Abstract
The success or failure of interspecific crosses is vital to evolution and to agriculture, but much remains to be learned about the nature of hybridization barriers. Several mechanisms have been proposed to explain postzygotic barriers, including negative interactions between diverged sequences, global genome rearrangements, and widespread epigenetic reprogramming. Another explanation is imbalance of paternally and maternally imprinted genes in the endosperm. Interspecific crosses between diploid Arabidopsis thaliana as the seed parent and tetraploid Arabidopsis arenosa as the pollen parent produced seeds that aborted with the same paternal excess endosperm phenotype seen in crosses between diploid and hexaploid A. thaliana. Doubling maternal ploidy restored seed viability and normal endosperm morphology. However, substituting a hypomethylated tetraploid A. thaliana seed parent reestablished the hybridization barrier by causing seed abortion and a lethal paternal excess phenotype. We conclude from these findings that the dominant cause of seed abortion in the diploid A. thaliana × tetraploid A. arenosa cross is parental genomic imbalance. Our results also demonstrate that manipulation of DNA methylation can be sufficient to erect hybridization barriers, offering a potential mechanism for speciation and a means of controlling gene flow between species.
INTRODUCTION
A variety of mechanisms have been described that minimize gene flow between species and contribute to their reproductive isolation. These may be prezygotic mechanisms, which reduce the frequency at which gametes combine to form a zygote, or postzygotic mechanisms, which reduce the viability or reproductive potential of the hybrid. In flowering plants, prezygotic barriers include different flowering times, attractiveness to different pollinators, failure of the pollen to adhere to the stigma surface, and abnormal growth of the pollen tube. Postzygotic barriers take effect after successful fertilization and include seed abortion and weakness or sterility of F1 hybrids and later generations (Stebbins, 1958; Coyne and Orr, 1998; Rieseberg and Carney, 1998; Tiffin et al., 2001).
Several mechanisms have been proposed to explain the operation of postzygotic barriers. One is the Dobzhansky-Muller model, in which genes from one species interact negatively with genes from another species, causing inviability or sterility in the hybrid offspring (reviewed by Coyne and Orr, 1998; Rieseberg and Carney, 1998). Another is “genome shock,” in which hybridization elicits restructuring of the genome, such as through changes to chromosomal organization or mobilization of repetitive sequences (McClintock, 1984). More recent studies indicate that genome shock could be associated with epigenetic effects such as the gene silencing triggered by homologous gene–gene interactions (Meyer and Saedler, 1996; Matzke and Matzke, 1998). In support of this hypothesis, it has been found that in allopolyploids the hybridization event is accompanied by changes in gene expression patterns, chromatin structure, and DNA methylation as well as sequence elimination (Henikoff and Matzke, 1997; Leitch and Bennett, 1997; Henikoff and Comai, 1998; Rieseberg and Noyes, 1998; Comai et al., 2000; Pikaard, 2001; Rieseberg, 2001; Madlung et al., 2002).
Although the success or failure of crosses between related plant species has been the subject of many studies (Thompson, 1930; Tiffin et al., 2001), the mechanisms that govern the outcome remain elusive. However, endosperm breakdown is cited frequently as the cause of seed failure in both interploidy crosses within a species and interspecific crosses (Watkins, 1932; Brink and Cooper, 1947; Stebbins, 1958; Haig and Westoby, 1991). The endosperm is the second fertilization product in flowering plant reproduction and develops after the fusion of the two polar nuclei (sister nuclei to the egg) with a sperm nucleus. Its role in seed development has long been debated, but it is generally considered to be involved in nutrition of the embryo (Lopes and Larkins, 1993). In Arabidopsis thaliana, as in most other dicots, the endosperm does not persist in the mature seed, and there is evidence that it plays a role in the transient storage of nutrients before delivery to the embryo (Hirner et al., 1998).
Whatever the function of the endosperm, its successful development appears crucial for the production of viable seeds; therefore, its failure in some interspecific crosses could represent a postzygotic hybridization barrier that operates early in the life cycle. Reproductive isolation caused by endosperm abortion has been attributed to disruption of the balance between maternal (m) and paternal (p) genomes (Johnston et al., 1980; Johnston and Hanneman, 1982). Normal development of endosperm usually requires a parental genomic ratio of 2m:1p, and deviations from this ratio, achieved by crossing plants of the same or related species but different ploidy levels, often are associated with seed abortion (Lin, 1984; Haig and Westoby, 1991). Maternal excess (a ratio > 2m:1p) generally is correlated with inhibited proliferation of the endosperm, and paternal excess generally is correlated with overgrowth (Haig and Westoby, 1991; Scott et al., 1998).
It is widely believed that the mechanism underlying the requirement for a specific parental genomic ratio in the endosperm is parental imprinting. This term refers to differential gene expression in offspring depending on the parent of origin, mediated via epigenetic modifications of a gamete's genome. Parental imprinting has been reported mainly in placental mammals (Moore and Haig, 1991; Bartolomei and Tilghman, 1997) and flowering plants (Kermicle and Alleman, 1990; Haig and Westoby, 1991; Scott et al., 1998). The major model for its evolution is the parental conflict theory, which presents imprinting as a struggle between maternally and paternally derived genomes over resource allocation from mother to offspring (Haig and Westoby, 1989, 1991). According to this model, there is selection pressure for growth promoters active during resource transfer to offspring (i.e., during endosperm development in flowering plants) to be expressed when they are inherited from the father but silenced when they are inherited from the mother, whereas growth inhibitors are selected for maternal expression and paternal silencing. Therefore, extra doses of maternal genomes provide extra active copies of growth inhibitors, resulting in small endosperm and seeds, whereas extra doses of paternal genomes provide extra active copies of growth promoters, resulting in an overgrowth phenotype. DNA methylation is an essential component of the imprinting mechanism in mammals (Tilghman, 1999) and also is involved in imprinting in maize (Lund et al., 1995; Finnegan et al., 1998). Adams et al. (2000) showed that reciprocal crosses in A. thaliana, in which one parent was hypomethylated by a METHYLTRANSFERASE1 (MET1) antisense transgene, phenocopy the effects of interploidy crosses. They concluded that DNA methylation plays an important role in parental imprinting in A. thaliana.
A. thaliana has been used to investigate the consequences of interspecific hybridization (Chen et al., 1998; Comai et al., 2000; Nasrallah et al., 2000; Madlung et al., 2002). Comai et al. (2000) performed a cross between diploid A. thaliana (2n = 2x = 10) as the seed parent and A. arenosa (also known as Cardaminopsis arenosa), which is widely regarded as a tetraploid (2n = 4x = 32), as the pollen parent. This cross was found to be unsuccessful as a result of a postzygotic hybridization barrier that caused the cessation of seed development when the embryo reached the globular stage. By contrast, these researchers found that substituting diploid with tetraploid A. thaliana in the A. thaliana × A. arenosa ([At × Aa]) cross resulted in a small proportion of viable seeds that germinated to produce a fertile hybrid, synthetic Arabidopsis suecica (SAS) (Chen et al., 1998), which resembled the natural allotetraploid A. suecica (2n = 4x = 26). Phenotypic instability and epigenetic gene silencing were significant features of SAS, with ∼0.4% of the genes in the allopolyploid subject to silencing (Comai et al., 2000), and changes to methylation patterns also were observed (Madlung et al., 2002).
In the work cited above, seed development was not described in more detail than noting whether seeds were viable or inviable and the stage at which embryos arrested. Therefore, it was not possible to determine whether seed abortion in the hybrid is caused by an imprinting-based mechanism or perhaps by a particular sensitivity of the endosperm to genome shock or allelic incongruity. Here, we describe the analysis of endosperm development after interspecific crosses between diploid and tetraploid A. thaliana as the seed parent and A. arenosa as the pollen parent. We found that the [2xAt × 4xAa] cross (with 2x A. thaliana as the seed parent) generated severely paternalized (overgrown) endosperm that resembled those produced in [2x × 6x] crosses in A. thaliana (Scott et al., 1998). By contrast, a [4xAt × 4xAa] cross (with 4x A. thaliana as the seed parent) restored seed viability and normal endosperm morphology. We further found that if a hypomethylated tetraploid A. thaliana seed parent was crossed to A. arenosa, extensive endosperm overgrowth and seed abortion were observed, suggesting that the rescuing effect of increased ploidy on the ability of A. thaliana to hybridize with A. arenosa was mediated by parental imprinting. This evidence strongly suggests that the dominant cause of endosperm failure in the [2xAt × 4xAa] cross is genomic imbalance rather than the sensitivity of the endosperm to allelic incongruity or epigenetic gene silencing. Significantly, given concerns regarding the deployment of genetically modified crop plants, the ability to erect postzygotic hybridization barriers by manipulating DNA methylation offers a potential means to control horizontal gene flow.
