Abstract
We present a molecular model of a chromatophore vesicle from Rhodobacter sphaeroides. These vesicles are ideal benchmark systems for molecular and systemic simulations, because they have been well studied, they are small, and they are naturally separated from their cellular environment. To set up a photosynthetic chain working under steady-state conditions, we compiled from the experimental literature the specific activities and geometries that have been determined for their constituents. This data then allowed defining the stoichiometries for all membrane proteins. This article contains the kinetic part of the reconstructed model, while the spatial reconstruction is presented in a companion article. By considering the transport properties of the Cytochrome c2 and ubiquinone pools, we show that their size and oxidation states allow for an efficient buffering of the statistical fluctuations that arise from the small size of the vesicles. Stoichiometric and kinetic considerations indicate that a typical chromatophore vesicle of Rb. sphaeroides with a diameter of 45 nm should contain approximately five bc1 monomers.
INTRODUCTION
The last decades of molecular cell biology were immensely influenced by the determination of three-dimensional structures at atomic resolution for many of the important proteins in biological cells, thus making way for the understanding of the detailed microscopic processes inside these proteins. Nowadays the focus starts to shift toward a systems biology treatment of subsets of the functional units of the cell, i.e., interconnected systems of proteins working on the same task. The most important theoretical tool to investigate cells at this systemic level is to simulate them on a computer (1–3). These dynamic models then reveal whether the knowledge about a particular system is both sufficiently complete and consistent and they help to state and test new hypotheses.
The photosynthetic apparatus of purple bacteria is an attractive model for such in silico approaches at the systemic or even molecular level. As this system is spatially confined to small vesicles, it only contains a manageably small number of molecules that belong to five integral membrane proteins and two electron carriers. Except for a few mechanistic details, the biological function of each macromolecule is known precisely. Moreover, the three-dimensional structures of all components could be determined in recent years: for the reaction center (RC) (4,5), the light harvesting complexes I and II (LH1 and LH2) (6–8), the F0F1–ATPase (9), and recently also for the Cytochrome bc1 complex (10). The electron carriers Cytochrome c2 (here abbreviated as c2) (11) and ubiquinone (Q) are well known, too. Consequently, the photosynthetic apparatus of purple bacteria may be considered a sufficiently well-studied and understood model system, ideally suited to develop new theoretical techniques. However, the wealth of published information about the photosynthetic apparatus of purple bacteria is spread out over literally hundreds of articles that appeared during at least three decades of research in a dozen or so journals. Additionally, not every part of the system is covered at the same level of detail. For example, the absorption of light by the bacteriochlorophylls of the light-harvesting complexes is understood at the level of quantum chemical calculations (8), whereas only an upper limit can be estimated for the diffusion coefficient of the quinone molecules in realistic protein-filled membranes from a single experiment (12). The first task of a systemic approach is therefore to compile a consistent set of data about the chromatophore vesicles that allows for a description of the important processes on equal or at least comparable footing.
Here we present a comprehensive model of a chromatophore vesicle from Rhodobacter (Rb.) sphaeroides working under steady-state conditions. Our incentive is to put together a biological benchmark system for dynamic molecular simulations, allowing us to study transport processes inside the model vesicle on a single molecule level, i.e., on a Brownian scale and resolution. Therefore, one needs dynamic data on protein diffusion inside the vesicle, how fast the proteins process photons, protons, etc., and how these reaction rates depend on external conditions. Most of these external conditions are concentrations or oxidation states of the other molecules in the vesicle. Details of the internal reactions of the proteins are considered only if they are relevant for their external behavior. Due to the amount of information, the material is split up into the kinetic process view, described in this article, and the spatial setup of the chromatophore vesicle, which is presented in the accompanying article (13).
Certainly, the two descriptions are closely related. Some kinetic arguments have to be considered in the spatial model and some spatial constraints affect the kinetics of photosynthesis. Consequently, this article also deals with certain aspects of the spatial arrangement of the transmembrane proteins, especially the number and placement of the bc1 complexes. In particular, recent atomic force and electron microscopy images of the membranes of chromatophore vesicles revealed how the light-harvesting complexes and reaction centers are arranged on the membrane (14–17). Surprisingly, the Cytochrome bc1 complex, without which photosynthesis cannot function, was not detected. This observation not only rekindled a long-going debate about whether the reaction centers form supercomplexes with the bc1s or not (18,19), but also presented a new challenge to reconstruct the actual geometry of such a chromatophore vesicle.
In this article, we employ the possibly simplistic assumption that the photosynthetic apparatus works as a linear conversion chain. So far, we do not account for additional control mechanisms, e.g., inhibitory feedback loops or controls as have been described for many important cellular processes (20). The reason for this is simply the lack of experimental evidence, for example whether proton pumping by the bc1 complex is shut off at too-low intravascular pH. As will be shown below, the kinetic model developed in this study provides stable steady-state solutions under typical physiological conditions. A short section at the end discusses possible control mechanisms. We hope that this work will contribute stimulating further experimental and theoretical work in this direction.
This article is organized as follows: the following section reviews the bacterial photosynthetic apparatus and introduces the concept of the process view. Then we collect the biochemical and biophysical data of the individual proteins before we assemble the photosynthetic conversion chain and determine the number of the bc1s. This completes the static view of the kinetic model. To incorporate dynamic effects, the transport capacities of the electron carriers are determined in the section Transport Processes and Reservoirs. Here, we also show how the size of their pools and their oxidation state is related to their kinetic buffering capacity against statistical fluctuations that stem from the photon capture rate and the small number of proteins on the vesicle.
THE SYSTEM
Bacterial photosynthesis
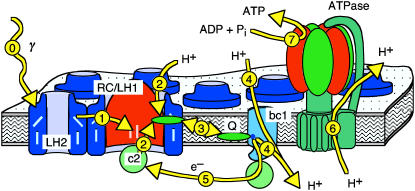
In text books, the photosynthetic system of the purple bacterium Rb. sphaeroides is usually depicted in schematic form as in Fig. 1. Shown is a part of a lipid bilayer, which contains one copy of each of the proteins plus the transport processes (denoted by arrows). In purple bacteria such as Rb. sphaeroides, the photosynthetic apparatus is mainly located on the so-called chromatophore vesicles, small specialized lipid vesicles of 30–60-nm diameter (21). These vesicles are densely packed with light-harvesting complexes (LHC) of types I and II (LH1, LH2). These collect the incident photons (process 0 in Fig. 1) and hand their energy on to the RCs (process 1) in the form of electronic excitations. The RC passes this energy on to a waiting ubiquinone (Q) in the form of an electron-proton pair, where the proton (H+) is taken up from the cytoplasm (process 2). Later, this ubiquinone, which has become a ubiquinol (QH2) by the uptake of a second of these pairs, unbinds from the RC and diffuses inside the membrane (process 3) to deliver its freight to the bc1 complex. According to the Q-cycle (22), the bc1 complex releases the protons to the inside of the vesicle and the stored energy is used to pump two further protons across the membrane (process 4). The electrons are then shuttled back to the RC by the water-soluble electron carrier protein Cytochrome c2 (process 5), while the proton gradient is the driving force for the synthesis of ATP from ADP and inorganic phosphate in the F0F1–ATP synthase (ATPase) (processes 6 and 7). For a more detailed description of some of these processes, see, e.g., the reviews of Schulten and co-workers (6,8).
