T. (1997). The 14-3-3 protein binds its target proteins with a common site located towards the C-terminus. FEBS Lett. 413, 273–276. [DOI] [PubMed] [Google Scholar]
- Imam, S.H., Buchanan, M.J., Shin, H.-C., and Snell, W.J. (1985). The Chlamydomonas cell wall: Characterization of the wall framework. J. Cell Biol. 101, 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan, D., Akita, M., and Keegstra, K. (2001). Molecular chaperones involved in chloroplast protein import. Biochim. Biophys. Acta 1541, 102–113. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., and Soll, J. (2002). Toc, Tic, and chloroplast import. Biochim. Biophys. Acta 1590, 177–189. [DOI] [PubMed] [Google Scholar]
- Jeanclos, E.M., Lin, L., Treil, M.W., Rao, J., DeCoster, M.A., and Anand, R. (2001). The chaperone protein 14-3-3η interacts with the nicotinic acetylcholine receptor α4 subunit: Evidence for a dynamic subunit stabilization. J. Biol. Chem. 276, 28281–28290. [DOI] [PubMed] [Google Scholar]
- Jones, D.H.A., Martin, H., Madrazo, J.K.A., Robinson, K.A., Nielson, P., Roseboom, P.H., Patel, Y., Howell, S.A., and Aitken, A. (1995). Expression and structural analysis of 14-3-3 proteins. J. Mol. Biol. 245, 375–384. [DOI] [PubMed] [Google Scholar]
- Kieliszewski, M.J., and Lamport, D.T.A. (1994). Extensin: Repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5, 157–172. [DOI] [PubMed] [Google Scholar]
- Knox, J.P. (1995). The extracellular matrix in higher plants. 4. Developmentally regulated proteoglycans and glycoproteins of the plant cell surface. FASEB J. 9, 1004–1012. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lamport, D.T.A. (2001). Life behind cell walls: Paradigm lost, paradigm regained. Cell. Mol. Life Sci. 58, 1363–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebich, I., and Voigt, J. (1995). A Chlamydomonas homologue to the 14-3-3 proteins: cDNA and deduced amino acid sequence. Biochim. Biophys. Acta 1263, 79–85. [DOI] [PubMed] [Google Scholar]
- Loppes, R., and Deltour, R. (1975). Changes in phosphatase activity associated with cell wall defects in Chlamydomonas reinhardtii. Arch. Microbiol. 103, 247–250. [DOI] [PubMed] [Google Scholar]
- Lu, G., DeLisle, A.J., De Vetten, N.C., and Ferl, R.J. (1992). Brain proteins in plants: An Arabidopsis 14-3-3 protein homologue to neurotransmitter pathway activators is part of a DNA-binding complex. Proc. Natl. Acad. Sci. USA 89, 11490–11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara, S., and Hase, E. (1975). Studies on the vegetative life cycle of Chlamydomonas reinhardtii Dangeard in synchronous culture. III. Some notes on the process of zoospore liberation. Plant Cell Physiol. 16, 371–375. [Google Scholar]
- Monk, B.C., Adair, W.S., Cohen, R.A., and Goodenough, U.W. (1983). Topography of Chlamydomonas: Fine structure and polypeptide components of the gametic flagellar membrane surface and the cell wall. Planta 158, 517–533. [DOI] [PubMed] [Google Scholar]
- Moore, B.W., and Perez, V.J. (1967). Specific acidic proteins of the nervous system. In Physiological and Biochemical Aspects of Nervous Integration, F.D. Carlson, ed (Woods Hole, MA: Prentice Hall), pp. 343–359.