RESULTS
Parental Ploidy Affects Seed Size and Viability in A. thaliana × A. arenosa Interspecific Crosses
To investigate the basis of the postzygotic hybridization barrier between A. thaliana and A. arenosa, we conducted a series of crosses between diploid (2x) or tetraploid (4x) A. thaliana (C24 ecotype) and 4x A. arenosa, with A. thaliana as the seed parent. Comai et al. (2000) performed similar crosses using the Landsberg erecta (Ler) ecotype of A. thaliana and reported that the [2xAt × 4xAa] cross produced seeds that invariably aborted at the globular stage of embryo development. By contrast, they found that the [4xAt × 4xAa] cross yielded a mixture of seeds, most of which aborted with embryos at more advanced stages of development than in the [2xAt × 4xAa] cross and a minority (5%) of which reached maturity and could be germinated to yield SAS. The cause of the difference in viability between the [2xAt × 4xAa] and [4xAt × 4xAa] crosses was not addressed directly, although the authors suggested that the crosses “can succeed when the parental genomic ratios are balanced.”
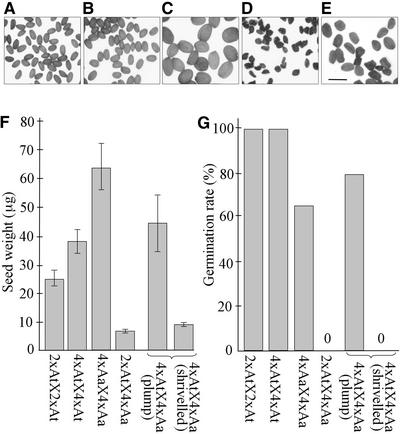
In our control crosses, we found that seeds produced by self-pollination of 2x A. thaliana, 4x A. thaliana, or 4x A. arenosa were plump and weighed a mean of 25.1, 38.3, and 65.4 μg, respectively (Figures 1A to 1C and 1F). The interspecific cross [2xAt × 4xAa] produced small, shriveled seeds (mean weight of 6.9 μg) (Figures 1D and 1F), which we later found to be aborted (see below). However, doubling the ploidy of the seed parent rescued approximately half of the progeny. Seeds from this cross, [4xAt × 4xAa], fell into two classes: 60% were relatively large and plump (mean weight of 44.1 μg) and 40% were smaller and shriveled (mean weight of 11.3 μg) (Figures 1E and 1F). Seeds from self-pollinated 2x and 4x A. thaliana germinated at 100% frequency, and seeds from selfed A. arenosa germinated at 66% frequency (Figure 1G). No [2xAt × 4xAa] seeds germinated, despite repeated cycles of stratification. By contrast, 80% of the plump seeds from the [4xAt × 4xAa] cross germinated to produce vigorous, healthy, and fertile hybrid plants. The shriveled seeds from this cross completely failed to germinate (Figure 1G).
Figure 1.
Seed Weight and Viability after Intraspecific and Interspecific Crosses between A. thaliana and A. arenosa.
(A) to (E) Mature seeds: [2xAt × 2xAt] (A), [4xAt × 4xAt] (B), [4xAa × 4xAa] (C), [2xAt × 4xAa] (D), and [4xAt × 4xAa] (E). Bar = 1 mm.
(F) Mean weight of desiccated mature seeds from (A) to (E). Seeds from the [4xAt × 4xAa] cross (E) were separated into two classes, plump and shriveled, which were weighed separately. Each bar represents the grand mean of several means each obtained by weighing seeds in groups of 10 and dividing by 10 (n = number of groups weighed). From left to right: 25.1 ± 2.8 μg (n = 5), 38.3 ± 4.4 (n = 4), 65.4 ± 8.1 (n = 7), 6.9 ± 0.6 (n = 9), 44.1 ± 10.1 (n = 7), and 11.3 ± 1.2 (n = 5).
(G) Germination frequencies of seeds from (A) to (E). From left to right: 100% (n = 22), 100% (n = 22), 66% (n = 25), 0% (n = 109), 52% (n = 138), and 0% (n = 45).
The ecotype of the seed parent appears to affect the proportion of viable seeds produced in the [4xAt × 4xAa] cross. We repeated the cross reported by Comai et al. (2000) to produce 5% viable seeds ([4xLerAt × 4xAa]) and also found lower viability (13%, n = 101) compared with when C24 was used as the seed parent.
Embryo Development in Intraspecific and Interspecific A. thaliana × A. arenosa Crosses
To investigate the underlying causes of the dramatic difference in seed viability between [2xAt × 4xAa] and [4xAt × 4xAa] crosses in the C24 ecotype, we compared embryo development in these crosses with that in self-pollinated 2x and 4x A. thaliana and 4x A. arenosa (Figure 2). Embryogenesis proceeded at similar rates in self-pollinated 2x and 4x A. thaliana, and embryos filled the entire volume of the seed cavity by 8 days after pollination (DAP) (Figures 2A and 2B). Embryo development in self-pollinated A. arenosa proceeded more slowly: embryos had reached only the globular stage by 8 DAP (Figure 2C), the heart stage by 13 DAP (Figure 2D), and filled the seeds at 17 DAP (data not shown).
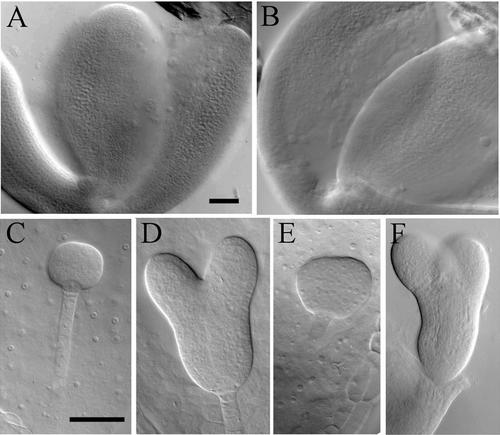
Figure 2.
Embryo Development in Seeds from Intraspecific and Interspecific Crosses between A. thaliana and A. arenosa.
(A) [2xAt × 2xAt] at 8 DAP.
(B) [4xAt × 4xAt] at 8 DAP.
(C) [4xAa × 4xAa] at 8 DAP.
(D) [4xAa × 4xAa] at 13 DAP.
(E) [2xAt × 4xAa] at 8 DAP.
(F) [4xAt × 4xAt] at 8 DAP.
Seeds were cleared and photographed using differential interference contrast optics. Bars = 50 μm for (A) and (B) and 50 μm for (C) to (F).
In [2xAt × 4xAa] seeds, embryos developed more rapidly than in self-pollinated A. arenosa, reaching the globular stage by 5 DAP (data not shown), but at 8 DAP, embryos still were at the globular–heart transition (Figure 2E), and seeds from 10 DAP onward usually were brown and collapsed. [4xAt × 4xAa] seeds fell into two categories: in ∼40% of the seeds, embryos arrested by the globular–heart transition as in [2xAt × 4xAa] (data not shown), but in the remainder of the seeds, embryos passed this stage, reaching the torpedo stage at 8 DAP (Figure 2F) and maximum size by 14 DAP (data not shown).
Endosperm Development in A. thaliana × A. arenosa Seeds: The Effect of Increasing Maternal Ploidy
Several studies have suggested that endosperm breakdown is the major cause of seed failure in interspecific crosses once successful fertilization has occurred, with embryo death as a secondary effect (Watkins, 1932; Brink and Cooper, 1947; Stebbins, 1958; Haig and Westoby, 1991). Therefore, we investigated endosperm development in seeds from [2xAt × 4xAa] and [4xAt × 4xAa] pollinations to determine whether it could be related to the contrasting outcomes of these crosses.
