Abstract
The S-layer of Bacillus sphaericus strain JG-A12, isolated from a uranium-mining site, exhibits a high metal-binding capacity, indicating that it may provide a protective function by preventing the cellular uptake of heavy metals and radionuclides. This property has allowed the use of this and other S-layers as self-assembling organic templates for the synthesis of nanosized heavy metal cluster arrays. However, little is known about the molecular basis of the metal-protein interactions and their impact on secondary structure. We have studied the secondary structure, protein stability, and Pd(II) coordination in S-layers from the B. sphaericus strains JG-A12 and NCTC 9602 to elucidate the molecular basis of their biological function and of the metal nanocluster growth. Fourier transform infrared spectroscopy reveals similar secondary structures, containing ∼35% β-sheets and little helical structure. pH-induced infrared absorption changes of the side-chain carboxylates evidence a remarkably low pK < 3 in both strains and a structural stabilization when Pd(II) is bound. The COO−-stretching absorptions reveal a predominant Pd(II) coordination by chelation/bridging by Asp and Glu residues. This agrees with XANES and EXAFS data revealing oxygens as coordinating atoms to Pd(II). The additional participation of nitrogen is assigned to side chains rather than to the peptide backbone. The topology of nitrogen- and carboxyl-bearing side chains appears to mediate heavy metal binding to the large number of Asp and Glu in both S-layers at particularly low pH as an adaptation to the environment from which the strain JG-A12 has been isolated. These side chains are thus prime targets for the design of engineered S-layer-based nanoclusters.
INTRODUCTION
The regularly structured paracrystalline surface layers (S-layers) are one of the most common surface structures found in bacteria and archaea (1,2). Most of the S-layers are composed of protein monomers with the ability to assemble in two-dimensional arrays exhibiting either oblique, square, or hexagonal lattice symmetry or other regular structures (3,4). They possess regularly arranged pores of identical size that enable the exchange of ions and small molecules between the living cells and their environment. The uranium-mining waste pile isolate Bacillus sphaericus JG-A12 is enveloped by a S-layer with a square symmetry, which is composed of identical protein monomers. The proteins are capable of selective and reversible binding of large amounts of metals (5), thus functioning probably as a barrier for toxic heavy metal ions in the environment (6,7). This has raised interest in their biotechnological applications for bioremediation (8) and for the generation of metal nanoclusters (9,10) as has also been shown for other S-layers (1,11,12). Here, we have studied metal-protein interactions in S-layers to understand both the molecular basis of a biological protection mechanism and the implications for technological applications.
The development of cluster-assembled materials with discrete, size-selected nanoparticles is of particular interest to enable the fine-tuning of the properties of nanoparticles (12,13). The latter usually differ significantly from those of the bulk material from which they are formed (12), allowing the generation of new materials (3). A promising approach to produce such nanoparticles is the use of self-assembling organic templates which allow the synthesis of a wide range of inorganic nanocrystal lattices (3,14–17). Due to the crystalline arrangement of the S-layer, functional groups are found in well-defined positions and orientations in the protein (3). S-layers have been shown to function as templates in natural mineralization processes (18–20) and have been used for the synthesis of CdS (21), Au (3,10), Pt, and Pd cluster arrays (22). In recent experiments, the S-layer of the closely related strain B. sphaericus NCTC 9602 has been successfully used to produce Pd-nanoclusters from bound Pd(II)-complexes by electron irradiation (22). In contrast to the low numbers of highly specific binding sites found in proteins that are conformationally regulated by metal-protein interactions, ultraviolet/VIS (visible) spectroscopy on S-layers has demonstrated the binding of 200–300 Pd complexes per S-layer monomer (23). Despite the correspondingly low specificity of the individual binding sites, which are also capable of binding a variety of different metals, their large number is unique for S-layers. This property must thus be linked to conserved patterns of metal-interacting amino acids and structural features that govern the accessibility of these groups. However, no information about the mechanism of the initial Pd(II) complexation, the impact on protein structure, and the functional groups involved in Pd(II)-complexation has been obtained. As the sites of nanocluster growth are determined by the metal-binding sites in the protein, their characterization and identification is of prime interest for the determination of the topology of metal nucleation within the protein matrix, as well as for the specific engineering of metal-binding sites through site-directed mutagenesis. Due to the lack of a high resolution x-ray structure of S-layers, spectroscopy is currently the most powerful approach to characterize metal binding.
In this study, extended x-ray absorption fine structure (EXAFS), x-ray absorption near edge structure (XANES), and Fourier transform infrared (FTIR) spectroscopy were used to analyze the general features of the complexation of Pd(II) within the large number of binding sites of the native S-layer proteins of the strains B. sphaericus JG-A12 and NCTC 9602. Specifically, the predominant chemical elements involved in the complexation are identified and their location within side-chain or backbone groups is addressed. In addition, the secondary structure and pH-dependent structural transitions of these proteins were studied because the organisms are specifically adapted to a low pH environment. Experiments were designed to investigate the function of carboxylates in metal-protein interactions in these S-layers, which are rich in Asp and Glu residues. The data evidence an acidic average pKa < 3 of the titratable carboxylic acids. The latter are specifically involved in metal-protein interactions resulting in the stabilization of the secondary structure of both S-layer proteins. Additional coordination by nitrogen is likely to also originate in side-chain interactions rather than coordination to the peptide backbone. The data reveal a relation between metal binding and S-layer stability at low pH, indicative of an adaptation to the specific environment from which the strain JG-A12 has been isolated. These data identify prime targets for the engineering of metal-binding sites in S-layer proteins.
MATERIALS AND METHODS
Isolation of the S-layer protein
Both B. sphaericus strains, JG-A12 and NCTC 9602, were grown in nutrient broth, containing 3 g/L meat extract (Merck KGaA, Darmstadt, Germany) and 5 g/L Bacto Peptone (Becton, Dickinson, Franklin Lakes, NJ). Cells were harvested at the late exponential growth phase by centrifugation at 10,000 × g for 20 min. Cell biomass was washed once, centrifuged, and resuspended in a buffer solution of 50 mM TRIS-HCl, 1 mM MgCl2 × 6 H2O, 3 mM NaN3, pH 7.5 (standard buffer). For the removal of the bacterial flagella from cells of B. sphaericus JG-A12, the cells were treated with a rotating-blade bender IKA T8 (IKA Labortechnik, Stauffen, Germany) at maximum speed for 10 min at 4°C. Flagella-free cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C. The biomass of both B. sphaericus strains was resuspended 1:1 in standard buffer, and a few crystals of DNase II and RNase were added. The cells were disintegrated using a high-shear fluid processor (M-110S Microfluidizer processor, Microfluidics, Newton, MA) at 4°C, a pressure of 960 bar, and three passes, which results in a desintegration rate of >99%. After washing the cell wall fragments two times in standard buffer, the plasma membranes were solubilized in 1% Triton X-100 in standard buffer for 10 min at room temperature. Remaining cell wall fragments were washed twice. Peptidoglycan was lysed by incubating the samples in a standard buffer containing 0.2 mg/ml lysozyme for 6 h at 30°C. The S-layer fraction was washed several times, resuspended in standard buffer, and stored at 4°C.
For further purification of the S-layer, protein suspensions were mixed with 6 M guanidine hydrochloride in 50 mM Tris, pH 7.2, until the solutions became clear. After stirring the solutions for 2 h at room temperature, nonprotein components were precipitated by centrifugation at 12,400 × g for 60 min at 4°C. The supernatants were dialyzed two times against 2 l 10 mM CaCl2, 3 mM NaN3 for 24 h at 4°C using dialysis tubings with a molecular weight cutoff of 50,000. Reassembled S-layers were harvested by centrifugation at 12,400 × g for 60 min at 4°C, resuspended in 10 mM CaCl2, 3 mM NaN3, and stored at 4°C until use. Protein concentrations were determined using the Protein Assay Kit (Sigma-Aldrich Chemie GmbH, Deisenhofen, Germany) according to the manufacturer's instructions.
Metalization of the S-layer
For sorption of Pd(II), the protein was dialyzed against H2O and 10 mg of it were incubated in 100 ml of a solution of 2 mM Na2PdCl4 (pH = 3.1), which was prepared 24 h before the use and kept in the dark. After 3 h of incubation at room temperature under shaking in the darkness, the sample was centrifuged and the pellet was resuspended in H2O. Residual salts were removed by dialysis of the metalized proteins against H2O. For EXAFS analysis, the protein samples were dried in a vacuum oven (48 h, 80°C) and pulverized. Previous controls done in our laboratory (C. Hennig, J. Raff, T. Reich, and S. Selenska-Pobell, unpublished data) revealed that the EXAFS spectra of Pd(II)-bound S-layers dried at 80°C and 30°C are almost superimposable, showing that the Pd(II) coordination is very little affected by the dehydration temperature. This is in agreement with the high stability of S-layer secondary structures (4).
