Abstract
Low glutelin content1 (Lgc1) is a dominant mutation that reduces glutelin content in rice grains. Glutelin is a major seed storage protein encoded by a multigene family. RNA gel blot and reverse transcriptase–mediated PCR analyses revealed that Lgc1 acts at the mRNA level in a similarity-dependent manner. In Lgc1 homozygotes, there is a 3.5-kb deletion between two highly similar glutelin genes that forms a tail-to-tail inverted repeat, which might produce a double-stranded RNA molecule, a potent inducer of RNA silencing. The hypothesis that Lgc1 suppresses glutelin expression via RNA silencing is supported by transgenic analysis using this Lgc1 candidate region, by reporter gene analysis, and by the detection of small interfering RNAs. In this context, Lgc1 provides an interesting example of RNA silencing occurring among genes that exhibit various levels of similarity to an RNA-silencing–inducing gene. Possible mechanisms for gene silencing of the glutelin multigene family by Lgc1 are discussed.
INTRODUCTION
Glutelin is a major seed storage protein, accounting for 60% of total endosperm protein in rice. It is encoded by a multigene family that consists of the GluA and GluB subfamilies (Takaiwa et al., 1991). Typically, they show 60 to 65% similarity to each other and >80% similarity within each subfamily. Glutelin is synthesized as a 57-kD precursor and then cleaved into a 37- to 39-kD acidic subunit and a 22- to 23-kD basic subunit. In a mutant line, Low Glutelin Content-1 (LGC-1), the content of glutelin is reduced and the contents of other seed storage proteins, including prolamin, are increased (Iida et al., 1993). Such upregulation is not specific to LGC-1 and is thought to be a kind of compensation for the reduction of glutelin. Although glutelin is accumulated in protein body II, prolamin is accumulated in protein body I (Tanaka et al., 1980). Because protein body I is barely digested in the human body (Ogawa et al., 1987), LGC-1 can be used as a low-protein cultivar. Thus, it is useful for patients who must restrict protein intake, such as kidney disease patients (Mochizuki and Hara, 2000). LGC-1 and some cultivars developed using LGC-1 as a cross parent are beginning to be used for this type of diet therapy.
This low-glutelin trait is conferred by a dominant mutation, Lgc1. On the other hand, several mutants affected in glutelin other than LGC-1 have been described (Kumamaru et al., 1988; Iida et al., 1993, 1997). The acidic subunits of glutelin appear as several spots in two-dimensional electrophoresis. glu1, a mutation deficient in spot 1a, is mapped on chromosome 2 (Iida et al., 1997). The glu2 and glu3 mutations, which are deficient in spots 2a and 3a, respectively, are mapped to chromosomes 10 and 1, respectively. They are recessive and are thought to be loss-of-function mutations of glutelin genes. Interestingly, in LGC-1, spot 1a is deleted and most glutelin proteins also appear to be reduced (Iida et al., 1993), which means that Lgc1 may be related in some way to glu1. We wished to understand the mechanism by which glutelin content is reduced in LGC-1.
RNA silencing is a post-transcriptional gene-silencing mechanism that is conserved among various organisms. It includes post-transcriptional gene silencing (PTGS) in higher plants and fungi (in which it is also called quelling) and RNA interference (RNAi) in animals (Voinnet, 2002). PTGS was found as a gene-silencing phenomenon of endogenous and exogenous homologous genes by sense transgenes. PTGS also can be induced by infection with a virus that has sequences homologous with those of the target genes. RNAi is a form of sequence-specific gene silencing induced by the injection of double-stranded RNA (dsRNA) and can be a useful tool in analyzing gene functions in nematodes, insects, and mammals. These RNA-silencing phenomena require common components, such as RNA-dependent RNA polymerases and eIF2C-like proteins (Cogoni and Macino, 1999; Tabara et al., 1999; Catalanotto et al., 2000; Dalmay et al., 2000b; Fagard et al., 2000; Mourrain et al., 2000; Smardon et al., 2000), and are associated with the generation of 21- to 25-nucleotide small interfering RNAs (siRNAs) (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Ketting et al., 2001; Catalanotto et al., 2002). dsRNA is thought to be involved in all RNA-silencing phenomena. In vitro RNAi systems have revealed the biochemical mechanism of RNA silencing. dsRNA is cleaved into 21-nucleotide double-stranded siRNAs by Dicer, which is a member of the RNase III family (Bernstein et al., 2001). The resultant siRNAs are incorporated in an RNA-induced silencing complex (RISC), which cleaves target RNA near the center of the region complementary to the antisense strand of siRNA (Hammond et al., 2000; Zamore et al., 2000; Elbashir et al., 2001). RNA-dependent RNA polymerase also is thought to function in the degradation of the target RNA (Lipardi et al., 2001; Sijen et al., 2001).
Here, we report the molecular cloning and functional analysis of Lgc1. Lgc1 is a structure harboring a tail-to-tail inverted repeat consisting of a functional GluB gene and a truncated GluB gene. This structure is thought to produce a GluB hairpin RNA with a dsRNA region, a potent inducer of RNA silencing. That is, RNA silencing is responsible for the low-protein trait in LGC-1, which is useful for patients on a restricted protein diet.
RESULTS
GluA and GluB Are Suppressed Differentially in LGC-1
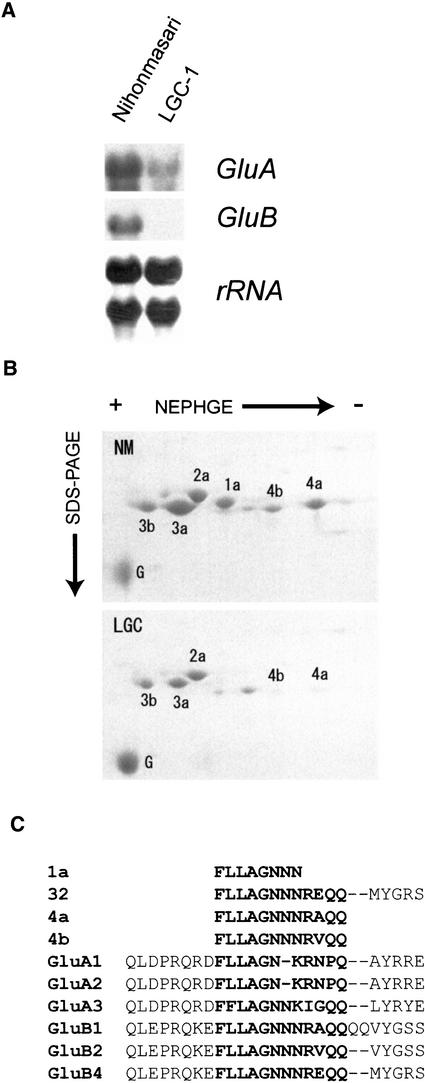
To characterize the mode of reduction of glutelin content in LGC-1, RNA gel blot analysis was performed (Figure 1A). Total RNAs of LGC-1 and its original cultivar, Nihonmasari, were extracted from developing seeds at 16 days after flowering (DAF), when glutelin genes are expressed abundantly. The steady state mRNA levels of both glutelin subfamilies were reduced in LGC-1, suggesting that these reductions are responsible for the low content of glutelin in LGC-1. However, the degree of suppression was different between GluA and GluB. The expression of GluA showed a severalfold reduction in LGC-1 compared with Nihonmasari, whereas GluB was suppressed more remarkably. Two-dimensional electrophoresis of rice seed proteins revealed that glutelin spot 1a was absent and the intensities of spots 4a and 4b were reduced severely, whereas all of the other glutelin spots were weakened (Figure 1B).
Figure 1.
The GluA and GluB Subfamilies Are Suppressed Differentially in LGC-1.
(A) RNA gel blot analysis of the GluA and GluB subfamilies in LGC-1. RNA samples were extracted from developing endosperm at 16 DAF. GluA1 and GluB1 cDNA were used as probes. rRNA was visualized by methylene blue staining of the blotted nylon membrane.
(B) Two-dimensional electrophoresis of glutelin acidic subunits of Nihonmasari (NM) and LGC-1 (LGC). Eighty micrograms of protein prepared from mature seeds was subjected to nonequilibrium pH gradient gel electrophoresis (NEPHGE) and then subjected to SDS-PAGE. The arrows indicate the directions of electrophoresis. G indicates the 26-kD globulin.
