Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) possesses a large 29.7-kb positive-stranded RNA genome. The first open reading frame encodes replicase polyproteins 1a and 1ab, which are cleaved to generate 16 “nonstructural” proteins, nsp1 to nsp16, involved in viral replication and/or RNA processing. Among these, nsp10 plays a critical role in minus-strand RNA synthesis in a related coronavirus, murine hepatitis virus. Here, we report the crystal structure of SARS-CoV nsp10 at a resolution of 1.8 Å as determined by single-wavelength anomalous dispersion using phases derived from hexatantalum dodecabromide. nsp10 is a single domain protein consisting of a pair of antiparallel N-terminal helices stacked against an irregular β-sheet, a coil-rich C terminus, and two Zn fingers. nsp10 represents a novel fold and is the first structural representative of this family of Zn finger proteins found so far exclusively in coronaviruses. The first Zn finger coordinates a Zn2+ ion in a unique conformation. The second Zn finger, with four cysteines, is a distant member of the “gag-knuckle fold group” of Zn2+-binding domains and appears to maintain the structural integrity of the C-terminal tail. A distinct clustering of basic residues on the protein surface suggests a nucleic acid-binding function. Gel shift assays indicate that in isolation, nsp10 binds single- and double-stranded RNA and DNA with high-micromolar affinity and without obvious sequence specificity. It is possible that nsp10 functions within a larger RNA-binding protein complex. However, its exact role within the replicase complex is still not clear.
The severe acute respiratory syndrome coronavirus (SARS-CoV) is a positive-stranded RNA virus with a large 29.7-kb genome that encodes 14 open reading frames (ORFs) (26, 30). The first of these ORFs covers two-thirds of the genome and codes for the replicase polyproteins 1a and 1ab (pp1a and pp1ab) (30). pp1ab is formed by frame shifting of the ribosome into the −1 frame during translation, just prior to the pp1a stop codon (2, 30). The two polyproteins are cleaved into 16 “nonstructural” proteins, nsp1 to nsp16, by two viral cysteine proteases, a 3CL-like protease and a papain-like protease (10, 21, 22). These proteolytically generated proteins and/or their cleavage intermediates are involved in viral replication and/or generation of the nested subgenomic mRNAs required for expression of the downstream ORFs within the host cell (32). The remaining ORFs can be categorized into the structural proteins (those that are part of the virion) and the “accessory” proteins. Immunofluorescence microscopy studies in coronaviruses such as murine hepatitis virus (MHV) reveal that some, if not all, of the nsp proteins, including p15 (counterpart to SARS-CoV nsp10), assemble into distinct cytoplasmic, membrane-associated replicase complexes that actively perform viral RNA synthesis (21, 24).
We have recently undertaken an initiative to generate a structure-function-interaction map of the entire proteome of SARS-CoV and its interactions with the host cell, presenting a comprehensive set of targets for therapeutic intervention (http://sars.scripps.edu/). To this end, we are using a structural and functional proteomics approach employing bioinformatics, structural biology, genetic approaches, and macromolecular interaction studies. Our studies and those of other laboratories on the SARS-CoV replicase proteins are beginning to provide the first clues as to the complex replication, transcription, and RNA processing events unique to the life cycle of coronaviruses. We recently determined the nuclear magnetic resonance structure of nsp7 and showed that it adopts a novel fold consisting of five helices, three of them forming a sheet (20). Subsequently, Zhai et al. (31) determined the crystal structure of a supercomplex consisting of eight subunits each of nsp7 and nsp8. This hollow cylinder-like assembly allows the passage of RNA and likely forms the core framework of the replicase complex. The central channel of the cylinder has an internal diameter of ∼30 Å and is positively charged, favoring nucleic acid binding, implying a role in conferring processivity to the RNA-dependent RNA polymerase (31). It has been demonstrated that nsp8 interacts with nsp9 (27), and the structure of the latter has been independently determined by two groups (9, 27). nsp9 binds single-stranded RNA (ssRNA) and double-stranded DNA (dsDNA) without obvious sequence specificity, optimally binding 45-mers that likely wrap once around a dimer of the protein (9, 27).