X-ray absorption spectroscopy
Palladium K-edge x-ray absorption spectra were collected at the Rossendorf Beamline located at the European Synchrotron Radiation Facility (ESRF), Grenoble, France (24), using a Si(111) double-crystal monochromator and Si-coated mirrors for focusing and rejection of higher harmonics. Samples were cooled to 30 K in a closed-cycle He cryostat, and data were collected either in transmission mode or in fluorescence mode using an Ar-flushed ionization chamber or a 13-element Ge detector, respectively. The energy was calibrated by measuring the Pd K-edge transmission spectrum of a palladium foil and defining the first inflection point as 24350 eV. The Pd-loaded samples were measured as dry samples. The EXAFS oscillations were isolated from the raw, averaged data by removal of the preedge background, approximated by a first-order polynomial, followed by μ0-removal via spline fitting techniques and normalization using a Victoreen function. Dead-time correction was applied to fluorescence data. The theoretical scattering phase and amplitude functions used in data analysis were calculated using FEFF8 (25). The amplitude reduction factor was held constant at 1.0 for the FEFF8 calculation and EXAFS fits. The shift in threshold energy, ΔE0, was varied as a global parameter in the fits. A metallic Pd foil; powder form of PdO, PdCl2, and [Pd(NH3)4]Cl2; and a solution of 2 mM Na2PdCl4 (pH 3.1) were used as reference compounds. For the Pd-loaded S-layer protein spectrum, data for phase shifts and backscattering-amplitudes were obtained from the PdO reference compound (Pd-O and Pd-Pd scattering).
Proteolysis of S-layer proteins
The endoproteinase Glu-C recognizes and cleaves specifically -Glu-P1′- and -Asp-P1′- bonds at pH = 7.8 in phosphate buffer. For digestion with the endoproteinase Glu-C (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany), 200 μg of the purified S-layer proteins and of the metalized proteins were resuspended in 32 μl of a 50 mM KH2PO4/Na2HPO4 buffer (pH = 7.8). After addition of 2 μg of the endoproteinase Glu-C, the solution was incubated for 24 h at 37°C. The resulting fragments were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis with a 4% stacking gel and a 10% or 12.5% separation gel as described (26) using the Mini-PROTEAN II (Bio-Rad GmbH, Munich, Germany). After electrophoresis, gels were stained with Coomassie brilliant blue R 250 or with Sypro-Ruby (Bio-Rad) and visualized with a VersaDoc Imaging System (Bio-Rad). The densitometric analysis of the gels was carried out with the software RFLPscan (Scanalytics, BD Biosciences, Rockville, MD).
FTIR
Spectra were recorded with a Vector22 equipped with a dialysis-coupled internal reflectance unit Bio-ATR-II (Bruker Optics GmbH, Ettlingen, Germany) and a liquid nitrogen-cooled mercury-cadmium-telluride detector. A total of 50 μl of suspensions of S-layers at a concentration of 40–50 mg/ml were spread on the ATR-crystal (Si). A total of 10 mM Tris-Cl buffer, pH 7, was layered on top of the sample, which was then sealed with a dialysis membrane (10 kD MW cutoff). Lowering of the pH was carried out by extensive buffer exchange through tubings connected to the dialysis volume and a peristaltic pump. Interferograms were recorded at 2 cm−1 resolution and coadded for the calculation of absorption spectra. Reference spectra were recorded for each measured pH with the identical buffer solutions, and water absorption was corrected in the final spectra by minimizing the broad absorption band of water at 2000 cm−1. For experiments in D2O, 2 ml of S-layer suspension were spun 15 min at 12,000 × g (MiniSpin table centrifuge, Eppendorf, Westbury, NY) and the pellet resuspended in D2O sample. The procedure was repeated three times with 1 h waiting between spins. The pH was adjusted using microliter amounts of NaOD and DCl in otherwise unbuffered solution. The sample was kept overnight in D2O at room temperature. Spectra were recorded the next day immediately after a final D2O exchange and transfer of the pellet to the sealed ATR-cell. All measurements were done at room temperature. The amide I and II absorption was fitted by Gaussian/Lorentzian lines using the spectrometer software OPUS. Positions of the main components in the amide I range around 1630–1640, 1650–1660, and 1680–1690 cm−1 and in the amide II band were determined from the negative peaks in the second derivative spectrum and fixed in an initial fit of band shapes and widths. In the final fit, all parameters were free to vary.
RESULTS
Secondary structure and stability of native S-layer proteins
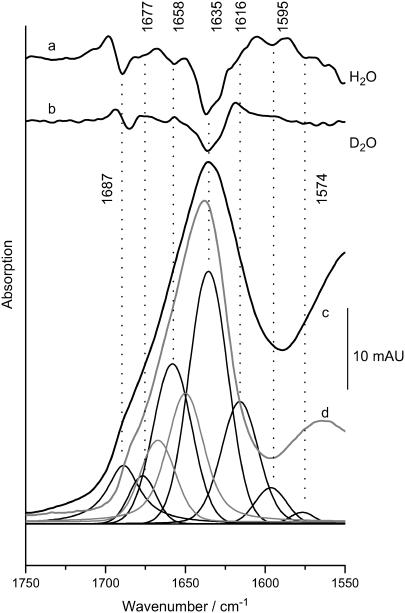
We have studied by FTIR the secondary structure of S-layers of B. sphaericus strains NCTC 9602 and JG-A12 as a function of pH and Pd(II) binding. The vibrational modes of the peptide backbone give rise to amide I (peptide C=O stretching) and amide II (peptide C-N stretching coupled to NH bending) absorptions in the 1620–1690 and 1540–1560 cm−1 range, respectively. The amide I absorption frequency depends on protein secondary structure allowing us to assess relative amounts of secondary structure based on the intensity of corresponding spectral components (27–29). Here, we have used attenuated total reflectance (ATR) FTIR-spectroscopy (30,31) coupled to dialysis (32), which allows us to record infrared (IR)-spectra in bulk aqueous phase at varying pH. Fig. 1 c shows the absorption spectrum of strain JG-A12 in the amide I and II range at pH 6. The amide I peak is found at 1636 cm−1. Individual spectral components centered at the negative peaks in the second derivative spectrum (Fig. 1 a) were fitted (see Materials and Methods). The main component at 1635 cm−1 is indicative of β-structure as well as the band at 1685 cm−1. The integral intensity of these bands suggests that ∼35% of the peptide backbone forms β-sheets. The frequency of the second-largest component at 1658 cm−1 lies in a region of strong overlap of α-helical structure (1650–1656 cm−1) disordered peptide backbones (1645–1657 cm−1) and turns (typically absorbing above 1660 cm−1). It is thus likely that several structures contribute to the 1658-cm−1 -absorption. The lack of the 1658-cm−1 shoulder in the second derivative spectrum when measured in D2O (Fig. 1 b) particularly argues for a large contribution of disordered structure, typically exhibiting a ∼10-cm−1 downshift in D2O, whereas the amide I modes of helices and turns respond less to the isotope exchange (33). The smallest component at 1677 cm−1 can be assigned to β-turns (5% integral intensity) typically absorbing above 1660 cm−1. Table 1 summarizes the amide I band assignments for both strains. The similar spectral properties of both strains imply very similar secondary structure compositions. The IR-based assessment of β-sheet is in agreement with secondary structure predictions, whereas the amount of α-helices is <20%, in contrast to up to 50% predicted by the algorithms listed in Table 1. The low helical amount is confirmed by the analysis of the amide I′ mode in D2O (Fig. 1 d) where the expected downshift of the high frequency band of the β-sheet to 1682 cm−1 is observed. The band analysis resolves a 1649 cm−1-absorbing component, i.e., at lower frequency than the 1651–1653 cm−1 range typical of helices. This indicates that the 1649 cm−1 band accounts mainly for absorption by random structure and cannot be fully assigned to helices. There is also no indication of a component at 1630 cm−1, a frequency typical of strongly solvent-interacting α-helices. In the region of turns, however, the D2O measurements provide a more accurate description, because overlapping contributions from random structures (mainly accounted for by the 1658 cm−1 in H2O) are downshifted and allow the appearance of a clear 1667-cm−1 component in the fit. This band is typical of the absorption by turns and probably describes the high frequency part of the amide I′ band more realistically than the strong overlapped fitted bands at 1677 and 1658 cm−1 in H2O. Therefore, the contribution of turns shown in Table 1 is taken from the D2O experiment, whereas the amount of β-sheet determined from the H2O experiments is fully reproduced with the spectra obtained in D2O.
