Abstract
The rate of synthesis of cytochrome f is decreased ∼10-fold when it does not assemble with the other subunits of the cytochrome b6f complex in Chlamydomonas reinhardtii chloroplasts. This assembly-mediated regulation of cytochrome f synthesis corresponds to a regulation of petA mRNA initiation of translation. Here, we demonstrate that cytochrome f translation is autoregulated by its C-terminal domain. Five cytochrome f residues conserved throughout all chloroplast genomes—residue Gln-297 in the transmembrane helix and a cluster of four amino acids, Lys-Gln-Phe-Glu, at positions 305 to 308, in the stromal extension—participate in the formation of a translation repressor motif. By contrast, positively charged residues in the stromal extension have little influence on the autoregulation process. These results do not favor a direct interaction between the repressor motif and the petA 5′ untranslated region but suggest the participation of a membrane-bound ternary effector.
INTRODUCTION
Energy-transducing systems, whether located in the inner mitochondrial membrane or in the chloroplast thylakoid membrane, are made of several multimeric proteins that are comprised of nucleus- and organelle-encoded subunits. How the various subunits, encoded by two distinct genetic compartments, are synthesized in the stoichiometry required for their functional assembly is a key issue in the study of organelle biogenesis.
Photosynthesis mutants bearing a genetic lesion for the expression of a single subunit often display a pleiotropic loss in the entire set of subunits from the same chloroplast protein complex: the various subunits of each photosynthetic protein accumulate through some concerted process (reviewed by Wollman et al., 1999; Choquet and Vallon, 2000). The efficient post-translational degradation of unassembled subunits represents a major contribution to this concerted accumulation. However, the rate of translation of some chloroplast-encoded subunits, rather than their half-life, also is controlled by protein assembly, a phenomenon that we have defined as a control by epistasy of synthesis (CES) process (Wollman et al., 1999; Choquet and Vallon, 2000). Therefore, protein assembly in the chloroplast can be regarded as the result of a hierarchical expression of the constitutive subunits: the CES subunits are those whose rates of synthesis are decreased in the absence of their assembly partners, the latter being defined as dominant subunits. Many studies of photosynthesis mutants from Chlamydomonas reinhardtii have shown that each protein complex of the thylakoid membrane contains at least one CES subunit (Choquet et al., 1998; Wollman et al., 1999; Choquet and Vallon, 2000).
To date, cytochrome f, a subunit of the cytochrome b6f complex that is encoded by the chloroplast petA gene, is the best-characterized CES protein. It associates with cytochrome b6 and subunit IV (SUIV), the products of the chloroplast petB and petD genes, respectively, with the iron-sulfur protein coded by the nuclear petC gene and with four mini-proteins of <4 kD (reviewed by Wollman, 1998). Cytochrome f is synthesized as a precursor protein with a 31-residue lumen-targeting peptide that drives the translocation of most of the protein through the thylakoid membrane. Once the transit peptide is cleaved, a large N-terminal heme binding domain (residues 32 to 282) remains in the lumenal space. This globular domain is tethered to the membrane by a 20–amino acid transmembrane helix (residues 283 to 302) that ends up in the stroma by a short stretch of 15 residues (positions 303 to 317). Together, the transmembrane helix and the stromal extension form the C-terminal domain.
Cytochrome f is a CES protein because its rate of synthesis is regulated by the availability of its assembly partners. In mutants that lack dominant subunits from the cytochrome b6f complex, such as cytochrome b6 and SUIV, the rate of synthesis of cytochrome f decreases to ∼10% of that observed in the wild type. However, this residual cytochrome f is inserted properly in the membrane and displays the same proteolytic resistance as in the wild type: unassembled cytochrome f is a poor substrate for chloroplast proteases (Kuras and Wollman, 1994).
The assembly-mediated control of cytochrome f synthesis probably originates from the regulation of the initiation of translation of petA mRNA. Indeed, cytochrome f does not behave as a CES subunit when the coding region of the petA gene is translated under the control of the unrelated atpA 5′ untranslated region (UTR): its rate of synthesis no longer is regulated by the presence of its assembly partners (Choquet et al., 1998). We observed the same loss of CES behavior in two other instances in which cytochrome f nevertheless was translated under the control of its regular petA gene 5′ UTR. In one instance, non-CES cytochrome f corresponded to a site-directed truncation of the C-terminal domain of the protein. The N-terminal domain, no longer tethered to the membrane, accumulated as a soluble protein in the thylakoid lumen (Kuras et al., 1995b). In the other instance, non-CES cytochrome f corresponded to a short-lived apoprotein form that resulted from the site-directed destruction of the residues required for covalent binding of the c-type heme (Kuras et al., 1995a). Both mutant strains still expressed the entire set of cytochrome b6f subunits in an unassembled state, but the rate of synthesis of the non-CES cytochrome f variants increased three times with respect to that in the wild type independent of the presence or absence of their assembly partners (Kuras et al., 1995b; Choquet et al., 1998). We reasoned that this paradoxical behavior of cytochrome f synthesis could be explained in one of two ways: by a transactivation hypothesis, in which some other cytochrome b6f subunit, when not fully assembled, acts as an activator of cytochrome f translation; or by an autoregulation hypothesis, in which the unassembled C-terminal domain of cytochrome f exposes a translational repressor motif. This motif would be shielded upon assembly with the other subunits, allowing further translation of the petA mRNA.
Here, we test these hypotheses. First we used a chloroplast reporter protein whose level of expression can be assessed quantitatively in vivo to show that the 5′ UTR of the petA mRNA is sufficient to confer cytochrome b6f–dependent CES behavior to an unrelated protein. Then we performed an extensive site-directed mutagenesis study of the C-terminal domain of cytochrome f that demonstrates its role as a repressor motif for cytochrome f translation and shows which residues contribute to this motif. A subset of these site-directed mutations was presented in a preliminary form at the 673rd meeting of the Biochemical Society (Choquet et al., 2001). From the structural and physicochemical properties of the repressor motif, we conclude that the CES process for cytochrome f expression most likely requires interaction with a membrane-bound translational activator.
RESULTS
The petA 5′ UTR Is Sufficient to Confer the CES Behavior of Cytochrome f to a Reporter Protein
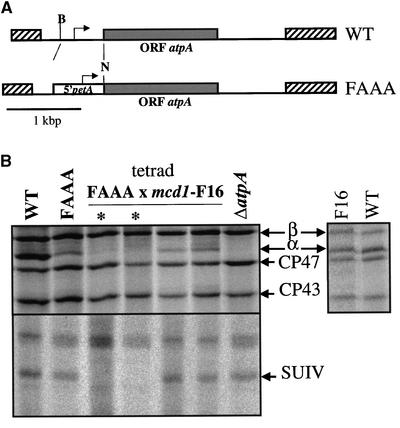
Using antibiotic resistance as a reporter function driven by the petA 5′ UTR, we had proposed previously that the 5′ UTR of the petA mRNA controlled the CES behavior of cytochrome f (Choquet et al., 1998). This conclusion is substantiated by the results shown in Figure 1. In this experiment, we quantitatively assessed the expression in vivo of a reporter protein driven by the petA 5′ UTR in conditions in which the assembly of cytochrome b6f complexes was prevented or allowed.
Figure 1.
Synthesis of the α-Subunit Translated under the Control of the petA 5′ UTR Is Regulated by the Presence of Cytochrome f Assembly Partners.