In this paper, we describe the structure of nsp10, a protein located in the SARS-CoV proteome just downstream of key replicase complex proteins: the nsp7-nsp8 supercomplex as well as the RNA-binding nsp9 on one end and the RNA polymerase (nsp12) on the other (26). The intervening nsp11 is a short 13-residue peptide (Fig. 1). Homologues of nsp10 are found in all coronaviruses (Fig. 1); nsp10 is among the more conserved of the SARS-CoV nsp proteins. nsp10 has been shown to play a critical role in the replicase complex, as a temperature-sensitive mutation of this protein in MHV prevented minus-strand RNA synthesis at the nonpermissive temperature (25). The structure of SARS-CoV nsp10 was solved to a resolution of 1.8 Å by single-wavelength anomalous dispersion (SAD) on a hexatantalum dodecabromide (Ta6Br122+)-derivatized crystal. nsp10 contains two Zn2+-binding sites, an N-terminal subdomain rich in helices, and a C-terminal domain consisting of β-strands and loops. The topological arrangement seen in this structure has not, to the best of our knowledge, been previously observed and can therefore be regarded as a new protein fold. The unique arrangement of residues that coordinate the first Zn2+ ion (involving a histidine and three cysteines) defines a new class of Zn fingers, while the second Zn finger has features of the gag-knuckle fold group (14). The physical location of nsp10 in the SARS-CoV proteome within a cluster of proteins which interact with RNA (26) as well as the occurrence of two Zn fingers and a well-defined positively charged patch on the protein surface hinted at a possible RNA-binding function. However, RNA-binding experiments indicate that while nsp10 binds to both single-stranded and double-stranded DNA and RNA, it does so with only high-micromolar affinity. This does not rule out the possibility that it may bind specifically and/or with greater affinity to nucleic acid as part of a larger complex that confers processivity during replication of the genomic or subgenomic RNA of the virus.
FIG. 1.
Sequence of SARS-CoV nsp10. Shown is a schematic of the SARS-CoV genome depicting the location of the replicase polyprotein and the different structural and accessory genes. The immediate neighborhood of nsp10 within pp1a/1ab is expanded in the inset, under which a sequence alignment of different homologues of nsp10 in coronaviruses is shown. RdRp, RNA-dependent RNA polymerase; TGEV, porcine transmissible gastroenteritis virus; IBV, avian infectious bronchitis virus; Hel, helicase; ExoN, exonuclease; Nendoll, endonuclease; 2′-o-MT, 2′-o-methyltransferase; EDV, epidemic diarrhea virus.
MATERIALS AND METHODS
Construct design, cloning, expression, and purification.
The predicted full-length sequence of nsp10 (gi:29837503; NP_828868) extends from nucleotides 12955 to 13371 of the SARS-CoV genome, corresponding to residues Ala4231 to Gln4369 of pp1a/pp1ab (26). A construct corresponding to residues 10 to 139 of nsp10 (truncating the first nine residues predicted to be random coiled) was amplified by PCR from genomic cDNA of the SARS-CoV Tor-2 strain using Taq polymerase and primer pairs encoding the 5′ and 3′ ends (forward, 5′-ATGAATTCAACTGTGCTTTCCTTCTGTGCTAGGGCCGGCCGGG-3′; reverse, 5′-CTGCATCAAGGGTTCGCGGACCCGGCCGGCCCTA-3′). The PCR product was cloned into the expression vector pMH1F, a derivative of pBAD (Invitrogen). Expression in pMH1F is driven by the araBAD promoter, and the recombinant protein is produced with an N-terminal Thio6His6 tag (MGSDKIHHHHHH).
A sequence-verified clone was transformed into the Escherichia coli methionine-auxotrophic strain DL41. An overnight culture from a fresh transformant was used to inoculate flasks of 2XYT-ampicillin media. The culture was grown at 37°C with vigorous shaking to an optical density at 600 nm of 0.6 to 0.7, induced with 0.2% (wt/vol) l-arabinose, and grown at 14°C for 8 h. The cells were harvested by centrifugation and lysed by sonication in buffer containing 50 mM potassium phosphate, pH 7.8, 300 mM NaCl, 10% glycerol, 5 mM imidazole, 0.5 mg/ml lysozyme, 100 μl/liter benzonase, and EDTA-free protease inhibitor (one tablet per 50 ml buffer; Roche). The lysate was clarified by ultracentrifugation at 100,000 × g for 45 min at 4°C, and the soluble fraction was applied onto a metal chelate column (Talon resin charged with cobalt; Clontech). The column was washed with 20 mM Tris, pH 7.8, 300 mM NaCl, 10% glycerol, 5 mM imidazole and eluted with 25 mM Tris, pH 7.8, 300 mM NaCl, 150 mM imidazole. The eluate was then purified by anion exchange on a Poros HQ column using a linear gradient of NaCl (0 to 1 M) in 25 Tris-HCl, pH 8.0, followed by size exclusion on a Superdex 75 column equilibrated with 10 mM Tris, pH 7.8, 150 mM NaCl. Pure fractions of nsp10 were concentrated to 3.5 mM and either used immediately for crystallization trials or flash frozen in liquid nitrogen for later use.