FIGURE 1.
IR absorption in the amide spectral range of a suspension of S-layer from strain JG-A12. The structure-sensitive amide I mode (c) was approximated by fitted bands whose frequencies where determined from negative peaks in the second derivative of the spectrum measured in H2O (a). Original data and the sum of the fitted components are superimposed. The integral intensity of the bands above 1600 cm−1 was used to estimate the type and amount of secondary structure (summarized in Table 1). Disappearance of the shoulder at 1658 cm−1 in D2O (b) argues for a large contribution from turns and unordered structure rather than α-helices (52). (d) Absorption in D2O (shaded spectrum), where a better distinction between turns and random structure is achieved as evident from the larger separation of the two fitted bands (shaded) at 1667 and 1649 cm−1, respectively (for clarity only these two peaks in the amide I′ range are shown. Spectra are scaled to the 1635 cm−1 peak, which is barely affected in D2O as is typical of β-sheet absorption).
TABLE 1.
Amide and carboxylate IR-absorptions for S-layers of the B. sphaericus strains JG-A12 and NCTC 9602
| Amide I components (cm−1) | Integral band intensity (%)
|
|
|---|---|---|
| JG-A12 | NCTC 9602 | |
| 1687 | 11 | 6 |
| 1676 | 5 | 5 |
| 1658 | 25 | 25 |
| 1635 | 39 | 48 |
| 1616 | 20 | 16 |
| Structure assignment | % | % |
| Helix | <18 (10–45) | <18 (9–44) |
| β-Strand | 33 (25–50) | 36 (24–52) |
| Turns | 10–20 (0–40) | 10–20 (0–40) |
| Random | >30 (0–40) | >30 (0–40) |
| Carboxylate vibrations | Native | Pd(II)-bound | Native | Pd(II)-bound |
| s COO− (cm−1) | 1400 | 1386−1410 | 1400 | 1386−1410 |
| as COO− (cm−1) | 1570 | 1560 | 1573 | 1572 |
| C=O stretch | 1720 | 1720 | 1720 | 1720 |
Secondary structure estimation is based on published assignments (27,29,53) as reviewed in Barth and Zscherp (52). The range of results of secondary structure predictions from primary structure-based algorithms (54–56) is shown in brackets. Integral intensities were transformed into relative amounts of secondary structure using absorptivity ratios of 0.6:1:1.5 for random coils and turns: α-helices, β-sheet (57).
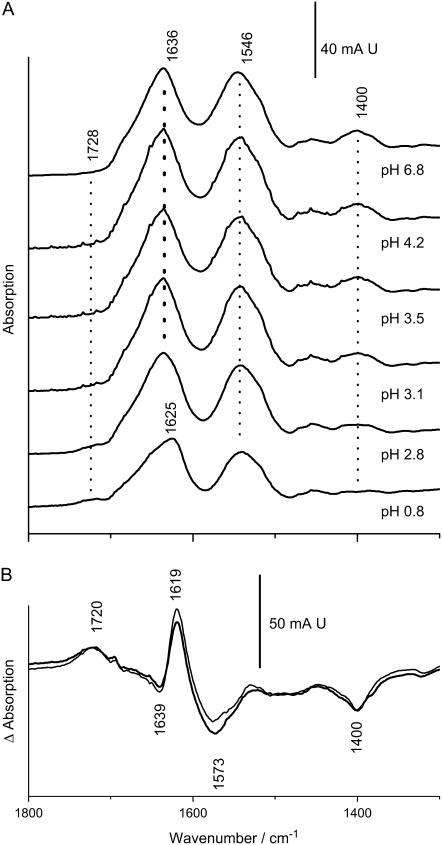
The secondary structure of both strains shows little dependence on pH down to pH 2.5 as seen for JG-A12 in Fig. 2 A. Below this pH, the protein denatures and the amide I band broadens and shifts irreversibly to 1625 cm−1. In addition, acidification reduces the IR absorption of the symmetric and antisymmetric COO−-stretching modes at 1400 and in the 1560–1580 cm−1 range, respectively, because these groups become protonated. Correspondingly, the absorption by the C=O-stretching mode of protonated carboxylic acids at ∼1720 cm−1 increases with acidification. These changes can be visualized by the subtraction of a spectrum recorded at neutral pH from that recorded at pH 0.8 (Fig. 2 B). The pH dependence of the 1400 cm−1 band reveals an unusual low pK of 2.8–3.0 at which half-maximal reduction of the COO− absorption occurs. Both strains exhibit virtually undistinguishable responses to pH as is evident from the superposition of their pH-induced difference spectra.
FIGURE 2.
(A) pH-dependent IR-absorption of the S-layer of strain JG-A12. A suspension of S-layers (20–30 μL) was measured in attenuated total reflection on a Si-crystal (Bio-ATR-II) and was successively exposed to solutions of different pH (10 mM sodium phosphate) by dialysis. The protonation of carboxylic acids causes the reduction of the COO−-stretching modes, which have lost half of their initial intensity at pH 3 (symmetric COO− at 1400 cm−1). Irreversible denaturation is evidenced by the downshift of the amide I absorption at pH 0.7. (B) pH-induced absorption changes (spectra at pH 0.8 minus spectra at pH 7) for S-layers of JG-A12 (thick line) and NCTC 9602 (thin line) visualize both the carboxylate and amide I absorption change in the pH 7 to pH 0.7 interval.
Secondary structure and stability of Pd(II)-bound S-layers
Fig. 3 A shows the pH-dependent IR-absorption of Pd(II)-bound S-layers from strain JG-A12. At neutral pH, the amide I mode is downshifted by 4 cm−1 versus the native protein. The amide II peak appears broadened and upshifted in frequency. However, acidification shows that the actual amide II band is superimposed with a broad absorption at 1562 cm−1 (compare Figs. 2 A and 3 A). Based on its pH sensitivity (as is obvious in the difference spectrum in Fig. 3 B), we assign the large integral intensity at ∼1562 cm−1 to the antisymmetric COO−-stretching vibrations in the Pd(II)-bound S-layer. The center frequency of these vibrations is downshifted by 10 cm−1 versus the native S-layer of JG-A12. The unobscured amide II mode is seen at 1533 cm−1 in the acidified sample at pH 0.7 when the COO− groups are replaced by COOH. In addition, metal binding causes splitting of the symmetric COO−-stretching mode into two components at ∼1410 and 1386 cm−1. The metal-induced appearance of absorption of the symmetric COO− stretch above 1400 cm−1 has been also observed with Ca2+ binding to carboxylates in proteins (34) strongly suggesting that carboxylates coordinate the metal ion also in the S-layers. The metal-induced spectral features are identical in both B. sphaericus strains except for the Pd(II)-dependent downshift of the antisymmetric COO−-stretching vibration, which is not seen in the reference strain NCTC 9602. Remarkably, acidification below pH 1 of the metal-bound S-layers does not generate the 1625 cm−1 amide I absorption of the denatured state, revealing an increased resistance of the Pd(II)-complexed proteins to acidic pH.
FIGURE 3.
(A) pH-dependent IR-absorption of Pd(II)-bound S-layer of strain JG-A12. (B) pH-induced absorption changes in S-layers from strain JG-A12 (thick line) and NCTC 9602 (thin line) calculated as in Fig. 2 show the carboxylate absorption changes, whereas amide absorption changes are largely blocked. The asymmetric COO−-stretching modes are enhanced versus the native proteins and their average frequency is downshifted by 10 cm−1 in JG-A12, whereas it is not altered in NCTC 9602. Experimental conditions as in legend to Fig. 2.