(A) Scheme of wild-type (WT) atpA and chimeric FAAA genes. Relevant restriction sites are as follows: B, BseRI; N, NcoI. ORF, open reading frame.
(B) Newly synthesized cytochrome f and SUIV detected by pulse-labeling experiments in the wild-type and ΔatpA control strains, in the parental strain FAAA, and in the four progeny of a representative tetrad from the cross FAAA (mt+) × mcd1-F16 (mt−). Asterisks indicate members of the tetrad that are incapable of phototrophic growth, lack SUIV, and show repressed translation of the α-subunit. The gel at right shows that the expression of the α-subunit is not affected in the original mcd1-F16 strain (Drager et al., 1998).
We used the chimeric gene FAAA, in which translation of the unrelated α-subunit of the ATP synthase complex, encoded by the atpA gene, is driven by the petA 5′ UTR (Figure 1A). The rate of translation of this chimeric atpA gene can be determined accurately by pulse-labeling experiments in the transformed strain FAAA, in which it replaces the endogenous atpA gene. The chimeric gene allowed only limited expression of the α-subunit, ∼40% of that observed in the wild type. However, this level of expression was sufficient to sustain photosynthetic growth on minimal medium (MM) (Drapier et al., 2002).
We then investigated whether the synthesis of the α-subunit, now translated under the control of the petA 5′ UTR, was regulated by the availability of cytochrome f assembly partners. We crossed the strain FAAA (mt+) to a mcd1-F16 (mt−) strain lacking SUIV as a result of a nuclear mutation that prevents the stable accumulation of the petD mRNA (Drager et al., 1998). In that cross, the four daughter cells of the tetrad inherited the chloroplast FAAA gene. By contrast, the nuclear mutation mcd1 was transmitted to only half of the progeny. Indeed, two members of each tetrad were unable to grow on MM because they lacked SUIV, whereas the other two were phototrophic and expressed SUIV (Figure 1B). We compared the rate of translation of the α-subunit among the progeny that expressed or did not express SUIV by performing a 5-min pulse labeling of whole cells in the presence of 14C-acetate and cycloheximide, an inhibitor of cytoplasmic translation (Figure 1B). The rate of synthesis of the α-subunit in the two members of the tetrad expressing SUIV was similar to that observed in the parental strain FAAA. By contrast, the synthesis of the α-subunit was barely detectable in the progeny that inherited the mcd1 mutation and lacked SUIV. Note the unaltered rate of synthesis of the α-subunit when expressed from the resident atpA gene in an mcd1-mutated context (Figure 1B, gel at right).
Thus, we conclude that the petA 5′ UTR is able to confer extensive cytochrome b6f–dependent CES behavior to an unrelated reporter protein. These cis-acting signals control the rate of translation of a downstream coding region by sensing the assembly status of cytochrome f in the thylakoid membranes. We then wondered how the cytochrome f protein participated in this regulation and performed site-directed mutagenesis to identify which protein motifs were involved in the sensing mechanism.
Mapping the C-Terminal End of the Regulatory Motif
We first mapped the C-terminal end of the putative regulatory motif of cytochrome f by constructing a set of C-terminal truncations of increasing length (Table 1). Mutated petA genes were inserted in the chloroplast genome of Chlamydomonas in place of the resident petA gene. The resulting transformants are referred to hereafter as fmut, where mut stands for one of the mutations listed Table 1. The same mutations also were associated, in plasmids pfmutΔpetD, with a deletion of the petD gene that encodes SUIV, a major assembly partner of cytochrome f, located a few kilobases downstream of petA (see supplemental data online). Transformation of the chloroplast genome with these plasmids yielded transformed strains designated fmutΔpetD. These transformants are incapable of phototrophic growth, whereas the mutations introduced in the C-terminal domain of fmut-transformed strains may or may not impair the function of the cytochrome b6f complex. Thus, all mutations were associated with the aadA cassette that confers spectinomycin resistance (Goldschmidt-Clermont, 1991) inserted at either a neutral EcoRV site to select fmut transformants or in place of the petD gene for the recovery of fmutΔ petD strains. Transformed strains were selected on the basis of their ability to grow on spectinomycin-supplemented medium.
Table 1.
Phenotypes of Cytochrome f Truncations
| C-Terminal Amino Acid Sequencea | Strain | CESb | Phototrophic Growthc |
|---|---|---|---|
| VLLTQVLLVLKKKQFEKVQLAEMNF | Wild type | + | + |
| VLLTQVLLVLKKKQFEKVQ* | f312St | + | + |
| VLLTQVLLVLKKKQFEK* | f310St | ± | + |
| VLLTQVLLVLKKKQ* | f307St | − | + |
| VLLTQVLLVLKKKQFEKF* | f310F311St | + | + |
Amino acid sequence of the end of the transmembrane helix (underlined) and of the stromal extension in the wild-type and mutant strains listed in the second column. Asterisks indicate premature stop codons. Residues mutated with respect to the wild-type sequence are shown in boldface in the first column.
+ indicates that cytochrome f is still controlled by epistasy of synthesis in that particular strain; − indicates a strain in which cytochrome f synthesis escapes the CES control; ± correspond to a situation in which the CES control is reduced but not abolished completely.
+ indicates that the strains are capable of phototrophic growth on MM.
To determine whether these petA mutations impaired the CES process, we compared the rates of synthesis of the mutated versions of cytochrome f in pairs of strains expressing or lacking SUIV. When petA mutations do not impair the CES process, the rates of cytochrome f synthesis should display a 10-fold decrease in the absence of SUIV. Conversely, if the CES process is impaired, the rate of cytochrome f synthesis should be similar in the presence or absence of SUIV. In these cases, we even expected higher rates of synthesis than in the wild type, based on our previous observations drawn from the expression of a non-CES soluble version of cytochrome f (Kuras et al., 1995b).
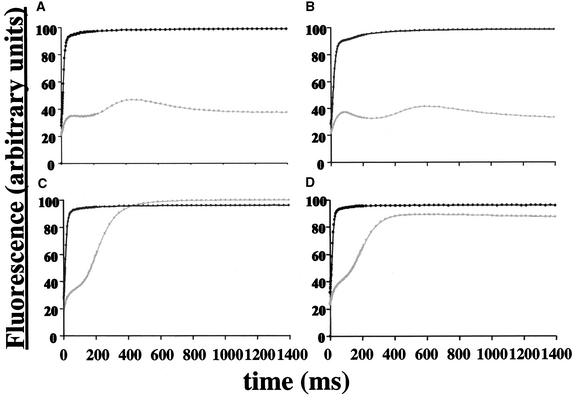
We studied three mutated versions of cytochrome f that lacked the last 6, 8, or 11 amino acids of the protein. The corresponding transformed strains, f312St, f310St, and f307St, were phototrophic, as determined from their ability to grow on MM under high light. Accordingly, their fluorescence induction kinetics were typical of a wild-type pattern (Zito et al., 1997), as illustrated in Figure 2 for the f307St strain. After dark adaptation, the fluorescence yield of f307St cells placed under continuous illumination showed transient phases of increase and decline until it reached a stationary level that was approximately three times lower than the maximum fluorescence level measured with the same strain in the presence of 10 μM DCMU (FmaxDCMU), an inhibitor of photosystem II. This low stationary level is a signature of active electron flow through the photosynthetic apparatus and demonstrates that cytochrome b6f complexes still were assembled and functional in the strains that expressed a truncated form of cytochrome f. Although highly conserved, the last 11 residues of the protein were not required for its function. As expected from the deletion of the petD gene that prevents the assembly of cytochrome b6f complexes, strains f312StΔpetD, f310StΔpetD, and f307StΔpetD, which express these truncated versions of cytochrome f in the absence of SUIV, showed fluorescence induction kinetics typical of mutants deficient in cytochrome b6f complex activity: their fluorescence yield increased steadily up to a stationary level that was similar to the FmaxDCMU level (Figure 2, f307StΔpetD).