FIGURE 1.
Artistic textbook-style rendering of the photosynthetic apparatus of purple bacteria. The inside of the chromatophore vesicle is below the membrane. Please see text for abbreviations and a description of the processes (yellow arrows) taking place at or around the lipid membrane that contains the transmembrane proteins.
The process view
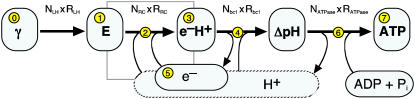
Although the textbook-style picture sketched above is well suited to explain qualitatively the successive steps in photosynthesis, it tends to neglect or even obscure the quantitative aspects. As will be explained in more detail below, the photosynthetic apparatus is not only a set of (transmembrane) proteins and reactions, but also a conversion chain where light energy is converted via some intermediate forms into chemical energy. This notion of a photosynthetic chain implies that the different proteins involved are chained to each other functionally (see Fig. 2). All the spatial information contained in Fig. 1 about, for example, the dimensions of the proteins is omitted here. The thick arrows indicate the central conversion path from light energy over excitons (E), electron proton pairs, and the transmembrane proton gradient into chemical energy. The thin arrows denote the auxiliary reactions. The network view of Fig. 2 is the complementary picture to the spatial model of Fig. 1. The complete model of the vesicle must incorporate both these views, since either one of them alone is clearly not sufficient. However, both Figs. 1 and 2 omit a lot of internal details of the proteins that perform the respective conversion steps.
FIGURE 2.
Schematic view of the photosynthetic apparatus as a conversion chain. The thick arrows denote the path through which the photon energy is converted into chemical energy stored in ATP via the intermediate stages (rounded rectangles). Each conversion step takes place in proteins that work in parallel. Their number N times the conversion rate of a single protein R determines the total throughput of this step. The microscopic reactions taking place in the RC (denoted by the shaded box), which are here summarized as a single thick arrow, are detailed in Fig. 3. The abbreviations used are γ for the incoming photons collected in the LHCs, E for the excitons in the LHCs and in the RCs, e−H+ for the electron-proton pairs stored on the quinols, e− for the electrons on the Cytochrome c2, ΔpH for the transmembrane proton gradient, and H+ for the protons outside of the vesicle (broken outline of the respective reservoir). Compare this representation to the spatially focused textbook-style view of Fig. 1.
Two interwoven cycles can be identified from Fig. 2:
The electrons cycle between the RCs and the bc1s in a closed loop and
The protons enter the vesicle via the RCs and bc1s and leave it again through the ATPase.
The rather unspecific return path of the protons via the cytoplasm is denoted in Fig. 2 with a broken outline of the proton reservoir. When adopting this process view, one needs to consider the quantitative aspects of a balanced chain. The process view consequently requires compiling the technical data of the proteins of the photosynthetic chain: numbers of individual proteins, throughputs with respect to the driving forces, relative placements on the vesicle, spatial and dynamical limits, kinetic rates of the physical and chemical processes, time constants of association and dissociation, diffusion times, etc.
Under constant external conditions the whole chain will be in equilibrium, where the output of each stage equals the input of the subsequent stage. Although each of these conversion processes has a different mechanism and, consequently, different turnovers with different dependencies on external conditions, it may be safely assumed that ATP synthesis works best when the stoichiometries of the chain links Ni are inversely related to their respective throughputs Ri:
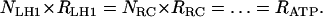 |
(1) |
In addition to these stoichiometric properties we can expect that the photosynthetic vesicles are built according to two familiar contrasting design principles that we know from our daily life's experience, expressed there rather vaguely as robustness (23) and efficiency (6). In our context this means that the photosynthetic machinery is robust with respect to the external conditions, especially against too much or too little light. As lipid membranes can only withstand a limited pH gradient corresponding to a transmembrane voltage of ≈200 mV, the composition of the vesicle and the interplay of the proteins should not allow for too many protons inside the vesicle even for unexpectedly high light intensities. Another aspect is that a malfunction of a photosynthetic vesicle should not endanger the cell. As there exists an upper bound to the total number of proteins imposed by the size of the vesicle, the stoichiometries should be optimized to make best use of the volume and the resources of a vesicle.
The following section starts with separately discussing each of the proteins of the chromatophore vesicles from Rb. sphaeroides. The chain segments are then linked according to their specific activities to build up the integrative overview from the conventional view, the process view, and the spatial constraints. During the setup of this model chromatophore it will become apparent that, although the photosynthesis of purple bacteria is thoroughly studied at the level of the individual proteins, some important technical data are still missing.
BIOCHEMICAL AND BIOPHYSICAL DATA FOR THE INDIVIDUAL COMPONENTS
In this section the necessary geometric and dynamic data are compiled from experimental studies of the individual proteins of the photosynthetic chain without relating them yet. Certainly, experimental data should always contain error bars, and these should be a critical part of the following discussion. However, experimental error estimates are not always given in the primary literature and are hard to estimate by outsiders. At this preliminary stage, we will therefore proceed with the bare numbers.
Capturing the photons: the light-harvesting complexes
As the photosynthetic chain is triggered by the captured photons, it cannot run faster than the total rate of photon capture that can be computed from the photon capture rate per LHC and the total number of LHCs per vesicle. There are two different types of LHCs, the smaller LH2 and the larger LH1, which always encircle an RC. We first determine the respective photon capture rates for both types and get back to their total numbers when discussing the RCs in the next section.
The capture rate of the LHCs at a particular wavelength is obtained as the product of the spectral density of incident photons, nγ(λ), times the wavelength-dependent absorption cross section of the LHCs, σLHC(λ). In the LHCs, the carotenoids and bacteriochlorophylls absorb the incident photons with a combined absorption spectrum as given, for example, in Cogdell et al. (24). These absorption spectra are normalized using the extinction coefficient of Bchl of ∼140 1/(mM cm) reported in Francke and Amesz (25), which translates into an effective photon capture cross section of σBchl = 2.32 Å2 per Bchl molecule at the wavelength of the Bchl absorption maximum. This value, measured in vivo, i.e., in the native environment, includes all effects from the environment of the Bchls in the LHCs and also from their averaged directional sensitivity. Each α–β-dimer of the LHCs containing a total of three Bchls, consequently has an absorption cross section per dimer σαβ = 3σBchl. Using the appropriate number of α–β-dimers per LHC gives the normalized absorption cross section of the LHCs, denoted by σLH1(λ) and σLH2(λ), respectively.