Digestion with Glu-C
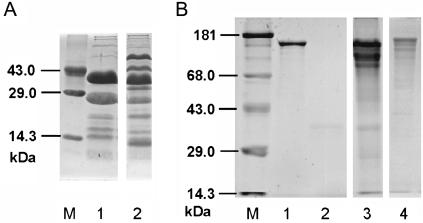
The FTIR results strongly indicate complexation of Pd(II) by Asp and Glu side chains and a concomitant stabilization of the secondary structure. The spectroscopically derived involvement of carboxylic acids in Pd(II) binding is strongly supported by the digestion of the S-layer proteins with the endoproteinase Glu-C. At pH = 7.8, the enzyme cleaves specifically -Glu-P1′- and -Asp-P1′- bonds (presumably located on the surface of the protein). Proteolytic digestion of the purified recrystallized S-layer protein of strains JG-A12 and NCTC 9602 with the Glu-C yields five and nine fragments, respectively (Fig. 4 A). The S-layer protein of JG-A12 (1207 aa) contains 63 Glu and 62 Asp residues, whereas that of NCTC 9602 (1197 aa) possesses 64 Glu and 77 Asp residues (35). In both cases the Glu and Asp residues are regularly distributed in the amino acid sequences. Thus most of the cleavage sites were buried inside the folded protein and the difference has to be explained by both deviations in the primary structure, especially in the C-terminal parts (35), and differences in the protein conformation. However, the densitometric analysis demonstrates that the endoproteinase only digests <5% of the Pd(II)-complexed S-layer protein of JG-A12 (Fig. 4 B, lane 3). Additionally occurring bands were the result of a slight fragmentation caused by the metalization procedure or the metalization itself (Fig. 4 B, lane 4). In summary, these results strongly support the suggested interaction of Pd(II) with carboxyl groups, which, thereby, become blocked for proteolytic attack by Glu-C.
FIGURE 4.
SDS-gelelectrophoresis. (A) The S-layer proteins of B. sphaericus JG-A12 (lane 1) and NCTC 9602 (lane 2) were digested with Glu-C and separated using a 12.5% gel. M, marker. (B) Native and Pd(II)-complexed S-layer proteins of B. sphaericus JG-A12 were digested with Glu-C and analyzed using a 10% gel. Lane 1: purified S-layer protein. Lane 2: Glu-C. Lane 3: purified S-layer protein with complexed Pd(II) after digestion with Glu-C. Lane 4: S-layer protein with complexed Pd(II); M, marker.
Pd(II) coordination studied by XANES spectroscopy
Pd(II)-protein interactions were further addressed by XANES spectroscopy. XANES provides information on the average oxidation state and local coordination environment of metals such as Pd (36). Small shifts (a few eV) in XANES absorption edge energies can occur when a metal changes its average oxidation state. Just above the absorption edge, light elements such as C, N, and O produce a strong signal (white line), rendering XANES particularly useful to detect Pd-O and Pd-N shells as their contribution to EXAFS amplitudes is very weak. We have based the distinction between Pd-O, Pd-N, Pd-Cl, or Pd-Pd bonds in metal-bound S-layers on the reference compounds PdO, [Pd(NH3)4]Cl2, PdCl2, Na2PdCl4, and Pd foil.
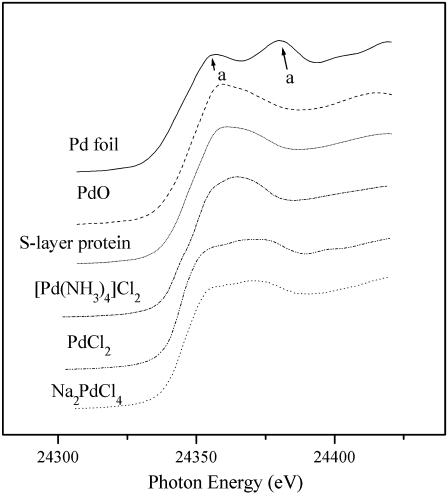
Fig. 5 shows the XANES regions of the XAS spectrum obtained with the Pd(II)-bound S-layer from strain JG-A12 and for reference compounds containing two oxidation states of palladium: Pd(II) (PdO, [Pd(NH3)4]Cl2, PdCl2, Na2PdCl4) and metallic Pd (0.025-mm thick palladium foil). Comparison of the experimental spectrum to the reference spectra clearly shows that Pd is present as Pd(II) in the Pd-loaded S-layer protein sample because the two absorption maxima (∼24360 and ∼24380 eV) characteristics of metallic Pd (feature marked a in Fig. 5) are absent. The fine structure of XANES of the Pd-loaded S-layer resembles that of [Pd(NH3)4]Cl2 and PdO, indicating that Pd-O and Pd-N are the predominant bonds that contribute to the metal-protein binding.
FIGURE 5.
Pd K-edge XANES region of EXAFS spectra of Pd-loaded S-layer protein of B. sphaericus JG-A12 and reference compounds (Pd foil, PdO, Na2PdCl4, [Pd(NH3)4]Cl2, and PdCl2).
Pd(II) coordination studied by EXAFS spectroscopy
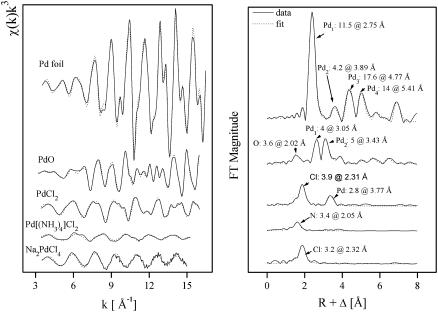
EXAFS spectroscopy samples the local structure around an atom (37). It provides element-specific, short-range structural and chemical information on Pd coordination, the identities, coordination numbers of the neighboring atoms, and their bond distances. Here, we have used this technique to address the coordination environment of Pd and to verify its chemical identity in the palladium-loaded S-layer protein of B. sphaericus JG-A12 sample. We have studied reference compounds to identify possible contributions from multiple coordination modes of Pd(II) when bound to S-layers. The data ranges of Δk and ΔR used in the fits of the EXAFS spectra are presented in Table 2. The k3 weighted EXAFS spectra of the reference compounds: Pd foil, PdO, PdCl2, [Pd(NH3)4]Cl2, and Na2PdCl4, and their corresponding Fourier transforms (FT) are shown in Fig. 6. Table 3 summarizes the fit parameters. It is well established that the FT of χ(k) over a finite k range is a radial structure function exhibiting a series of peaks whose positions and magnitudes are related to the interatomic distances and the number of atoms in the different coordination shells, respectively. FT peak distances are reported in units of angstroms and are uncorrected for scattering phase shift, i.e., R + ΔR. In the case of Pd foil, the FT peaks of metallic Pd were attributed to six Pd-Pd shells with distances of 2.75, 3.89, 4.77, 5.41, 6.15, and 7.34 Å. The major peak corresponds to ∼12 Pd atoms at a Pd-Pd interatomic distance of 2.75 ± 0.01 Å as reported (38) and in agreement with x-ray diffraction studies. However, in the case of the PdO spectrum, the FT reveals several shells containing four oxygen atoms at 2.02 Å, consistent with x-ray diffraction studies (2.018 Å) (39), four palladium atoms at 3.05 Å, and five palladium atoms at 3.43 Å. Additional Pd-Pd contributions appear beyond 3.5 Å, corresponding to higher coordination shells. The Na2PdCl4 and [Pd(NH3)4]Cl2 FT spectra indicate that palladium is coordinated to 3–4 chloride atoms at a distance of 2.32 Å and 3–4 nitrogen atoms at 2.05 Å, respectively. The nearest environment of Pd in PdCl2 consist of 4 Cl (2.31 Å) and 2–3 Pd (3.77 Å). The latter results are in good agreement with those obtained with x-ray diffraction (40).
TABLE 2.
Data range of Δk and ΔR used to fit the EXAFS spectra of the S-layer samples and the reference compounds
| Sample | Δk | ΔR |
|---|---|---|
| Pd foil | 3.5–16.5 | 2–7.2 |
| PdO | 3.3–16 | 1–7 |
| PdCl2 | 3.5–16 | 1–3.8 |
| Pd[(NH3)4]Cl2 | 3.5–15 | 1–2 |
| Na2PdCl4 | 3.5–15 | 1–2.3 |
| S-layer | 3.3–15 | 1–3.5 |
FIGURE 6.
Pd K-edge k3-weighted EXAFS spectra (left) and the corresponding FT (right) of the Pd reference compounds.
TABLE 3.
