Abstract
The foamy virus (FV) glycoprotein precursor gp130Env undergoes a highly unusual biosynthesis, resulting in the generation of three particle-associated, mature subunits, leader peptide (LP), surface (SU), and transmembrane (TM). Little structural and functional information on the extracellular domains of FV Env is available. In this study, we characterized the prototype FV (PFV) Env receptor-binding domain (RBD) by flow cytometric analysis of recombinant PFV Env immunoadhesin binding to target cells. The extracellular domains of the C-terminal TM subunit as well as targeting of the recombinant immunoadhesins by the cognate LP to the secretory pathway were dispensable for target cell binding, suggesting that the PFV Env RBD is contained within the SU subunit. N- and C-terminal deletion analysis of the SU domain revealed a minimal continuous RBD spanning amino acids (aa) 225 to 555; however, internal deletions covering the region from aa 397 to 483, but not aa 262 to 300 or aa 342 to 396, were tolerated without significant influence on host cell binding. Analysis of individual cysteine point mutants in PFV SU revealed that only most of those located in the nonessential region from aa 397 to 483 retained residual binding activity. Interestingly, analysis of various N-glycosylation site mutants suggests an important role of carbohydrate chain attachment to N391, either for direct interaction with the receptor or for correct folding of the PFV Env RBD. Taken together, these results suggest that a bipartite sequence motif spanning aa 225 to 396 and aa 484 to 555 is essential for formation of the PFV Env RBD, with N-glycosylation site at position 391 playing a crucial role for host cell binding.
Viral envelope glycoproteins initiate entry of membrane-enveloped viruses into cells by binding to cell surface receptors, followed by conformational changes leading to membrane fusion and delivery of the genome containing viral capsid to the cytoplasm (7). The envelope (Env) glycoproteins of foamy viruses (FVs) are no exception and mediate attachment to host cells through binding to yet unknown cellular receptor molecules. Viral particles are then taken up by endocytosis, and a pH-controlled glycoprotein-mediated fusion of viral and cellular lipid membranes leads to the release of FV capsids into the cytoplasm of the host cell (35).
The principal domain structure of the prototype FV (PFV) Env, comprising an 18-kDa N-terminal signal or leader peptide (LP), a central 80-kDa surface (SU) subunit, and a C-terminal 48-kDa transmembrane (TM) subunit, is similar to other retroviral glycoproteins, although the 126-amino-acid (aa) LP is unusually long (reviewed in reference 24). Biosynthesis and maturation of the FV glycoprotein precursor gp130Env is also unusual in several aspects. First, gp130Env is translated as a full-length precursor protein into the rough endoplasmic reticulum, where it initially adopts a type III membrane topology with both its N and C termini located intracytoplasmically (12, 25). Second, only during its transport to the cell surface is it posttranslationally processed by cellular, most likely furin-like, proteases and not by the signal peptidase complex. Processing results in formation of at least three subunits (10, 11). In the heterotrimeric FV glycoprotein complex, the N-terminal LP has a type II membrane topology, whereas the C-terminal TM subunit has a type I membrane topology. The internal SU subunit presumably associates with extracellular domains of TM on the luminal side (25, 42). Processing of the SU/TM but not the LP/SU cleavage site is essential for generation of infectious viral particles in the supernatant (1, 36). Third, all three subunits are incorporated into FV particles, and interactions of the gp18LP subunit with the viral capsid are essential for FV budding and particle release (25, 42).
Image reconstruction analysis from electron micrographs of negatively stained virions revealed the characteristic, prominent Env spike structures on FV particles, indicating that the FV Env glycoprotein, similar to other viral glycoproteins, forms trimeric complexes containing three copies of each of the three individual subunits (41). However, additional high-resolution structural information of FV glycoprotein subunits is not available and little functional analysis of the extracellular domains has been performed. In addition, the cellular receptor molecules utilized by FV particles for attachment to host cells have not been identified, largely because the extremely broad host range of FVs precludes classic expression cloning approaches. In this study we describe the characterization of the receptor-binding domain (RBD) of the PFV envelope glycoprotein using a flow cytometric assay and recombinant PFV Env immunoadhesins.
MATERIALS AND METHODS
Cells.
The human kidney cell line 293T (9), the canine thymus cell line Cf2Th (33), the canine osteosarcoma cell line D17 (39), the baby hamster kidney fibroblast cell line BHK-21 (30), the feline kidney cortex cell line CRFK (5), and the human fibrosarcoma cell line HT1080 (37), as well as the variants thereof stably expressing an enhanced green fluorescent protein zeocin (EGZ) fusion protein (HT1080 EGZ) or, in addition, the wild-type PFV Env protein (HT1080 Env) as described previously (4), were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics. The human kidney cell line 293F (Invitrogen) was cultivated in 293 SFMII (Invitrogen) serum-free medium supplemented with Glutamax and antibiotics.
Expression constructs.