As a first approximation to the photon spectrum nγ(λ), the spectrum of the sunlight filtered through the earth's atmosphere is used, i.e., the spectrum that reaches the ground (see, e.g., (26)). Taking the growth conditions for Rb. sphaeroides given in Feniouk et al. (21) as representative for their native environment, the power spectrum Eγ(λ)nγ(λ) is normalized to the total light intensity of 18 W/m2. Eγ(λ) is the energy of a photon of wavelength λ. The total photon absorption rate RLH1 of a closed LH1 ring of 16 α–β dimers at this light intensity is then
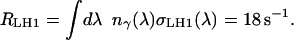 |
(2) |
This rate applies to circular, closed LH1 rings of 16 α–β–subunits each. In Rb. sphaeroides, the native form of the LH1 rings are homodimers of an RC plus an incomplete LH1 ring of ∼12 subunits (see, e.g., (25) and (15)). Consequently the total capture rate per LH1/RC ring reduces to 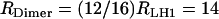 s−1. The same procedure being applied to the smaller LH2 rings, which consist of eight instead of 16 subunits and have a slightly different absorption spectrum σLH2(λ), yields a total photon capture rate of RLH2 = 10 s−1. The largest contribution to the uncertainty of the capture rates RDimer and RLH2 is due to the photon spectrum taken as the sun spectrum filtered only by the atmosphere, while the bacteria normally live in an aqueous environment. However, it will be shown below that the absolute values of RDimer and RLH2 are not critical, as there are more LHCs present on the vesicle than necessary at this light intensity. Hence, we will proceed with the rates estimated above.
s−1. The same procedure being applied to the smaller LH2 rings, which consist of eight instead of 16 subunits and have a slightly different absorption spectrum σLH2(λ), yields a total photon capture rate of RLH2 = 10 s−1. The largest contribution to the uncertainty of the capture rates RDimer and RLH2 is due to the photon spectrum taken as the sun spectrum filtered only by the atmosphere, while the bacteria normally live in an aqueous environment. However, it will be shown below that the absolute values of RDimer and RLH2 are not critical, as there are more LHCs present on the vesicle than necessary at this light intensity. Hence, we will proceed with the rates estimated above.
Fixating the light energy: the reaction center
The photons harvested in the LHCs are funneled as electronic excitations (excitons) to the special pair Bchls of the RC where charge separation takes place. An electron is then transferred onto a quinone molecule waiting at the Qb site of the RC. Concurrently, a proton is taken up from the outside of the vesicle and finally transferred onto the quinone. Once the quinone has taken up two of these electron-proton pairs, it is released again into the inner membrane space and diffuses to the bc1 complex.
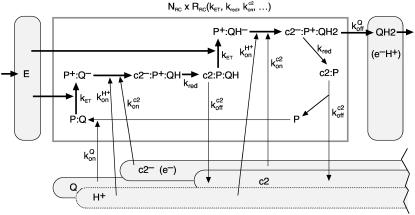
The charge separation and energy transfer onto the quinone take place in several distinct steps, as depicted in Fig. 3. The overall throughput of a single RC, RRC, is determined from the combination of all these individual reactions. However, for our rate consideration it is sufficient to know which of them is the rate-limiting one. Apart from the light intensity, two candidates are given in the literature: Gerencsér et al. (27) identified the unbinding of the oxidized Cytochrome c2 from the RC as the bottleneck, limiting the turnover to <800 electrons per second. A smaller value of 270 c2 per second and RC was reported by Paddock et al. (28). But even this slower rate is much higher than another limit imposed by the unbinding of the quinol from the RC, which was determined by Milano et al. (29) to take ∼25 ms. As this time covers only the longest out of a handful of steps, a conservative estimate would be approximately twice that duration. Therefore, we assume a total time to load a quinone with two electron-proton pairs of some 50 ms or, correspondingly, a total output of ∼20 quinols per second.
FIGURE 3.
Detailed view of the processes that take place inside the reaction center. This network of reactions belongs in the boxed area of the overview figure (Fig. 2). All these individual reactions with their individual rates k together determine the overall conversion rate RRC of a single RC. As in Fig. 2, the thick arrows denote the flow of the energy from the excitons through the cyclic charge-state changes of the special pair Bchl (P) of the RC. Again, as in Fig. 2, the rounded rectangles denote the reservoirs, which are here labeled with the respective transporter molecule. The abbreviations are the same as used throughout the text and in Fig. 2.
A slightly faster turnover of the RC was determined by Barz et al. (30): during a train of short saturating light pulses spaced 20-ms apart, ∼80% of dark-adapted, wild-type RCs underwent a full QH2-to-Q exchange cycle. The main difference between these experiments and our estimate, based on the considerations of Milano et al. (29), is that we are interested in the case of continuous illumination. Then, as will be explained below, the quinones are mostly reduced and there are only a few Qs available to replace the QH2s in the RCs. The number of Qs is much higher in a dark adapted cell which reduces the time needed for rebinding to the Qb site of the RC.
As each quinol carries two electrons and the RC has an efficiency of close to 1 (8), the RC can process up to ∼40 photons per second captured by the LHCs. With the photon capture rates RDimer and RLH2 calculated above, one RC can handle the one LH1 surrounding it plus an additional three LH2s. This ratio is compatible with a recent AFM observation of native chromatophore membranes of Rb. sphaeroides (16). We will therefore proceed with a typical unit of one RC directly surrounded by one LH1 and connected to another three LH2s which load the RC with a rate of excitons of RRC = RDimer + 3RLH2 = 44 s−1. The energy from these captured photons is then transferred onto 22 QH2 molecules per second. These considerations, i.e., the typical photon flux, the capture cross section of the LHCs and the rate-limiting processes in the RCs, show that the RCs work close to their maximal turnover.
For higher light intensities, the ratio of LH2s per LH1/RC should decrease to avoid overloading of the RC. Actually, variations in both of the stoichiometries between LH1s and LH2s and of the absolute absorption cross section have been observed for bacteria grown in low- or high-light conditions (31,32). Alternatively, the light intensity at which the surrounding LH1 alone would be able to load the RC can be estimated as ∼50 W/m2. This corresponds to 1/20th of the intensity of the sunlight at noon on a sunny summer day (26).
Pumping protons: the bc1 complex
The energy of the photons, which was loaded onto the quinol molecules at the RCs as two electron-proton pairs per quinol, is used in the bc1 complex according to the so-called Q-cycle (22): the quinol docks to the Qo site of the bc1 and the protons are released into the interior of the vesicle; one of the electrons is passed directly onto a waiting c2, while the other, via the Qi site of the bc1, is passed onto another quinone, together with another proton from the outside of the vesicle. The overall balance is that for each quinol starting from the RC, four protons are pumped into the vesicle against the proton gradient. For each quinol, two c2s are required to return the electrons to the RC.
Although the duration of the processes in the bc1 complex is not fully known yet, an estimate sufficient for our purpose can be determined from the measured overall enzymatic activity of the bc1s 2.5 μMol c2 are reduced per minute by 1 nMol of bc1 (33), i.e., 21 c2s per second by every single bc1. In vivo, the bc1 always occurs as a homodimer. Correspondingly, at each bc1 dimer, 42 quinol molecules coming from the RC induce the release of 168 protons/s into the vesicle. This measured rate was determined without a proton gradient, whereas in the vesicle the bc1s have to pump protons against such a gradient. Certainly, there should be some maximal proton gradient that the bc1s can cope with and the activity of the bc1s will slow down with increasing ΔpH across the membrane. However, this dependence has not been measured so far.