(C) Alignment of internal amino acid sequences of glutelin spots. The region from 191 to 217 amino acids of GluB1 is shown. Internal amino acid sequences of 1a, 4a, and 4b were determined in this work. The sequence of spot 32 was determined by Komatsu et al. (1993). Other sequences are deduced from genomic or cDNA clones (Takaiwa et al., 1991). GluB4 corresponds to cDNA clone RG21 (Masumura et al., 1989). The region of amino acid sequences determined in this work is shown in boldface.
Spots 1a, 4a, and 4b are thought to be GluB proteins because GluB gene expression was suppressed more strongly, as shown in the RNA gel blot analysis. To confirm this notion, internal amino acid sequences of glutelin spots 1a, 4a, and 4b were determined by the Cleveland method using digestion with Staphylococcus aureus V8 protease, because the N termini of the glutelin proteins are blocked. An S. aureus V8 protease fragment of spot 1a has the amino acid sequence FLLAGNNN (Figure 1C). Spots 4a and 4b had the same sequence as spot 1a in this region. All of the deduced amino acid sequences of the GluB genes reported to date have the same sequence in this region, whereas those of the GluA genes do not. These results suggest that spots 1a, 4a, and 4b are GluB protein subunits. Consistent with this finding, Komatsu et al. (1993) reported that spot 1a (designated spot 32 in their report) is a GluB protein subunit and spots 2a, 3a, and 3b are GluA protein subunits. The internal amino acid sequence of spot 1a/32 was identical to the deduced amino acid sequence of GluB4. The internal amino acid sequences of spots 4a and 4b were identical to the deduced amino acid sequences of GluB1 and GluB2, respectively.
Lgc1 Does Not Affect the Promoter Activity of GluB
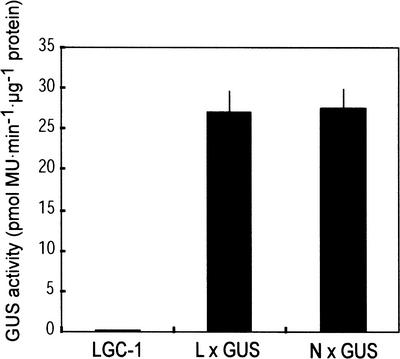
We then used reporter gene analysis to determine whether GluB is suppressed transcriptionally or post-transcriptionally (Figure 2). Pollen from a transgenic line with a β-glucuronidase (GUS) reporter gene driven by the −573-bp region of the GluB1 promoter, which is sufficient to confer full endosperm-specific expression (Wu et al., 1998), was crossed with LGC-1. The Lgc1 gene that is transmitted from an egg cell, as well as the Lgc1 gene that is transmitted from pollen, is active in F1 endosperm. Indeed, we confirmed the Lgc1 phenotype in hybrid seeds of this cross (data not shown). GUS activity in developing seeds of F1 hybrids between the GluB1:GUS transgenic line and LGC-1 (26.2 ± 4.2 pmol methylumbelliferone·min−1· μg−1 protein) was similar to that in developing seeds of F1 hybrids between the transgenic line and Nihonmasari (27.5 ± 4.8 pmol methylumbelliferone·min−1·μg−1 protein). This finding suggests that Lgc1 does not influence the activity of this GluB1 promoter fragment. Given that this promoter fragment can confer the full expression of GluB, it is likely that GluB is suppressed in a post-transcriptional manner in LGC-1.
Figure 2.
The Promoter Activity of GluB1 Is Not Affected by Lgc1.
GUS activity in developing endosperm at 19 DAF was measured. LGC-1, nontransgenic LGC-1 seeds; L × GUS, F1 seeds between a GluB1:GUS transgenic plant and LGC-1; N × GUS, F1 seeds between a GluB1:GUS transgenic plant and nontransgenic Nihonmasari. Pollen from the transgenic plant was crossed with LGC-1 and Nihonmasari. Each column represents the average GUS activity of 5 (LGC-1 and N × GUS) or 10 (L × GUS) samples. Standard deviations are shown as bars. MU, methylumbelliferone.
Molecular Cloning of a Candidate Region for Lgc1
To genetically map Lgc1, restriction fragment length polymorphism (RFLP) analysis was performed using an F2 population between LGC-1 and an indica cultivar, Kasalath. Preliminary mapping using RFLP makers G365 and XNpb243 suggested that Lgc1 is located near glu1 on chromosome 2 (data not shown). The fact that both LGC-1 and a glu1 homozygote lack spot 1a prompted us to perform further mapping. Thus, the protein profiles of 160 F2 seeds between LGC-1 and the glu1 homozygote were examined using one-dimensional SDS-PAGE. All of the F2 seeds showed either the Lgc1 or glu1 phenotype, and no seed with a wild-type band pattern was obtained, suggesting that Lgc1 and glu1 are tightly linked.
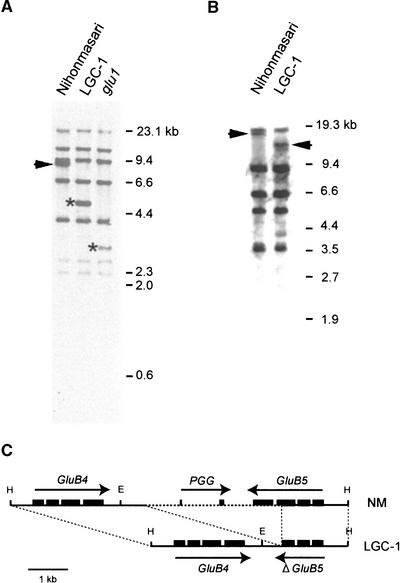
Because this result raises the possibility that Lgc1 is a mutation of a member of the GluB subfamily, DNA gel blot analysis was performed using a GluB1 cDNA clone as a probe. In the DNA gel blot analysis, an 8.5-kb band present in Nihonmasari (Figure 3A, arrowhead) was not observed in LGC-1, whereas a band of 5.5 kb was present specifically in LGC-1 (Figure 3A, top asterisk). There was no cross-hybridization between the GluA and GluB subfamilies under this condition. The presence of the 5.5-kb band was correlated perfectly with the Lgc1 phenotype in 78 F2 seeds between LGC-1 and Nihonmasari (data not shown). Also in the glu1 homozygote, the 8.5-kb band was not observed and a 3.0-kb glu1-specific band was detected. This finding suggests that Lgc1 and glu1 are mutations of the same GluB gene that encodes spot 1a.
Figure 3.
Molecular Cloning of a Candidate Region for Lgc1.
(A) LGC-1 and a glu1 homozygote have a mutation in the same band of GluB. Genomic DNA was digested with HindIII and run on a 0.8% agarose gel. A blotted membrane was probed with a GluB1 cDNA clone and washed with 1× SSC with 0.1% SDS at 65°C. The band indicated by the arrowhead in Nihonmasari disappeared, and the bands indicated by asterisks were detected in LGC-1 and glu1 homozygotes.
(B) The bands exhibiting polymorphism between Nihonmasari and LGC-1 in EcoRI digestions (arrowheads) were cloned and analyzed. The conditions for DNA gel blot analysis were as described for (A).
(C) Structures of a candidate region for Lgc1 and the corresponding region in Nihonmasari (NM). The HindIII fragments corresponding to the bands indicated by the arrowhead in Nihonmasari and by the asterisk in LGC-1 described in (A) are shown. Black boxes and arrows indicate exons and directions of transcription, respectively. The region shown by a dashed line in Nihonmasari is deleted in LGC-1. ΔGluB5 is the truncated GluB5 gene, and PGG is a putative gene predicted by GeneScan. E, EcoRI; H, HindIII.