Crystallization and data collection.
nsp10 was crystallized using the nanodroplet vapor diffusion method (23). Drops (100 nl 3.5 mM protein plus 100 nl crystallant) were dispensed into 96-well low-profile Greiner plates using an Innovadyne liquid-handling robot. After 21 days at 4°C, harvestable cubic crystals (∼50 μm by ∼50 μm by ∼50 μm) were obtained in 2.0 M ammonium formate, 0.1 M HEPES, pH 7.5. A native 1.8-Å data set (at a wavelength of 1.0 Å) was collected on beamline 8.2.1 at the Advanced Light Source (Berkeley, CA). Native crystals were derivatized by being soaked for 3 days in a solution of the Ta6Br122+ cluster compound (1 mM) in cryoprotectant (30% glycerol in reservoir solution). The use of Ta6Br122+ in the derivatization of crystals of large macromolecules has been described earlier (1). The cluster is a regular octahedron of six Ta atoms with 12 bridging Br atoms at the edges of the octahedron. Both Ta and Br atoms exhibit large anomalous dispersion signals at their absorption edges of 9,918.74 eV (1.25 Å) and 13,476.54 eV (0.92 Å), respectively. Single-wavelength anomalous diffraction (SAD) data were collected to 2.11 Å on a derivatized crystal at the same beam line at a wavelength of 1.2553 Å, corresponding to the inflection point of the Ta L-III absorption edge. Reflections of both native and SAD datasets were indexed in I-centered cubic lattices (space group I23), integrated, and scaled using HKL2000 (19). The Laue group was later identified to be I213.
Structure determination and refinement.
Initial phases were obtained by SAD phasing with the 2.11-Å data set at the Ta inflection wavelength using the program SOLVE (29). While six Ta sites (corresponding to a single Ta6 cluster) were obtained in both space groups I23 and I213, an interpretable map was obtained only in I213. The resulting phases had a figure of merit of 0.68 after density modification procedures using RESOLVE (28). The resultant SAD phases were merged, improved, and extended for using the 1.8-Å native data set using the programs CAD and DM as implemented in the CCP4 package, assuming 1 monomer in the asymmetric unit with a Matthews coefficient of 3.4 and solvent content of 63.7% (7). Automated model building using Arp/wARP (15) traced ∼85% of the backbone and docked 65% of the sequence, including the side chains. The rest of the sequence was manually built into the density using “O” (13) and refined against the high-resolution native data to 1.8 Å with iterative rounds of model building and refinement using Refmac5 (18) of CCP4 (6). A summary of data collection and refinement statistics is shown in Table 1. The stereochemical quality of the final refined model was checked using Procheck (16), and ribbon diagrams were made using Pymol (8). The coordinates and the structure factors have been deposited in the Protein Data Bank (PDB).
TABLE 1.
Data collection and refinement statisticsd
| Statistical elemente | Crystal type
|
|
|---|---|---|
| TaBr (infection wavelength) | Native | |
| Data collection statistics | ||
| Space group | I213 | I213 |
| Cell dimensions | a = b = c = 108.302 Å | a = b = c = 108.874 Å |
| Wavelength (Å) | 1.2553 | 1.0000 |
| Resolution range (Å) | 50.0-2.1 | 50.0-1.8 |
| No. of observations | 252, 264 | 316, 394 |
| No. of unique reflections | 1,2407 | 19,035 |
| Completeness (%) | 99.8 (99.3) | 99.97 (100.0) |
| Redundancy | 9.2 | 5.7 |
| Mean I/σ(I) | 12.85 (2.5) | 22.96 (2.36) |
| Rsyma on I | 0.110 (0.685) | 0.048(0.600) |
| Highest resolution shell (Å) | 2.18-2.10 | 1.86-1.80 |
| Refinement statistics | ||
| Rworkb | 0.196 (0.27) | |
| Rfreec | 0.233 (0.298) | |
| Protein atoms (avg. B factor) | 961 (24.8) | |
| Solvent atoms (avg. B factor) | 188 (38.5) | |
| Hetero atoms (avg. B factor) | 8 (37.8) | |
| RMSDf bond length | 0.014 Å | |
| RMSD bond angle | 1.49 ° | |
| Stereochemical validation | ||
| Most favored (non-Gly, non-Pro) | 93 (86.9%) | |
| Generously allowed | 14 (13.1%) | |
| Additionally allowed | 0 | |
| Disallowed | 0 | |
Rsym = Σhkl[(ΣjIj − <I>)/ΣjIj].
Rwork = ΣhklFo − Fc/ΣhklFo, where Fo and Fc are the observed and calculated structure factors, respectively.