EXAFS structural parameters of the Pd reference compounds
| Sample | Shell | N* | R[Å]† | σ2 [Å2]‡ | ΔE[eV] |
|---|---|---|---|---|---|
| Pd foil | Pd-Pd1 | 11.5 ± 0.4 | 2.75 | 0.0021 | −18.0 |
| Pd-Pd2 | 4.2 ± 0.9 | 3.89 | 0.0019 | ||
| Pd-Pd3 | 17.6 ± 2 | 4.77 | 0.0026 | ||
| Pd-Pd4 | 14.0 ± 2 | 5.41 | 0.0013 | ||
| PdO | Pd-O1 | 3.6 ± 0.3 | 2.02 | 0.0020 | −10.9 |
| Pd-Pd1 | 4.0 ± 0.3 | 3.05 | 0.0026 | ||
| Pd-Pd2 | 5.0 ± 0.4 | 3.43 | 0.0018 | ||
| [Pd(NH3)4]Cl2 | Pd-N | 3.4 ± 0.2 | 2.05 | 0.0019 | −14.5 |
| PdCl2 | Pd-Cl | 3.9 ± 0.2 | 2.31 | 0.0020 | −15.6 |
| Pd-Pd | 2.8 ± 0.3 | 3.77 | 0.0027 | ||
| Na2PdCl4 | Pd-Cl | 3.2 ± 0.2 | 2.32 | 0.0010 | −12.2 |
Errors in coordination numbers are ±25% and standard deviations as estimated by EXAFSPAK.
Errors in distance are ±0.02 Å.
Debye-Waller factor.
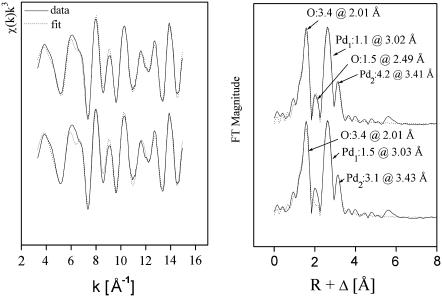
The Pd K-edge EXAFS spectrum of the palladium species formed on S-layer protein from B. sphaericus strain JG-A12 and its corresponding FT are shown in Fig. 7 as calculated with two models (see below). The data exhibit an excellent signal/noise ratio allowing analysis up to 15 Å−1 as summarized in Table 4. Four peaks are found at bond distances of 2.01, 2.49, 3.02, and 3.41 Å corresponding to Pd-O, Pd-O, Pd-Pd, and Pd-Pd bonds, respectively. The distances were identified using Pd-O and Pd-Pd backscattering phase and amplitude functions obtained from atomic coordinates of PdO using the FEFF 8 program. The first shell can be fitted to ∼3–4 oxygen atoms at a distance of 2.01 Å. This is in good agreement with the crystal structure of palladium acetate where each palladium is surrounded by four bridging acetate ligands with Pd-O distances in the range of 1.973–2.014 Å (41). These results strongly support the assignment to oxygen atoms particularly from the carboxyl groups of aspartic and glutamic acids demonstrated by FTIR.
FIGURE 7.
Pd K-edge k3-weighted EXAFS spectra (left) and the corresponding FT (right) of the Pd complexes formed by S-layer of B. sphaericus JG-A12 according to model a lower traces and model b upper traces.
TABLE 4.
EXAFS structural parameters of the palladium complexes formed by the S-layer protein of B. sphaericus JG-A12 according to the models a and b
| Sample | Shell | N* | R[Å]† | σ2 [Å2]‡ | ΔE[eV] | Error§ |
|---|---|---|---|---|---|---|
| S-layer protein (model a) | Pd-O | 3.4 ± 0.1 | 2.01 | 0.0037 | −12.1 | 0.29 |
| Pd-Pd1 | 1.5 ± 0.1 | 3.03 | 0.0027 | |||
| Pd-Pd2 | 3.1 ± 0.5 | 3.43 | 0.0078 | |||
| S-layer protein (model b) | Pd-O1 | 3.4 ± 0.1 | 2.01 | 0.0039 | −14.15 | 0.26 |
| Pd-O2 | 1.5 ± 0.3 | 2.49 | 0.0050 | |||
| Pd-Pd1 | 1.1 ± 0.1 | 3.02 | 0.0019 | |||
| Pd-Pd2 | 4.2 ± 0.6 | 3.41 | 0.0094 |
Errors in coordination numbers are ±25% and standard deviations as estimated by EXAFSPAK.
Errors in distance are ±0.02 Å.
Debye-Waller factor.
Error is given as the normalized fit error ∑(χdata(k)k3 − χfit(k)k3)2/(P − F). (P number of data points, F number of variables.)
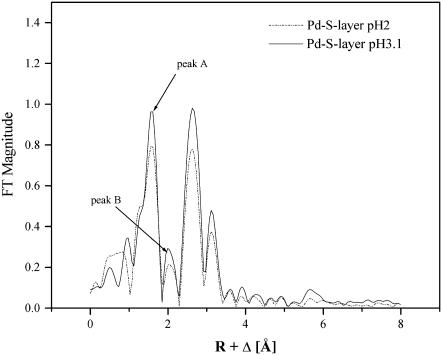
The second FT shell at 2.49 Å does not correspond to a peak in the model compound spectra. It may be due to a side-lobe peak, originating from a truncation effect due to the limited reciprocal space integrated in the FT (in the following designated model a) or may have a structural origin (model b). A similar FT peak at 2.1 Å (at a distance of 2.49 Å) is present also in the Pd reference sample PdO (Fig. 6). Fig. 8 compares the EXAFS amplitudes of two Pd-S-layers prepared and measured under identical conditions at pH 3.1 and pH 2. According to model a, peak B may be a side-lobe of peak A. The intensities of both peaks change proportionally with the change in pH as expected for side lobes in a given Fourier-integration range. On the other hand, this is also true for the intensities of the peaks at larger distances. Thus, the assignment to a side lobe is ambiguous.
FIGURE 8.
FT of the EXAFS spectra of Pd-treated S-layers at pH 2.0 and pH 3.1. Note the constant ratio of the amplitudes of the peak A and peak B indicating that the latter may be a side lobe of peak A. See text for details.
In model b, the shell at distance 2.49 Å may originate in the contribution from a light atom (oxygen or nitrogen as was demonstrated by XANES and FTIR) since this bond distance is too short for a Pd-Pd bond length. EXAFS does not allow us to distinguish between light elements. The structural parameters of the Pd complexes formed by the S-layer protein of B. sphaericus JG-A12 according to the two models are presented in Table 4. Both models are statistically equivalent best-fit models.
DISCUSSION
It has been shown that the heavy-metal-binding capability of the S-layer of B. sphaericus strain JG-A12 may provide a biological barrier for the entry of uranium into the cell (5–7). This discovery has prompted biotechnological investigations of the use of these proteins for the bioremediation of uranium-containing environmental sites (8). Alternatively, these S-layers have been used as organic templates for the generation of nanostructured heavy metal clusters (7,10,42) as reviewed recently (9). The biological function as well as the technological applications are based on a primary interaction between a metal cation and the protein matrix. Here, we have addressed the molecular details of metal binding and its impact on protein secondary structure and stability using IR and x-ray absorption spectroscopic (XAS) methods. XAS can be performed under nondestructive conditions. This agrees with our previous work on the uranium LIII edge XAS characterization of the U complexes formed by cells and isolated S-layer protein from B. sphaericus (7). In addition, combined XAS and near IR absorption spectroscopy has been applied to the complex of human serum transferrin with Np (IV), where also no irradiation damage was observed (43). The investigations reported here have revealed four salient properties of S-layer proteins of the two B. sphaericus strains JG-A12 and NCTC 9602: 1), predominance of β-sheet secondary structure over helical structure (for quantitation see Table 1), 2), an average pKa near pH 3 of the solvent-accessible carboxylates, 3), the involvement of carboxyl groups in structure-stabilizing metal-protein interactions, and 4), coordination of Pd in oxidation state II by oxygen and nitrogen. The large contribution of β-sheets deduced from the low amide I frequency in both strains is in general agreement with structure predictions for S-layer proteins (44). In contrast, the amount of predicted α-helices depends strongly on the prediction method, whereas the IR data presented here indicate an upper limit of 18% α-helical structure.