In developing seeds of self-pollinated A. thaliana, the endosperm proliferates as a syncytium until the embryo reaches the heart stage (∼5 DAP) and then begins to cellularize from the micropylar pole (Mansfield and Briarty, 1990). Several days before cellularization, three domains of endosperm are evident: central peripheral endosperm (PE), which is composed of regularly spaced nuclei with associated cytoplasm lining the central region of the embryo sac; micropylar PE, which consists of nuclei embedded in a common cytoplasm surrounding the suspensor; and chalazal endosperm (ChE), a dense multinucleate tissue at the chalazal pole. After cellularization of the PE, the embryo grows and gradually consumes the endosperm; only one or two layers of the endosperm (the aleurone) persist in the mature seed. Endosperm development is modified after interploidy crosses, with reciprocal crosses between parents of different ploidies having opposite effects (Scott et al., 1998). In seeds generated by the cross [2x × 6x], which produces an endosperm with the genomic ratio of 2m:3p, the endosperm (especially the ChE) overproliferates and usually fails to cellularize. [6x × 2x] crosses, which generate 6m:1p endosperm, display complementary phenotypes (underproliferation and early cellularization). Seeds invariably abort in crosses between 2x and 6x plants. [2x × 4x] and [4x × 2x] crosses, which generate 2m:2p and 4m:1p endosperm, respectively, show similar parent-of-origin effects on endosperm development, but these are less severe and do not cause seed lethality.
For the present study, developing seeds from control [2xAt × 2xAt] and [4xAt × 4xAt] crosses, and interspecific [2xAt × 4xAa] and [4xAt × 4xAa] crosses, were examined using confocal and light microscopy of whole-mount seeds, with particular emphasis on endosperm cellularization (Figure 3A) and proliferation (Figures 3B to 3J). (Self-pollinated A. arenosa was not included because the seeds were too large to image using these techniques.) In self-pollinated 2x and 4x A. thaliana, we observed cellularized endosperm in seeds at 5 DAP containing heart-stage embryos. In the interspecific [2xAt × 4xAa] cross, embryos did not pass the globular–heart transition and the endosperm was not observed to cellularize. In the [4xAt × 4xAa] cross, cellularization was delayed both in absolute time and with respect to embryogenesis, occurring at 8 DAP, when the embryo had reached the torpedo stage (Figure 3A).
Figure 3.
Embryo and Endosperm Development in Seeds from Intraspecific and Interspecific Crosses between A. thaliana and A. arenosa.
(A) Stages of embryogenesis from 2 to 8 DAP. Shaded boxes indicate that the endosperm is cellularizing. CC, curled cotyledon; D, dermatogen; G, globular; H, heart; LC, linear cotyledon; T, torpedo; Z, zygote.
(B) Endosperm proliferation in developing seeds. Central PE nuclei were counted in serial optical sections through whole-mount Feulgen-stained seeds using confocal microscopy. The mean number of nuclei (±se, n = 3 to 7) is plotted for each cross from 2 to 7 or 8 DAP.
(C) to (J) Endosperm development at the chalazal pole. (C) to (F) show confocal micrographs of Feulgen-stained seeds at 6 DAP. (G) to (J) show cleared whole-mount seeds at 8 DAP photographed using differential interference contrast optics. The ChE is marked with arrows in (G) and (H) and with dashed lines in (I) and (J). Cn, cotyledon. Bars = 50 μm.
(C) and (G) [2xAt × 2xAt].
(D) and (H) [4xAt × 4xAt].
(E) and (I) [2xAt × 4xAa].
(F) and (J) [4xAt × 4xAa].
Counts of the numbers of PE nuclei at different stages were made using serial optical sections (Figure 3B). In A. thaliana, the rate of endosperm proliferation reaches a maximum just before cellularization (Scott et al., 1998), although the cellular PE continues to divide as it fills the seed cavity (Brown et al., 1999). In the present study, the endosperm in seeds from 2x A. thaliana began to cellularize at 5 DAP (Figure 3A), when the PE contained 433 ± 1.7 (mean ± se) nuclei (Figure 3B). Beyond this stage, the number of nuclei increased only slightly, to a maximum of 485 ± 15.2 at 7 DAP. Seeds from 4x A. thaliana followed a similar pattern, with cellularization occurring at the same time (Figure 3A) and the rate of PE proliferation peaking slightly later (Figure 3B).
In the interspecific [2xAt × 4xAa] cross, the endosperm was not observed to cellularize (Figure 3A) and the PE continued to proliferate steadily after division in the A. thaliana endosperm had ceased (Figure 3B). At 8 DAP, the last time point at which nuclei were counted, PE from [2xAt × 4xAa] seeds had reached a mean of 615 ± 2.5 nuclei (Figure 3B). Endosperm proliferation in the [4xAt × 4xAa] cross was less extreme and more closely followed the pattern seen in A. thaliana seeds. The endosperm cellularized later (Figure 3A), and the peak rate of mitosis also was delayed (Figure 3B), but the rate decreased at the time of cellularization. The maximum number of PE nuclei counted in [4xAt × 4xAa] seeds was 484 ± 28.0 at 8 DAP, at which stage the PE began to cellularize.
Representative images of the ChE at 6 and 8 DAP are shown in Figures 3C to 3J, and area measurements are given in Table 1. In self-pollinated 2x and 4x A. thaliana, the size of the ChE decreased between 6 and 8 DAP as the endosperm cellularized and was consumed by the embryo (Figures 3C, 3D, 3G, and 3H, Table 1). At 6 DAP, the size of the ChE in the [2xAt × 4xAa] cross did not differ significantly from that in [2xAt × 2xAt] seeds (Figure 3E, Table 1). However, as the interspecific seeds continued to develop, the ChE increased dramatically in volume (Figure 3I, Table 1). In [4xAt × 4xAa] seeds, the ChE was slightly smaller than that in [2xAt × 4xAa] seeds at 6 DAP, but by 8 DAP, [4xAt × 4xAa] ChE showed a much smaller increase in size than was seen in the [2xAt × 4xAa] cross (Figures 3F and 3J, Table 1). Unlike the case for embryos, which either arrested at the globular–heart transition or continued to develop, endosperm in the [4xAt × 4xAa] cross did not fall into two obvious phenotypic classes.
Table 1.
Size of the ChE in Intraspecific and Interspecific Crosses between A. thaliana and A. arenosa
| Cross | DAP | ChE, Maximum Cross-Sectional Area (μm2, mean ± se) |
Comparison | P Value a |
|---|---|---|---|---|
| 2xAt × 2xAt | 6 | 1,251 ± 66 (n = 5) | ||
| 8 | 603 ± 122 (n = 4) | |||
| 4xAt × 4xAt | 6 | 2,012 ± 55 (n = 8) | ||
| 8 | 761 ± 109 (n = 4) | |||
| 2xAt × 4xAa | 6 | 1,563 ± 167 (n = 10) | 2xAt × 2xAt at 6 DAP | 0.059 |
| 8 | 13,470 ± 658 (n = 6) | 2xAt × 2xAt at 8 DAP | <0.000b | |
| 4xAt × 4xAa | 6 | 1,194 ± 138 (n = 9) | 4xAt × 4xAt at 6 DAP | <0.000b |
| 2xAt × 4xAa at 6 DAP | 0.011b | |||
| 8 | 3,583 ± 246 (n = 16) | 4xAt × 4xAt at 8 DAP | <0.000b | |
| 2xAt × 4xAa at 8 DAP | <0.000b | |||
| 4xMET1a/sAt × 4xMET1a/sAt |
6 | 2,312 ± 62 (n = 8) | ||
| 8 | 821 ± 89 (n = 4) | |||
| 4xMET1a/sAt × 4xAa |
6 | 4,089 ± 457 (n = 10) | 4xMET1a/sAt × | 0.002b |
| 4xMET1a/sAt at 6 DAP | ||||
| 8 | 28,100 ± 5,549 (n = 5) | 4xMET1a/sAt × | 0.008b | |
| 4xMET1a/sAt at 8 DAP |
Probability of the null hypothesis (no difference) in a two-tailed t test.
Significant at the 95% confidence level.