Five percent (1,023 reflections) of randomly chosen reflections was used in the calculation of Rfree.
Values in parentheses are for data corresponding to the outermost shell.
For the phasing step, the following values were obtained: figure of merit SOLVE, 0.36; RESOLVE, 0.68; DM, 0.76.
RMSD, root mean square deviation.
Electrophoretic mobility shift assay.
Gel mobility shift assays were used to measure the affinity of nsp10 binding to nucleic acid. RNA and DNA oligomers designed to generate double-stranded hairpins, dimers, and concatemer conformations (5′-GTTAAGGCCTTAAGGCCTTTAAAGGCCTTAAGGCCTTAAC-3′) were synthesized commercially. RNA oligonucleotides were heated to 65°C for 5 min and cooled slowly to room temperature, while DNA oligonucleotides were boiled for 5 min and cooled similarly to generate double-stranded nucleic acid. Single-stranded RNA (5′-AUAUGGAAGAGCCCUA-3′) was also synthesized commercially. Nucleic acid substrates were resuspended at 1 mg/ml in water containing 0.1% sodium dodecyl sulfate to prevent RNase degradation. Binding mixtures of 5 μg nucleic acid (80 to 100 pmol) were incubated with different concentrations of nsp10 at 37°C for 1 h. Binding mixtures were separated by native polyacrylamide gel analysis on precast 6% acrylamide DNA retardation gels (Invitrogen). Nucleic acid was detected by staining with SYBR-gold stain (Molecular Probes), which is suitable for dsDNA, dsRNA, and ssRNA detection. SYBR-gold fluorescence was imaged on a UV light source equipped with a digital camera. Protein was then detected by SYPRO-ruby poststain according to the manufacturer's protocol (Molecular Probes).
Protein structure accession number.
The coordinates and the structure factors have been deposited in the PDB under accession number 2FYG.
RESULTS
Structure determination by Ta SAD phasing.
Crystals of SARS-CoV nsp10 soaked in Ta6Br122+ were used to obtain phases for structure determination. Characteristic of tantalum bromide derivatization (1), large dispersive anomalous differences were observed in the low-resolution shells (>5.0 Å), which diminished from 4.5 to 3.0 Å and then increased again from 3.0 to 2.1 Å in a highly redundant single-wavelength anomalous data set collected at the L-III absorption edge of Ta. Five of the six sites had reasonable occupancies (0.2 to 0.8) and B (temperature) factors (0.67, 0.63, 0.24, 0.22, 0.21). The Ta6Br122+ cluster was found in the interstitial bulk solvent space near one of the threefold axes. This region was encased by six protein monomers that formed two distinct types of trimers. The N-terminal His tag residues from one trimer and two polar loops from the other trimer aided in encasing the cluster. The final model (Fig. 2a) contains amino acids 10 to 132 of SARS-CoV nsp10, along with five residues at the N terminus contributed by the purification tag (6× His tag) of the construct. We did not see density for five residues at the C terminus. The final R and Rfree values are 19.5 and 23.0%, respectively. Data collection and refinement statistics as well as stereochemical parameters are listed in Table 1.
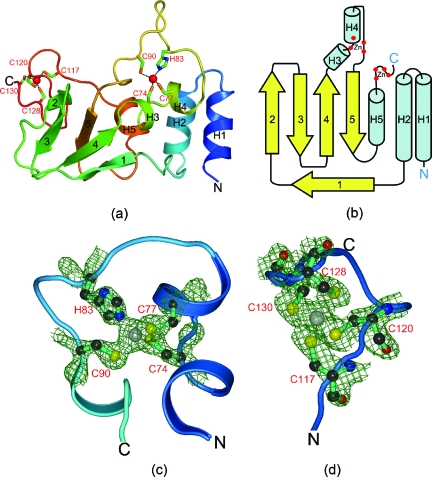
FIG. 2.
Structure of SARS-CoV nsp10. (a) Ribbon diagram of SARS-CoV nsp10 showing the arrangement of helices and strands. The secondary structures are colored from blue (N terminus) to red (C terminus) and are numbered from H1 to H5 for helices and 1 to 5 for the β-strands. (b) Topology diagram showing the connectivities between the secondary structural elements in the nsp10 structure. Helices are in cyan and strands are in yellow, with the same numbering scheme as that described for panel a. (c) Electron density observed at the first Zn2+-binding site. The residues coordinating the Zn2+ ion are shown as balls and sticks. (d) Electron density observed at the second Zn2+-binding site. The four cysteine residues coordinating the metal ion at the second Zn2+ ion near the protein C terminus are shown as balls and sticks. The 2Fo-Fc maps are contoured at 1.0 σ, where Fo and Fc are the observed and calculated structure factors, respectively.
nsp10 adopts a new fold.