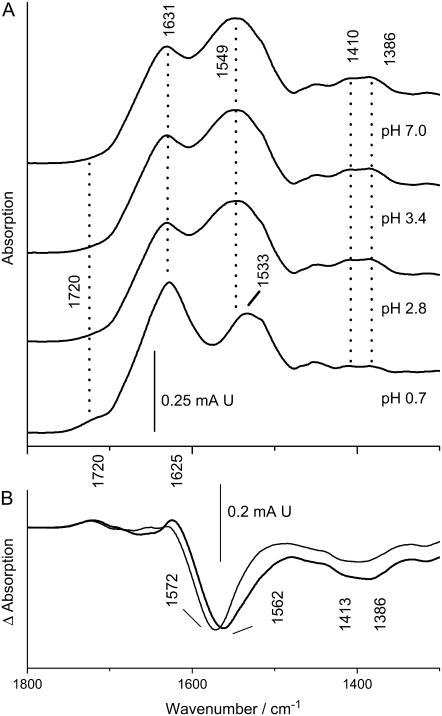
Unexpectedly, Pd(II) binding stabilizes the S-layer secondary structure against acidification. The amide I frequency of the Pd (II)-bound samples shows less response to acidification than the native proteins (2 cm−1 vs. 11 cm−1 pH-induced downshift), resulting in an amide I mode that is 4 cm−1 higher in the acidified metal-bound state versus the irreversibly denatured metal-free state. Stabilization may be mediated by metal-side-chain and metal-backbone interactions. The data strongly argue for metal-carboxylate interactions as an important mode of Pd(II) binding and thus of structural stabilization. Biochemical evidence for coordination by carboxylates is provided by the fact that the Pd(II)-bound S-layer of B. sphaericus JG-A12 becomes fully protected against proteolytic attack by the -Asp-P1′- and -Glu-P1′-specific protease Glu-C. Although the S-layer protein of B. sphaericus JG-A12 possesses 125 possible cleavage sites regularly distributed within the primary structure of the protein, only four of these can be attacked by Glu-C. This indicates the inaccessibility of most of the carboxylates due to their structural seclusion and/or the large size of the enzyme. The Pd(II)-dependent blockade of the few accessible sites fully supports the role of Asp and Glu in Pd(II) complexation. However, a much larger number of sites is involved in complexation than only those that affect Glu-C digestion. From preliminary inductive coupled plasma mass spectroscopy (ICP-MS) we have obtained an estimate of more than 400 mol Pd bound per mol S-layer protein of B. sphaericus JG-A12, in agreement with the several hundreds of metal atoms bound per monomer of other S-layers (23). Spectroscopic evidence is based on the IR absorption of side-chain carboxylates. S-layers from both strains exhibit an increase in the absorption of the antisymmetric COO−-stretching modes in the 1550–1580 cm−1 range and a broadening of the symmetric COO−-stretching absorption. In principle, amide II modes may contribute to the Pd(II)-induced absorption increase in this range. However, measurements in D2O, where the amide II absorption shifts to ∼1450 cm−1 (C-N stretching uncoupled from N-H bending upon H/D exchange) reveals that the intensity increase and the pH sensitivity between 1560 and 1580 cm−1 must be solely attributed to carboxylates (free from overlap with amid II′ in D2O), whereas no pH sensitivity is observed in the amide II′ modes (free from overlap with carboxylates). Thus, at neutral pH, the antisymmetric COO−-stretching absorption in D2O is larger in the Pd(II) -bound state in the S-layer of JG-A12 than in the metal-free state (Fig. 9, a and b). The assignment to carboxylates is confirmed by the fact that the 1560–1580 cm−1 bands vanish completely upon acidification when the COO− groups are transformed into COOH groups (Fig. 9, c and e, for JG-A12, Fig. 9, d and f, for NCTC 9602). The broad C=O absorption of the COOH groups in the acidified samples and its low frequency (see also Figs. 2 and 3) evidence a broad distribution of H-bond strengths typical of solvent-accessible protonated carboxyl groups.
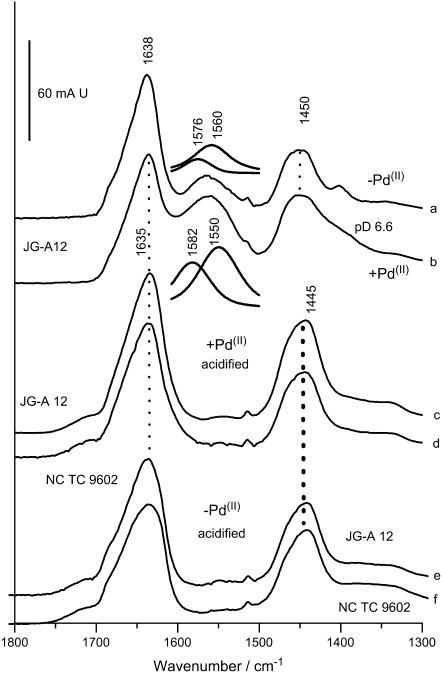
FIGURE 9.
FTIR spectra of S-layers of B. sphaericus strain JG-A12 and NCTC 9602 measured in D2O. (a) Spectrum of JG-A12 at pH 7, metal free. (b) Spectrum of JG-A12 at pH 7, Pd(II) bound. Note the broadening of the 1400 cm−1 absorption (symmetric COO− stretching) and the increase of the ∼1560 cm−1 absorption (antisymmetric COO−- stretching) as compared to a. The antisymmetric COO−-stretching modes where fitted by two components absorbing at 1576 and 1560 cm−1 (inset). Besides the strong absorption enhancement, the salient effect of Pd(II) binding is the 10 cm−1 downshift of the 1560 cm−1 absorption, whereas the upshift of the high frequency part is less pronounced. (c) Absorption of JG-A12 in the Pd(II)-bound state acidified with DCl. (d) Absorption of NCTC 9602 in the Pd(II)-bound state acidified with DCl. Note the absence of residual absorption in the 1560–1570 cm−1 range and the lack of pH sensitivity in the amide II′ mode (1450 cm−1) in c and d as compared to a and b. (e) Absorption of JG-A12 in the metal-free state acidified with DCl. (f) Absorption of NCTC 9602 in the metal-free state acidified with DCl. Note the correspondence of the amide II′ modes in c and e and in d and f.
The spectra further show that Pd(II) binding shifts the center of the antisymmetric stretching frequency from 1567 to 1558 cm−1 in the S-layer of B. sphaericus JG-A12 (Fig. 9, a and b). Correspondingly, the average frequency difference, Δ, between antisymmetric and symmetric COO−-stretching modes is reduced. In model compounds, this has been found to be characteristic of carboxylates that chelate a metal ion rather than coordinating it in unidentate fashion, where an upshift of the antisymmetric COO−-stretching mode and an increase of Δ is typically expected (45). The relation between carboxylate vibrations and the type of metal ion coordination in proteins has been established in studies of various Ca2+-binding proteins (46,47). Again, a downshift of the antisymmetric COO− stretch was found to be correlated with bidentate coordination. Thus, bidentate coordination appears to be the predominant Pd(II) binding mode in the S-layer of strain JG-A12, whereas unidentate coordination seems to be less abundant. The lack of a significant shift of the antisymmetric COO−-stretching mode in NCTC 9602 suggests that a bridging coordination is more prevalent as this binding mode has little effect on this mode in Asp and Glu side chains in solution. The magnitude of the described effects on the COO− absorption implies that the vast majority of carboxylates is affected by Pd(II) binding. For example, the IR-spectral changes caused by Ca2+ binding to the four carboxylate-containing binding sites in calmodulin (47) containing 148 amino acids would be barely visible on the scale that covers the amide I absorption of the ∼10-fold larger S-layer proteins. The IR data are thus consistent with a multitude of carboxylate- (and nitrogen-) containing Pd(II)-binding sites found earlier for other S-layers (36) and supported by preliminary ICP-MS results of the strains studied here.
The XANES data indicate that oxygen and nitrogen are the predominant groups that coordinate Pd(II) in the S-layer of JG-A12. Their relative contributions were assessed by the iterative target test factor analysis as described (48). The calculation reveals a mixture containing 55% of Pd-O and 45% of Pd-N bonding. EXAFS analysis indicated the presence of an additional shell at a distance of 2.49 Å, which was not found in PdO. This shell may be due to a truncation effect by the limited reciprocal space integrated in the FT (model a) or may have a structural origin (model b). The presence of such unphysical shells is well documented. A weak peak between the main Oax and Cl peaks in EXAFS spectra of uranyl chloride complexes has been interpreted as an “overlap effect” (49). A similar effect was observed with the U(VI) aquo chloro complexes, and an assignment to a coordination shell was ruled out by factor analysis (50). With respect to model b, a survey of the interatomic palladium-oxygen (nitrogen) distances in the Cambridge Structure database indicated that 2.49 Å is too long to correspond to a Pd-O or Pd-N distance. It could be a Pd….O “contact” but the origin of this oxygen atom is unknown. A shell at the same distance for the Pd coordination sphere of the precatalytic solution of the phosphine-free Heck reaction using energy dispersive EXAFS (EDE) has been reported (51) but could not be fitted to carbon, oxygen, or even phosphorus. The authors reported that the shell may indicate the presence of a lighter element, but there is no definitive information about its nature. Although the origin of the peak at 2.49 Å cannot be clarified, our data interpretation is not impeded by this ambiguity because there are also no significant differences between the structural parameters of the S-layer-bound Pd complexes determined with either model (Table 4).