- Muslin, A.J., Tanner, J.W., Allen, P.M., and Shaw, A.S. (1996). Interaction of 14-3-3 with signalling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Oecking, C., Eckerskorn, C., and Weiler, E.W. (1994). The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic proteins. FEBS Lett. 352, 163–166. [DOI] [PubMed] [Google Scholar]
- Perkins, D.N., Creasy, D.M., and Cottrell, J.S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Petosa, L., Masters, S.C., Bankstone, L.A., Pohl, J., Wang, B., Fu, H., and Liddington, R.C. (1998). 14-3-3 ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 273, 16305–16310. [DOI] [PubMed] [Google Scholar]
- Rausch, H., Larsen, N., and Schmitt, R. (1989). Phylogenetic relationship of the green alga Volvox carteri deduced from small-subunit ribosomal RNA comparison. J. Mol. Evol. 29, 255–265. [DOI] [PubMed] [Google Scholar]
- Roberts, K. (1974). Crystalline glycoprotein cell walls of algae: Their structure, composition and assembly. Philos. Trans. R. Soc. Lond. B 268, 129–146. [DOI] [PubMed] [Google Scholar]
- Roberts, K., Grief, C., Hills, G.J., and Shaw, P.J. (1985). Cell wall glycoproteins: Structure and function. J. Cell Sci. 2 (suppl.), 105.–127. [DOI] [PubMed] [Google Scholar]
- Roberts, M.R., Salinas, J., and Collinge, D.B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50, 1031–1039. [DOI] [PubMed] [Google Scholar]
- Schlösser, U.G. (1966). Enzymatisch gesteuerte Freisetzung von Zoosporen bei Chlamydomonas reinhardtii Dangeard in Synchronkultur. Arch. Microbiol. 54, 129–159. [Google Scholar]
- Sehnke, P.C., DeLille, J.M., and Ferl, R.J. (2002). Consummating signal transduction: The role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14 (suppl.), S339.–S354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Soll, J. (2002). Protein import into the chloroplasts. Curr. Opin. Plant Biol. 5, 529–535. [DOI] [PubMed] [Google Scholar]
- Todd, A., Cossons, N., Aitken, A., Price, G.B., and Zannis-Hadjopoulos, M. (1998). Human cruciform binding protein belongs to the 14-3-3 family. Biochemistry 37, 14317–14325. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heusden, G.P.H., Griffith, D.J.F., Ford, J.C., Chin-A-Woeng, T.F.C., Schrader, P.A.T., Carr, A.M., and De Steensma, H.M. (1995). The 14-3-3 proteins encoded by the Bmh1 and Bmh2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 229, 45–53. [PubMed] [Google Scholar]
- Vogeler, H.-P., Voigt, J., and König, W.A. (1990). Polypeptide pattern of the insoluble wall component of Chlamydomonas reinhardtii and its variation during the vegetative cell cycle. Plant Sci. 71, 119–128. [Google Scholar]
- Voigt, J. (1984). A study of cross-links between the glycoprotein subunits within the insoluble inner cell wall layer of Chlamydomonas reinhardtii. Mitt. Staatsinst. Allg. Bot. Hambg. 19, 83–98. [Google Scholar]
- Voigt, J. (1985). Macromolecules released into the culture medium during the vegetative cell cycle of the unicellular green alga Chlamydomonas reinhardtii. Biochem. J. 226, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, J. (1986). Biosynthesis and turnover of cell wall glycoproteins during the vegetative cell cycle of Chlamydomonas reinhardtii. Z. Naturforsch. 41c, 885–896. [Google Scholar]
- Voigt, J. (1988). The LiCl-soluble cell wall layers of Chlamydomonas reinhardtii contain several immunologically related polypeptides. Planta 173, 373–384. [DOI] [PubMed] [Google Scholar]
- Voigt, J., Hinkelmann, B., and Harris, E.H. (1997). Production of cell wall polypeptides by different cell wall mutants of the unicellular green alga Chlamydomonas reinhardtii. Microbiol. Res. 152, 189–198. [DOI] [PubMed] [Google Scholar]
- Voigt, J., Liebich, I., Hinkelmann, B., and Kiess, M. (1996). Immunological identification of a putative precursor of the insoluble glycoprotein framework of the Chlamydomonas cell wall. Plant Cell Physiol. 37, 91–101. [DOI] [PubMed] [Google Scholar]
- Voigt, J., Liebich, I., Kiess, M., and Frank, R. (2001). Subcellular distribution of 14-3-3 proteins in the unicellular green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 268, 6449–6457. [DOI] [PubMed] [Google Scholar]
- Voigt, J., and Münzner, P. (1987). The Chlamydomonas cell cycle is regulated by a light/dark-responsive cell-cycle switch. Planta 172, 463–472. [DOI] [PubMed] [Google Scholar]
- Voigt, J., Münzner, P., and Vogeler, H.-P. (1991). The cell wall glycoproteins of Chlamydomonas reinhardtii: Analysis of the in vitro translation products. Plant Sci. 75, 129–142. [Google Scholar]
- Waffenschmidt, S., Kusch, T., and Woessner, J.P. (1999). A transglutaminase immunologically related to tissue transglutaminase catalyzes cross-linking of cell wall proteins in Chlamydomonas reinhardtii. Plant Physiol. 121, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt, S., Spessert, R., and Jaenicke, L. (1988). Oligosaccharide side chains of wall molecules are essential for cell wall lysis in Chlamydomonas reinhardtii. Planta 175, 513–519. [DOI] [PubMed] [Google Scholar]
- Waffenschmidt, S., Woessner, J.P., Beer, K., and Goodenough, U.W. (1993). Isodityrosine cross-linking mediates insolubilization of cell walls in Chlamydomonas. Plant Cell 5, 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., and Shakes, D.C. (1996). Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 43, 384–398. [DOI] [PubMed] [Google Scholar]
- Woessner, J.P., and Goodenough, U.W. (1994). Volvocine cell walls and their constituent glycoproteins: An evolutionary perspective. Protoplasma 181, 245–258. [Google Scholar]
- Yaffe, M.B. (2002). How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53–57. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Wang, H., Liu, D., Liddington, R.C., and Fu, H. (1997). Raf-1 kinase and exoenzyme S interact with 14-3-3 ζ through a common site involving lysine 49. J. Biol. Chem. 272, 13717–13724. [DOI] [PubMed] [Google Scholar]