To determine whether this polymorphism is involved in the Lgc1 phenotype, a polymorphic EcoRI fragment in LGC-1 and the corresponding Nihonmasari fragment (Figure 3B) were isolated. Physical mapping and DNA sequencing revealed that the EcoRI fragment contains a member of the GluB subfamily (GluB5) and that a 3.5-kb fragment is deleted from the third exon to the downstream region in LGC-1 (Figure 3C). Construction of λ phage contigs revealed that GluB4 is located only 3.8 kb downstream of GluB5 in a tail-to-tail direction. GluB4 in LGC-1 has a sequence identical to that in Nihonmasari from at least 0.58 kb upstream of the initiation codon to ∼1.0 kb downstream of the termination codon, suggesting that GluB4 in LGC-1 is functional. GluB4 shares 99.8% identity with GluB5 in the coding region: three nucleotide substitutions are in the fourth exon and two are in the third intron (see supplemental data online). The promoter region is identical at least up to −587 bp. A search of the rice EST database in the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/homology) revealed that both genes are expressed in endosperm. The amino acid sequence of the acidic subunit of GluB4 is identical to that of GluB5 and to the internal amino acid sequence of spot 1a in two-dimensional electrophoresis. This sequence identity, together with the observation that glu1, a recessive mutation deficient in spot 1a, also has a mutation in the 8.5-kb HindIII band containing both GluB4 and GluB5 (Figure 3C), suggest that GluB4 and GluB5 encode spot 1a and that glu1 has a mutation involving both GluB4 and GluB5.
GeneScan with maize matrix (http://genes.mit.edu/GENSCAN.html) predicted a gene in the region between GluB4 and GluB5 (see supplemental data online). This gene, designated PGG (Predicted Gene by GeneScan), consists of two exons separated by one intron of 971 bp and encodes a protein of 25 amino acids. No homolog of PGG was found in plants or other organisms.
Expression of Members of the Glutelin Multigene Family in LGC-1
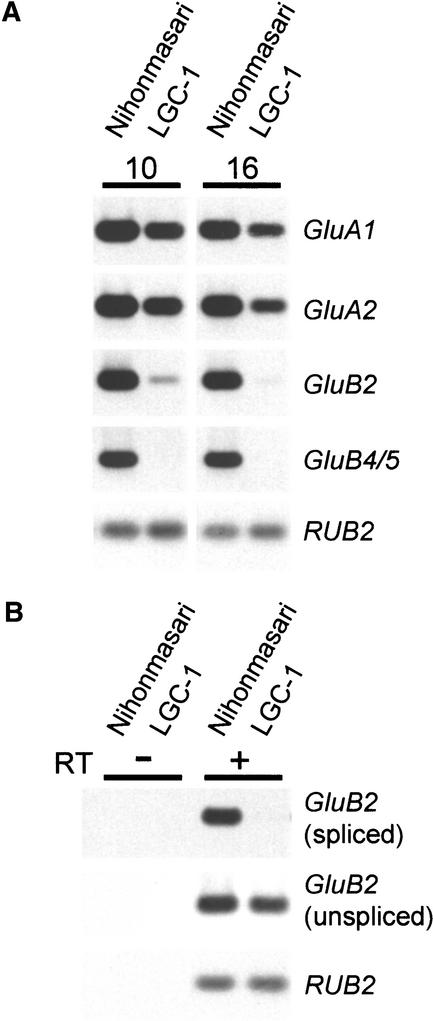
To analyze the expression of some members of the GluA and GluB subfamilies separately, quantitative reverse transcriptase–mediated (RT) PCR was performed (Figure 4A). Total RNA was extracted from developing seeds at 10 and 16 DAF. Because of the high similarity between GluB4 and GluB5, expression of GluB4 could not be distinguished from that of GluB5. Therefore, the RT-PCR product represents both GluB4 and GluB5 mRNAs. No signal for GluB4/5 was detected at either 10 or 16 DAF in LGC-1, indicating that GluB4/5 were suppressed severely. However, an increased number of PCR cycles enabled the detection of signals for GluB4/5 in LGC-1, suggesting that GluB4/5 are expressed at very low levels in LGC-1 (data not shown). These results are consistent with the observation that spot 1a encoded by GluB4 and GluB5 appears deficient in LGC-1 (Iida et al., 1993) (Figure 1B). GluB2 is 83.2% similar to GluB4 and likely encodes spot 4b. The expression of GluB2 in LGC-1 was ∼1/10th of that in Nihonmasari at 10 DAF and ∼1/30th of that at 16 DAF, which is consistent with the presence of a faint signal for spot 4b (Figure 1B, bottom).
Figure 4.
Expression of the glutelin Multigene Family during Seed Development.
(A) Expression of members of the glutelin multigene family was detected by quantitative RT-PCR using primer sets specific to each member. RT-PCR products were transferred onto nylon membranes and detected with specific probes. Endosperms were collected at 10 and 16 DAF. GluB4/5 represents both GluB4 and GluB5 expression because they cannot be distinguished as a result of their very high similarity. RUB2 is the ubiquitin gene, which is a reference for the amount of cDNA used for PCR.
(B) Unspliced and spliced GluB2 were detected by quantitative RT-PCR. One set of primers was designed for the spliced GluB2 in the first and third exons, and another set of primers was designed for the unspliced GluB2 in the first and second introns. Reverse transcriptase–treated (RT+) and untreated (RT−) RNAs, which were prepared from the developing endosperm at 16 DAF, were used as templates for PCR.
GluA genes also were suppressed, although less severely. A GluA gene, P0460E08.38, is the only functional glutelin gene on chromosome 1 (http://rgp.dna.affrc.go.jp). Because glu3, a recessive mutation deficient in spot 3a, is mapped very near P0460E08.38 on chromosome 1 (Iida et al., 1997), this glutelin gene is thought to encode spot 3a. Another GluA gene, OSJNBa0050N08.16, is found on BAC OSJNBa0050N08, which contains RFLP marker sequences close to glu2 such as C961, suggesting that the BAC clone is located near glu2 on chromosome 10. Because glu2 is a recessive mutation deficient in spot 2a, OSJNBa0050N08.16 likely encodes spot 2a. According to the currently available rice genome sequence (http://rgp.dna.affrc.go.jp), P0460E08.38 and OSJNBa0050N08.16 are thought to be GluA1 and GluA2, respectively. The similarities of GluA1 and GluA2 to GluB4 are 69.1 and 60.0%, respectively. The degree of suppression of GluA1 in LGC-1 is very similar to that of GluA2: approximately one-half at 10 DAF and approximately one-fifth at 16 DAF. These results suggest that the degree of suppression of glutelin genes in LGC-1 depends on their similarities to GluB4/5 and becomes more severe during seed development. Furthermore, the observation that Lgc1 suppresses GluA genes on chromosomes 1 and 10 but Lgc1 is located on chromosome 2 indicates that Lgc1 acts in trans.
As another approach to examine the post-transcriptional suppression of the glutelin genes in LGC-1, RNA levels of spliced and unspliced glutelin products were examined (Figure 4B). Although the RT-PCR product corresponding to the spliced GluB2 product was reduced significantly in LGC-1, the RT-PCR product corresponding to the unspliced GluB2 product was largely unaffected in LGC-1. Because only mature mRNA is exported from the nucleus to the cytoplasm, this observation suggests that cytoplasmic degradation of GluB2 mRNA occurs in LGC-1. Collectively, Lgc1 suppresses the expression of the glutelin multigene family in similarity-dependent, trans, and post-transcriptional manners. These characteristics are common to RNA silencing.
An Inverted Repeat Consisting of a Functional GluB and a Truncated GluB Confers the Lgc1 Phenotype
GluB5 in LGC-1 lacks a transcription termination signal, because the deletion includes a putative poly(A) signal and the poly(A) site predicted from an EST for GluB5 (Rothnie, 1996). As a consequence of the absence of a transcription termination signal, transcription from the GluB5 promoter may generate a read-through product containing a sense-truncated GluB5, an intergenic region, and an antisense GluB4, which can form a hairpin RNA with the GluB4/5 dsRNA regions. Because dsRNA suppresses the expression of homologous genes, which is referred to as RNAi, it is hypothesized that GluB4/5 dsRNA may suppress glutelin expression in LGC-1.