Figure 2.
Fluorescence Induction Kinetics of Representative Transformed Strains.
Fluorescence induction curves of dark-adapted cells from wild-type (A), f307St (B), f307StΔpetD (C), and fΔK (D) (see Table 3 and Figure 6) strains in the presence (black symbols and lines) or absence (gray symbols and lines) of 10 μM DCMU. Dots represent individual time-point measurements. a.u., arbitrary units.
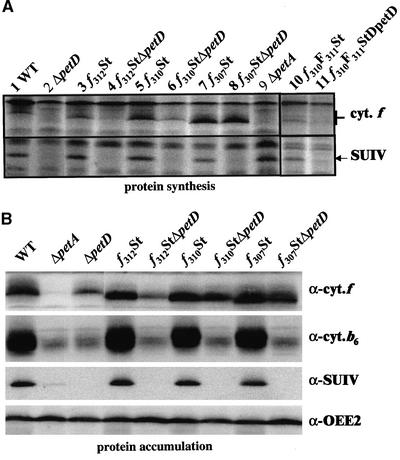
The CES behavior of these truncated forms of cytochrome f then was assessed by comparing their rates of synthesis in pulse-labeled strains expressing or not expressing SUIV. Because the accumulation of petA mRNA was similar in all strains (data not shown), variations in the rate of cytochrome f synthesis should result only from variations in the efficiency of the CES process. As a result of the C-terminal truncations, the three cytochrome f variants displayed greater electrophoretic mobility than their wild-type counterparts (Figure 3A). The rate of synthesis of cytochrome f in strain f312St was similar to that of wild-type cytochrome f (cf. lanes 1 and 3) and was reduced ∼10-fold in the absence of SUIV (lane 4), as was observed for wild-type cytochrome f (lane 2) in strain ΔpetD. Therefore, the CES process was not affected by the removal of the last six amino acids of the polypeptide.
Figure 3.
Expression of Cytochrome b6f Complex Subunits in Strains Expressing Truncated Cytochromes f.
(A) Synthesis rates of cytochrome f and SUIV determined by pulse-labeling experiments in the wild type (WT), ΔpetD, ΔpetA, and strains expressing truncated cytochrome f versions (see list in Table 1).
(B) Immunodetection with specific antibodies of cytochrome b6f subunit accumulation in the same strains. Accumulation of the OEE2 subunit from the photosystem II complex provided a loading control.
By contrast, the rate of cytochrome f synthesis was increased threefold in the f307St strain (lane 7) and remained at the same high level when cytochrome b6f assembly was impaired because of the absence of SUIV in strain f307StΔpetD (lane 8). Therefore, the 11-residue-truncated cytochrome f escaped the CES process, even though it still was assembled in a functional cytochrome b6f complex. Thus, we conclude that some residues involved specifically in the CES process are located between positions 307 and 312. Cytochrome f truncated by only eight residues exhibited intermediate behavior (Figure 3). In the presence of SUIV, its rate of synthesis was increased slightly with respect to that in the wild type (143 ± 39%; lane 5), whereas in the absence of SUIV, it still was synthesized to 46 ± 19% of the wild-type level (lane 6). Thus, in the absence of SUIV, cytochrome f synthesis still was repressed (143 versus 46%), but the efficiency of the CES process was decreased significantly. We noted that cytochrome f truncated by seven residues only but carrying the additional substitution Val-311→Phe for the last residue of the truncated protein [strains f310F311St(/ΔpetD); Figure 3A, lanes 10 and 11] still was regulated by CES. Thus, residues 310 and 311 are not required as such for the CES process. Possibly, the decreased efficiency of the CES regulation in strain f310St is attributable to the vicinity of the negative charge carried by the C terminus of the protein, which is shifted by one residue in strain f310F311St. Together, these experiments confirm that residues critical for the CES process are restricted to amino acids upstream of position 310.
The accumulation of the truncated cytochromes f in the various strains detected in immunoblot experiments with a specific antibody raised against the entire protein (Figure 3B) closely paralleled the rate of cytochrome f synthesis: the truncated versions of cytochrome f were as resistant to proteolytic degradation as regular cytochrome f. Accumulation of the other cytochrome b6f subunits was not altered significantly in these strains compared with their wild-type or ΔpetD counterparts. In the f307St strain, which accumulated three times the wild-type amount of cytochrome f, cytochrome b6 accumulation was increased slightly (117 ± 10.9% of the wild-type level), whereas the accumulation of SUIV remained unchanged (93 ± 7.3% of the wild-type level). Thus, overexpression of cytochrome f did not result in increased accumulation of the assembled cytochrome b6f complex.
Phylogenetic Conservation of the C-Terminal Domain of Cytochrome f
Because these experiments stressed the importance for the CES process of residues from the stromal extension up to position 309, we examined the phylogenetic conservation of the C-terminal domain of cytochrome f. The sequence from Chlamydomonas was aligned with sequences available in the EMBL database on July 30, 2002 (Figure 4). The stromal extension was highly conserved, with residues 303 to 306, 308, 310, and 311 invariant in all species. Special attention should be paid to residue 307, a Phe residue in all chloroplast cytochrome f sequences, which is replaced by aliphatic residues (Val or Ile) in cyanobacteria. Phe also was substituted (by a Tyr) in cytochrome f from Euglena gracilis, which is encoded by a nuclear gene (D. Gonzalez-Halphen, personal communication).
Figure 4.
Phylogenetic Conservation of the C-Terminal Domain from Cytochrome f.
The amino acid sequence of the C-terminal domain from Chlamydomonas was aligned with sequences from other species. Sequences in the first group are from land plants, and the second and third groups correspond to algae and cyanobacteria sequences, respectively. Residues from the transmembrane helix are boxed. Only amino acids that differ from those found at the same positions in Chlamydomonas are indicated, in lightface for conservative substitutions or in boldface for more drastic substitutions. Unchanged residues are symbolized by dots. At bottom, asterisks indicate stress residues invariant in all species, and the circled x indicates residues conserved in all chloroplast cytochrome f but not in cyanobacterial cytochrome f. The thick black line over the Chlamydomonas sequence delineates the region involved in the CES process identified by deletion analysis (residues 307 to 309), whereas residues critical for the formation of the repressor motif, identified by point mutations, are underlined.
By contrast, the sequence of the transmembrane helical domain was less conserved through evolution. Only two residues, Phe-289 and Gln-297, were invariant in all species. The stromal end of the helix (Leu-Val-Leu at positions 300 to 302) still was fairly conserved, with few substitutions, most often conservative in some algae species, whereas the rest of the helix was highly variable.