Hypomethylation of 4x A. thaliana Raises a Hybridization Barrier in the [4xAt × 4xAa] Cross
The data presented above suggest that the success or failure of crosses between A. thaliana and A. arenosa may be determined by the ratio of maternal to paternal genomes in the endosperm. Lethality in the [2xAt × 4xAa] cross appeared to be attributable to paternal excess causing endosperm overproliferation, which in the [4xAt × 4xAa] cross was ameliorated by doubling the maternal contribution. The ability to reduce endosperm proliferation by increasing the maternal genomic contribution suggests the operation of parental imprinting (Haig and Westoby, 1991; Scott et al., 1998). We demonstrated previously that parental imprinting in A. thaliana involves the activity of the DNA MET1 gene (Adams et al., 2000). A. thaliana plants transformed with an antisense MET1 construct (MET1 a/s) have hypomethylated genomes (Finnegan et al., 1996) and show alterations in the behavior of their gametes in crosses with wild-type plants. For example, a [2x × 4x] cross in the C24 background normally produces moderate endosperm overproliferation and viable seeds, but substitution of the seed parent with a C24 2x MET1 a/s plant causes massive endosperm overproliferation and seed lethality of the type usually associated with a [2x × 6x] cross (Adams et al., 2000). The phenotype is consistent with a model in which hypomethylation blocks parental imprinting during the production of the maternal gametes (polar nuclei), causing their “paternalization” (Adams et al., 2000; Spielman et al., 2001). By analogy with the experiment described above, if parental imprinting is a factor in the outcome of A. thaliana × A. arenosa crosses, substitution of a normally methylated seed parent in the cross [4xAt × 4xAa] with a hypomethylated 4x A. thaliana seed parent should result in a paternal excess phenotype.
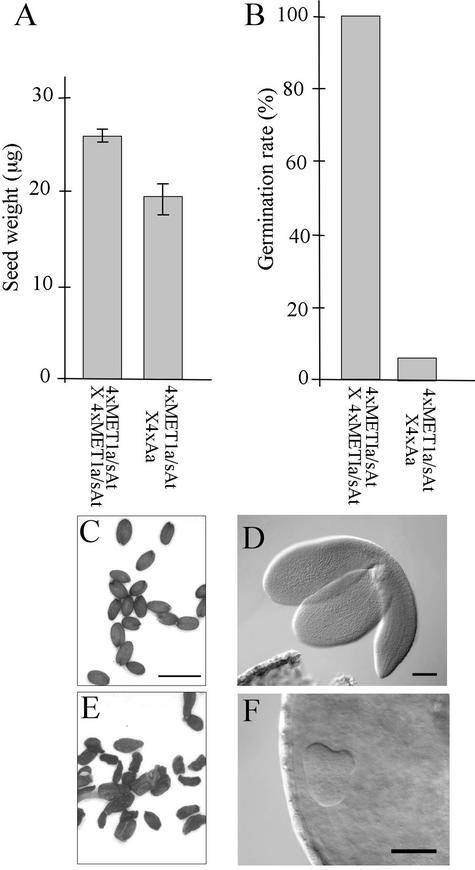
Accordingly, we replaced 4x A. thaliana seed parents with 4x MET1 a/s A. thaliana and analyzed the resulting seeds for viability and diagnostic endosperm phenotypes. The control [4xMET1a/sAt × 4xMET1a/sAt] seeds were plump, with a mean weight of 26.9 μg, and germinated at a frequency of 100% (Figures 4A to 4C). By contrast, seeds from the [4xMET1a/sAt × 4xAa] cross were invariably shriveled, with a mean weight of 19.6 μg, and had a germination frequency of only 7% (Figures 4A, 4B, and 4E). The small number of hybrids produced from this cross either died soon after germination or grew slowly into small plants that first flowered after 7 months (data not shown). Embryos in the [4xMET1a/sAt × 4xMET1a/sAt] cross grew to fill the seeds (Figure 4D), but the majority of [4xMET1a/sAt × 4xAa] embryos arrested at the globular–heart transition (Figure 4F), the same developmental stage at which embryos ceased to develop in [2xAt × 4xAa] seeds (Figure 2E).
Figure 4.
Embryo and Seed Development in Crosses Using Hypomethylated 4x A. thaliana.
(A) Mean weight of desiccated mature seeds, calculated as for Figure 1F. From left to right: 26.9 ± 0.8 μg (n = 4) and 19.6 ± 2.9 μg (n = 9).
(B) Germination rate. From left to right: 100% (n = 28) and 7% (n = 88).
(C) to (F) Comparisons of seed development. (C) and (E) show mature seeds. Bar = 1 mm. (D) and (F) show embryos at 8 DAP, either extruded (D) or in cleared seeds (F), photographed using differential interference contrast optics. Bars = 100 μm (D) and 50 μm (F).
(C) and (D) [4xMET1a/sAt × 4xMET1a/sAt].
(E) and (F) [4xMET1a/sAt × 4xAa].
Endosperm phenotypes in the [4xMET1a/sAt × 4xMET1-a/sAt] cross were similar to those seen in self-pollinated 4x A. thaliana, with the rate of PE mitosis peaking at 6 DAP (Figure 5A) and ChE decreasing in size between 6 and 8 DAP (Figures 5B and 5D, Table 1). In the [4xMET1a/sAt × 4xAa] cross, both the PE and the ChE overproliferated relative to [4xMET1a/sAt × 4xMET1a/sAt] seeds (Figures 5A, 5C, and 5E, Table 1). However, unlike the case in the [2xAt × 4xAa] cross, the endosperm cellularized in approximately half of the seeds by 8 DAP (data not shown).
Figure 5.
Endosperm Development in Crosses Using Hypomethylated 4x A. thaliana.
(A) Central PE nuclei, counted as for Figure 3B. The mean number of nuclei (±se, n = 3 to 4) is plotted for each cross from 2 to 7 or 8 DAP.
(B) to (E) Endosperm development at the chalazal pole. (B) and (C) show confocal micrographs of Feulgen-stained seeds at 6 DAP. (D) and (E) show cleared whole-mount seeds at 8 DAP photographed using differential interference contrast optics. The ChE is marked with an arrow in (D) and with a dashed line in (E). Cn, cotyledon; R, radicle. Bars = 50 μm.
(B) and (D) [4xMET1a/sAt × 4xMET1a/sAt].
(C) and (E) [4xMET1a/sAt × 4xAa].
Reciprocal Parent-of-Origin Phenotypes in Crosses with SAS
Although the data presented above are consistent with the hypothesis that parental imprinting is involved in the hybridization barrier between 2x A. thaliana and 4x A. arenosa, they are derived from crosses in only one direction, with A. thaliana as the seed parent. Our attempts to perform the reciprocal cross were unsuccessful. A. arenosa carries a self-incompatibility system that prevents most self-fertilization (Comai et al., 2000). This phenomenon often is associated with unilateral incompatibility, which also prevents successful pollination of self-incompatible plants by pollen from related species (deNettancourt, 1997). We found that the majority of A. thaliana pollen failed to germinate on A. arenosa stigmas, and the few pollen grains that did germinate produced short pollen tubes that did not penetrate the stigma surface (data not shown). We were unable to overcome this barrier by bud pollination or pretreatment of stigmas with salt solutions, both of which are known to compromise self-incompatibility systems (Hiscock and Dickinson, 1993). Consequently, we abandoned attempts to perform the reciprocal of the [2xAt × 4xAa] cross directly and sought an alternative, albeit indirect, method to test complementary parent-of-origin effects in crosses involving the two Arabidopsis species.
The approach we developed used SAS (2n = 4x = 26), which carries one genome from A. thaliana and one from A. arenosa. Comai et al. (2000) reported that pollination of A. thaliana or A. arenosa by SAS produced seeds, as did pollination of SAS by either parent. In the present study, pollen from SAS plants (which were generated by our [4xAt × 4xAa] cross) also germinated successfully on stigmas of 2x and 4x A. thaliana and 4x A. arenosa, and seeds were produced in all crosses. Interestingly, reciprocal crosses between SAS and A. arenosa produced seeds that were not significantly different in weight from selfed seeds of the maternal parent (data not shown). This finding suggests that the presence of the A. arenosa genome in SAS has much the same effect on seed weight as an A. arenosa parent alone. Therefore, the use of SAS allowed us to cross A. thaliana and A. arenosa genomes in both directions and produce seeds in which to investigate parent-of-origin effects on endosperm development after hybridization.