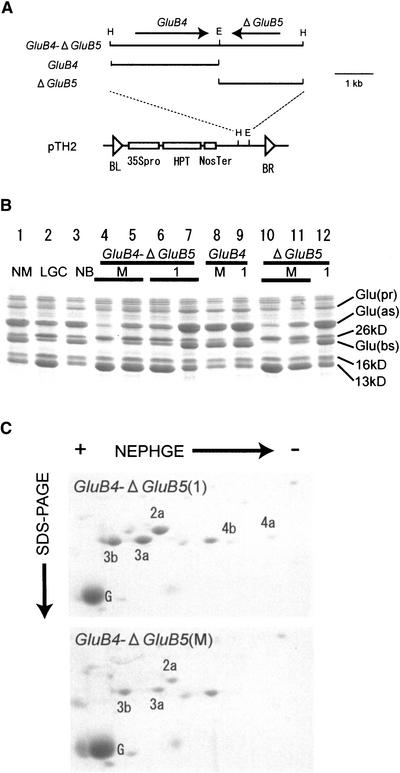
To assess this hypothesis, transformation analysis was performed. A rice cultivar, Nipponbare, was transformed with a HindIII fragment containing both GluB4 and the truncated GluB5 gene (ΔGluB5). This fragment was designated GluB4-ΔGluB5 (Figure 5A). Twenty independent transformants (T0) were obtained, and the phenotypes of T1 seeds were analyzed (Figure 5B). The endosperm half of a seed was used for analysis of protein profiles by SDS-PAGE, and the embryo half was grown to isolate genomic DNA of T1 plants. Nineteen of 20 T0 transformants produced at least one T1 seed with the Lgc1 phenotype: low glutelin and high prolamin contents. The one remaining T0 transformant did not produce T1 progeny with the Lgc1 phenotype (Figure 5B, lane 7, Table 1). This T1 plant had only one copy of the transgene. Thus, in the GluB4-ΔGluB5 transgene, the efficiency of conferring the Lgc1 phenotype was 95%. Sense transgenes are known to occasionally suppress the expression of homologous genes (Vaucheret et al., 1998). This phenomenon is called cosuppression. Therefore, the suppression of glutelin expression by the GluB4-ΔGluB5 transgene could be attributable to cosuppression.
Figure 5.
The GluB4-ΔGluB5 Inverted Repeat Confers the Lgc1 Phenotype Efficiently.
(A) Structures of constructs used for transformation experiments. GluB4-ΔGluB5 represents a HindIII fragment containing the GluB4-ΔGluB5 inverted repeat from LGC-1. GluB4 represents a HindIII-EcoRI fragment containing only GluB4. ΔGluB5 represents an EcoRI-HindIII fragment containing only ΔGluB5. These fragments were inserted into HindIII or HindIII-EcoRI sites in the pTH2 binary vector.
(B) Analysis of protein profiles of transformants by SDS-PAGE. Only representative profiles are shown. NM, Nihonmasari; LGC, LGC-1; NB, Nipponbare. M and 1 represent transformants with multiple copies and one copy of the transgene, respectively. Glu(pr), glutelin precursors; Glu(as), glutelin acidic subunits; Glu(bs), glutelin basic subunits; 26kD, 26-kD globulin; 16kD, 16-kD prolamin; 13kD, 13-kD prolamin.
(C) Two-dimensional nonequilibrium pH gradient gel electrophoresis (NEPHGE) of glutelin acidic subunits in GluB4-ΔGluB5 transformants. The arrows indicate the direction of electrophoresis. GluB4-ΔGluB5(1) represents a GluB4-ΔGluB5 transformant with one copy of the transgene showing modest reduction of glutelin; GluB4-ΔGluB5(M) represents a GluB4-ΔGluB5 transformant with multiple copies of the transgene showing severe reduction of glutelin. G indicates the 26-kD globulin.
Table 1.
Efficiency of Conferring the Lgc1 Phenotype
| Constructs | Total | One Copya | More Than One Copy |
|---|---|---|---|
| GluB4-ΔGluB5 | 19 of 20 | 6 of 7 | 13 of 13 |
| ΔGluB5 | 8 of 16 | 0 of 6 | 8 of 10 |
| GluB4 | 0 of 20 | 0 of 8 | 0 of 12 |
Values shown represent number of T0 transformants producing seeds with the Lgc1 phenotype per total number of T0 transformants.
One band was observed in DNA gel blot analyses using the 35S promoter of Cauliflower mosaic virus as a probe and HindIII or XhoI as a restriction enzyme.
To answer this question, a HindIII-EcoRI fragment containing only GluB4, which has a sequence identical to that of the corresponding region in Nihonmasari, was transformed (Figure 5A). In all 20 independent T0 transformants obtained, no T1 seeds with the Lgc1 phenotype were obtained (Figure 5B, lanes 8 and 9, Table 1). This finding indicates that the GluB4-ΔGluB5 transgene causes the Lgc1 phenotype at a much higher frequency than the GluB4 transgene.
Because PGG is deleted in LGC-1, it also is possible that a reduced dose of PGG has a dominant effect in Lgc1 heterozygotes. However, the GluB4-ΔGluB5 transgene, which lacks PGG and does not change the copy number of PGG in the genome, conferred the Lgc1 phenotype at a very high frequency. This finding suggests that this putative gene is not involved in the Lgc1 phenotype.
The GluB4-ΔGluB5 transgene conferred the Lgc1 phenotype at a high frequency, suggesting that this region contains an element responsible for the Lgc1 phenotype. On the other hand, the inability of the GluB4 transgene to confer the Lgc1 phenotype suggests that the HindIII-EcoRI GluB4 fragment is not involved in conferring or is not sufficient to confer the Lgc1 phenotype. Thus, an EcoRI-HindIII fragment containing ΔGluB5 but not GluB4 was introduced into Nipponbare (Figure 5A). Of 16 independent T0 transformants, 8 plants produced T1 seeds with the Lgc1 phenotype and the remaining 8 plants did not (Figure 5B, Table 1). This result suggests that the suppression of glutelin genes in LGC-1 is partly attributable to ΔGluB5. However, of seven GluB4-ΔGluB5 transformants with a single transgene band in DNA gel blot analysis, six showed the Lgc1 phenotype, whereas of six ΔGluB5 transformants with a single transgene band, none showed the Lgc1 phenotype (Table 1). Lgc1 suppresses glutelin genes in F1 hybrids between LGC-1 and Nihonmasari, which have only one copy of ΔGluB5. This finding suggests that GluB4-ΔGluB5, and not ΔGluB5 alone, is responsible for the Lgc1 phenotype in LGC-1. Thus, the results of these transgenic experiments are consistent with the hypothesis that the expression of glutelin genes is suppressed by a hairpin RNA produced from the GluB4-ΔGluB5 inverted repeat in LGC-1.
The two-dimensional protein profiles of the transformants with glutelin content similar to that in LGC-1 (Figure 5B, lanes 5, 6, and 11) were similar to the two-dimensional protein profile of LGC-1. For example, in a GluB4-ΔGluB5 transformant with a single transgene band, GluB proteins (spots 1a, 4a, and 4b) were weakened severely, whereas GluA proteins (spots 2a, 3a, and 3b) were reduced less severely [Figure 5C, GluB4-ΔGluB5(1)]. On the other hand, some GluB4-ΔGluB5 and ΔGluB5 transformants had lower glutelin contents than LGC-1 (Figure 5B, lanes 4 and 10). In a GluB4-ΔGluB5 transformant with a glutelin content lower than that of LGC-1, GluA proteins also were reduced severely [Figure 5C, GluB4-ΔGluB5(M)], suggesting that this is the main cause of the lower glutelin contents in these transformants, although GluB proteins also were suppressed more severely than they were in LGC-1. Similar results were obtained in a ΔGluB5 transformant with a glutelin content lower than that in LGC-1. Interestingly, these transformants have multiple transgenes. It is possible that the more severe Lgc1 phenotype is related to a greater dose of the transgene. Consistent with this possibility, a slight gene dose effect was observed among LGC-1 and F1 hybrids between LGC-1 and Nihonmasari (see supplemental data online).