An Amino Acid Cluster Centered on Residue Phe-307 Contributes to the Repressor Motif
Using a similar strategy, we then performed a systematic analysis of the contribution of each amino acid from the region included between the end of the transmembrane helix at position 303 and residue 309. Four positively charged Lys residues (positions 303 to 305 and 309), present in the stromal extension and conserved through evolution, could be involved in electrostatic interactions with the petA gene 5′ UTR. Therefore, each Lys residue was substituted by an uncharged Met residue. Substitutions in two other positions, Gln-306→Ala and Glu-308→Phe, also decreased polarity, whereas a third substitution, Phe-307→Ser, greatly reduced bulkiness. None of these mutations affected the accumulation of the petA mRNA (data not shown). All substitutions and their effects on the CES process and phototrophic growth are summarized in Table 2. With the exception of the strain carrying the substitution Lys-303→Met, all substitution mutants were phototrophic and indistinguishable from the wild type in terms of growth rates and fluorescence induction kinetics (data not shown). The accumulation of SUIV, which reflects the amount of assembled cytochrome b6f complex (Kuras and Wollman, 1994), was similar to that observed in the wild type, which is indicative of a fully assembled cytochrome b6f complex (data not shown). By contrast, strain f303M grew poorly on MM under dim light (8 μE· m−2·s−1), died when grown on MM under high light (80 μE· m−2·s−1), and accumulated only 15 to 20% of the assembled cytochrome b6f complex (data not shown).
Table 2.
Phenotypes of the Mutations Introduced in the Stromal Extension of Cytochrome f
| C-Terminal Amino Acid Sequence | Strain | CES | Phototrophic Growtha |
|---|---|---|---|
| VLLTQVLLVLKKKQFEKVQLAEMNF | Wild type | + | + |
| VLLTQVLLVLKKKQFEMVQLAEMNF | f309M | + | + |
| VLLTQVLLVLKKKQFFKVQLAEMNF | f308F | ± | + |
| VLLTQVLLVLKKKQSEKVQLAEMNF | f307S | − | + |
| VLLTQVLLVLKKKAFEKVQLAEMNF | f306A | ± | + |
| VLLTQVLLVLKKMQFEKVQLAEMNF | f305M | ± | + |
| VLLTQVLLVLKMKQFEKVQLAEMNF | f304M | + | + |
| VLLTQVLLVLMKKQFEKVQLAEMNF | f303M | − | ∓ |
Same conventions as in Table 1.
+ indicates that the strains are capable of phototrophic growth on MM; ∓ indicates that the strain grows poorly or not at all on MM, even though a small residual electron flow at the level of the cytochrome b6f complex still is observed in fluorescence induction kinetics (see text).
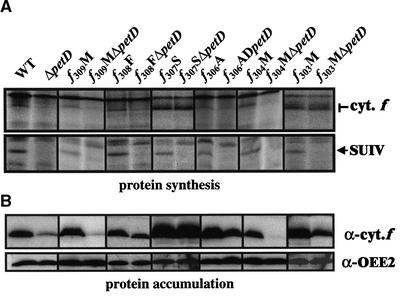
The rate of synthesis of cytochrome f was studied in these mutants as described above. The pattern of cytochrome f synthesis in the pairs of strains f309M(/ΔpetD) and f304M(/ΔpetD) was identical to that observed in the wild-type/ΔpetD strains (Figure 5A). Thus, the CES process was not affected by these substitutions. Three substitutions, Lys-305→Met (data not shown), Gln-306→Ala, and Glu-308→Phe (Figure 5A, Table 2), led to a partially deregulated CES phenotype, similar to what we observed in the f310St(/ΔpetD) strains (Figure 3). Cytochrome f showed a slight overexpression in the presence of SUIV and a moderate repression in the absence of the dominant subunit.
Figure 5.
Cytochrome f Expression in Representative Strains Carrying Mutations in the Stromal Extension.
Synthesis (A) and accumulation (B) of cytochrome f in the wild type (WT), in the control ΔpetD strain, and in the mutant strains f309M(/ΔpetD), f308F(/ΔpetD), f307S(/ΔpetD), f306A(/ΔpetD), f304M(/ΔpetD), and f303M(/ΔpetD). Translation of SUIV is shown in (A), and OEE2 accumulation provided a loading control in (B).
By contrast, residue Phe-307, which is typical of chloroplast-encoded cytochromes f because Val or Ile is found in that position in cyanobacteria, was critical for the regulation of CES. When Phe-307 was substituted by a Ser residue, we observed a complete loss of the CES process and a marked overexpression of cytochrome f, irrespective of the presence or absence of SUIV (Figure 5A). A complete loss of the CES process also was observed in strains carrying the substitution Lys-303→Met (Figure 5A).
With the exception of the Lys-303→Met substitution, these mutations did not decrease the stability of cytochrome f, as shown by immunodetection of the protein, which remained proportional to its rate of synthesis (Figure 5B). In the case of the Lys-303→Met substitution, a decreased proteolytic resistance of the protein was observed, in that cytochrome f accumulated to only 220 ± 14% and 80 ± 10% of the wild-type level in strains f303M and f303MΔpetD, respectively, although its rate of synthesis was increased three times in both strains (Figures 5A and 5B).
This series of point mutations led us to identify a cluster of four amino acids, centered on the Phe-307 residue and specific to chloroplastic cytochrome f, that provides an essential contribution to the formation of the repressor motif.
The Topology but not the Sequence of the Junction between the Transmembrane Helix and the Stromal Extension Is Crucial for the CES Process
The loss of the CES process for cytochrome f upon the removal of a positively charged residue at position 303 called for further investigation. This residue could contribute directly to the folding of the downstream cluster. However, this residue, being the first residue of the “stop-transfer” signal of the stromal extension, also could be critical for the junction between the transmembrane helix and the stromal extension. Its substitution by a hydrophobic residue could tilt the C-terminal exit of the helix at the membrane surface and alter the topology of the Phe-307 cluster with respect to the plane of the thylakoid membrane. All of the mutations listed in Table 3 preserve the primary sequence of the stromal extension but modify its junction with the transmembrane helix by interspacing one additional hydrophobic (Ile or Val) or positively charged (Lys or Arg) residue between residues Leu-302, the last residue of the transmembrane helix, and Lys-303, the first amino acid of the stromal extension. We also deleted one of the three Lys residues at the beginning of the stromal extension. None of the transformants was able to grow on MM under high light.
Table 3.
Phenotypes of Strains with Amino Acid Sequence Alterations at the Junction between the Transmembrane Helix and the Stromal Extension
| C-Terminal Amino Acid Sequence | Strain | CES | Phototrophic Growth |
|---|---|---|---|
| RIQGLLVFFSFVLLTQVLLVLKKKQFEKV | Wild type | + | + |
| RIQGLLVFFSFVLLTQVLLVLIKKKQFEKV | f302::I | − | ∓ |
| RIQGLLVFFSFVLLTQVLLVLVKKKQFEKV | f302::V | − | ∓ |
| RIQGLLVFFSFVLLTQVLLVLRKKKQFEKV | f302::R | − | ∓ |
| RIQGLLVFFSFVLLTQVLLVLKKKKQFEKV | f302::K | − | ∓ |
| RIQGLLVFFSFVLLTQVLLVLΔKKQFEKV | fΔK | − | ∓ |
Same conventions as in Table 2. Δ indicates the deletion of the corresponding amino acid from the wild-type sequence.