Using the protons: the ATP synthase
The last stage of the photosynthetic chain is the F0F1–ATP synthase (ATPase), which uses the proton gradient built up by the bc1s to synthesize ATP from ADP and inorganic phosphate. Ten to fourteen protons are required to drive one rotation during which three ATPs are synthesized, i.e., on average four protons per ATP molecule (34). The throughput of the ATPase has a sigmoidal characteristic. It increases exponentially with small pH gradients over the membrane, but is then limited to a maximal value for transmembrane voltages of >200 mV. This maximal conversion rate was determined as ∼400 ATP per second for ATPase from spinach chloroplasts and as ∼100 ATP per second for bacterial ATPase from Escherichia coli (35,36). Due to the lack of data for ATPase from Rb. sphaeroides, we assume the same maximal throughput of ∼100 ATP per second as the bacterial ATPase from E. coli, requiring 400 protons per second being pumped into the vesicle.
CONSTRAINTS FOR THE COMBINED PHOTOSYNTHETIC APPARATUS
After having collected the necessary data for each of the proteins in the previous chapter we can now link the segments of the conversion chain. To arrive at a consistent picture, several constraints have to be satisfied. When the vesicle operates at steady state, i.e., under constant illumination, the output of one stage is the input of the next stage. These balance constraints together with the rates of the individual rates fix the relative stoichiometries of the proteins and carriers. The absolute stoichiometries are then obtained by combining these values with the limited space available on the vesicle surface, which limits the total number of proteins but not their stoichiometries. Obviously, the model vesicle should also obey all experimental observations, as long as they are not explicitly contradicting each other. Fortunately, we did not meet such conflicts during this work. In the case of different possible solutions to the above constraints (see below), we suggest those solutions to be favored that adhere to the general principles of efficiency and stability. The composition of the model vesicle developed in this subsection is summarized in Table 1.
TABLE 1.
Overview over the collected dynamical data of the proteins and their stoichiometries
| Protein | Throughput per protein [1/s] | H+ equivalentsper protein [1/s] | Total number per avg. vesicle of 45-nm diameter | Rate determined from: |
|---|---|---|---|---|
| LH2 | 10 γ | 20 | 60 | Absorption spectra + |
| LH1 dimer | 2 × 14γ | 56 | 10 | + light intensity of 18 W/m2 |
| RC | 22 QH2 | 88 | 20 | QH2 (un)binding |
| bc1 dimer | ≤2 × 42 c2 | 168 | 3 (…10) | Measured activity at ΔpH = 0 |
| ATPase | ≤100 ATP | 400 | 1 | Measured throughput |
| Cytochrome c2 | 80 e− | 160 | 20 | (Un)binding at the bc1 |
| Ubiquinone | 10 × 2(e−H+) | 40 | 100 | (Un)binding at the RC and the bc1 |
The proton equivalents are calculated according to the reactions 2γ ⇒ 1 QH2 at the RC, 1 QH2 ⇒ 4H+ at the bc1 and 4H+ ⇒ 1 ATP at the ATPase.
In applying the constraints we again start from the LHCs, which are the largest objects on the vesicle and cover most of its surface. One LH1/RC dimer occupies an area of 234 nm2 (17); hexagonally positioned circular LH1 complexes need an area of 146 nm2 each. An LH2 monomer, with eight instead of 16 subunits, consequently should cover only one-quarter of this area, 37 nm2. One unit of one LH1/RC dimer plus six LH2s—three per RC (see above)—occupies an area of 456 nm2. A small vesicle of 30-nm diameter can accommodate four of these units, which still leaves some space for the bc1s and the ATPase. A large vesicle of 60-nm diameter provides space for ∼20 of these RC/LH1/LH2 units, i.e., for 40 single RC/LH1s and 120 LH2 rings. A midsize vesicle with an outer diameter of 45 nm is just big enough for 11 of these units. When leaving some space for bc1s and an ATPase it can accommodate 20 RCs, which can supply up to ∼440 quinols per second at the chosen light intensity of 18 W/m2.
Because the position and number of proteins of the next step of the conversion chain, the bc1 complex, is unclear from the available experimental observations (see (13)), we continue at the last stage of the photosynthetic chain, the ATPase.
The previous subsection mentioned the experiments concerning the characteristic of the ATPase. Their number, on the other hand, was determined by Feniouk et al. (21) as one ATPase per vesicle on average. To keep this single ATPase running most efficiently, ∼400 protons per second have to be pumped into the vesicle (see Using the Protons: the ATP Synthase). Since the ATPase's throughput cannot increase beyond this value even for dangerously high proton gradients, however, the supply of protons should not exceed this maximal turnover—or some kind of safety-valve would have to discard the excess protons without making any use of them.
Now, knowing the total output of the 20 RCs of an average-size vesicle—440 quinols per second, i.e., the equivalent of 1760 protons—and the input necessary to drive the ATPase most efficiently—400 protons per second—we come back to the bc1 complex. Obviously, the numbers do not match by a factor of ∼4. The question is whether the number of bc1s matches the larger output of the RCs, or the smaller input of the ATPase.
The measured activity per bc1 dimer (up to 168 protons per second) is approximately twice the output of one RC (22 quinols per second for the assumed typical illumination). If the total number of bc1s were related to the output of the RCs, an average-sized vesicle would consequently contain ∼10 bc1 dimers, the same number as LH1/RC dimers. This is more than the ratio of one bc1 per two RCs measured earlier (37). However, to keep the one ATPase per vesicle running, only 400 protons per second are required on average, which can already be provided by five bc1 monomers. Consequently, at the typical illumination, only five RCs of 20 are required to keep the ATPase running at full speed.
Because the ATPase has a fixed maximal turnover, determined by the association and dissociation times of the reactants, a stable solution can only be obtained by linking the number of bc1s to the ATPase and, consequently, assigning the bc1s as the kinetic bottleneck for the whole conversion chain. This prevents the interior of the chromatophore vesicles from being filled with too many protons, i.e., that it becomes too acidic, finally putting even the whole vesicle at risk (see, however, Possible Control Mechanisms). On the other hand, an electronic excitation decaying in the RC, because there are not enough bc1s to keep up with the photon capture rate, does not cause any harm to the vesicle (8). We therefore suggest, based on the rates of the ATPase and the bc1s, that a chromatophore vesicle contains only two or three bc1 dimers.
When determining the capture rate of the LHCs in Capturing the Photons: the Light-Harvesting Complexes, a light intensity of 18 W/m2 had been assumed leading to the above mismatch between the output of the RCs and the input of the ATPase. Obviously, this mismatch decreases with decreasing light intensity. Already at a reduced light intensity of 4.5 W/m2, the photon capture rate and the maximal throughput of the ATPase would match. The bacteria can consequently grow well under reduced light intensity, too, a situation that is encountered by them at least twice a day during twilight. This consideration favors a setup where the antenna part, i.e., the number of LHCs and RCs, of the photosynthetic chain is oversized compared to the subsequent steps, which are then less affected by changes of the light intensity. However, even so, it remains unclear why there are so many LH1/RC dimers. The same total absorption cross section could be accomplished by fewer LH1/RCs and more LH2s.