Suppression of Glutelin Expression in LGC-1 Is Related to RNA Silencing
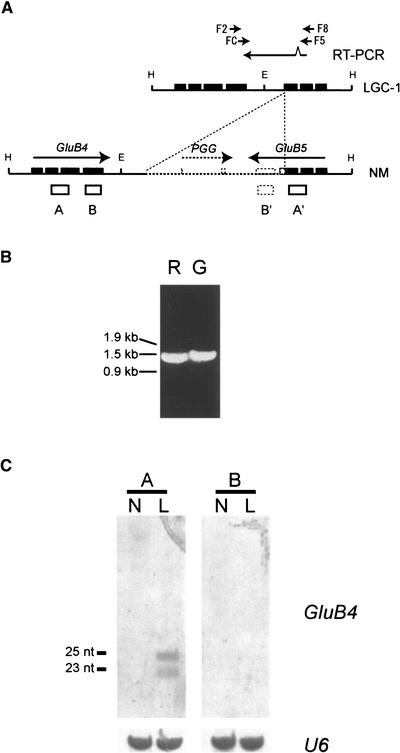
The hypothesis that GluB4/5 dsRNA suppresses glutelin expression in LGC-1 is based on the assumption that a read-through occurs in LGC-1. To confirm this hypothesis, an attempt was made to detect a read-through product by nested RT-PCR using RNA prepared from developing LGC-1 seeds at 16 DAF. PCR primers designed in the fourth exon of GluB4 and the second exon of GluB5 detected a read-through product from GluB5 to GluB4 containing the intergenic region (Figures 6A and 6B). The RT-PCR product was slightly smaller than a genomic PCR product. Sequencing of the RT-PCR product revealed that the second intron was spliced out precisely, suggesting that the RT-PCR product was obtained from RNA transcribed from the GluB5 promoter. An RT-PCR product corresponding to this product was not observed in Nihonmasari (data not shown).
Figure 6.
Detection of a Read-Through Product and siRNA.
(A) Structure of a read-through product and positions of probes used to detect siRNA. The top scheme shows the structure of the read-through product detected by nested RT-PCR. The positions of the primers used in the first-round PCR (F2 and F8) and second-round PCR (FC and F5) are shown. The bottom scheme shows the positions of probes used to detect siRNA (open boxes). Region A in GluB4 is located in the possible dsRNA-forming region and has a corresponding region in GluB5 of LGC-1 (A′). Region B in GluB4 is located in the possible dsRNA-forming region, and the corresponding region in GluB5 (B′) is deleted in LGC-1. The region indicated by the dashed lines is deleted in LGC-1. E, EcoRI; H, HindIII; NM, Nihonmasari.
(B) Amplification of a read-through product from LGC-1. Lane M, molecular mass markers; lane R, the nested RT-PCR product indicated in (A); lane G, the PCR product amplified from LGC-1 genomic DNA with the primer set FC and F5.
(C) Detection of siRNA. Two kinds of GluB4 probes (A and B; see [A]) were used in RNA gel blot analyses of siRNA. Regions A and B correspond to 534 to 1037 bp and 1429 to 1819 bp, respectively, relative to the initiation codon of the GluB4 genomic clone. Two siRNAs of different sizes, which probably correspond to long and short siRNAs, were detected in LGC-1 when region A was used as a probe. The same membrane was probed with the U6 small nuclear RNA probe. L, LGC-1; N, Nihonmasari. The positions of 23- and 25-nucleotide (nt) RNAs are shown at left as a reference.
Furthermore, RNA gel blot analysis was performed to detect siRNA, a hallmark of RNA silencing. When region A, containing parts of the second and third exons of GluB4 (Figure 6A), which is within the possible dsRNA-forming region, was used as a probe, two bands with sizes of 21 to 25 nucleotides were detected specifically in LGC-1 (Figure 6C). Because plants often have two distinct classes of siRNAs, short siRNAs of ∼21 to 22 nucleotides and long siRNAs of ∼24 to 26 nucleotides (Hamilton et al., 2002), these bands are thought to be siRNAs. This observation suggests that Lgc1 acts through RNA silencing. Although no RNA molecule with a dsRNA region was amplified in our RT-PCR analysis (data not shown), this is probably because a double-stranded molecule is not an efficient template for reverse transcription or PCR as a result of the fact that the antisense strand inhibits primer binding (Siebert et al., 1995). Thus, these findings support the involvement of RNAi in the suppression of glutelin genes in LGC-1.
When a part of the fourth exon (Figure 6A, region B) of GluB4, which is deleted in ΔGluB5, was used as a probe, no signal for siRNA was detected. In in vitro RNAi systems, spreading of RNA targeting is reported to occur only in the 3′→5′ direction, because it is thought to be a primer-dependent process (Lipardi et al., 2001; Sijen et al., 2001). However, in plants, spreading of RNA targeting occurs in both the 3′→5′ and 5′→3′ directions (Voinnet et al., 1998; Vaistij et al., 2002), probably because SGS2/SDE1 RNA-dependent RNA polymerase does not require primers to synthesize RNA, as does the fungi RNA-dependent RNA polymerase QDE-1 (Makeyev and Bamford, 2002). The failure to detect siRNA for this region suggests that spreading may not occur in LGC-1. Interestingly, virus-induced gene silencing of some endogenous genes, such as phytoene desaturase and ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit, does not cause spreading of RNA targeting, because the endogenous GluB4 gene does not cause spreading in LGC-1 (Jones et al., 1999; Thomas et al., 2001; Vaistij et al., 2002).
DNA Methylation of Glutelin Genes in LGC-1
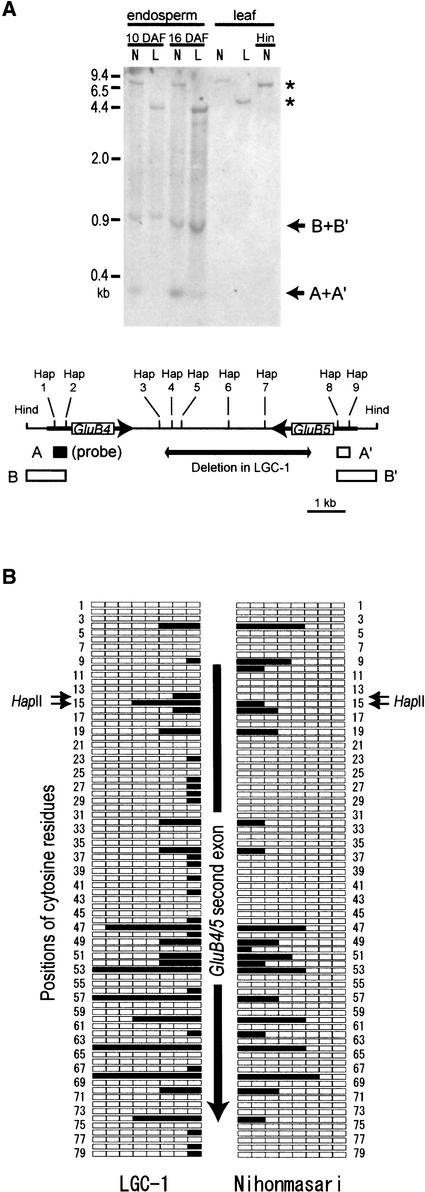
RNA silencing often is associated with DNA methylation. Genomic DNAs from mature leaves and developing endosperm were digested with HindIII and the methylation-sensitive restriction enzyme HapII and subjected to DNA gel blot analysis using a HapII fragment of GluB4 (Figure 7A, fragment A) as a probe. Because GluB4 and GluB5 are highly similar to each other, they sometimes cannot be distinguished in this analysis. For example, the 0.3-kb band indicated by A+A′ contains both fragment A and fragment A′ and the 0.9-kb band indicated by B+B′ contains both fragment B and fragment B′, because the sizes of the HapII fragments to which the fragment A probe hybridizes are identical. The HapII sites in GluB4/5 structural genes—Hap 1, Hap 2, Hap 8, and Hap 9—are in the possible dsRNA-forming region. In mature leaves, most of the HapII sites are thought to be methylated, because only one band corresponding to the HindIII fragment was detected in both Nihonmasari and LGC-1 and no fragment resulting from HapII digestion was detected in either Nihonmasari or LGC-1. In developing endosperm, some HapII-digested fragments were detected, indicating that there is less DNA methylation in endosperm. The intensity of the band corresponding to fragments A and A′ in LGC-1 is weaker than that in Nihonmasari, suggesting that the Hap 1, Hap 2, Hap 8, and Hap 9 sites are methylated more strongly in LGC-1. However, the bands corresponding to B and B′, the presence of which indicates unmethylated Hap 2 and Hap 8 sites, were observed as major bands in Nihonmasari and LGC-1, suggesting that only a modest increase of DNA methylation occurs at these sites.
Figure 7.
DNA Methylation of the glutelin Genes in Developing Endosperm.