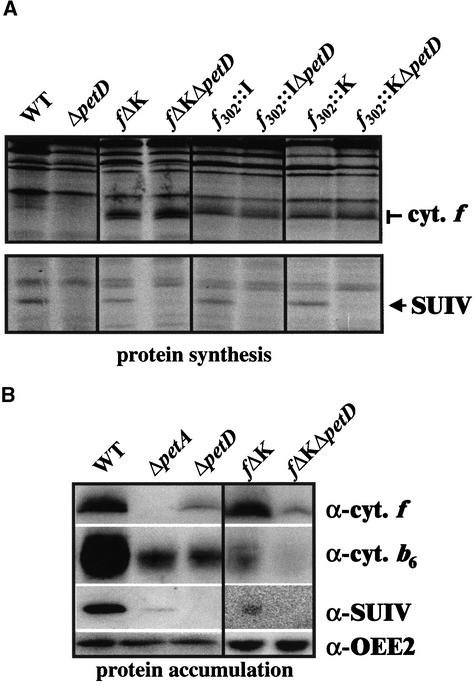
As illustrated in Figure 6B for strain fΔK, the accumulation of SUIV or cytochrome b6 was reduced significantly in these transformants, ranging from 15 to 70% of the wild-type amount depending on the strains. This was indicative of a defect in cytochrome b6f complex assembly that was substantiated by the pattern of fluorescence induction, which is typical of an impairment of electron flow at the level of the cytochrome b6f complex (Figure 2, fΔK): the fluorescence yield increased steadily to a stationary level, instead of showing a transient increase followed by a decline phase, as in photosynthetically active strains. However, the stationary level of fluorescence observed in the mutants remained below the FmaxDCMU level. Thus, electron transfer through the cytochrome b6f complex, although hampered severely, still occurred in these strains. Further functional characterization of cytochrome b6f activity in strains f302::I, f302::K, and fΔK demonstrated a strongly reduced amount of functional complex (G. Finazzi and R. Barbagallo, personal communication).
Figure 6.
Expression of Cytochrome b6f Complex Subunits in Strains Altered in the Junction between the Transmembrane Helix and the Stromal Extension.
(A) Rates of cytochrome f and SUIV synthesis in fΔK(/ΔpetD), f302::I(/ΔpetD), and f302::K(/ΔpetD) strains compared with those in the wild type (WT) and ΔpetD.
(B) Cytochrome b6f subunit accumulation in control and fΔK(/ΔpetD) strains analyzed by immunoreaction with specific antibodies. Accumulation of the OEE2 protein is shown as a loading control.
The synthesis of cytochrome f escaped the CES process in all of these strains [Table 3, Figure 6A for strains fΔK(/ΔpetD), f302::I(/ΔpetD), and f302::K(/ΔpetD)]: the mutated cytochromes presented a threefold increased rate of synthesis compared with wild-type cytochrome f, regardless of whether SUIV was synthesized or not. Insertions of an extra hydrophobic residue at the C-terminal end of the helix in strain f302::I should rotate by 100° the orientation of the hydrophilic stretch that protrudes at the stromal side of the thylakoid membranes. Mutations fΔK and f302::K did not modify the amino acid sequence of the protein up to the first two and three amino acids of the stromal extension. These observations strongly suggest that the CES process for cytochrome f synthesis requires a precise topology, rather than a precise primary sequence, at the exit of the transmembrane helix of the polypeptide chain. However, the spacing between the Phe-308 cluster and the transmembrane helix was also modified by these insertions. This finding prompted us to determine whether some residues within the transmembrane helix also contribute to the formation of the regulatory motif.
Importance of Residues from the Transmembrane Helix for the CES Process
The mutations performed within the helical domain are listed in Table 4. Two cytochrome f variants, f298::S and f295::A, bear one amino acid insertion one or two helical turns, respectively, before the C-terminal end of the transmembrane helix. We also substituted by a Cys the two residues, Phe-289 and Gln-297, conserved in all species. The additional mutations Gln-284→ Cys, Gly-285→Cys, Leu-299Leu-302→AlaAla, and Lue-302→ Val substituted amino acids that are often found in petA sequences from other species at both ends of the transmembrane helix. Here again, the accumulation of petA mRNA was similar in all mutant strain (data not shown).
Table 4.
Characterization of the Mutation Introduced in the Transmembrane Helix
| C-Terminal Amino Acid Sequence | Strain | CES | Phototrophic Growth |
|---|---|---|---|
| RIQGLLVFFSFVLLTQVLLVLKKKQFEKV | Wild type | + | + |
| RIQGLLVFFSFVLLTQVSLLVLKKKQFEK | f298::S | − | ∓ |
| RIQGLLVFFSFVLLATQVLLVLKKKQFEK | f295::A | − | + |
| RIQGLLVFFSFVLLTQVLLVVKKKQFEKV | f302V | + | + |
| RIQGLLVFFSFVLLTQVAAVLKKKQFEKV | f299A300A | + | + |
| RIQGLLVFFSFVLLTCVLLVLKKKQFEKV | f297C | − | + |
| RIQGLLVCFSFVLLTQVLLVLKKKQFEKV | f289C | + | + |
| RIQCLLVFFSFVLLTQVLLVLKKKQFEKV | f285C | + | + |
| RICGLLVFFSFVLLTQVLLVLKKKQFEKV | f284C | + | + |
Same conventions as in Table 2.
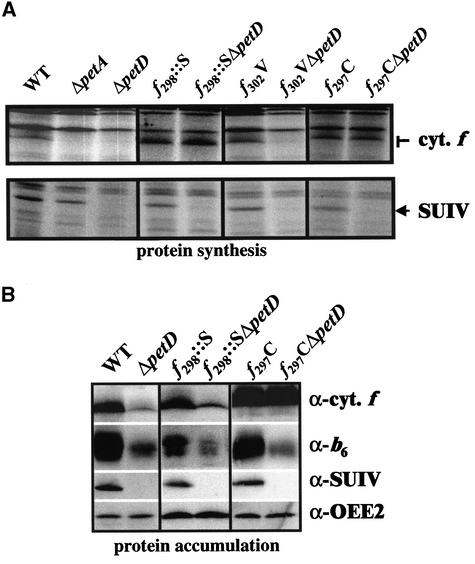
Mutant f298::S displayed a fluorescence phenotype and a pleiotropic loss in cytochrome b6f subunits typical of an impairment of protein assembly (Figure 7B), whereas mutant f295::A and the other strains carrying a substitution within the transmembrane helix were capable of phototrophic growth and behaved like the wild type in terms of growth rate, fluorescence kinetics, and cytochrome b6f assembly (data not shown).
Figure 7.
Expression of Cytochrome b6f Complex Subunits in Strains Bearing Mutations in the Transmembrane Helix of Cytochrome f.
(A) petA and petD genes translated in three representative strains carrying a mutation within the transmembrane helix alone or associated with a deletion of the petD gene. WT, wild type.
(B) Accumulation of cytochrome b6f subunits in strains f298::S(/ΔpetD) and f297C(/ΔpetD) probed with specific antibodies.