Our cautious estimate of only two or three bc1 dimers per vesicle is partly driven by the lack of detailed knowledge about the bc1 characteristic, i.e., whether and how the above cited enzymatic activity changes with respect to a proton gradient. So far we assumed the simplest case, where the activity is independent of ΔpH. If, however, the activity decreases with increasing ΔpH, a higher number of bc1s may be accommodated, bringing their number closer to the ratio of one bc1 for every two RCs determined earlier. Then the vesicle would contain some five bc1 dimers, which together cover approximately the same area as a single LH1/RC dimer.
TRANSPORT PROCESSES AND RESERVOIRS
Apart from the conversion rates of the transmembrane proteins of the photosynthetic chain, which are limited by internal conformational changes (e.g., movement of the Rieske protein of the bc1 complex) or transfer rates, the stationary overall rate could be diffusion-limited, too. Therefore, we now look at the connectors between the transmembrane proteins, the diffusing transport molecules ubiquinone and Cytochrome c2, and discuss whether their transport capacities impose any constraints on the locations of the bc1 complexes.
Diffusion of the electron carriers Cytochrome c2 and ubiquinone
The electrons cycle between the RC and the bc1 via two carriers, the water-soluble Cytochrome c2 and the membrane-bound quinone (Q) (see Figs. 1 and 2). These carriers move diffusively between their respective docking sites on the proteins. Their overall transport capacity is consequently determined not only by the binding dynamics at the docking sites but also by their numbers and the times they need to diffuse from the RC to the bc1 and back again.
The c2 molecules, which each transfer one electron from the bc1 complex to the RC, are confined to the inside of the vesicle. They have a diameter of 3.3 nm, i.e., ∼1/10th of the inner vesicle diameter. Even without knowing exactly how long the c2s take to bind to or unbind from both the RC and the bc1 it is possible to derive upper limits for the overall process from the turnovers of the proteins explained in the previous section. The enzymatic activity of the bc1 of 42 c2s per second means that, on average, one c2 is loaded every 23 ms. During this time the c2 binds to the bc1, takes up an electron, and dissociates again, making way for the next c2. Actually, the bc1 occurs as a dimer where a c2 bound to one of the monomers blocks the binding site of the other monomer (38). Therefore, a c2 can spend, at most, 11.5 ms at the bc1. The time that the c2 spends at the RC was determined as 1.25 ms. Consequently, the c2 spends some 13 ms per roundtrip associated with one of the membrane proteins.
The time spent with free diffusion can be estimated by the following argument: in the most extreme case, i.e., when the bc1 and the RC are located as far apart as possible, the c2 has to diffuse diametrically through the vesicle along the inner diameter and back again, before it can pick up the next electron. The distance to be covered in an average-size vesicle is therefore, at most, Lc2 ≈ 37 nm. With the (free) diffusion coefficient of Dc2 = 1.5 × 102 nm2/μs (39), this corresponds to a diffusion time of 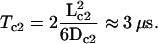 This is ∼4 orders of magnitude shorter than the times spent at the RC and bc1. Although this formula describes free unbounded three-dimensional diffusion, it gives a reasonable estimate for the particular confined geometry of when the c2 will have explored the whole inside of the vesicle and found a corresponding reaction partner.
This is ∼4 orders of magnitude shorter than the times spent at the RC and bc1. Although this formula describes free unbounded three-dimensional diffusion, it gives a reasonable estimate for the particular confined geometry of when the c2 will have explored the whole inside of the vesicle and found a corresponding reaction partner.
Summing up all contributions we find that one round-trip of the c2 from the RC to the bc1 and back amounts to ∼13 ms, or, correspondingly, that one c2 can shuttle at least 80 electrons per second. For every electron taken up at the bc1, two protons are released into the vesicle, so this rate is equivalent to ∼160 protons pumped into the vesicle. As one ATPase has a maximal throughput of some 400 protons per second, three c2s per vesicle would be more than enough to shuttle all electrons necessary for one ATPase. Now, in a real vesicle there is approximately one c2 per RC (40), i.e., by a factor-of-10 more than what seems necessary. Consequently, the diffusion and also the (un)binding of the c2 are not limiting the overall performance of the photosynthetic chain. The main contribution to the round-trip time of the c2 is due to the association with the bc1. As there are far more c2s available than necessary and, furthermore, as the c2s seem to spend much less time at one of the many RCs than at the few bc1s, there should nearly always be an oxidized c2 waiting at one of the bc1 dimer's binding sites to receive the electron from an arriving quinol. Therefore, the c2 pool of the vesicle is expected to be mainly oxidized under steady-state conditions.
A similar conclusion—that transport is not limiting the overall rate—is reached for the other electron carrier, the membrane-bound ubiquinone, as well. For the quinol an association time of 46 ms is obtained at the bc1, twice the time than for the c2, as for every QH2 arriving, two c2 have to take up the electrons. For our treatment of the QH2 binding the dimeric nature of the bc1 is not important. Every single Q actually has only one-half of that timespan available for unloading its electron proton freight: one of the electrons is not returned directly to the RC but stored away temporarily to a quinone via the Qi site. This storage-quinone has to unload its load, too: for every QH2 starting at the RC, two QH2 arrive at the bc1 Qo site. However, for our purpose it makes no difference whether the Qo site of the bc1 is occupied half of the time by QH2s coming from the bc1 or whether the binding of the QH2s from the RC takes twice as long. Above, we estimated that the Q-binding dynamics limits the overall throughput of the RCs to one Q every 50 ms. During every round-trip the quinone consequently spends up to ∼100 ms associated with the two transmembrane proteins.
For the diffusion time, we again consider the extreme case, where the RC and the bc1 are located on opposite sides of the vesicle. Then, for a 45-nm-diameter vesicle, a Q has to cover a distance of Lq = 65 nm from the RC to the bc1 and the same distance back again, diffusing along the circumference of the vesicle in the middle of the membrane. The time required for two-dimensional diffusion is 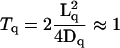 ms with the quinone's free diffusion coefficient of Dq = 2 nm2/μs (12). This time fits well to an observed lag of 1 ms between the onset of illumination and the onset of observable reduction of the bc1 in dark-adapted cultures of Rb. sphaeroides (30,41). Again, the diffusion time is much smaller than the total time necessary for binding, charge transfer, and unbinding. One complete roundtrip lasts for ∼100 ms. As explained above, each quinol is worth four pumped protons. Consequently, 10 quinols per vesicle are enough to supply the 400 protons per second that the ATPase can handle. This number is much smaller than the typical quinone pool of at least 10 Qs per RC, i.e., of >200 Qs per vesicle (40).
ms with the quinone's free diffusion coefficient of Dq = 2 nm2/μs (12). This time fits well to an observed lag of 1 ms between the onset of illumination and the onset of observable reduction of the bc1 in dark-adapted cultures of Rb. sphaeroides (30,41). Again, the diffusion time is much smaller than the total time necessary for binding, charge transfer, and unbinding. One complete roundtrip lasts for ∼100 ms. As explained above, each quinol is worth four pumped protons. Consequently, 10 quinols per vesicle are enough to supply the 400 protons per second that the ATPase can handle. This number is much smaller than the typical quinone pool of at least 10 Qs per RC, i.e., of >200 Qs per vesicle (40).