(A) DNA methylation in GluB4 and GluB5. Genomic DNA from mature leaves and developing endosperm was digested with the methylation-sensitive restriction enzyme HapII together with HindIII, run on a 1.5% agarose gel, and subjected to DNA gel blot analysis. The scheme at bottom shows the positions of HapII and HindIII sites in the GluB4-GluB5 region. Fragment A in GluB4 (black box) was used as a probe. Under stringent washing conditions, the GluB4 probe did not hybridize to other members of the glutelin multigene family except GluB5. As a result of the very high similarity between GluB4 and GluB5, fragments A and A′ (open box A′) and fragments B and B′ (open boxes B and B′) were detected as the same bands (arrows A+A′ and B+B′). Asterisks indicate bands that correspond to HindIII fragments, although the sizes of the bands are different between Nihonmasari and LGC-1 as a result of the deletion in LGC-1. Hap, HapII site; Hind, HindIII site; L, LGC-1; N, Nihonmasari.
(B) Methylation pattern of the top strand of the second exon of GluB4/5 determined by the bisulfite-mediated genomic sequencing method. Closed and open boxes indicate the numbers of methylated and unmethylated cytosine residues, respectively, in eight clones. The vertical arrow shows the position and direction of the second exon of GluB4/5. Numerals indicate the positions of cytosine residues. Positions 14 and 15 (HapII) correspond to the Hap 2 and Hap 8 sites in (A).
To investigate the methylation pattern of GluB4/5 in detail, the top strand of the region that includes the second exon was analyzed using the bisulfite-mediated genomic sequencing method (Figure 7B). A mixture of genomic DNA from rice endosperm at 10 DAF and from Arabidopsis leaf was used. Complete conversion of unmethylated cytosine to uracil was verified using the Arabidopsis ASA1 region, which is known to be unmethylated (Jeddeloh et al., 1998). Because GluB4 and GluB5 are highly similar to each other, they cannot be distinguished in this analysis. A significant increase in methylation was observed at positions 14 and 15, which correspond to the Hap 2 and Hap 8 sites in Figure 7A. This observation is consistent with the results of the DNA gel blot analysis described above. The average percentage of cytosine residues that are methylated in Nihonmasri is 10.9%, whereas this percentage is 15.8% in LGC-1. The majority of cytosine residues were equally or more strongly methylated in LGC-1, but some were not. Both the DNA gel blot analysis and the bisulfite-mediated genomic sequencing experiments suggest that a modest increase of DNA methylation occurs in LGC-1 endosperm.
DISCUSSION
The low-glutelin cultivar LGC-1 is beginning to be used as low-protein rice in diet therapy for patients with kidney disease. This trait is conferred by a single dominant mutation, Lgc1. Here, we report the molecular cloning of Lgc1 and reveal the mechanism of reduction of glutelin content by Lgc1.
Lgc1 Is a Possible Hairpin RNA–Producing Structure
Lgc1 has characteristics common to RNA silencing. (1) Lgc1 does not affect the promoter activity of GluB, and cytoplasmic degradation of GluB RNA is thought to occur in LGC-1, suggesting that the suppression is likely post-transcriptional. (2) Lgc1 suppresses glutelin genes on chromosomes 1 and 10, whereas Lgc1 is located on chromosome 2, suggesting that Lgc1 acts in trans. (3) The degree of suppression of glutelin genes in LGC-1 depends on the similarity with GluB4/5, suggesting that Lgc1 suppresses the glutelin multigene family in a similarity-dependent manner. (4) siRNA for GluB4/5, which is a hallmark of RNA silencing, was detected in LGC-1. The structure of the Lgc1 candidate region, together with these characteristics, suggests that GluB5 in LGC-1 lacks a transcription termination signal and produces a read-through product from GluB5 to GluB4. This could form a hairpin RNA with the GluB4/5 dsRNA-forming region, leading to the suppression of glutelin genes by RNAi. In fact, a read-through product from GluB5 to GluB4 was detected in LGC-1. Consistent with this hypothesis, the transgenic experiments suggest that the GluB4-ΔGluB5 inverted repeat, rather than ΔGluB5 alone, is responsible for suppression of the glutelin genes. These results indicate that Lgc1 is a “structure” that produces a GluB hairpin RNA rather than a gain-of-function mutation of a GluB5 structural gene.
In higher plants, RNAi often is achieved by transgenes producing a hairpin RNA (Waterhouse et al., 1998). The efficiency of suppression by the transgene is ∼60%, whereas it increases to nearly 100% if the transgene contains an intron (Smith et al., 2000). In the case of the GluB4-ΔGluB5 transgene, the efficiency of conferring the Lgc1 phenotype is 95%. This high efficiency is thought to be caused by introns in ΔGluB5. In fact, the second intron of GluB5 was spliced out precisely in the read-through product detected by RT-PCR.
Although one copy of the ΔGluB5 transgene was not able to suppress glutelin gene expression in the transformation experiments, relatively frequent suppression was observed when more than one copy of the ΔGluB5 transgene was integrated into the rice genome. A possible explanation for this observation is that, if two copies of ΔGluB5 are integrated as an inverted repeat, they will efficiently produce a hairpin RNA because of the lack of transcription termination signals. If they are integrated as a direct repeat, as a result of the lack of transcription termination signals, they will efficiently produce a direct repeat RNA, which is another inducer of RNA silencing (Ma and Mitra, 2002). This hypothesis also could explain why multiple copies of the GluB4 transgene, which is thought to have a transcription termination signal, were not able to induce RNA silencing, at least efficiently.
Endogenous mutations that cause gene silencing often are associated with duplications. For example, duplication of chalcone synthase genes in soybean results in a trans-dominant suppression of homologous genes (Todd and Vodkin, 1996). Duplications of phosphoribosylanthranilate isomerase (PAI) genes in Arabidopsis (Bender and Fink, 1995) and the R gene family in maize (Ronchi et al., 1995) induce DNA methylation of homologous genes and cause gene silencing. Lgc1 is not a mutation that involves duplication, but it has a structure similar to that of some mutations that do involve duplication. Particularly, the structure of PAI1 to PAI4 duplication is very similar to that of Lgc1. The duplication is composed of a tail-to-tail inverted repeat of highly similar genes with promoters. However, the action of the GluB4-ΔGluB5 inverted repeat may be different from that of the PAI1 to PAI4 inverted repeat because the former appears to increase the DNA methylation level of GluB4/5 only modestly, whereas the latter appears to increase DNA methylation of PAI genes drastically (Luff et al., 1999). If so, it will be interesting to determine what differentiates their action. Although these mutations were analyzed in detail, it remains unclear whether their mode of action is transcriptional or post-transcriptional. Lgc1 represents a specific case in which the involvement of RNA silencing was demonstrated.
Function of Short and Long siRNAs
In higher plants, two major siRNA molecules have been reported: short and long siRNAs (Hamilton et al., 2002). Llave et al. (2002) cloned siRNAs in Arabidopsis and revealed that they consist of 21- and 24-nucleotide molecules, which likely represent short and long siRNAs. This is in contrast to the siRNA of fruit flies, which has a length of 21 nucleotides. The short siRNA in plants is expected to have the same function as the siRNA in fruit flies (i.e., it is incorporated into RISC and serves as a guide for sequence-specific mRNA cleavage). This view is supported by the observation that short siRNA is correlated with the degradation of target mRNAs (Hamilton et al., 2002). Furthermore, Hamilton et al. (2002) suggested that long siRNA is correlated with the systemic silencing and DNA methylation of target genes. In LGC-1, both short and long siRNAs were detected. This observation is consistent with short siRNA having a role in RNA degradation and long siRNA having a role in DNA methylation. However, Lgc1 behaves as a Mendelian factor in the panicles of F1 hybrids, suggesting that the Lgc1 phenotype is not transmitted in the panicle, at least not efficiently. This may mean that long siRNA is not involved in systemic RNA silencing. Alternatively, a surveillance system that prevents the transmission of the RNA-silencing signal exists in spikelets as well as in the shoot apex (Foster et al., 2002). Another possibility is that most glutelin has been accumulated in rice grain before systemic RNA silencing is established, although long siRNA can be detected at 10 DAF, a relatively early stage of seed development (data not shown).