The nsp10 monomer seen in the asymmetric unit has a single domain that can be roughly segregated into three regions: a largely helical subdomain at the N terminus, an irregular β-sheet region, and a C-terminal region consisting mostly of loops (Fig. 1). The helical region encompasses two antiparallel helices at the N terminus (H1 and H2) along with three other small helices, H3, H4, and H5. This is followed by a large loop that connects the helical hairpin to an irregularly curved β-sheet that resembles an imperfect β-barrel with strands in the order 2, 3, 1, 4, 5. The hydrogen bonding pattern is broken at multiple regions between the strands of the barrel, and the first β-strand is considerably bent compared to the other four. The middle of the polypeptide consists of three small helices and a large loop insertion. The C-terminal region of the polypeptide (residues 112 to 131) is largely unstructured, with irregular loops. A schematic of the topology of the nsp10 fold is shown in Fig. 2b.
We observed clear electron densities suggesting two bound ions in the structure (Fig. 2c and d). An X-ray absorption spectroscopy experiment yielded a strong peak at 9,658 eV corresponding to the K edge of Zn (data not shown), indicating that nsp10 contains Zn. That both electron densities correspond to Zn2+ ions was confirmed by analyzing a Zn anomalous difference map (data not shown). We therefore modeled two Zn2+ ions into the electron densities; when refined with full occupancies, these have B factors (21.24 and 36.42) that are comparable to those of the rest of the protein molecule (24.8).
Structure comparison using the entire polypeptide chain against known structures in the Protein Data Bank (PDB) (3) by DALI (11) did not yield any hits with significant Z scores (>2.0). DALI searches with the individual subdomains (with five helices as one and the β-sheet as the other) also did not yield any structural homologues, and neither did various combinations of loop inclusions/deletions with these two domains, indicating that nsp10 adopts a novel fold. Sequence homologues of SARS-CoV nsp10 are found only in related coronaviruses (Fig. 1) and are notably absent among prokaryotes or eukaryotes. None of these homologues have been structurally characterized; the SARS-CoV nsp10 structure is the first structural representative of this family of proteins.
nsp10 is a Zn finger protein.
The two Zn fingers present in the structure of SARS-CoV nsp10 were compared with known Zn2+-binding motifs (14). Several unique conformational features are seen in both Zn fingers. The first Zn finger is present in the middle of the polypeptide chain, where the metal ion is coordinated by three cysteines and a histidine residue (C74, C77, H83, and C90; Fig. 2c). Of these, C77 lies in a short, single-turn α-helix (H4), while the other three are in loops. The conformation of protein residues in the vicinity of the Zn2+ ion are such that this site cannot be placed in any of the known classes of Zn2+-binding sites (14). The only group in which it can be placed is the loosely defined category of “Zn2+-binding loops” (14). The second Zn2+ finger is coordinated by four cysteine residues at the C-terminal tail of the protein (C117, C120, C128, and C130; Fig. 2d). Conformational comparison of the protein backbone as well as the side chains of the four cysteines in the vicinity of this Zn2+ finger suggests this motif is a member of the gag-knuckle fold group (14). Specifically, it belongs to the polymerase gag-knuckle family, members of which include one of the Zn fingers of the large subunit of yeast RNA polymerase (residues 63 to 82 of PDB no. 1I3Q) and the dsRNA-binding protein sigma3 of the reovirus outer capsid (residues 48 to 77 of PDB no. 1FN9). The primary function of a majority of Zn fingers of this family is to bind single- and/or double-stranded RNA; however, it may also be a protein-protein interaction module (14). Surface charge distribution indicates that this Zn finger lies in a negatively charged region that is clustered with Asp and Glu residues (Fig. 3b). It is noteworthy that while many of the residues near these two metal ions are exposed to the solvent, the Zn2+ ions are themselves buried.
FIG. 3.
Analysis of electrostatic charge distribution and symmetry mates of nsp10. (a) The surface of one of the nsp10 monomers is shown with electrostatic potential colored from blue (positive) to red (negative) in the range of +4.1 kT to −4.1 kT. Important residues that contribute to the positive charge are shown as balls and sticks over a semitransparent protein surface. The two metal ions are shown as spheres. (b) Two symmetry-related dimers observed in the crystal. The two zinc atoms Zn1 and Zn2 are shown. (c) Modeled effect of a Q65E mutation on the surface charge of nsp10. A large contiguous negatively charged surface patch is generated which may unfavorably alter a critical binding interface with an interacting protein and/or RNA. This mutation causes a temperature-sensitive defect in minus-strand RNA synthesis in MHV (25). WT, wild type.