As shown by pulse-labeling experiments, the CES process was unaffected in strain f302V(/ΔpetD) (Figure 7) as well as in strains f299A300A(/ΔpetD), f289C(/ΔpetD), f285C(/ΔpetD), and f284C(/ΔpetD) (data not shown). By contrast, the CES process was abolished completely in strains f298::S and f297C (Figure 7A) as well as in strain f295::A (data not shown): the rates of synthesis of cytochrome f were increased threefold with respect to the wild type, irrespective of the presence or absence of SUIV. In strains f297C(/ΔpetD), cytochrome f accumulation was increased three times, irrespective of the presence or absence of SUIV (Figure 7B). Thus, the Gln-297→Cys substitution did impair the CES process without altering the stability of the mutant cytochrome f. By contrast, cytochrome f was strongly destabilized in strain f298::S(/ΔpetD) (Figure 7B): despite the threefold increased rate of synthesis, cytochrome f accumulated only to 115 ± 10% and 20 ± 7% of the wild-type level in the presence or absence of SUIV.
DISCUSSION
Cytochrome f Synthesis Is Autoregulated through a C-Terminally Located Repressor Motif
We observed previously that cytochrome f synthesis is reduced in mutant strains defective in cytochrome b6f complex assembly because of the absence of another subunit of the same protein complex (Kuras and Wollman, 1994). This downregulation of cytochrome f synthesis, which we named a CES process (Wollman et al., 1999; Choquet and Vallon, 2000) is mediated by the 5′ UTR of the petA mRNA, showing that it results from an actual regulation of cytochrome f translation initiation. This is illustrated in the present study by the ability of the 5′ UTR of the petA mRNA to confer a cytochrome b6f assembly–dependent rate of translation to the unrelated α-subunit of the ATP synthase complex. This result, obtained with an endogenous reporter gene, confirms quantitatively the qualitative observation made previously with the heterologous AadA reporter protein (Choquet et al., 1998).
However, we also had observed an opposite effect in two other mutants defective in cytochrome b6f assembly: a threefold increase in the rate of cytochrome f synthesis in strain f283St (Kuras et al., 1995b) that lacked the C-terminal transmembrane anchor and the stromal extension of the protein as well as in strain f52L55V that expressed a very unstable hemeless version of cytochrome f (Choquet et al., 1998). Both mutant strains failed to assemble cytochrome b6f complexes. We started the present study by performing truncations of various length at the C terminus of cytochrome f and found that residues from the stromal extension at positions 307 to 310 played a critical role in the regulation of cytochrome f translation. Cytochrome f escaped the CES process in strains such as f307St that still display a functional assembly of cytochrome b6f complexes: its rate of synthesis was higher than in the wild type and independent of the presence or absence of an assembly partner such as SUIV. Thus, we can exclude the transactivation hypothesis, which postulates that some other cytochrome b6f subunits, when not assembled fully, can act as activators for cytochrome f translation. We conclude that cytochrome f translation actually is autoregulated via some motif that extends in the short stromal stretch at the C-terminal part of the protein. The phenotype of f307St also offers a unique opportunity to assess the physiological contribution of the CES process to the fitness of photosynthesis in Chlamydomonas grown under various environmental conditions. Preliminary experiments in which the CES mutant and a wild type were grown in phototrophic conditions in a 5% CO2 atmosphere under 125 μE· m−2·s−1 illumination showed slower rates in the exponential phase of growth of the mutant. Further experiments using a variety of growth conditions are required before any definite conclusions can be drawn regarding the importance of the CES process in the physiology of Chlamydomonas.
Characteristics of the Repressor Motif
Further mutational analysis stressed the CES contribution of five residues that are conserved strictly in all chloroplast-encoded cytochromes f: a four–amino acid cluster—Lys-Gln-Phe-Glu at positions 305 to 308 from the stromal extension—and residue Gln-297 in the transmembrane helix. The repressor motif also depends strictly on the position of the stromal cluster with respect to the membrane-embedded Gln-297, because insertions or deletions of one residue in the intervening sequence fully abolished the CES process and provoked a stimulation of cytochrome f synthesis. Thus, the topology as well as the sequence of the C-terminal domain determines the efficiency of the negative feedback. These characteristics have precluded, until now, the successful overexpression of the repressor motif fused to a carrier protein.
On the basis of our C-terminal truncations, we can exclude any further CES contribution from the very last eight residues of the C-terminal stromal extension of cytochrome f. However, we cannot exclude the possibility that other substitutions, harsher than those that we have used, would reveal additional contributions from some residues upstream of position 305. Within the membrane itself, the deleterious effect for the CES process of an Ala insertion at position 295, close to the critical Gln-297 residue, could be indicative of a structural contribution of some unidentified upstream intramembrane residue, but the Ala insertion could merely cause a steric reorientation of Gln-297 with respect to the axis of the transmembrane helix. Unfortunately, the structure of the C-terminal domain of cytochrome f remains unknown, although the lumenal N-terminal domain has been solved at atomic resolution (Martinez et al., 1994; Chi et al., 2000). The crystal structure of the homologous bc1 complex cannot be used in that respect because sequence homology is restricted to the core of the complex, which consists of cytochrome b and the Rieske protein, whereas cytochromes f and c1 have widely different sequences.
A Ternary Effector Is Needed for the CES Process
In most instances in which an autoregulation of translation has been documented, the autoregulated protein has an RNA binding function that mediates its interaction with its own transcript (Portier et al., 1990; Mosner et al., 1995; Wu and Bag, 1998; Hendrick et al., 2001). Here, we found no evidence for such a direct interaction. First, the critical residue Gln-297, buried in the membrane, is unlikely to interact with a stromally located mRNA. Second, we examined the possible contribution of the four Lys residues in the stromal extension of cytochrome f that were reasonable candidates for electrostatic interactions with RNA molecules. Substitution of Lys-305 altered the CES process, but this residue is part of the critical tetrapeptide CES cluster mentioned above. Most importantly, the cluster of three positive charges at the exit of the transmembrane helix is not critical for the formation of the repressor motif, because substitutions of either Lys-304 or Lys-309 by a Met had no effect on the CES process. Lys-303, whose substitution by a Met abolished the CES process, is more likely to play an indirect role, because this substitution moves the stop-transfer signal of the transmembrane helix one residue downstream in the polypeptide sequence; therefore, it should modify the position of the tetrapeptide CES cluster with respect to the membrane plane.
As an alternative, the interaction between unassembled cytochrome f and the petA 5′ UTR should rely on a ternary effector associated with the membrane and able to interact with the cytochrome f domain extending from the membrane-buried Gln-297 residue to the cluster Lys-Gln-Phe-Glu at positions 305 to 308. In this view, the CES effector would be able to modulate the translation of cytochrome f depending on its binding to the repressor motif exposed by unassembled cytochrome f. The CES effector could be either a translational repressor that is activated upon binding to the repressor motif or a translational activator that is trapped upon binding to the repressor motif. It would become available to drive cytochrome f synthesis only when released from cytochrome f (i.e., when cytochrome b6f assembly resumes). In the latter view, the CES effector would act as a gene-specific translational activator capable of competitive binding to the 5′ petA mRNA and to the regulatory motif within the C-terminal domain of cytochrome f. We favor this model because the requirement for nucleus-encoded activators is a common feature of chloroplast gene expression (for recent reviews, see Barkan and Goldschmidt-Clermont, 2000; Zerges, 2000).