Possible Mechanism of Suppression of the Glutelin Multigene Family by Lgc1
The absence of spreading of RNA targeting in LGC-1 suggests that siRNAs from the possible dsRNA-forming region in GluB4/5 are involved directly in the degradation of mRNA of glutelin genes other than GluB4 and GluB5. A RISC containing the siRNA from GluB4/5 may cleave mRNAs at a site with a sequence complementary to a sequence in the target glutelin gene. Elbashir et al. (2001) demonstrated that one nucleotide mismatch in the central region of the 19-nucleotide double-stranded region in siRNA abolishes RNAi, whereas nucleotide mismatches are more permissive in the 2-nucleotide 3′ overhang and near the end of the 19-nucleotide region. GluB2 shares 82.9% identity with GluB4 in the possible dsRNA-forming region. GluB2 and GluB4/5 share three perfect matching regions of at least 21 nucleotides (two are 21 nucleotides and one is 23 nucleotides in length) in this region, which suggests that GluB2 mRNA could be cleaved by a RISC containing the GluB4/5 siRNA. On the other hand, GluA1 and GluA2, which share 66.4 and 65.0% identity, respectively, with GluB4/5 in the possible dsRNA-forming region, have no sequences that perfectly match GluB4/5 with 19 or more nucleotides in this region. Both have a 20-nucleotide sequence with only one mismatch that could be located at the fifth nucleotide from the end of the 19-nucleotide double-stranded region in an siRNA. Given the fact that mismatches near the end region of siRNA can induce weak RNAi, these siRNAs may be able to induce weak RNAi against GluA1 and GluA2. In addition, it is thought that plant micro RNAs (single-stranded siRNA-related small RNAs that can cleave mRNA as a component of RISC) can cleave mRNA in spite of a small number of mismatches to the target mRNA (Tang et al., 2003). The difference in the degree of suppression among the glutelin multigene family in LGC-1 is thought to be caused by differences in the effectiveness with which the GluB4/5 siRNAs cleave their targets.
Some GluB4-ΔGluB5 and ΔGluB5 transformants have glutelin contents that are lower than the glutelin content of LGC-1 (Figure 5B, lanes 4 and 10). An interesting feature of these transgenic lines is that they could be used to produce “super low-protein rice.” This rice may be more beneficial than LGC-1 for kidney disease patients.
METHODS
Plant Materials
Rice (Oryza sativa) LGC-1, the glu1 homozygote, and the GluB1:GUS transformants were described by Iida et al. (1993), Iida et al. (1997), and Wu et al. (1998), respectively.
Protein Analysis
One-dimensional SDS-PAGE was performed according to Iida et al. (1997). Two-dimensional electrophoresis was performed essentially as described by Iida et al. (1997) except that the pH gradient gel contained 2.5% Ampholine, pH 3.5 to 10, 2.5% Pharmalite, pH 8.5 to 10, and 8 M urea and that SDS-PAGE was performed according to Komatsu et al. (1993). Internal amino acid sequences were determined as described by Komatsu et al. (1993).
Plasmid Constructions
Genomic DNA from Nihonmasari and LGC-1 was digested with EcoRI and run on a gel. Gel fragments corresponding to the band showing a polymorphism in DNA gel blot analysis were cut out. Eluted DNA from the gel fragments was inserted into the EcoRI site of λDash II (Stratagene, La Jolla, CA), and phage libraries were constructed. Genomic DNA partially digested with MboI also was used to construct λDash II phage libraries. These libraries were screened with GluB1 cDNA as a probe, and phage contigs were constructed. HindIII (GluB4-ΔGluB5), HindIII-EcoRI (GluB4), and EcoRI-HindIII (ΔGluB5) fragments from λ clones were inserted into the HindIII site (GluB4-ΔGluB5) or the HindIII-EcoRI sites (GluB4 and ΔGluB5) in a binary vector, pTH2 (Figure 1B). pTH2 was constructed by introducing the 35S promoter of Cauliflower mosaic virus–hygromycin phosphotransferase gene–nopaline synthase terminator cassette into pTRA415(R)-delNTP (Fukuoka et al., 2000).
DNA and RNA Gel Blot Analyses
Leaf genomic DNA was prepared from mature leaves and developing seeds according to Rogers and Bendich (1985). Developing seeds were ground in a mortar and extracted with 10× TE (100 mM Tris-HCl and 10 mM EDTA, pH 7.5) and phenol:chloroform:isoamyl alcohol (25:24:1). Genomic DNA, total RNA, and small RNA from endosperm were isolated simultaneously from the extract using a DNA/RNA kit (Qiagen, Hilden, Germany). Total RNA for the RNA gel blot analysis was extracted by the method of Takaiwa et al. (1987). DNA and RNA gel blot analyses were performed according to Sambrook et al. (1989). For the DNA gel blot analysis, 5 μg of DNA was used for each digestion. Labeling of probes with digoxigenin and detection of signal were performed according to the instructions of the manufacturer (Roche Diagnostics, Mannheim, Germany). CDPstar (Roche Diagnostics) was used as a substrate for alkaline phosphatase. In the RNA gel blot analysis, 10 μg of total RNA was subjected to electrophoresis on a 1.2% agarose gel and transferred to a nylon membrane. mRNA detection was performed with the enhanced chemiluminescence system (Amersham Pharmacia Biotech, Buckinghamshire, UK) under stringent washing conditions (0.1× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] and 0.4% SDS containing 6 M urea at 42°C). Small interfering RNA was detected according to Dalmay et al. (2000a) except for the use of the digoxigenin-labeled PCR product as a probe. Each lane was loaded with 20 μg of small RNA. The oligonucleotide used to detect U6 small nuclear RNA was 5′-GCTAATCTT-CTCTGTATCGTTCCAATTTTATCGG-3′. The oligonucleotide was labeled with a digoxigenin oligonucleotide tailing kit (Roche Diagnostics).
Reverse Transcriptase–Mediated PCR
For nested reverse transcriptase–mediated (RT) PCR, cDNA was made from total RNA of endosperm at 16 days after flowering using the SuperScript first-strand synthesis system (Gibco BRL, Gaithersburg, MD) and random hexamers as primers. The first-round PCR was performed with the primer set F8 (5′-ATGAGCACCAAAAGATCCAC-3′) and F2 (5′-GTTTTGGAACGTTAATGCCCATAG-3′). The second-round PCR was performed with the primer set F5 (5′-AGTTGTTGCTCTATATGTCTT-CGACT-3′) and FC (5′-CTCCTAGATATCAACAACAGAC-3′). PCR conditions were as follows: 94°C for 2 min of denaturalization, 35 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 3 min, and a final extension at 72°C for 5 min. ExTaq (Takara, Tokyo, Japan) was used as the DNA polymerase. For quantitative RT-PCR, cDNA was synthesized from DNase I–treated total RNAs of endosperm using the SuperScript first-strand synthesis system (Gibco BRL) and oligo(dT) as a primer. The PCR conditions were as described above except that the number of cycles was 13. The reduced number of PCR cycles enables the quantification of mRNA. For analysis of unspliced products, cDNA was synthesized using a mixture of the gene-specific primers GluB2R1 and RUB2R1. The number of PCR cycles was 13 for GluB2 and RUB2 and 23 for the unspliced GluB2. The PCR product was run on a gel together with PCR products from a dilution series of standard RNA as a reference. The gel was blotted to a membrane and hybridized with specific probes. Signals were detected with the enhanced chemiluminescence system (Amersham Pharmacia Biotech).
The primers were as follows: 5′-CGAACGTAAATCCATGGCACAAC-3′ (GluB4/5F1) and 5′-CCTCACTTAGTAGCTCATTGTT-3′ (GluB4/5R1) for GluB4/5; 5′-CCTTTTGGTACCTCGATATAGC-3′ (GluB2F1) and 5′-AAG-CCTCACTTAGTAGCTCAGT-3′ (GluB2R1) for GluB2; 5′-GAAAACTCT-ATCAATGATACTAGG-3′ (GluB2intF1) and 5′-TTTTAAATATGAATTTAATGTGAAAAC-3′ (GluB2intR1) for the unspliced GluB2; 5′-TTCTTCGATGTCTCTAATGAGCAA-3′ (GluA1F1) and 5′-AGCTAACAAGAAATC-CCTTTGCCT-3′ (GluA1R1) for GluA1; 5′-TTCTTCGATGTCTCTAATGAGTTG-3′ (GluA2F1) and 5′-AGCTAACAAGAAATCCCTCTGTCG-3′ (GluA2R1) for GluA2; and 5′-AATCAGCCAGTTTGGTGGAGCTG-3′ (RUB2F1) and 5′-ATGCAAATGAGCAAATTGAGCACA-3′ (RUB2R1) for the 3′ untranslated region of RUB2, a ubiquitin gene (Wang et al., 2000). Specificities of the primer sets were verified by sequencing the RT-PCR products.