The crystallographically determined nonbonded Pd-Pd distances in Pd acetate in the region of 3.105–3.203 Å were also found in the EXAFS spectrum of the Pd-loaded S-layer, but the first shell could also equally well be fitted to a contribution from nitrogen since EXAFS spectroscopy cannot distinguish between neighboring elements in the periodic table. Therefore, the question of coordination by nitrogen was further addressed by the FTIR measurements in D2O. Fig. 9 shows the shifted amide II′ absorption at 1450 cm−1, which is free from overlap with carboxylate contributions. Pd(II) binding affects the amide II′ modes neither at neutral pH (amide II′ at 1450 cm−1, independent of Pd(II), Fig. 9, a and b) nor in acidified samples (amide II′ at 1445 cm−1, independent of Pd(II), Fig. 9, c and e, for strain JG-A12, Fig. 9, d and f, for strain NCTC 9602). Thus, the predominant effect of Pd(II) is on carboxylate vibrations, rather than C-N-stretching modes of the peptide backbone. This indicates that the possible contributions of nitrogen to Pd(II) complexation are likely to result from side chains. Based on the evidence for carboxylates as Pd(II)-coordinating groups, we have analyzed possible relations between carboxyl- and nitrogen-bearing amino acids in the primary structure. We find that 42% and 47% of the Asp and Glu residues in the S-layer proteins from strain NCTC 9602 and JG-A12, respectively, follow or precede a nitrogen-bearing amino acid. This is ∼1.5-fold more often than expected from the amino acid composition of both strains containing 12% carboxyl- and 20% nitrogen-bearing amino acids, typical also of S-layers from other phylogenetic branches. Among the nitrogen-bearing residues, Lys and Asn are the most frequent amino acids found next to a carboxylate. The possible functional role of these relations needs further investigation. In combination with the XANES data, however, nitrogens from amino acid side chains are likely to participate in Pd(II) coordination in addition to carboxylates of neighboring Asp and Glu residues. The IR absorption frequency of the C-N stretches of nitrogen-bearing amino acid side chains in H2O is typically found below 1500 cm−1 and is generally low in intensity (52). Thus, the lack of clear Pd(II)-dependent effects in the amide II range is consistent with contributions from side-chain nitrogens but argues against the participation of backbone nitrogen in Pd(II) coordination. The location of positively charged side chains next to carboxylates could also cause the lowering of the pK of Asp and Glu side chains by charge stabilization through salt bridges. Such a topology may provide a molecular adaptation of the S-layers of B. sphaericus to an environment where a high heavy-metal-binding capacity must be maintained at low pH.
In summary, we have shown that carboxylates of Asp and Glu residues exhibit an unusually low pK and are coordination sites for Pd(II) in the two related B. sphaericus strains JG-A12 and NCTC 9602, which become structurally stabilized by the heavy metal. Acidic amino acids and probably nitrogen-bearing side chains are thus prime targets for the site-directed modification of S-layer properties such as metal-binding capacity and secondary structure stability, which are relevant to metalization-based biotechnological applications.
Acknowledgments
We are grateful to A. C. Scheinost, A. Rossberg, and H. Funke for their assistance in the XAS measurements and to Jenny Philipp for her biochemical support. We also thank A. Rossberg for his support in using the Iterative Target Test Factor analysis.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG, Bonn, Germany) grant SE 671/7-2, the Saxon Ministry of Science and the Fine Arts (Dresden, Germany) grant 7531.50-03.0370-01, and by the European Community (EC, Brussels, Belgium) grant GRD1-413 2001-40424.
References
- 1.Sleytr, U. B., and M. Sara. 1997. Bacterial and archaeal S-layer proteins: structure-function relationships and their biotechnological applications. Trends Biotechnol. 15:20–26. [DOI] [PubMed] [Google Scholar]
- 2.Beveridge, T. J. 1994. Bacterial S-layers. Curr. Opin. Struct. Biol. 4:204–212. [Google Scholar]
- 3.Dieluweit, S., D. Pum, and U. B. Sleytr. 1998. Formation of a gold superlattice on an S-layer with square lattice symmetry. Supramolecular Sci. 5:15–19. [Google Scholar]
- 4.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276–302. [DOI] [PubMed] [Google Scholar]
- 5.Selenska-Pobell, S., P. Panak, V. Miteva, G. Bernhard, and H. Nitsche. 1999. Selective accumulation of heavy metals by three indigenous Bacillus strains, B. cereus, B. megaterium and B. sphaericus, from drain waters of a uranium waste pile. FEMS Microbiol. Ecol. 29:59–67. [Google Scholar]
- 6.Raff, J. 2002. Wechselwirkungen der Hüllproteine von Bakterien aus Uranabfallhalden mit Schwermetallen. Thesis FZR-358, Forschungszentrum Rossendorf.
- 7.Merroun, M., J. Raff, A. Rossberg, C. Hennig, T. Reich, and S. Selenska-Pobell. 2005. Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl. Environ. Microbiol. 71:5532–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raff, J., U. Soltman, S. Matys, M. Schnorpfeil, H. Böttcher, W. Pompe, and S. Selenska-Pobell. 2002. Bacterial-based bioremediation of uranium mining waste waters by using sol-gel ceramics. In Uranium in the Aquatic Environment. B. J. Merkel, B. Planer-Friedrich, and C. Wolkersdorfer, editors. Springer, Berlin. 615–622.
- 9.Pollmann, K., J. Raff, M. Merroun, K. Fahmy, and S. Selenska-Pobell. 2006. Metal binding by bacteria from uranium mining waste piles and its technological applications. Biotechnol. Adv. 24:58–68. [DOI] [PubMed] [Google Scholar]
- 10.Merroun, M., A. Rossberg, C. Hennig, A. Scheinost, and S. Selenska-Pobell. 2006. Spectroscopic characterization of gold nanoparticles formed by cells and S-layer protein of Bacillus sphaericus JG-A12. Mater. Sci. Eng. C. In press.