A distinct positively charged patch is also noticeable on one side of the protein surface (Fig. 3a). Extending to almost 30 Å along one direction, this positive patch encompasses the first Zn finger (Zn atom labeled Zn1 in Fig. 3a) along with several residues that are conserved across the different viral nsp10s. A few important residues that contribute to this positive charge include Lys93, Arg78, Ser11, and Asn40. Such a large positive surface coupled with the observation of two Zn fingers points to the possibility of an RNA-binding function for this protein. The charge distribution as seen in two different dimers composed of symmetry-related monomers as observed from crystal packing is shown in Fig. 3b. The second Zn finger (Zn atom labeled Zn2) appears to play a critical role in both structural integrity of the C-terminal half of the structure as well as its oligomerization, as evidenced by the close proximity and its interaction in the two symmetry-related molecules (Fig. 3b). Analytical size exclusion chromatography indicates that nsp10 is a dimer in solution (data not shown). The high symmetry of the I-centered cubic lattice makes it difficult to predict which of the several symmetry-related oligomers is physiologically relevant. Two of these are shown in Fig. 3b.
Several proteins in the SARS-CoV proteome possess one or more Zn2+-binding motifs: nsp10, the papain-like protease domain of nsp3, the N-terminal domain of nsp13 (helicase/NTPase), and nsp14 (exonuclease) (26). nsp2 and the N-terminal domain of nsp12 (RNA-dependent RNA polymerase) also contain cysteine-rich regions that may harbor Zn fingers. Zn fingers are found in proteins that perform diverse functions in various cellular processes: replication, repair, transcription, translation, metabolism, signaling, cell proliferation, and apoptosis (see reference 14 for a review). They function as interaction modules and bind to a wide variety of ligands including proteins and small molecules, but most often they bind nucleic acids.
Physiological role of nsp10.
nsp10 is found in all coronaviruses, and its sequence is highly conserved (between 51 and 56% identity) across the entire length of the protein (Fig. 1), suggesting its importance in the coronavirus life cycle. In MHV, there is evidence that nsp10 plays a critical role in the replicase complex: a temperature-sensitive Gln65Glu mutation caused a defect in minus-strand RNA synthesis at the nonpermissive temperature of 40°C, while plus-strand synthesis appeared to be unaffected (25). Gln65 is absolutely conserved in all coronavirus nsp10 proteins. When modeled on the SARS-CoV nsp10 structure, mutation of Gln65 to a Glu residue generates a large contiguous negatively charged surface patch (Fig. 3c) which may unfavorably alter a critical binding interface with an interacting protein and/or RNA. Immunofluorescence studies using confocal microscopy in MHV-infected cells have demonstrated that p15 (MHV counterpart of SARS-CoV nsp10) colocalizes with p28, p65, 3CL protease, p10, p22, p12, the RNA-dependent RNA polymerase, the helicase, and the nucleocapsid protein (counterparts of SARS-CoV nsp1, nsp2, nsp5, nsp7, nsp8, nsp9, nsp12, nsp13, and sars9a, respectively) at the site of replication complexes (4, 5). Further, specific interactions between MHV p15, p28, and p10 were seen in yeast two-hybrid and coimmunoprecipitation experiments, suggesting a direct role of nsp10 in the replicase complex (5).
In the pp1ab polyprotein, SARS-CoV nsp10 is preceded by three proteins demonstrated to interact with RNA (nsp7, nsp8, and nsp9) (9, 27, 31) and is followed by the suite of putative viral RNA-modifying enzymes (nsp12 to nsp16) (26). The proximity of nsp10 to viral RNA-binding and RNA-modifying proteins, the presence of two coordinated Zn2+ ions in the crystal structure, and the presence of the highly conserved positively charged face (Fig. 3a) suggested that nsp10 could play a role in viral RNA synthesis by binding directly to RNA. The affinity of nsp10 for nucleic acid was estimated by electrophoretic mobility shift assays (Fig. 4). nsp10 displayed only micromolar affinity for single-stranded and double-stranded nucleic acids and did not exhibit a preference for RNA over DNA. Nucleic acid binding was not diminished by the presence of up to 250 mM sodium chloride in the binding buffer. The nsp9 RNA binding protein (9) also exhibited micromolar affinities for nucleic acid with low substrate preference, characteristic of nonspecific binding. While the affinities presented here for nsp10 and previously for nsp9 would generally be considered insufficient to describe specific RNA-binding proteins, we are unable to rule out the possibility that nsp10 contributes to a larger RNA-binding protein complex.
FIG. 4.