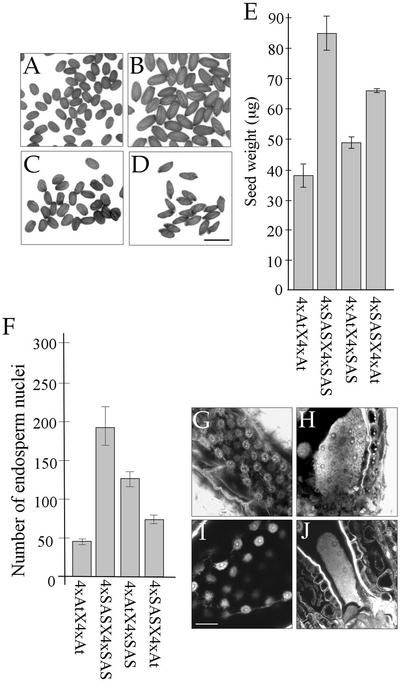
Seeds from self-pollinated SAS were larger than those from 4x A. thaliana, and more than double the weight (Figures 6A, 6B, and 6E). A comparison of [4xAt × 4xAt] and [4xAt × 4xSAS] seeds showed that substituting an SAS pollen parent for 4x A. thaliana increased seed size (Figure 6C) and increased weight by 28% (Figure 6E). In the reciprocal cross, [4xSAS × 4xAt] seeds were smaller and weighed 22% less than [4xSAS × 4xSAS] seeds (Figures 6D and 6E). Endosperm proliferation and morphology showed the same trends. Counting PE nuclei in seeds with an SAS parent at later times after pollination proved not feasible because of their size; therefore, we counted nuclei at the late globular stage only (Figure 6F). [4xSAS × 4xSAS] endosperm contained 315% more nuclei than [4xAt × 4xAt] endosperm at this stage. [4xAt × 4xSAS] seeds contained 77% more PE nuclei than [4xSAS × 4xAt] seeds (Figures 6F, 6G, and 6I) and a relatively large ChE (Figures 6H and 6J). [4xAt × 4xSAS] endosperm had 174% more nuclei than [4xAt × 4xAt] endosperm, whereas [4xSAS × 4xAt] endosperm had 63% fewer nuclei than [4xSAS × 4xSAS] endosperm. The timing of endosperm cellularization also showed parent-of-origin effects, with PE in [4xSAS × 4xAt] seeds beginning to cellularize at 7 DAP and those in [4xAt × 4xSAS] seeds beginning to cellularize at 10 DAP (data not shown).
Figure 6.
Seed and Endosperm Development in Intraspecific and Interspecific Crosses between A. thaliana and SAS.
(A) to (D) Mature seeds: [4xAt × 4xAt] (A), [4xSAS × 4xSAS] (B), [4xAt × 4xSAS] (C), and [4xSAS × 4xAt] (D). Bar = 1 mm.
(E) Mean weight of desiccated mature seeds from (A) to (D), calculated as for Figure 1F. From left to right: 38.3 ± 4.4 μg (n = 4), 84.2 ± 6.4 μg (n = 5), 49.0 ± 1.8 μg (n = 6), and 65.9 ± 0.2 μg (n = 3).
(F) Central PE nuclei, counted as for Figure 3B. Each bar represents the mean number of endosperm nuclei in seeds at the mid-globular stage of embryogenesis (n = 3). From left to right: 46 ± 4.9, 191 ± 28, 126 ± 9.3, and 71 ± 3.5.
(G) to (J) Confocal micrographs of Feulgen-stained seeds at 6 DAP showing the proliferation of PE ([G] and [I]) and ChE ([H] and [J]). Bar = 50 μm.
(G) and (H) [4xAt × 4xSAS].
(I) and (J) [4xSAS × 4xAt].
Reciprocal crosses between 2x A. thaliana and SAS might be expected to show the effects of a greater imbalance between parental genomes than crosses between 4x A. thaliana and SAS; indeed, crosses resulted in shriveled nonviable seeds in both directions (data not shown). Endosperm in [2xAt × 4xSAS] seeds exhibited a paternalized phenotype (overproliferation and late cellularization), suggesting that in this case, as in [2xAt × 4xAa] seeds, the increased parental imbalance had a lethal outcome. Additionally crosses between SAS and 4x MET1 a/s A. thaliana showed an enhanced parent-of-origin effect on seed weight compared with crosses with normally methylated 4x A. thaliana. The [4xMET1a/sAt × 4xSAS] cross produced seeds that weighed 40.2 μg, an increase of 49% compared with seeds from self-pollinated 4x MET1 a/s A. thaliana. The reciprocal cross [4xSAS × 4xMET1a/sAt] produced seeds of 20.4 μg, 76% lighter than [4xSAS × 4xSAS] seeds.
Together, the evidence presented here suggests that SAS has a paternalizing influence when used as a pollen parent in crosses with A. thaliana and a maternalizing influence when used as a seed parent.
DISCUSSION
Factors in the Hybridization Barrier between A. thaliana and A. arenosa
Interspecific crosses between A. thaliana as the seed parent and A. arenosa as the pollen parent failed when A. thaliana was in its natural diploid state, but doubling the maternal contribution produced viable seeds. This outcome was reported by Comai et al. (2000), but we have extended the analysis and examined endosperm and seed development in these crosses. The [2xAt × 4xAa] cross yielded small, shriveled seeds containing embryos that arrested by the globular–heart transition (Figures 1 and 2). By contrast, 60% of seeds from our [4xAt × 4xAa] cross were plump and contained full-grown embryos, and of these, 80% germinated (Figure 1). The success of the [4xAt × 4xAa] cross was expected because the naturally occurring species A. suecica is thought to have arisen from such an ancestral hybridization (Kamm et al., 1995; Mummenhoff and Hurka, 1995; Okane et al., 1996). However, the ecotype of the seed parent appears to affect the proportion of viable seeds produced. Using C24, we found ∼50% viable seeds in the [4xAt × 4xAa] cross; by contrast, Comai et al. (2000) found only 5% viable seeds when using the Ler ecotype as the seed parent. We also found a lower viability (13%, n = 101) with Ler than when C24 was used as the seed parent. Interecotype crosses between Ler and C24 did not reflect this difference, showing no parent-of-origin phenotypes in reciprocal crosses.
We have repeated the ancestral [4xAt × 4xAa] cross to yield SAS. The formation of this hybrid shows that increasing maternal dosage in the interspecific cross between A. thaliana and A. arenosa rescues seeds from abortion. Here, we aimed to explore the mechanism of this phenomenon.
Interspecific seeds incorporate genomes that differ in two ways, species of origin and parent of origin. Interaction between genomes in the offspring may be affected by either or both of these differences. Seed abortion in interspecific crosses could be attributable to (1) allelic incongruity (negative interactions among the products of diverged gene sequences), (2) genome shock (widespread preprogrammed changes to genomic structure or gene expression), or (3) parental imprinting (because of ploidy imbalance or divergence in expression patterns of imprinted genes, or both). The first two models can explain the failure of hybridization at any stage of development from zygote formation onward. The last can account only for the failure of a stage at which imprinting operates, which in the case of flowering plants is mainly if not exclusively endosperm development.
Several predictions can be formulated to distinguish between the two classes of models. If parental imprinting is involved in interspecific seed failure, we would expect the relative ploidy of the parents to affect the outcome and also that reciprocal crosses would produce complementary phenotypes in interspecific endosperm. Conversely, the allelic-incongruity and genome-shock hypotheses are not necessarily ploidy dependent, and these phenomena would not be expected to cause reciprocal endosperm phenotypes. Furthermore, if the two species' genomes are incompatible, or if there is widespread disruption to genomic organization or gene expression after hybridization, we would expect postgermination hybrids to exhibit a variety of morphological abnormalities, although it has been proposed that endosperm can act as a filter for offspring with low fitness (Ehlenfeldt and Ortiz, 1995), suggesting that seeds containing the most abnormal offspring could abort.
Our results show that in the [2xAt × 4xAa] cross, the endosperm appeared to be the major site of abnormalities before seed abortion (Figure 3, Table 1). [2xAt × 4xAa] endosperm displayed a dramatic overgrowth phenotype, as described previously for the severe paternal genomic excess generated by [2x × 6x] crosses in A. thaliana (Scott et al., 1998). Several features of endosperm growth exhibit similarities in the two crosses: the increased mitotic rate resulting in overproliferated PE, the absence of endosperm cellularization, and the development of a large ChE. These phenotypes support the hypothesis that seed lethality in the [2xAt × 4xAa] cross is caused by lethal paternalization as a result of an imbalance in active copies of maternally and paternally imprinted genes contributed by 2x A. thaliana and 4x A. arenosa. Further support for the involvement of imprinting in the hybridization barrier between A. thaliana and A. arenosa is provided by the observation that doubling maternal ploidy to make the [4xAt × 4xAa] cross resulted in a dramatic reduction in endosperm proliferation, restoration of PE cellularization, and rescue of seed abortion (Figures 1 to 3, Table 1). Seeds from the [2xAt × 6xAt] cross also were rescued by doubling maternal ploidy: the [4xAt × 6xAt] cross produced mainly viable seeds (our unpublished data).