Whereas the quinone diffusion coefficient used above was measured in a lipid membrane without any transmembrane proteins, the membrane of the chromatophore vesicle is densely loaded with obstacles for quinone diffusion. Consequently, the effective diffusion coefficient through the gaps between the LHCs will be smaller. However, the population of the quinone pool would even allow for round-trip times of close to 1 s, i.e., under steady-state conditions the effective diffusion coefficient could be smaller by nearly three orders of magnitude before the quinone diffusion comes close to limiting the overall performance. Consequently, the electron carriers do not impose any constraint on the positions of the bc1 on the vesicle: the diffusion times—which depend on the protein placement—are only a small contribution to the transport capacities of the c2s and of the Qs.
On the numbers of the electron carriers
What is the reason for the 10-fold abundance of the electron carriers? The chromatophore vesicles are small and therefore only contain a handful of proteins in each of the subsequent conversion steps. Consequently, the output of each stage of the photosynthetic chain will show relatively large fluctuations. The bc1s and the ATPase in particular have a maximal turnover. Whereas their throughput will follow any smaller than average input with smaller turnovers, it cannot catch up during a temporarily increased output of the previous step. In the absence of buffers the average total throughput of the whole chain is limited to the maximal turnover of the slowest link minus the average width of the fluctuations of the previous step. With buffers present between the conversion steps, each protein is fed from an essentially static reservoir and can work at its maximal turnover all the time.
In the photosynthetic chain there are two buffers. One is the closed electron cycle between the RCs and the bc1s and the other one is implemented via the open proton cycle between the bc1s and the ATPase as the proton buffering capacity of the vesicle. The specific proton buffering capacity of chromatophores from Rb. capsulatus (42) implies that there are eight titratable groups of pKa = 8 per RC. Unfortunately it is not feasible to estimate the total number of protons inside a running vesicle from this value determined at a specific pH. One would need to know the full dependence of ΔpH versus the proton number for the whole range of pH gradients. Nevertheless, this specific proton buffering capacity allows one to estimate that roughly nine out of ten protons inside the vesicle are bound. Ignoring the buffering, 60 protons inside a midsize vesicle are enough to create a ΔpH of 3.4, corresponding to a transmembrane voltage of 200 mV. Correspondingly, at least 10 times as many protons can be expected inside a real working vesicle. This number is enough to gap statistical fluctuations in the proton supply from the bc1s to the ATPase of up to one-second length.
The other buffer contains ∼100 quinols, which supply the electrons to drive the bc1s. They can bridge a second's shortage from the RCs as well. But this number is misleading as it was pointed out above that the electron cycle is closed. Consequently, every electron released from a quinol at the bc1 has to be stored onto a c2, and for every QH2 two oxidized c2s are necessary. Thus, when the c2 pool is initially completely oxidized, the 20 c2s can take up electrons from no more than 10 QH2s. This is enough to supply the 2–3 bc1s for some 100 ms. So, if there are no photons during a 10th of a second, the bc1s and the ATPase still keep on running pumping protons. However, to achieve this buffering time the c2 pool has to be completely oxidized, which again requires that the bc1 complexes reduce the c2 more slowly than they can be oxidized at the RCs (see previous section). This is one reason why it could be advantageous for the bacteria to have more RCs than would be needed from the rate considerations: this ensures an oxidized c2 pool, which ensures a maximal buffering capacity against statistical fluctuations of the photon rate. This becomes even more important at low light conditions, when the lower photon rate implies larger relative fluctuations. These considerations may explain the larger-than-necessary c2 pool, but still, 90% of the quinone pool seems to be superfluous. One may suspect that their high number might be necessary due to a greatly reduced diffusion coefficient through the transmembrane proteins. This, however, would result in a much larger lag between the onset of illumination and bc1 activity than was observed.
We consequently suggest two reasons why the bc1 should be the bottleneck of the photosynthetic chain:
To prevent too-high proton concentrations inside the vesicle as explained in the section on the Constraints for the Combined Photosynthetic Apparatus.
To ensure that the electron carriers are in the correct oxidation state to act as a buffer against a fluctuating photon rate.
Positions of the bc1s
The evidence collected so far for the proteins on and inside of the vesicle, gives hardly any clue as to where on the vesicle the bc1 complexes should be placed. Essentially all of the dynamical data, including the diffusion of the electron carriers, is compatible with any setup ranging from super complexes, where the bc1s are attached to the LH1/RC units, to a strict separation between the bc1s on the one hand and LH1/RCs on the other hand. There is only one experimental observation giving an indirect indication that the bc1s are spatially separated from the RCs, which is the observed lag between the onset of light and the activity of the bc1 (see Diffusion of the Electron Carriers Cytochrome c2 and Ubiquinone). This lag can naturally be explained with a setup where the RCs and the bc1 are separated by a distance comparable to the vesicle diameter, i.e., as far away as possible on the vesicle. The accompanying article (13) on the putative vesicle geometry uses further arguments to place the bc1s at the vesicle pole near the ATPase.
Possible control mechanisms
As mentioned in the Introduction, the discussion of the photosynthetic machinery did not account for possible feedback mechanisms. Such mechanisms are often the basis for robustness of cellular behavior (23) or for adaptation (20). Unfortunately, there is hardly any experimental evidence on control mechanisms in chromatophore vesicles. Above, we noted the saturating production of the F0F1–ATP synthase at increasing membrane potential. To our knowledge, corresponding data is neither available for the pumping rate of the bc1 complex against an increasing intravesicular proton concentration nor for the quinone-loading capacity of RCs at different values of pH. Protonation reactions may also affect the conformations of individual LH rings or even the shape of the entire vesicle, thus affecting the diffusion behavior of quinone carriers that need to penetrate the LH rings. Much work, both experimental and theoretical, is needed to fully understand the behavior at a molecular scale of such a simple prototype of a cellular organelle.
SUMMARY AND CONCLUSIONS
This article presented the reconstruction of a putative molecular model of a chromatophore vesicle from Rb. sphaeroides. These vesicles are specialized to hold the simple photosynthetic apparatus of the bacteria. They appear an ideal model system for dynamic simulations on a systemic or molecular level, because they are small, naturally isolated from their cellular environment, and contain only six different proteins with known structure. This prompted us to compile the relevant kinetic and geometric information about all the parts of the photosynthetic apparatus from Rb. sphaeroides and to try to assemble from it a complete vesicle. To this effect, we used the notion of the photosynthetic apparatus as a conversion chain, which converts light energy via some intermediate steps into the chemically stored energy of the synthesized ATP molecules. This process view allowed us to relate the numbers of the proteins to their respective activities which, in turn, allows for an estimate of how many bc1 complexes per vesicle can be expected.