Nucleic acid binding by nsp10. Binding mixtures containing (A) 80 pmol double-stranded RNA, (B) 82 pmol double-stranded DNA, (C) 100 pmol single-stranded RNA, or (D) no nucleic acid were incubated with various concentrations of nsp10. Binding mixtures shown in lanes 0 to 7 contained 0, 10, 20, 40, 80, 160, 320, and 800 μM nsp10, respectively. Free and bound nucleic acids were detected by SYBR-gold staining (Invitrogen). A double-stranded DNA ladder (1 Kb Plus; Invitrogen) was included as a marker (lane M). The positions of free nucleic acid (F), shifted nucleic acid-nsp10 complexes (S), and supershifted multiplexes (O) on the gels are indicated at the left. The shifted and supershifted nucleic acid peaks coincide with SYPRO-ruby protein stain in each gel (not shown).
nsp10 also appears to contribute significantly to the cytopathogenic effects observed in SARS infection in lung tissue by interacting with the oxidoreductase system in mitochondria (17). Yeast two-hybrid experiments screening a human embryo lung cDNA library for interactions, using SARS-CoV nsp10 as bait, revealed two subunits of cellular RNA polymerase B complex (BTF3 and ATF5), the NADH 4L subunit, and cytochrome oxidase II as hits (17). Specific interaction with cytochrome oxidase II was supported by a glutathione S-transferase pull-down assay and Western blotting (17). Interestingly, transient expression of SARS-CoV nsp10 in KMB-17 cells inhibited replication of human coronavirus (HCoV) 229E (17), perhaps by competitively interfering with the HCoV replicase complex.
DISCUSSION
The SARS-CoV proteome contains a number of proteins with very low sequence similarity to other proteins. Even at the structural level, SARS-CoV proteins are proving to yield a significant number of new folds. To date, out of the nine proteins with available three-dimensional structural information (the ADP ribose 1-phosphatase [ADRP] domain of nsp3, nsp5, nsp7, nsp8, nsp9, nsp10, sars2, and sars7a and the N-terminal domain of sars9a), five contain new folds. Both in isolation (20) and within the nsp7-nsp8 hexadecamer supercomplex (31), nsp7 adopts a novel fold consisting of four and three helices, respectively. In the supercomplex, nsp8 also consists of a new fold (31). It occurs in two distinctly different conformations: a “golf club”-like structure (composed of an N-terminal “shaft” domain and a C-terminal “head” domain) and a “bent golf club” conformation, with the long helical shaft bent in the middle into two shorter helices linked by a coil (31). Certain features of the nsp9 fold were found to bear some similarity to four different existing folds (trypsin-like proteases, the C-terminal domain of μ-transposase, α- and β-subunits of F1-ATP synthase-like, and small protein B) (9, 27). However, there are sufficient differences for it to be defined as a unique nucleic acid-binding fold. Finally, the nuclear magnetic resonance structure of the N-terminal domain of sars9a (nucleocapsid protein) revealed a five-stranded β-sheet topology not found in other RNA-binding proteins (12). As more structures of the SARS-CoV proteome and those of other viruses are structurally characterized, it will be interesting to see if the preponderance of uncommon folds is unique to coronaviruses or true of viruses in general.
Conclusions.
This study reports the first structural characterization of SARS-CoV nsp10, a protein critical to viral replication that is highly conserved in coronaviruses. It plays a vital but as-yet uncharacterized role in negative-strand RNA synthesis by the replicase complex (25). nsp10 adopts a tertiary fold that, to our knowledge, has not been previously observed. It contains two Zn fingers, the first of which belongs to a new Zn finger family. The presence of the two Zn fingers, a large conserved positively charged patch on the surface of the protein, as well as its physical location within a cluster of RNA-binding/RNA-modifying proteins in the replicase polyprotein all seem to suggest an RNA-binding function. While the gel-shift assays reported here confirm the ability of purified nsp10 to bind RNA, the observed interaction is weak and not sequence specific. Previous immunolocalization and interaction studies have indicated that in vivo, nsp10 acts in concert with numerous other viral proteins, counterparts of SARS-CoV nsp1, nsp2, nsp5, nsp7, nsp8, nsp9, nsp12, nsp13, and sars9a (4, 5). It is in the context of these other components of the replicase complex that the physiological role of nsp10 is likely to become clearer. While speculative, it is highly likely that nsp9 and nsp10 might in fact bind to the hexameric nsp7-nsp8 supercomplex as part of a larger structure that directs double-stranded RNA through its central channel towards the RNA polymerase for genome replication. While still in their infancy, experiments designed to study replicase assembly and function in SARS-CoV and related coronaviruses hold promise to elucidate this complex process in the viral life cycle.