Among the nuclear factors that control chloroplast gene expression, MCA1 (Gumpel et al., 1995) and TCA1 (Wostrikoff et al., 2001) are required specifically for the expression of petA. MCA1 is involved in the post-transcriptional stabilization of petA mRNA, binds to its 5′ UTR (S. Purton, Y. Choquet, and D.B. Stern, unpublished data), and is predicted to be a transmembrane protein (S. Purton, unpublished data). It could interact with the membrane-buried cytochrome f residue Gln-297 that plays a critical role in trapping the ternary effector. TCA1, which controls the translation of petA mRNA, also acts on its 5′ UTR, as expected for the ternary CES factor. Although we have not cloned TCA1 and therefore still lack direct sequence evidence for its association with the thylakoid membranes, we note that translational activators in yeast mitochondria (McMullin and Fox, 1993; Green-Willms et al., 2001) are membrane bound. Cloning of the TCA1 gene and further biochemical studies of MCA1 and TCA1 should provide molecular information on their possible roles in the CES process.
Generality of the Regulatory Motif
There are only a few examples of protein assembly control of translation. The assembly of the bacterial flagellum (reviewed by Chilcott and Hughes, 2000). controls the expression of several components, most of them at the level of transcription, with the exception of the FlgM protein. FlgM is thought to be regulated at the level of translation or cotranslational secretion through some multistep regulation process (Karlinsey et al., 1998). In eukaryotes, the only example of protein assembly control of translation that does not result from a mere loss of mRNA or from the known mRNA binding function of the autoregulated protein is the post-transcriptional regulation of α-tubulin expression in Chinese hamster cells (Gonzalez-Garay and Cabral, 1996). The present CES regulation mechanism may be more common in organelle gene expression than was thought previously. It is a general feature of chloroplast gene expression in Chlamydomonas, because at least one CES subunit is found in each main photosynthetic complex (Choquet and Vallon, 2000). Also, there is indirect evidence for CES behavior by some chloroplast subunits in tobacco or barley (Gamble and Mullet, 1989; Kim et al., 1994; Rodermel et al., 1996), including cytochrome f (Barkan et al., 1994; Fisk et al., 1999; Monde et al., 2000). Cytochrome f is not expected to be under CES control in cyanobacteria, given the absence of these ternary effectors, which belong to the class of specific translational activators that are observed in organelles only. Indeed, in contrast to Chlamydomonas, high levels of unassembled and unprocessed pre-apocytochrome f were reported to accumulate in the thylakoid of Synechococcus elongatus, with no indication of an autocontrol of synthesis (Tichy and Vermaas, 1999). It is noteworthy that one of the critical CES residues identified during the course of this work, the aromatic Phe-307 residue, is a conserved signature of chloroplast cytochrome f, whereas aliphatic residues (Val or Ile) are found at similar positions in cyanobacterial sequences. In cyanobacteria, the major cytochrome b6f complex subunits are encoded by the petC/petA and petB/petD operons (Schneider et al., 2000), in which the stable petA gene product should be translated only after the petC gene product is made. This operon organization has been lost in chloroplasts. Thus, the CES process could be regarded as a translational regulation mechanism specific to the organelle, which has to cope with the splitting of operons and the migration to the nucleus of some of the structural genes that encode subunits of organelle-located oligomeric proteins.
METHODS
Strains, Media, and Genetic Methods
A wild-type Chlamydomonas reinhardtii strain (mt+) derived from strain 137C was used as a control strain and for chloroplast transformation experiments. The other strains used in this work were the deletion strains ΔpetD and ΔpetA (Kuras and Wollman, 1994), the chloroplast transformant FAAA (Drapier et al., 2002), and the nuclear mutant mcd1-F16 (Drager et al., 1998). Wild-type, mutant, and transformed strains were grown on Tris-acetate-phosphate medium, pH 7.2, at 25°C under dim light (5 to 6 μE·m−2·s−1) (Harris, 1989). Crosses were performed as described by Harris (1989).
Nucleic Acid Manipulations
Plasmid pWFΔpetD, bearing the wild-type petA gene associated with deletions of the petA-petD intergenic region and of the petD gene, replaced by the aadA cassette (see supplemental data online), was constructed from pWFA (Kuras et al., 1997) using the strategy described previously (Kuras et al., 1995b).
Plasmid pFCmut was created by cloning the 876-bp BstEII-EcoRV fragment from plasmid pWF (Kuras and Wollman, 1994) into the vector pBKS− digested by EcoRV and XhoI; both fragments were first blunt-ended by treatment with Klenow. pFCmut, digested by EcoRI and SacI, treated with Klenow, and religated on itself, yielded plasmid pFCter, which contains the last 189 codons and the 3′ untranslated region of the petA gene but lacks most of the restriction sites from the pBKS− polylinker.
Plasmid pFCter was used as a template in PCR using primers FHIS6 and FHISStop (see supplemental data online). The 3.8-kb amplified product was digested by PstI and ligated on itself to yield plasmid pFCterH6, which contained the 3′ part of the petA coding sequence fused at its C-terminal end to a six-His tag.
To generate the cytochrome f versions mutated in the C-terminal domain (see supplemental data online), plasmid pFCter (or pFCterH6) was used as a template in PCR using appropriate pairs of partially overlapping oligonucleotides as primers. PCR products then were digested with the single cutter enzymes, whose restriction sites were introduced together with the mutations and religated on themselves to yield the various mutated plasmids pCterM. These plasmid were sequenced to check for the presence of the correct mutation. The 554-bp AccI-EcoRV fragments excised from these plasmids then were ligated with the 8.5- and 6.7-kb fragments from plasmids pWFA and pWFΔpetD digested with the same enzyme to yield plasmids pfM and pfMΔpetD, respectively.
Transformation of Chlamydomonas and Characterization of the Transformed Strains
Wild-type cells were transformed by tungsten particle bombardment as described previously (Kuras and Wollman, 1994) with a helium particle gun. Transformants, which contained the aadA cassette, were selected on Tris-acetate-phosphate-spectinomycin–containing plates (100 μg/mL) and subcloned on spectinomycin-containing plates until they reached homoplasmy, as determined by DNA filter hybridization or restriction fragment length polymorphism analysis of specific PCR products. Characterization of cytochrome b6f mutant phenotypes was performed using their fluorescence induction kinetics according to Zito et al. (1997). Strains transformed by plasmids pfMΔpetD were screened by fluorescence for defective cytochrome b6f activity. At least three independent transformants were analyzed for each construct. Phototrophic growth was tested by plating the cells on minimal medium under dim light (5 to 8 μE·m−2·s−1) or high light (80 μE·m−2·s−1).
Pulse-labeling experiments, protein isolation, separation, and immunoblot analysis using 125I-protein A were performed as described by Kuras and Wollman (1994). All pulse-labeling experiments were repeated twice and performed on three independent transformants. Cell extracts were loaded on an equal chlorophyll basis. Quantification of the rate of translation of the chloroplast-encoded polypeptides and of the accumulation of cytochrome b6f subunits was performed on PhosphorImager scans using the program ImageQuant (Molecular Dynamics, Sunnyvale, CA). Values for the rates of synthesis of cytochrome f and the apoprotein of CP43 were corrected for background by measuring in each lane an empty window of the same area below cytochrome f. Accumulation of cytochrome b6f subunits was normalized to that of the OEE2 protein from the photosystem II complex. The rates of translation of cytochrome f variants or of the chimeric α-subunit from the ATP synthase complex were normalized to that of the apoprotein of CP43 from photosystem II to correct for variations in the uptake and incorporation of radiolabeled acetate in the various strains.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The GenBank accession number for MCA1 is AF330231.