The dynamical data about the throughputs and characteristics of the proteins are not complete, but appear sufficient to assemble the chain. The numbers of light-harvesting complexes and reaction centers are easily determined from the available membrane area and the observed maximal speed of the reaction centers as ∼10 LH1/RC dimers for a midsize vesicle with a diameter of 45 nm. Feniouk et al. (21) showed experimentally that each vesicle contains a single ATPase. The number of bc1 complexes, however, is unclear. When their number is related to the output of the RCs we expect approximately one bc1 per RC, i.e., ∼10 bc1 dimers per vesicle. When their number is matched to the throughput of the ATPase, approximately five monomers are already enough to keep the vesicle running. Both these estimates are either a factor of two higher or lower, respectively, than the experimentally determined stoichiometry. The safest setup is to have as few bc1 complexes as possible, so that their total throughput is the bottleneck for the whole photosynthetic chain. Then the interior of the vesicle cannot become too acidic and possibly be damaged. This is also the overall most efficient setup where most of the vesicle surface is used for capturing photons, keeping ATP synthesis running even at low light conditions. Additionally, with only a few bc1s the pools of the electron carriers Cytochrome c2 and ubiquinone are mainly oxidized and reduced, respectively, i.e., in an oxidation state that allows for a maximal buffering against statistical fluctuations of the photon rate. By these arguments and also by considering the diffusive transport capacities of Cytochrome c2 and ubiquinone, the number of bc1 complexes can be estimated in good agreement with experimental evidence. Still, these considerations give us no clue as where to position them on our model vesicle because the diffusional processes involving Cytochrome c2s and quinones/quinols appear not rate-limiting.
In this work, we set up a steady-state picture of the photosynthetic apparatus. The model is currently used in our laboratory as a basis for stochastic dynamic simulations of a complete vesicle at the molecular level. First of all, such simulations will provide a consistency test of the experimental data available. For example, most of the rate constants used in our model were derived from quite different experiments, often performed under different conditions. These numbers should all be treated with appropriate error estimates. The whole-vesicle simulation would then test how well these values fit together and test their relations and dependencies. The simulation will also reveal which of the parameters are sensitive to changes of the external conditions, whether all important processes and parameters were included, and whether the geometries used are valid.
The systemic approach of this study is, of course, not limited to photosynthesis itself but could be applied to other functional units of the cell as well. With the field of molecular whole system simulations still being vastly unexplored, however, bacterial photosynthesis is an ideal benchmark system to learn how to set up such simulations.
Acknowledgments
We thank Armen Mulkidjanian and Carola Hunte for insightful discussions and the anonymous reviewers for helpful comments.
References
- 1.Ishii, N., M. Robert, Y. Nakayama, A. Kanai, and M. Tomita. 2004. Toward large-scale modeling of the microbial cell for computer simulation. J. Biotechnol. 113:281–294. [DOI] [PubMed] [Google Scholar]
- 2.Price, N. D., J. L. Reed, and B. O. Palsson. 2004. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat. Rev. Microbiol. 2:886–897. [DOI] [PubMed] [Google Scholar]
- 3.Papin, J., T. Hunter, B. O. Palsson, and S. Subramanian. 2005. Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. Mol. Cell Biol. 6:99–111. [DOI] [PubMed] [Google Scholar]
- 4.Allen, J. P., G. Feher, T. O. Yeates, H. Komiya, and D. C. Rees. 1987. Structure of the reaction center from Rhodobacter sphaeroides R-26: the protein subunits. Proc. Natl. Acad. Sci. USA. 84:6162–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermler, U., G. Fritzsch, S. K. Buchanan, and H. Michel. 1994. Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 Å resolution: cofactors and protein-cofactor interactions. Structure. 2:925–936. [DOI] [PubMed] [Google Scholar]
- 6.Hu, X., A. Damjanović, T. Ritz, and K. Schulten. 1998. Architecture and mechanism of the light harvesting apparatus of purple bacteria. Proc. Natl. Acad. Sci. USA. 95:5935–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koepke, J., X. Hu, C. Muenke, K. Schulten, and H. Michel. 1996. The crystal structure of the light-harvesting complex II (B800–850) from Rhodospirillum molischianum. Structure. 4:581–597. [DOI] [PubMed] [Google Scholar]
- 8.Hu, X., T. Ritz, A. Damjanović, F. Authenrieth, and K. Schulten. 2002. Photosynthetic apparatus of purple bacteria. Q. Rev. Biophys. 35:1–62. [DOI] [PubMed] [Google Scholar]
- 9.Abrahams, J. P., A. G. W. Leslie, R. Lutter, and J. E. Walker. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 370:621–628. [DOI] [PubMed] [Google Scholar]
- 10.Berry, E. A., L.-S. Huang, L. K. Saechao, N. G. Pon, M. Valkova-Valchanova, and F. Daldal. 2004. X-ray structure of Rhodobacter capsulatus Cytochrome bc1: comparison with its mitochondrial and chloroplast counterparts. Photosynth. Res. 81:251–275. [DOI] [PubMed] [Google Scholar]
- 11.Axelrod, H. L., E. C. Abresch, M. Y. Okamura, A. P. Yeh, D. C. Rees, and G. Feher. 2002. X-ray structure determination of the Cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides. J. Mol. Biol. 319:501–515. [DOI] [PubMed] [Google Scholar]
- 12.Marchal, D., W. Boireau, J. M. Laval, J. Moiroux, and C. Bourdillon. 1998. Electrochemical measurement of lateral diffusion coefficients of ubiquinones and plastoquinones of various isoprenoid chain lengths incorporated in model bilayers. Biophys. J. 74:1937–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geyer, T., and V. Helms. 2006. A spatial model of the chromatophore vesicles of Rhodobacter sphaeroides and the position of the Cytochrome bc1 complex. Biophys. J. 91:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jungas, C., J.-L. Ranck, J.-L. Rigaud, P. Joliot, and A. Verméglio. 1999. Supramolecular organization of the photosynthetic apparatus of Rhodobacter sphaeroides. EMBO J. 18:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheuring, S., F. Francia, J. Busselez, B. A. Melandri, J.-L. Rigaud, and D. Lévy. 2004. Structural role of PufX in the dimerization of the photosynthetic core complex of Rhodobacter sphaeroides. J. Biol. Chem. 279:3620–3626. [DOI] [PubMed] [Google Scholar]