- 11.Mertig, M., R. Kirsch, W. Pompe, and H. Engelhardt. 1999. Fabrication of highly oriented nanocluster arrays by biomolecular templating. Eur. Phys. J. D. 9:45–48. [Google Scholar]
- 12.Seifert, G. 2004. Nanocluster magic. Nat. Mater. 3:77–78. [DOI] [PubMed] [Google Scholar]
- 13.Andres, R. P., J. D. Bielefeld, J. I. Henderson, D. B. Janes, V. R. Kolagunta, C. P. Kubiak, W. J. Mahoney, and R. G. Osifchin. 1996. Self-assembly of a two-dimensional superlattice of molecularly linked metal clusters. Science. 273:1690–1693. [Google Scholar]
- 14.Patolsky, F., Y. Weizmann, and I. Willner. 2004. Actin-based metallic nanowires as bio-nanotransporters. Nat. Mater. 3:692–695. [DOI] [PubMed] [Google Scholar]
- 15.McMillan, R. A., C. D. Paavola, J. Howard, S. L. Chan, N. J. Zaluzec, and J. D. Trent. 2002. Ordered nanoparticle arrays formed on engineered chaperonin protein templates. Nat. Mater. 1:247–252. [DOI] [PubMed] [Google Scholar]
- 16.Braun, E., Y. Eichen, U. Sivan, and G. Ben Yoseph. 1998. DNA-templated assembly and electrode attachment of a conducting silver wire. Nature. 391:775–778. [DOI] [PubMed] [Google Scholar]
- 17.Mertig, M., R. Kirsch, and W. Pompe. 1998. Biomolecular approach to nanotube fabrication. J. Appl. Physiol. A66:723–727. [Google Scholar]
- 18.Schultze-Lam, S., G. Harauz, and T. J. Beveridge. 1992. Participation of a cyanobacterial S layer in fine-grain mineral formation. J. Bacteriol. 174:7971–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultze-Lam, S., and T. J. Beveridge. 1994. Nucleation of celestine and strontianite on a cyanobacterial S-layer. Appl. Environ. Microbiol. 60:447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown, D. A., T. J. Beveridge, W. Keevil, and B. L. Sherriff. 1998. Evaluation of microscopic techniques to observe iron precipitation in a natural microbial biofilm. FEMS Microbiol. Ecol. 26:297–310. [Google Scholar]
- 21.Shenton, W., D. Pum, U. B. Sleytr, and S. Mann. 1997. Synthesis of cadmium sulphide superlattices using self-assembled bacterial S-layers. Nature. 389:585–587. [Google Scholar]
- 22.Wahl, R., M. Mertig, J. Raff, S. Selenska-Pobell, and W. Pompe. 2001. Electron-beam induced formation of highly ordered palladium and platinum nanoparticle arrays on the S-layer of Bacillus sphaericus NCTC 9602. Adv. Mater. 13:736–740. [Google Scholar]
- 23.Wahl, R., H. Engelhardt, W. Pompe, and M. Mertig. 2005. Multivariate statistical analysis of two-dimensional metal cluster arrays grown in vitro on a bacterial surface layer. Chem. Mater. 17:1887–1894. [Google Scholar]
- 24.Matz, W., N. Schell, G. Bernhard, F. Prokert, T. Reich, J. Claußner, W. Oehme, R. Schlenk, S. Dienel, H. Funke, F. Eichhorn, M. Betzel, U. Strauch, G. Hüttig, H. Krug, W. Neumann, V. Brendler, P. Reichel, M. A. Denecke, and H. Nitsche. 1999. ROBL—a CRG beamline for radiochemistry and material research at the ESRF. J. Synchrotron. Radiat. 6:1076–1085. [Google Scholar]
- 25.Ankudinov, A. L., B. Ravel, J. J. Rehr, and S. D. Conradson. 1998. Real-space multiple-scattering calculation and interpretation of x-ray absorption near-edge spectra. Phys. Rev. B. 58:7565–7575. [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- 27.Surewicz, W. K., H. H. Mantsch, and D. Chapman. 1993. Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry. 32:389–394. [DOI] [PubMed] [Google Scholar]
- 28.Arrondo, J. L., A. Muga, J. Castresana, and F. M. Goni. 1993. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 59:23–56. [DOI] [PubMed] [Google Scholar]
- 29.Dong, A., P. Huang, and W. S. Caughy. 1990. Protein secondary structures in water from second-derivative amide I infrared spectra. Biochemistry. 29:3303–3308. [DOI] [PubMed] [Google Scholar]
- 30.Hofer, P., and U. P. Fringeli. 1979. Structural investigation of biological material in aqueous environment by means of infrared-ATR spectroscopy. Biophys. Struct. Mech. 6:67–80. [DOI] [PubMed] [Google Scholar]
- 31.Goormaghtigh, E., V. Raussens, and J.-M. Ruysschaert. 1999. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim. Biophys. Acta. 1422:105–185. [DOI] [PubMed] [Google Scholar]
- 32.Fahmy, K. 2001. Application of ATR-FTIR spectroscopy for studies of biomolecular interactions. Recent Res. Devel. Biophys. Chem. 2:1–17. [Google Scholar]
- 33.Goormaghtigh, E., V. Cabiaux, and J.-M. Ruysschaert. 1994. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. III. Secondary structures. Subcell. Biochem. 23:405–450. [DOI] [PubMed] [Google Scholar]
- 34.Mizuguchi, M., M. Nara, Y. Ke, K. Kawano, T. Hiraoki, and K. Nitta. 1997. Fourier-transform infrared spectroscopic studies on the coordination of the side-chain COO- groups to Ca2+ in equine lysozyme. Eur. J. Biochem. 250:72–76. [DOI] [PubMed] [Google Scholar]
- 35.Pollmann, K., J. Raff, M. Schnorpfeil, G. Radeva, S. Selenska-Pobell. 2005. Novel surface layer protein genes in Bacillus sphaericus associated with unusual insertion elements. Microbiology. 151:2961–2973. [DOI] [PubMed]
- 36.Davis, R. J., and M. Boudart. 1994. Structure of supported PdAu clusters determined by x-ray absorption spectroscopy. J. Phys. Chem. 98:5471–5477. [Google Scholar]
- 37.Koningsberger, D. C., and R. Prins. 1988. X-Ray Absorption. Wiley, New York.
- 38.Polizzi, S., P. Riello, A. Alerna, and A. Eneditti. 2001. Nanostructure of Pd/SiO2 supported catalysts. Phys. Chem. Chem. Phys. 3:4614–4619. [Google Scholar]
- 39.Wasser, J., H. A. Levy, and S. W. Peterson. 1953. The structure of PdO. Acta Crystallogr. 6:661–663. [Google Scholar]
- 40.Wells, A. F. 1938. Crystal structures of palladous chloride PdCl2. Z. Kristallogr. 100:189. [Google Scholar]
- 41.Skapski, A. C., and M. L. Smart. 1970. The crystal structure of Trimeric palladium(II) acetate. J. Chem. Soc. D11:658b–659. [Google Scholar]
- 42.Merroun, M., K. Pollmann, J. Raff, A. Scheinost, and S. Selenska-Pobell. 2003. EXAFS studies of palladium nanoclusters formed at the cells and S-layers of Bacillus sphaericus JG-A12. FZR-Report 400:25. [Google Scholar]
- 43.Llorens, I., C. Den Auwer, Ph. Moisy, E. Ansoborlo, C. Vidaud, and H. Funke. 2005. Neptunium uptake by serum transferrin. FEBS Lett. 272:1739–1744. [DOI] [PubMed] [Google Scholar]
- 44.Sara, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deacon, G. B., and R. J. Phillips. 1980. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 33:227–250. [Google Scholar]
- 46.Nara, M., M. Tasumi, M. Tanokura, T. Hiraoki, M. Yazawa, and A. Tsutsumi. 1994. Infrared studies of interaction between metal ions and Ca(2+)-binding proteins. Marker bands for identifying the types of coordination of the side-chain COO- groups to metal ions in pike parvalbumin (pI = 4.10). FEBS Lett. 349:84–88. [DOI] [PubMed] [Google Scholar]
- 47.Nara, M., M. Tanokura, T. Yamamoto, and M. Tasumi. 1995. A comparative study of the binding effects of Mg2+, Ca2+, Sr2+, and Cd2+ on calmodulin by Fourier-transform-infrared spectroscopy. Biospectroscopy. 1:47–54. [Google Scholar]
- 48.Rossberg, A., T. Reich, and G. Bernhard. 2003. Complexation of uranium(VI) with protocatechuic acid-application of iterative transformation factor analysis to EXAFS spectroscopy. Anal. Bioanal. Chem. 376:631–638. [DOI] [PubMed] [Google Scholar]
- 49.Servaes, K., C. Hennig, R. van Deun, and C. Gorller-Walrand. 2005. Structure of [UO2Cl4]2- in acetonitril. Inorg. Chem. 44:7705–7707. [DOI] [PubMed] [Google Scholar]
- 50.Hennig, C., J. Tutschku, A. Rossberg, G. Bernhard, and A. Scheinost. 2005. Comparative EXAFS investigation of uranium(VI) and -(IV) aqua chloro complexes in solution using a newly developed spectroelectrochemical cell. Inorg. Chem. 44:6655–6661. [DOI] [PubMed] [Google Scholar]
- 51.Evans, J., L. O'Neill, V. L. Kambhampati, R. Graham, S. Turin, A. Genge, A. J. Dent, and T. Neisus. 2002. Structural characterization of solution species implicated in the palladium-catalysed Heck reaction by Pd K-edge x-ray absorption spectroscopy: palladium acetate as a catalyst precursor. J. Chem. Soc., Dalton Trans. 10:2207–2212. [Google Scholar]
- 52.Barth, A., and C. Zscherp. 2002. What vibrations tell us about proteins. Q. Rev. Biophys. 35:369–430. [DOI] [PubMed] [Google Scholar]
- 53.Krimm, S., and J. Bandekar. 1986. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 38:181–364. [DOI] [PubMed] [Google Scholar]
- 54.King, R. D., M. Saqi, R. Sayle, and M. J. E. Sternberg. 1997. DSC: public domain secondary structure prediction. Comput. Appl. Biosci.. 13:473–474. [DOI] [PubMed] [Google Scholar]
- 55.Kneller, D. G., F. E. Cohen, and R. Langridge. 1990. Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 214:171–182. [DOI] [PubMed] [Google Scholar]
- 56.Meiler, J., M. Mueller, A. Zeidler, and F. Schmaeschke. 2001. Secondary structure prediction by JUFO. J. Mol. Model. (Online). 7:360–369. [Google Scholar]
- 57.de Jongh, H. H., E. Goormaghtigh, and J. M. Ruysschaert. 1996. The different molar absorptivities of the secondary structure types in the amide I region: an attenuated total reflection infrared study on globular proteins. Anal. Biochem. 242:95–103. [DOI] [PubMed] [Google Scholar]