Supplementary Material
Acknowledgments
We are grateful to D. Drapier, R. Kuras, and D.B. Stern for critical reading of the manuscript and/or stimulating discussion during the course of this work. We thank J.L. Santillan-Torres, A. Atteia, and D. Gonzalez-Halphen for the communication of the Euglena petA sequence before publication. This work was supported by the Centre National de la Recherche Scientifique (Unité Propre de Recherche 1261 and Unité Mixte de Recherche 7099) and by the Collège de France. K.W. was Attaché Temporaire d'Enseignement et de Recherche at the Collège de France.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011692.
Footnotes
Online version contains Web-only data.
References
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear gene in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Barkan, A., Walker, M., Nolasco, M., and Johnson, D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13, 3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y.I., Huang, L.S., Zhang, Z., Fernandez-Velasco, J.G., and Berry, E.A. (2000). X-ray structure of a truncated form of cytochrome f from Chlamydomonas reinhardtii. Biochemistry 39, 7689–7701. [DOI] [PubMed] [Google Scholar]
- Chilcott, G.S., and Hughes, K.T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., Stern, D.B., Wostrikoff, K., Kuras, R., Girard-Bascou, J., and Wollman, F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95, 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., and Vallon, O. (2000). Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82, 615–634. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., Wostrikoff, K., Rimbault, B., Zito, F., Girard-Bascou, J., Drapier, D., and Wollman, F.A. (2001). Assembly-controlled regulation of chloroplast gene translation. Biochem. Soc. Trans. 29, 421–426. [DOI] [PubMed] [Google Scholar]
- Drager, R.G., Girard-Bascou, J., Choquet, Y., Kindle, K.L., and Stern, D.B. (1998). In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 13, 85–96. [DOI] [PubMed] [Google Scholar]
- Drapier, D., Girard-Bascou, J., Stern, D.B., and Wollman, F.A. (2002). A dominant nuclear mutation in Chlamydomonas identifies a factor controlling chloroplast mRNA stability by acting on the coding region of the atpA transcript. Plant J. 31, 687–697. [DOI] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble, P.E., and Mullet, J.E. (1989). Translation and stability of proteins encoded by the plastid psbA and psbB genes are regulated by a nuclear gene during light-induced chloroplast development in barley. J. Biol. Chem. 264, 7236–7243. [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19, 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garay, M.L., and Cabral, F. (1996). α-Tubulin limits its own synthesis: Evidence for a mechanism involving translational repression. J. Cell Biol. 135, 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green-Willms, N.S., Butler, C.A., Dunstan, H.M., and Fox, T.D. (2001). Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem. 276, 6392–6397. [DOI] [PubMed] [Google Scholar]
- Gumpel, N.J., Ralley, L., Girard-Bascou, J., Wollman, F.A., Nugent, J.H., and Purton, S. (1995). Nuclear mutants of Chlamydomonas reinhardtii defective in the biogenesis of the cytochrome b6f complex. Plant Mol. Biol. 29, 921–932. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Hendrick, J.L., Wilson, P.G., Edelman, I.I., Sandbaken, M.G., Ursic, D., and Culbertson, M.R. (2001). Yeast frameshift suppressor mutations in the genes coding for transcription factor Mbf1p and ribosomal protein S3: Evidence for autoregulation of S3 synthesis. Genetics 157, 1141–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey, J.E., Tsui, H.C., Winkler, M.E., and Hughes, K.T. (1998). Flk couples flgM translation to flagellar ring assembly in Salmonella typhimurium. J. Bacteriol. 180, 5384–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Klein, P.G., and Mullet, J.E. (1994). Vir-115 gene product is required to stabilize D1 translation intermediates in chloroplasts. Plant Mol. Biol. 25, 459–467. [DOI] [PubMed] [Google Scholar]
- Kuras, R., Buschlen, S., and Wollman, F.A. (1995. a). Maturation of pre-apocytochrome f in vivo: A site-directed mutagenesis study in Chlamydomonas reinhardtii. J. Biol. Chem. 270, 27797–27803. [DOI] [PubMed] [Google Scholar]
- Kuras, R., de Vitry, C., Choquet, Y., Girard-Bascou, J., Culler, D., Buschlen, S., Merchant, S., and Wollman, F.A. (1997). Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 272, 32427–32435. [DOI] [PubMed] [Google Scholar]
- Kuras, R., and Wollman, F.-A. (1994). The assembly of cytochrome b6f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, R., Wollman, F.A., and Joliot, P. (1995. b). Conversion of cytochrome f to a soluble form in vivo in Chlamydomonas reinhardtii. Biochemistry 34, 7468–7475. [DOI] [PubMed] [Google Scholar]
- Martinez, S.E., Huang, D., Szczepaniak, A., Cramer, W.A., and Smith, J.L. (1994). Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure 2, 95–105. [DOI] [PubMed] [Google Scholar]
- McMullin, T.W., and Fox, T.D. (1993). COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J. Biol. Chem. 268, 11737–11741. [PubMed] [Google Scholar]
- Monde, R.A., Zito, F., Olive, J., Wollman, F.A., and Stern, D.B. (2000). Post-transcriptional defects in tobacco chloroplast mutants lacking the cytochrome b6f complex. Plant J. 21, 61–72. [DOI] [PubMed] [Google Scholar]
- Mosner, J., Mummenbrauer, T., Bauer, C., Sczakiel, G., Grosse, F., and Deppert, W. (1995). Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 14, 4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier, C., Dondon, L., and Grunberg-Manago, M. (1990). Translational autocontrol of the Escherichia coli ribosomal protein S15. J. Mol. Biol. 211, 407–414. [DOI] [PubMed] [Google Scholar]
- Rodermel, S., Haley, J., Jiang, C.Z., Tsai, C.H., and Bogorad, L. (1996). A mechanism for intergenomic integration: Abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc. Natl. Acad. Sci. USA 93, 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, D., Altenfeld, U., Thomas, H., Schrader, S., Muhlenhoff, U., and Rogner, M. (2000). Sequence of the two operons encoding the four core subunits of the cytochrome b6f complex from the thermophilic cyanobacterium Synechococcus elongatus. Biochim. Biophys. Acta 1491, 364–368. [DOI] [PubMed] [Google Scholar]
- Tichy, M., and Vermaas, W. (1999). Accumulation of pre-apocytochrome f in a Synechocystis sp. PCC 6803 mutant impaired in cytochrome c maturation. J. Biol. Chem. 274, 32396–32401. [DOI] [PubMed] [Google Scholar]
- Wollman, F.-A. (1998). The structure, function and biogenesis of cytochrome b6f complexes. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 459–476.
- Wollman, F.A., Minai, L., and Nechushtai, R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1411, 21–85. [DOI] [PubMed] [Google Scholar]
- Wostrikoff, K., Choquet, Y., Wollman, F.A., and Girard-Bascou, J. (2001). TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., and Bag, J. (1998). Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J. Biol. Chem. 273, 34535–34542. [DOI] [PubMed] [Google Scholar]
- Zerges, W. (2000). Translation of mRNAs encoded by chloroplast genomes. Biochimie 82, 583–601. [DOI] [PubMed] [Google Scholar]
- Zito, F., Kuras, R., Choquet, Y., Kossel, H., and Wollman, F.-A. (1997). Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol. 33, 79–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.