- 16.Bahatyrova, S., R. N. Frese, C. A. Siebert, J. D. Olsen, K. O. van der Werf, R. van Grondelle, R. A. Niederman, P. A. Bullough, C. Otto, and C. N. Hunter. 2004. The native architecture of a photosynthetic membrane. Nature. 430:1058–1062. [DOI] [PubMed] [Google Scholar]
- 17.Siebert, C. A., P. Qian, D. Fotiadis, A. Engel, C. N. Hunter, and P. A. Bullough. 2004. Molecular architecture of photosynthetic membranes in Rhodobacter sphaeroides: the role of PufX. EMBO J. 23:690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joliot, P., A. Verméglio, and A. Joliot. 1989. Evidence for supercomplexes between reaction centers, Cytochrome c2 and Cytochrome bc1 complex in Rhodobacter sphaeroides whole cells. Biochim. Biophys. Acta. 975:336–345. [Google Scholar]
- 19.Fernàndez-Velasco, J. G., and A. R. Crofts. 1991. Complexes or super complexes: inhibitor titrations show that electron transfer in chromatophores from Rhodobacter sphaeroides involves a dimeric UQH2:Cytochrome c2 oxidoreductase, and is delocalized. Biochem. Soc. Trans. 19:588–593. [DOI] [PubMed] [Google Scholar]
- 20.Tyson, J. L., K. C. Chen, and B. Novak. 2003. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15:221–231. [DOI] [PubMed] [Google Scholar]
- 21.Feniouk, B. A., D. A. Cherepanov, N. E. Voskoboynikova, A. Y. Mulkidjanian, and W. Junge. 2002. Chromatophore vesicles of Rhodobacter capsulatus contain, on average, one F0F1-ATP synthase each. Biophys. J. 82:1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crofts, A. R. 2004. The Q-cycle—a personal perspective. Photosynth. Res. 80:223–243. [DOI] [PubMed] [Google Scholar]
- 23.Alon, U., M. G. Surette, N. Barkai, and S. Leibler. 1999. Robustness in bacterial chemotaxis. Nature. 397:168–171. [DOI] [PubMed] [Google Scholar]
- 24.Cogdell, R. J., N. W. Isaacs, A. A. Freer, T. D. Howard, A. T. Gardiner, S. M. Prince, and M. Z. Papiz. 2003. The structural basis of light-harvesting in purple bacteria. FEBS Lett. 555:35–39. [DOI] [PubMed] [Google Scholar]
- 25.Francke, C., and J. Amesz. 1995. The size of the photosynthetic unit in purple bacteria. Photosynth. Res. 46:347–352. [DOI] [PubMed] [Google Scholar]
- 26.Gerthsen, C., H. O. Kneser, and H. Vogel. 1986. Physiks, 15th Ed. Springer-Verlag, Berlin, Heidelberg, New York, Tokyo.
- 27.Gerencsér, L., G. Laczkó, and P. Maróti. 1999. Unbinding of oxidized Cytochrome c from photosynthetic reaction center of Rhodobacter sphaeroides is the bottleneck of fast turnover. Biochemistry. 38:16866–16875. [DOI] [PubMed] [Google Scholar]
- 28.Paddock, M. L., S. H. Rongey, E. C. Abresch, G. Feher, and M. Y. Okamura. 1988. Reaction centers from three herbicide resistant mutants of Rhodobacter sphaeroides 2.4.1: sequence analysis and preliminary characterization. Photosynth. Res. 17:75–96. [DOI] [PubMed] [Google Scholar]
- 29.Milano, F., A. Agostiano, F. Mavelli, and M. Trotta. 2003. Kinetics of the quinone binding reaction at the Qb site of reaction centers from the purple bacteria Rhodobacter sphaeroides reconstituted in liposomes. Eur. J. Biochem. 270:4595–4605. [DOI] [PubMed] [Google Scholar]
- 30.Barz, W. P., A. Verméglio, F. Francia, G. Venturoli, B. A. Melandri, and D. Oesterhelt. 1995. Role of PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 2. PufX Is required for efficient ubiquinone/ubiquinol exchange between the reaction center Qb site and the Cytochrome bc1 complex. Biochemistry. 34:15248–15258. [DOI] [PubMed] [Google Scholar]
- 31.Reidl, H., J. R. Golecki, and G. Drews. 1983. Energetic aspects of photophosphorylation capacity and reaction center content of Rhodopseudomonas capsulata, grown in a turbidostat under different irradiances. Biochim. Biophys. Acta. 725:455–463. [Google Scholar]
- 32.Scheuring, S., J.-L. Rigaud, and J. N. Sturgis. 2004. Variable LH2 stoichiometry and core clustering in native membranes of Rhodospirillum photometricum. EMBO J. 23:4127–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao, K., L. Yu, and C.-A. Yu. 2000. Confirmation of the involvement of protein domain movement during the catalytic cycle of the Cytochrome bc1 complex by the formation of an intersubunit disulfide bond between Cytochrome b and the iron-sulfur protein. J. Biol. Chem. 275:38597–38604. [DOI] [PubMed] [Google Scholar]
- 34.Capaldi, R. A., and R. Aggeler. 2002. Mechanism of the F0F1–type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27:154–160. [DOI] [PubMed] [Google Scholar]
- 35.Junesch, U., and P. Gräber. 1991. The rate of ATP-synthesis as a function of ΔpH and Δψ catalyzed by the active, reduced H+-ATPase from chloroplasts. FEBS Lett. 294:275–278. [DOI] [PubMed] [Google Scholar]
- 36.Turina, P., B. A. Melandri, and P. Gräber. 1991. ATP synthesis in chromatophores driven by artificially induced ion gradients. Eur. J. Biochem. 196:225–229. [DOI] [PubMed] [Google Scholar]
- 37.van den Berg, W. H., R. C. Prince, C. L. Bashford, K.-I. Takamiya, W. D. Bonner, Jr., and P. L. Dutton. 1979. Electron and proton transport in the ubiquinone Cytochrome b-c2 oxidoreductase of Rhodopseudomonas sphaeroides. Patterns of binding and inhibition by antimycin. J. Biol. Chem. 254:8594–8604. [PubMed] [Google Scholar]
- 38.Hunte, C., S. Solmaz, and C. Lange. 2002. Electron transfer between yeast Cytochrome bc1 complex and Cytochrome c: a structural analysis. Biochim. Biophys. Acta. 1555:21–28. [DOI] [PubMed] [Google Scholar]
- 39.Eltis, L. D., R. G. Herbert, P. D. Barker, A. G. Mauk, and S. H. Northrup. 1991. Reduction of horse heart ferricytochrome c by bovine liver ferrocytochrome b5. Experimental and theoretical analysis. Biochemistry. 30:3663–3674. [DOI] [PubMed] [Google Scholar]
- 40.García, A. F., G. Venturoli, N. Gad'on, J. G. Fernández-Velasco, B. A. Melandri, and G. Drews. 1987. The adaptation of the electron transfer chain of Rhodopseudomonas capsulata to different light intensities. Biochim. Biophys. Acta. 890:335–345. [Google Scholar]
- 41.Crofts, A. R., and C. A. Wraight. 1983. The electrochemical domain of photosynthesis. Biochim. Biophys. Acta Rev. Bioenerg. 726:149–185. [Google Scholar]
- 42.Feniouk, B. A., M. A. Kozlova, D. A. Knorre, D. A. Cherepanov, A. Y. Mulkidjanian, and W. Junge. 2004. The proton-driven rotor of ATP synthase: Ohmic conductance (10 fS), and absence of voltage gating. Biophys. J. 86:4094–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]