We were unable to cross A. thaliana and A. arenosa in both directions because of the inability of A. thaliana pollen to germinate on A. arenosa stigmas. As an approximation, we performed reciprocal crosses between A. thaliana and SAS, which contains one diploid chromosome set from A. thaliana and one from A. arenosa. These crosses resulted in complementary endosperm phenotypes (Figure 6), as expected if parental imprinting affects endosperm development in interspecific crosses involving the A. thaliana and A. arenosa genomes.
Although our evidence supports the hypothesis that parental imprinting is an important factor in seed abortion in the [2xAt × 4xAa] cross, it is unlikely to be the only explanation, because we found that 40% of seeds from the rescuing [4xAt × 4xAa] cross still failed to complete development (Figure 1). We did not observe different classes of endosperm phenotypes in developing seeds, which we would expect if this 40% of seeds aborted as a result of lethal paternal excess. Therefore, allelic incongruity or genome shock could be responsible for the failure to rescue this proportion of the seeds. Hybrid offspring of the [4xAt × 4xAa] cross or their inbred progeny exhibited poor fertility that could not be explained fully by meiotic defects, as well as some morphological abnormalities, gene silencing, and changes to DNA methylation patterns, all of which support the hypothesis that genome shock could be an additional factor in the hybridization barrier between these species (Comai et al., 2000; Madlung et al., 2002).
The ability of the A. thaliana and A. arenosa genomes to function together to produce viable embryos in the [4xAt × 4xAa] cross and subsequently healthy, fertile hybrids suggests that there is no general incongruity between these two genomes caused by sequence divergence. Additionally, we found that the A. arenosa genome can complement two preglobular embryo-lethal mutants of 2x A. thaliana (emb9 and emb54 [Meinke, 1985]) (our unpublished data), advancing embryo development to the late globular stage typical of [2xAt × 4xAa] seeds.
However, allelic incongruity cannot be excluded as a cause of some degree of abortion, given the high level of polymorphism encountered in outbreeding species such as A. arenosa. It is possible that some A. arenosa genotypes in combination with A. thaliana could be lethal.
Hypomethylation Restores the Hybridization Barrier through Paternalization of Endosperm
Adams et al. (2000) showed that uniparental hypomethylation in A. thaliana mediated by the MET1 antisense construct (Finnegan et al., 1996) can be used to phenocopy ploidy imbalance in the endosperm, supporting the hypothesis that DNA methylation plays an important role in the imprinting mechanism of flowering plants. For example, a cross between a hypomethylated 2x seed parent and a wild-type 2x pollen parent produces seed and endosperm phenotypes similar to those produced by the wild-type [2x × 4x] cross. We interpret this finding as showing that the hypomethylated seed parent contributes ectopically active copies of paternal- as well as maternal-specific genes as a result of the loss of imprinting; therefore, it has a paternalizing effect on endosperm (Adams et al., 2000; Spielman et al., 2001). Replacing the wild-type seed parent with a hypomethylated seed parent in the [2x × 4x] cross tips the outcome from viable paternal excess to dramatic endosperm overgrowth and seed lethality (Adams et al., 2000), presumably as a result of this paternalizing effect. Similarly, in the present study, we found that replacing the wild-type 4x A. thaliana seed parent with a hypomethylated 4x plant in the [4xAt × 4xAa] cross resulted in the abortion of most of the resulting seeds with a strong paternal excess phenotype (Figure 4, Table 1).
Madlung et al. (2002) reported that treatment of SAS with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (azadC) enhanced the phenotypic instability of the hybrid plants as a result of changes in transcription patterns. However, this is unlikely to represent the same phenomenon that we reported above, for two reasons: (1) the azadC treatment caused variable degrees of abnormality associated with a general loss of the control of gene expression, such as fasciation and homeotic transformations, whereas changes to seed development mediated by uniparental hypomethylation had more precise and repeatable effects on endosperm morphology; and (2) the azadC treatment had comparatively little effect on A. thaliana, whereas uniparental hypomethylation had the same effect on seeds resulting from interspecific crosses between A. thaliana and A. arenosa as it had on A. thaliana seeds (Adams et al., 2000; this study). Therefore, we interpret the low success rate of the [4xMET1a/sAt × 4xAa] cross as being attributable mainly to lethal paternalization of the endosperm. However, the early death or slow growth of the small number of hybrids resulting from this cross could be the result of the effects of MET1 a/s–mediated hypomethylation on an already epigenetically compromised genome or of hypomethylation combined with allelic incongruity.
Imprinting, Hybridization Barriers, and Genomic Strength
We have shown that there is a hybridization barrier between 2x A. thaliana (when used as a seed parent) and 4x A. arenosa (when used as a pollen parent) that can be overcome by increasing maternal ploidy but restored by hypomethylation. This is significant for both understanding the nature of postzygotic hybridization barriers and manipulating them.
Interspecific crosses in several genera show a requirement for the correct balance of parental genomes in the endosperm that is not necessarily 2m:1p. To explain this observation, Johnston et al. (1980) proposed the endosperm balance number (EBN) hypothesis, according to which each species is assigned an EBN reflecting its effective ploidy, which is determined empirically by its behavior in interspecific crosses. In this system, it is a 2:1 ratio of maternal-to-paternal EBNs, rather than chromosome sets per se, that is required to ensure normal endosperm development. EBN and parental imprinting have been interpreted as reflecting the same phenomenon (Haig and Westoby, 1989, 1991). It has been observed previously that the EBN system could serve as a mechanism of reproductive isolation, because species with different EBNs cannot hybridize (Johnston et al., 1980; Johnston and Hanneman, 1982; Ehlenfeldt and Ortiz, 1995), presumably as a result of the dosage imbalance of imprinted genes in the endosperm. The abortion of [2xAt × 4xAa] seeds with a paternal excess phenotype indicates that 2x A. thaliana has a lower EBN than 4x A. arenosa, whereas the production of viable seeds with normal endosperm morphology by the [4xAt × 4xAa] cross shows that 4x A. thaliana has a similar EBN to 4x A. arenosa. Substitution of 4x MET1 a/s A. thaliana for the seed parent in the [4xAt × 4xAa] cross restored lethality and the paternal excess phenotype, suggesting that hypomethylation decreases EBN. Changing ploidy levels of one parent or its gametes has been proposed as a way to alter EBN and therefore the outcome of interspecific crosses, but our study documents an attempt to modify a hybridization barrier by manipulating the imprinting system itself.
EBNs generally are assessed on a single parameter, the ability of interspecific crosses to produce hybrid seeds. However, investigation of seed development in such crosses shows that there are more subtle effects than simple viability or inviability, such as changes to seed size (Ehlenfeldt and Ortiz, 1995). We found that reciprocal crosses between 4x A. thaliana and 4x SAS produced larger endosperm when SAS was the pollen parent than when it was the seed parent (Figure 6). Mature seed weight did not follow the same pattern, but this may be the result of maternal effects, because SAS self seeds were twice the weight of 4x A. thaliana seeds. Therefore, in assessing parent-of-origin effects on seed weight, we compared crosses in which the seed parent was held constant (i.e., [4xAt × 4xSAS] was compared with [4xAt × 4xAt] and [4xSAS × 4xAt] was compared with [4xSAS × 4xSAS]). By this method, we found that for both seed weight and endosperm proliferation, SAS had a growth-promoting effect when used as a pollen parent in crosses with A. thaliana plants at the same ploidy level and an inhibiting effect when used as a seed parent (Figure 6). This finding suggests that SAS, and by extension A. arenosa, has a paternalizing effect on interspecific seeds when used as a pollen parent in crosses with 4x A. thaliana and a maternalizing effect when used as a seed parent. One caveat is that the [4xAt × 4xAa] cross, in which both chromosome sets in the paternal genome are from A. arenosa, might be expected to produce larger seeds than [4xAt × 4xSAS], in which A. arenosa contributes only one paternal chromosome set; in fact, the seeds were of similar size and weight (Figures 1 and 6). However, it is possible that this finding reflects a limit to the resources that an A. thaliana parent can allocate to a seed, no matter how demanding the paternal genome.