Acknowledgments
We gratefully acknowledge Anna Tang, Christine Chen, Radin Aur, Thomas Clayton, and Amy Houle for technical assistance. Bioinformatics support for this project was provided by Enrique Abola, Anand Kolatkar, and Sophie Coon of The Scripps Research Institute and Weizhong Li and Adam Godzik of the Burnham Institute. We thank Bing Hao (Nikola Pavletich laboratory, Memorial Sloan-Kettering Institute, New York) for the kind gift of Ta6Br122+. We also acknowledge the helpful support of the beamline staff at Advanced Photon Source (GM/CA-CAT), Advanced Light Source (BL-8.2.1), and Stanford Synchrotron Radiation Laboratory (SSRL) (BL-11.1) synchrotron facilities for help in data collection. In particular, we thank Banumathi Sankaran (ALS) for valuable advice on data collection with the Ta6Br122+ soaks.
SSRL BL-11.1 is supported by the National Institutes of Health (NIH) National Center for Research Resources, NIH National Institutes of General Medical Sciences, Department of Energy, Office of Biological and Environmental Research, Stanford University, and The Scripps Research Institute. The General Medicine and Cancer Institutes Collaborative Access Team is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). This study was supported by National Institutes of Allergy and Infectious Disease/NIH Contract no. HHSN 266200400058C, “Functional and Structural Proteomics of the SARS-CoV,” to P.K.
Footnotes
TSRI manuscript 17990-CB.
REFERENCES
- 1.Banumathi, S., M. Dauter, and Z. Dauter. 2003. Phasing at high resolution using Ta6Br12 cluster. Acta Crystallogr. Sect. D. Biol. Crystallogr. 59:492-498. [DOI] [PubMed] [Google Scholar]
- 2.Baranov, P. V., C. M. Henderson, C. B. Anderson, R. F. Gesteland, J. F. Atkins, and M. T. Howard. 2005. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology 332:498-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, N. I. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockway, S. M., X. T. Lu, T. R. Peters, T. S. Dermody, and M. R. Denison. 2004. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 78:11551-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 7.Cowtan, K. 1994. DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31:34-38. [Google Scholar]
- 8.DeLano, W. L. 2002. The PyMOL Molecular Graphics System. http://pymol.sourceforge.net.
- 9.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsP9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, F., H. Y. Ou, L. L. Chen, W. X. Zheng, and C. T. Zhang. 2003. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 553:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holm, L., and C. Sander. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, Q., L. Yu, A. M. Petros, A. Gunasekera, Z. Liu, N. Xu, P. Hajduk, J. Mack, S. W. Fesik, and E. T. Olejniczak. 2004. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 43:6059-6063. [DOI] [PubMed] [Google Scholar]
- 13.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 14.Krishna, S. S., I. Majumdar, and N. V. Grishin. 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31:532-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamzin, V. S., A. Perrakis, and K. S. Wilson. 2001. The ARP/WARP suite for automated construction and refinement of protein models, p. 720-722. In M. G. Rossmann and E. Arnold (ed.), International tables for crystallography, vol. F. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 16.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 17.Li, Q., L. Wang, C. Dong, Y. Che, L. Jiang, L. Liu, H. Zhao, Y. Liao, Y. Sheng, S. Dong, and S. Ma. 2005. The interaction of the SARS coronavirus non-structural protein 10 with the cellular oxido-reductase system causes an extensive cytopathic effect. J. Clin. Virol. 34:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 20.Peti, W., M. A. Johnson, T. Herrmann, B. W. Neuman, M. J. Buchmeier, M. Nelson, J. Joseph, R. Page, R. C. Stevens, P. Kuhn, and K. Wuthrich. 2005. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 79:12905-12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 23.Santarsiero, B. D., D. T. Yegian, C. C. Lee, G. Spraggon, J. Gu, D. Scheibe, D. C. Uber, E. W. Cornell, R. A. Nordmeyer, W. F. Kolbe, J. Jin, A. L. Jones, J. M. Jaklevic, P. G. Schultz, and R. C. Stevens. 2002. An approach to rapid protein crystallization using nanodroplets. J. Appl. Crystallogr. 35:278-281. [Google Scholar]
- 24.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddell, S., D. Sawicki, Y. Meyer, V. Thiel, and S. Sawicki. 2001. Identification of the mutations responsible for the phenotype of three MHV RNA-negative ts mutants. Adv. Exp. Med. Biol. 494:453-458. [DOI] [PubMed] [Google Scholar]
- 26.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsP9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terwilliger, T. C. 2003. Statistical density modification using local pattern matching. Acta Crystallogr. Sect. D. Biol. Crystallogr. 59:1688-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D. Biol. Crystallogr. 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 31.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]