GUS Reporter Gene Assay
The GluB1:GUS transgenic plant was crossed with LGC-1 and its original cultivar, Nihonmasari, as a pollen donor. Developing seeds were harvested at 19 days after flowering and used to measure GUS activity as described by Wu et al. (1998). Each sample was a mixture of four seeds harvested from the same pollen-recipient plant.
Transformation Experiments and Analysis of Transformants
Transformation experiments were performed using the rice cultivar Nipponbare according to Fukuoka et al. (2000). Typically, three T1 seeds from each T0 plant were used for analysis. The endosperm half was used for SDS-PAGE analysis, and the embryo half was grown to isolate genomic DNA. Among the three T1 seeds, one seed exhibiting the Lgc1 phenotype was chosen and subjected to DNA gel blot analysis using the 35S promoter of Cauliflower mosaic virus as a probe to count the copy number of transgenes. When none of the three seeds showed the Lgc1 phenotype, one T1 plant with a transgene(s) (confirmed by PCR) was chosen and subjected to DNA gel blot analysis. In this case, more than three seeds were analyzed by SDS-PAGE to confirm that the transformant produced no seeds with the Lgc1 phenotype.
Bisulfite-Mediated Genomic Sequencing
The protocol was performed essentially as described by Paulin et al. (1998). Rice endosperm genomic DNA (140 ng) digested with StuI and PstI was mixed with 75 ng of leaf genomic DNA from Arabidopsis thaliana ecotype Columbia digested with DraI and HindIII. This mixture was subjected to the conversion reaction. The Arabidopsis ASA1 PCR product from the mixed DNA was used to verify the complete conversion of unmethylated cytosine to uracil (Kato et al., 2003). The second exon region of GluB4/5 was amplified with primers GluB45biF1 (5′-ATATATTGTTTATATGGGTATAAATTATGATT-3′) and GluB45biR1 (5′-TTTTACCACATAAACTACAACATATTCATA-3′) for the first round and GluB45biF2 (5′-TTATGTTATAATAATGTTGTTTTAGGTTATAT-3′) and GluB45biR2 (5′-CAACATATTCATATATATAACAATAAAAAACAAA-3′) for the second round.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
Accession numbers for the sequences cited in this article are as follows: D15618 and C98177, C961; AU094576, EST clone for GluB5; and AB093593, GluB4-ΔGluB5.
Supplementary Material
Acknowledgments
We thank F. Takaiwa for providing the glutelin cDNA clones and transgenic plants and I. Mitsuhara and Y. Ohashi for the pTH2 binary vector and helpful discussions. We are grateful to M. Kato and T. Kakutani for their help and for suggestions concerning the bisulfite-mediated genomic sequencing experiment. This work was supported by Ministry of Agriculture, Forestry and Fisheries Rice Genome Project Grants GS-1123 and PR-1203 and, in part, by the budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011452.
Footnotes
Online version contains Web-only data.
References
- Bender, J., and Fink, G.R. (1995). Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83, 725–734. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Catalanotto, C., Azzalin, G., Macino, G., and Cogoni, C. (2000). Gene silencing in worms and fungi. Nature 404, 245. [DOI] [PubMed] [Google Scholar]
- Catalanotto, C., Azzalin, G., Macino, G., and Cogoni, C. (2002). Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999). Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A.J., Mueller, E., and Baulcombe, D.C. (2000. a). Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12, 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Boulcombe, D.C. (2000. b). An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.-B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, T.M., Lough, T.J., Emerson, S.J., Lee, R.H., Bowman, J.L., Foster, R.L.S., and Lucas, W.J. (2002). A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14, 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka, H., et al. (2000). Agrobacterium-mediated transformation of monocot and dicot plants using the NCR promoter derived from soybean chlorotic mottle virus. Plant Cell Rep. 19, 815–820. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., Voinnet, O., Chappell, L., and Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Iida, S., Amano, E., and Nishio, T. (1993). A rice (Oryza sativa L.) mutant having a low content of glutelin and a high content of prolamine. Theor. Appl. Genet. 87, 374–378. [DOI] [PubMed] [Google Scholar]
- Iida, S., Kusaba, M., and Nishio, T. (1997). Mutants lacking glutelin subunits in rice: Mapping and combination of mutated glutelin genes. Theor. Appl. Genet. 94, 177–183. [Google Scholar]
- Jeddeloh, J., Bender, J., and Richard, E.J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12, 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. (1999). RNA–DNA interaction and DNA methylation in post-transcriptional gene silencing. Plant Cell 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., Miura, A., Bender, J., Jacobsen, S.E., and Kakutani, T. (2003). Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13, 421–426. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E.J., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H.A. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, S., Kajiwara, H., and Hirano, H. (1993). A rice protein library: A data-file of rice proteins separated by two-dimensional electrophoresis. Theor. Appl. Genet. 86, 935–942. [DOI] [PubMed] [Google Scholar]
- Kumamaru, T., Satoh, H., Iwata, N., Omura, T., Ogawa, M., and Tanaka, K. (1988). Mutants for rice storage proteins. Theor. Appl. Genet. 76, 11–16. [DOI] [PubMed] [Google Scholar]
- Lipardi, C., Wei, Q., and Paterson, B.M. (2001). RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107, 297–307. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff, B., Pawlowski, L., and Bender, J. (1999). An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3, 505–511. [DOI] [PubMed] [Google Scholar]
- Ma, C., and Mitra, A. (2002). Intrinsic direct repeats generate consistent post-transcriptional gene silencing in tomato. Plant J. 31, 37–49. [DOI] [PubMed] [Google Scholar]
- Makeyev, E.V., and Bamford, D.H. (2002). Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell 10, 1417–1427. [DOI] [PubMed] [Google Scholar]
- Masumura, T., Kidzu, K., Sugiyama, Y., Mitsukawa, N., Hibino, T., Tanaka, K., and Fujii, S. (1989). Nucleic sequence of a cDNA encoding a major rice glutelin. Plant Mol. Biol. 12, 723–725. [DOI] [PubMed] [Google Scholar]
- Mochizuki, T., and Hara, S. (2000). Usefulness of low protein rice on diet therapy in patients with chronic renal failure. Jpn. J. Nephrol. 42, 24–29. [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Kumamaru, T., Satoh, H., Iwata, N., Omura, T., Kasai, Z., and Tanaka, K. (1987). Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 28, 1517–1528. [Google Scholar]
- Paulin, R., Grigg, W., Davey, M.W., and Piper, A.A. (1998). Urea improves efficiency of bisulphate-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res. 26, 5009–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.O., and Bendich, A.J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Ronchi, A., Petroni, K., and Tonelli, C. (1995). The reduced expression of endogenous duplications (REED) in the maize R gene family is mediated by DNA methylation. EMBO J. 14, 5318–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnie, H.M. (1996). Plant mRNA 3′-end formation. Plant Mol. Biol. 32, 43–61. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23, 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H.A., and Fire, A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.-B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissan, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Takaiwa, F., Kikuchi, S., and Oono, K. (1987). A rice glutelin gene family: A major type of glutelin mRNAs can be divided into two classes. Mol. Gen. Genet. 208, 15–22. [Google Scholar]
- Takaiwa, F., Oono, K., Wing, D., and Kato, A. (1991). Sequence of three members and expression of a new major subfamily of glutelin genes from rice. Plant Mol. Biol. 17, 875–885. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Sugimoto, T., Ogawa, M., and Kasa, Z. (1980). Isolation and characterization of protein bodies in the rice endosperm. Agric. Biol. Chem. 44, 1633–1639. [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.L., Jones, L., Baulcombe, D.C., and Maule, A.J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25, 417–425. [DOI] [PubMed] [Google Scholar]
- Todd, J.J., and Vodkin, L.O. (1996). Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Jones, L., and Baulcombe, D.C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Béclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.-B., Mourrain, P., Palauqui, J.-C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2002). RNA silencing: Small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 5, 444–451. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Wang, J., Jiang, J., and Oard, J.H. (2000). Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.). Plant Sci. 156, 201–211. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.-B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.-Y., Suzuki, A., Washida, H., and Takaiwa, F. (1998). The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J. 14, 673–683. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.