The asymmetric effects of the SAS and A. thaliana genomes on seed development fit a model in which a paternally contributed A. arenosa genome within an endosperm can extract more resources from the A. thaliana mother in which the seed is developing than the maternal genome is able to withhold. This could operate, for example, by the A. arenosa paternal genome silencing more growth inhibitors, or repressing them for longer, than an A. thaliana paternal genome would. Conversely, a maternally contributed A. arenosa genome can withhold resources demanded by an A. thaliana paternal genome (e.g., by silencing more growth promoters than an A. thaliana maternal genome would). This phenomenon was predicted by the parental-conflict theory, according to which species evolve a balance between the activity of the male genome in acquiring resources for its offspring and the activity of the female genome in inhibiting resource acquisition (Haig and Westoby, 1989, 1991). In other words, each species reaches its own unique balance of conflict. However, when maternal and paternal genomes of two species with different “genomic strengths” are brought together in a hybrid seed, the dosage of expressed alleles of imprinted genes is likely to be unbalanced, resulting in complementary endosperm phenotypes and potentially in seed abortion. This finding suggests that instant speciation could be achieved by changing global methylation levels, for example, by mutation of a DNA methyltransferase or another component of the imprinting mechanism.
The parental-conflict model also predicts that imprinting should be relaxed in monogamous or inbreeding species, because the drive for conflict between the parents is reduced (Haig, 1992). The viable outcome of [2x × 4x] and [4x × 2x] crosses in A. thaliana (Scott et al., 1998), a predominantly inbreeding species, compared with the failure of [2x × 4x] and [4x × 2x] crosses in the closely related outbreeding species Brassica rapa (Håkansson, 1956) is in accord with this proposition. A. arenosa is an obligate outbreeder as a result of its self-incompatibility system; therefore, it would be predicted to have greater genomic strength than A. thaliana. This notion is supported by the reciprocal phenotypes in seed size and endosperm proliferation seen in the cross between 4x SAS and 4x A. thaliana, in which genomic strength was not masked by a ploidy imbalance.
Evolution of A. suecica
For many years, authors have suggested that A. thaliana and A. arenosa are the parents of A. suecica. Hylander (1957) proposed a cross between 2x A. thaliana and 2x A. arenosa followed by genome duplication. This scenario remains possible, but at present, 2x A. arenosa reportedly is restricted in distribution to Slovakia and Hungary, its ability to hybridize with A. thaliana has not been tested, and we were unable to obtain plants to perform this cross. Love (1961) suggested that a 4x A. arenosa seed parent was fertilized by an unreduced pollen grain of A. thaliana, but analysis of the A. suecica chloroplast genome has shown that A. thaliana was the ancestral seed parent (Mummenhoff and Hurka, 1995). Borgen (1987) proposed that the first step in hybridization was a cross between 2x A. thaliana and 4x A. arenosa, but Comai et al. (2000) and we have shown that seeds from this cross do not complete development. The data presented here and by Comai et al. (2000) suggest that the most plausible scenario for the evolution of A. suecica is fertilization of an unreduced A. thaliana embryo sac by a normally reduced sperm of 4x A. arenosa. Our results indicate that a cross between a reduced A. thaliana embryo sac and 4x A. arenosa would fail because of lethal paternal excess.
Conclusions
In this study, we found that parental imprinting most likely is the dominant mechanism for the postzygotic hybridization barrier between A. thaliana and A. arenosa. The cross can be rescued by doubling the ploidy of the seed parent, resulting in an endosperm with balanced maternal and paternal contributions. Additionally, we have shown that it is possible to revert to the nonviable state by hypomethylation of the seed parent, effectively raising a hybridization barrier. The ability to introduce a hybridization barrier may prove useful in the containment of genetically modified crops: genomic strength could be modified to ensure that they would be unable to produce viable seeds by hybridization with wild relatives. Our results also suggest that it would be possible to breach a hybridization barrier by changing the genomic strength, allowing normally isolated species to interbreed.
METHODS
Plant Material
Plants were grown for 3 to 4 weeks in a Fisons Fi-totron 600H growth cabinet (Bellevue, WA) at 26°C with a daylength of 16 h, transferred to a glasshouse, and grown in the same conditions. Arabidopsis thaliana plants used were C24 diploid (2x) hemizygous for an A9 barnase construct, which confers male sterility (Paul et al., 1992), and C24 tetraploid (4x) A9 barnase. Male-sterile segregants were used as seed parents in crosses, and their male-fertile siblings were used as pollen parents. Hypomethylated 4x C24 plants containing a MET1 antisense construct under the control of the 35S promoter (MET1 a/s) (Finnegan et al., 1996) were obtained by crossing 2x MET1 a/s plants with tetraspore (tes) mutants in the Ler background (Spielman et al., 1997). This cross resulted in progeny varying in ploidy from 2x to 5x. Tetraploid progeny from the cross were selected and screened for the presence of the MET1 a/s transgene (Adams et al., 2000). Backcrosses to 4x C24 for two generations were performed to cross out the tes mutation. The resulting plants were used as 4x MET1 a/s. Arabidopsis arenosa seeds were donated by Luca Comai (University of Washington, Seattle), and stocks were maintained by crossing within a small number of plants.
Artificial Pollination
For male-sterile seed parents (2x A9-barnase), open flowers were pollinated. For male-fertile plants, flower buds were emasculated 1 day before anthesis and pollinated 1 day later. For each cross, developing siliques were collected at 2 to 14 days after pollination and processed as described below. Mature seeds were collected from desiccated pods and weighed using a Mettler UMT2 microbalance (Mettler-Toledo, Leicester, UK).
Fertilization of A. arenosa by A. thaliana was attempted by emasculating and pollinating very young buds. Salt treatment was performed by applying a range of concentrations of NaCl (0.5 to 5% with 0.1% Tween 20) to the stigmas of open flowers, leaving them to dry for 1 h, and then pollinating them.
Confocal Laser Scanning Microscopy
Samples were prepared as described by Braselton et al. (1996) and imaged at the University of Bath using an Axiovert 100M Zeiss LSM510 laser scanning microscope (Jena, Germany). Feulgen-stained samples were excited using an argon ion laser at 488 nm and emissions detected at ≥515 nm. Images measuring 1024 × 1024 pixels were collected using a C-Apochromat 63×/1.2 water lens, saved in TIFF format, and processed using Adobe Photoshop 5.0 LE (Mountain View, CA). Peripheral endosperm nuclei were counted in serial optical sections throughout the depth of a Feulgen-stained seed. For larger seeds, peripheral endosperm nuclei were counted for one-half of the seed, and a total estimate was obtained by doubling this number. Measurements of chalazal endosperm area were taken using Zeiss LSM Image Browser software, version 3.1.0.99 (www.zeiss.de), or Scion Image (Frederick, MD), version 4.0.2 (www.scioncorp.com).
Clearing of Intact Seeds
To clear seeds for light microscopy, intact seed pods were placed in an 8:1 solution of ethanol:acetic acid at 4°C overnight. Pods were washed for 1 h in 100% ethanol, 90% ethanol, and 70% ethanol and stored at 4°C. Seeds were dissected from siliques in one drop of chloral hydrate fixative (chloral hydrate:glycerol:water, 8:1:3). Whole-seed preparations were examined with a Nikon Eclipse E800 microscope (Tokyo, Japan) using differential interference contrast optics. Images were made using a SPOT RT Color camera (Diagnostic Instruments, Sterling Heights, MI), saved as JPEG files, and processed using Adobe Photoshop 5.0 LE.
Upon request, all novel materials described in this article will be made available for noncommercial research purposes.
Acknowledgments
We thank Luca Comai for A. arenosa seeds and James Doughty for help with pollinations. C.B. was supported by a University of Bath studentship and by Ceres, Inc., and M.S. was supported by Sulis Innovation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010496.
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127, 2493–2502. [DOI] [PubMed] [Google Scholar]
- Bartolomei, M.S., and Tilghman, S.M. (1997). Genomic imprinting in mammals. Annu. Rev. Genet. 31, 493–525. [DOI] [PubMed] [Google Scholar]
- Borgen, L. (1987). Postglacial evolution in the Nordic flora: A review. Blyttia 45, 147–169. [Google Scholar]
- Braselton, J.P., Wilkinson, M.J., and Clulow, S.A. (1996). Feulgen staining of intact plant tissues for confocal microscopy. Biotech. Histochem. 71, 84–87. [DOI] [PubMed] [Google Scholar]
- Brink, R.A., and Cooper, D.C. (1947). The endosperm in seed development. Bot. Rev. 132, 423–541. [Google Scholar]
- Brown, R.C., Lemmon, B.E., Nguyen, H., and Olsen, O.A. (1999). Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12, 32–42. [Google Scholar]
- Chen, Z.J., Comai, L., and Pikaard, C.S. (1998). Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. USA 95, 14891–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L., Tyagi, A.P., Winter, K., Holmes-Davis, R., Reynolds, S.H., Stevens, Y., and Byers, B. (2000). Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12, 1551–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J.A., and Orr, H.A. (1998). The evolutionary genetics of speciation. Philos. Trans. R. Soc. Lond. B 353, 287–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deNettancourt, D. (1997). Incompatibility in angiosperms. Sex. Plant Reprod. 10, 185–199. [Google Scholar]
- Ehlenfeldt, M.K., and Ortiz, R. (1995). Evidence on the nature and origins of endosperm dosage requirements in Solanum and other angiosperm genera. Sex. Plant Reprod. 8, 189–196. [Google Scholar]
- Finnegan, E.J., Genger, R.K., Peacock, W.J., and Dennis, E.S. (1998). DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 223–247. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig, D. (1992). Genomic imprinting and the theory of parent-offspring conflict. Semin. Dev. Biol. 3, 153–160. [Google Scholar]
- Haig, D., and Westoby, M. (1989). Parent-specific gene expression and the triploid endosperm. Am. Nat. 134, 147–155. [Google Scholar]
- Haig, D., and Westoby, M. (1991). Genomic imprinting in endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B 333, 1–13. [Google Scholar]
- Håkansson, A. (1956). Seed development of Brassica oleracea after certain reciprocal pollinations. Hereditas 42, 373–396. [Google Scholar]
- Henikoff, S., and Comai, L. (1998). A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 149, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and Matzke, M.A. (1997). Exploring and explaining epigenetic effects. Trends Genet. 13, 293–295. [DOI] [PubMed] [Google Scholar]
- Hirner, B., Fischer, W.N., Rentsch, D., Kwart, M., and Frommer, W.B. (1998). Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 14, 535–544. [DOI] [PubMed] [Google Scholar]
- Hiscock, S.J., and Dickinson, H.G. (1993). Unilateral incompatibility within the Brassicaceae: Further evidence for the involvement of the self-incompatibility (S)-locus. Theor. Appl. Genet. 86, 744–753. [DOI] [PubMed] [Google Scholar]
- Hylander, N. (1957). Cardaminopsis suecica (Fr.) Hiit., a northern amphidiploid species. Bull. Jard. Bot. Etat. Brux. 27, 591–604. [Google Scholar]
- Johnston, S.A., den Nijs, T.P.M., Peloquin, S.J., and Hanneman, J.R.E. (1980). The significance of genic balance to endosperm development in interspecific crosses. Theor. Appl. Genet. 57, 5–9. [DOI] [PubMed] [Google Scholar]
- Johnston, S.A., and Hanneman, R.E. (1982). Manipulations of endosperm balance number overcome crossing barriers between diploid Solanum species. Science 217, 446–448. [DOI] [PubMed] [Google Scholar]
- Kamm, A., Galasso, I., Schmidt, T., and Heslop-Harrison, J.S. (1995). Analysis of a repetitive DNA family from Arabidopsis arenosa and relationships between Arabidopsis species. Plant Mol. Biol. 27, 853–862. [DOI] [PubMed] [Google Scholar]
- Kermicle, J.L., and Alleman, M. (1990). Gametic imprinting in maize in relation to the angiosperm life cycle. Dev. Suppl. 1990, 9–14. [PubMed] [Google Scholar]
- Leitch, I.J., and Bennett, M.D. (1997). Polyploidy in angiosperms. Trends Plant Sci. 2, 470–476. [Google Scholar]
- Lin, B.Y. (1984). Ploidy barrier to endosperm development in maize. Genetics 107, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1993). Endosperm origin, development, and function. Plant Cell 5, 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, A. (1961). Hylandra: A new genus of Cruciferae. Sven. Bot. Tidskr. 55, 211–217. [Google Scholar]
- Lund, G., Messing, J., and Viotti, A. (1995). Endosperm-specific demethylation and activation of specific alleles of alpha tubulin genes of Zea mays L. Mol. Gen. Genet. 246, 716–722. [DOI] [PubMed] [Google Scholar]
- Madlung, A., Masuelli, R.W., Watson, B., Reynolds, S.H., Davison, J., and Comai, L. (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129, 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1990). Endosperm cellularization in Arabidopsis thaliana. Arabidopsis Inf. Serv. 27, 65–72. [Google Scholar]
- Matzke, M.A., and Matzke, A.J.M. (1998). Epigenetic silencing of plant transgenes as a consequence of diverse cellular defence responses. Cell. Mol. Life Sci. 54, 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. (1985). Embryo-lethal mutants of Arabidopsis thaliana: Analysis of mutants with a wide range of lethal phases. Theor. Appl. Genet. 69, 543–552. [DOI] [PubMed] [Google Scholar]
- Meyer, P., and Saedler, H. (1996). Homology-dependent gene silencing in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 23–48. [DOI] [PubMed] [Google Scholar]
- Moore, T., and Haig, D. (1991). Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 7, 45–49. [DOI] [PubMed] [Google Scholar]
- Mummenhoff, K., and Hurka, H. (1995). Allopolyploid origin of Arabidopsis suecica (Fries) Norrlin: Evidence from chloroplast and nuclear genome markers. Bot. Acta 108, 449–456. [Google Scholar]
- Nasrallah, M.E., Yogeeswaran, K., Snyder, S., and Nasrallah, J.B. (2000). Arabidopsis species hybrids in the study of species differences and evolution of amphiploidy in plants. Plant Physiol. 124, 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okane, S.L., Schaal, B.A., and Al-Shehbaz, I.A. (1996). The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst. Bot. 21, 559–566. [Google Scholar]
- Paul, W., Hodge, R., Smartt, S., Draper, J., and Scott, R. (1992). The isolation and characterization of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol. Biol. 19, 611–622. [DOI] [PubMed] [Google Scholar]
- Pikaard, C.S. (2001). Genomic change and gene silencing in polyploids. Trends Genet. 17, 675–677. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H. (2001). Polyploid evolution: Keeping the peace at genomic reunions. Curr. Biol. 11, R925–R928. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H., and Carney, S.E. (1998). Plant hybridization. New Phytol. 140, 599–624. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H., and Noyes, R.D. (1998). Genetic map-based studies of reticulate evolution in plants. Trends Plant Sci. 3, 254–259. [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341. [DOI] [PubMed] [Google Scholar]
- Spielman, M., Preuss, D., Li, F.L., Browne, W.E., Scott, R.J., and Dickinson, H.G. (1997). TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development 124, 2645–2657. [DOI] [PubMed] [Google Scholar]
- Spielman, M., Vinkenoog, R., Dickinson, H.G., and Scott, R.J. (2001). The epigenetic basis of gender in flowering plants and mammals. Trends Genet. 17, 705–711. [DOI] [PubMed] [Google Scholar]
- Stebbins, G.L. (1958). The inviability, weakness and sterility of interspecific hybrids. Adv. Genet. 9, 147–215. [DOI] [PubMed] [Google Scholar]
- Thompson, W. (1930). Causes of difference in success of reciprocal interspecific crosses. Am. Nat. 64, 407–421. [Google Scholar]
- Tiffin, P., Olson, M.S., and Moyle, L.C. (2001). Asymmetrical crossing barriers in angiosperms. Proc. R. Soc. Lond. B Biol. Sci. 268, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman, S.M. (1999). The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell 96, 185–193. [DOI] [PubMed] [Google Scholar]
- Watkins, A. (1932). Hybrid sterility and incompatibility. J. Genet. 25, 125–162. [Google Scholar]