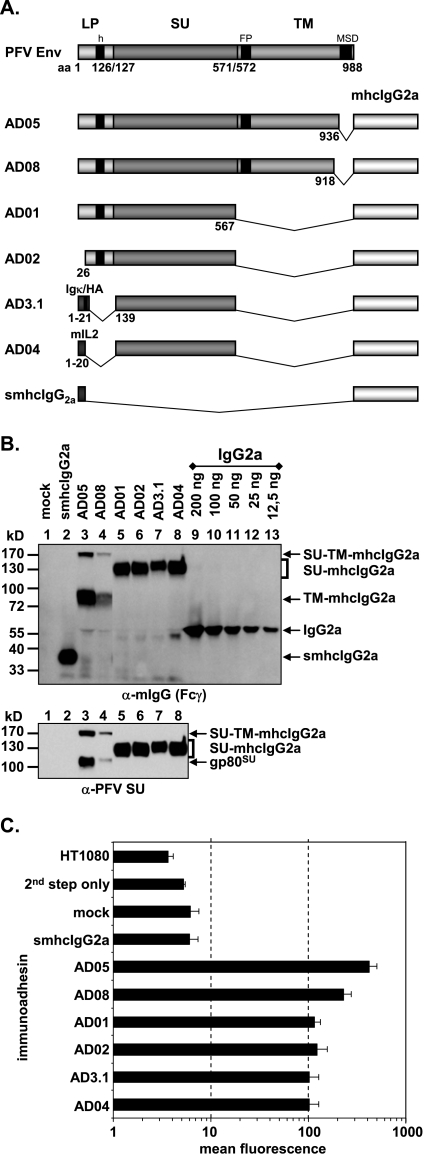
The PFV Env immunoadhesin constructs are based on the murine leukemia virus (MLV)-derived vector pczCFG5 IEGZ (8) containing a polylinker upstream of an encephalomyocarditis virus internal ribosomal entry site driving an EGZ fusion protein cassette. Besides the constant domains (hinge, CH2, CH3) of mouse immunoglobulin G2a (IgG2a), cloned by PCR from BALB/c total spleen mRNA, these immunoadhesins comprise various portions of the extracellular domains of the PFV Env (10). The individual cloning strategies and mutagenesis primers are available on request. All PCR-derived fragments were sequenced to confirm the desired mutations and to exclude further off-site mutations. Construct pAD05, serving as the basis of all other PFV Env immunoadhesins and containing the PFV Env extracellular domain aa 1 to 936, spanning the complete LP and SU domains as well as most of the extracellular domains of the TM subunit, has been described previously (10) (see Fig. 2A). Construct pAD08 harbors the extracellular domain aa 1 to 918 of PFV Env (Fig. 2A). In contrast, pAD01, pAD02, pAD3.1, and pAD04 lack C-terminal sequences containing the SU/TM cleavage site and the TM subunit (see Fig. 2A). Whereas pAD01 contains the full-length LP domain and the SU domain up to aa 567, the N-terminal first 25 aa of the LP were deleted in pAD02 (see Fig. 2A), a mutation that has been shown previously to enhance glycoprotein cell surface expression in context of the full-length Env protein (25). Constructs pAD3.1 and pAD04 have the complete PFV Env LP and part of the N terminus of SU (aa 1 to 138) replaced by the heterologous signal sequences of immunoglobulin kappa with a hemagglutinin tag or of mouse interleukin 2, respectively (see Fig. 2A). The control construct psmhcIgG2a, lacking any PFV Env sequences, is based on the SFG-sNefmCIgG2a retroviral vector and contains the signal sequence of mouse interleukin 2 fused to the constant domains of mouse IgG2a (see Fig. 2A).
FIG. 2.
Analysis of different subdomain constructs. (A) Schematic outline of the PFV Env domain structure and the PFV Env immunoadhesins containing different domains as indicated. LP, leader peptide; SU, surface; TM, transmembrane; h, hydrophobic domain of LP; FP, fusion peptide; MSD, membrane-spanning domain; mhcIgG2a, mouse IgG2a heavy chain constant domains; IgΚ, mouse IgG kappa light chain signal peptide; mIL2, mouse interleukin 2 signal peptide; HA, hemagglutinin A epitope tag. (B) Western blot analysis of 293T supernatants (100 μl) containing different PFV Env immunoadhesins, controls, or IgG2a standard for protein concentration determination as indicated using polyclonal anti-mouse IgG-Fcγ or monoclonal anti-PFV SU-specific antibodies. The identities of the individual proteins are given on the right. (C) Mean fluorescence and corresponding standard deviation (n = 3) of different immunoadhesins and controls on HT1080 target cells. Staining was done using 50 ng immunoadhesin or control in a volume of 830 μl.
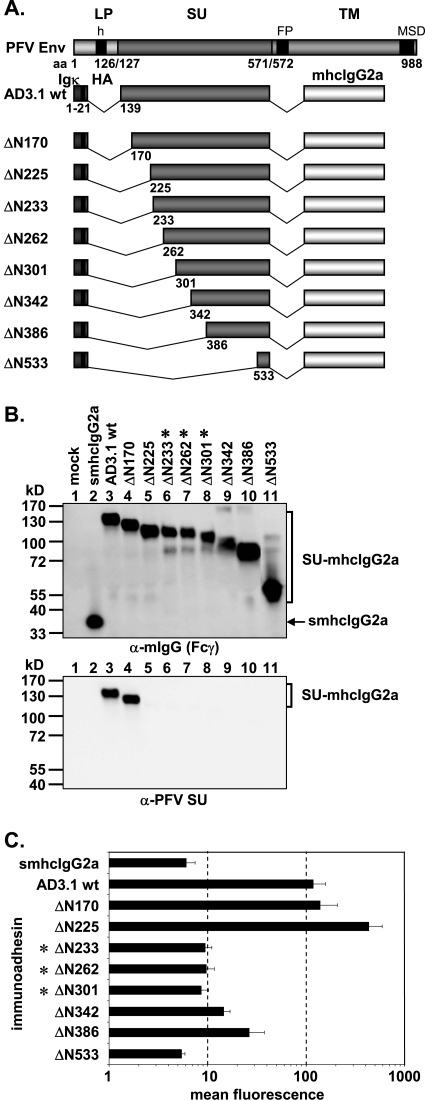
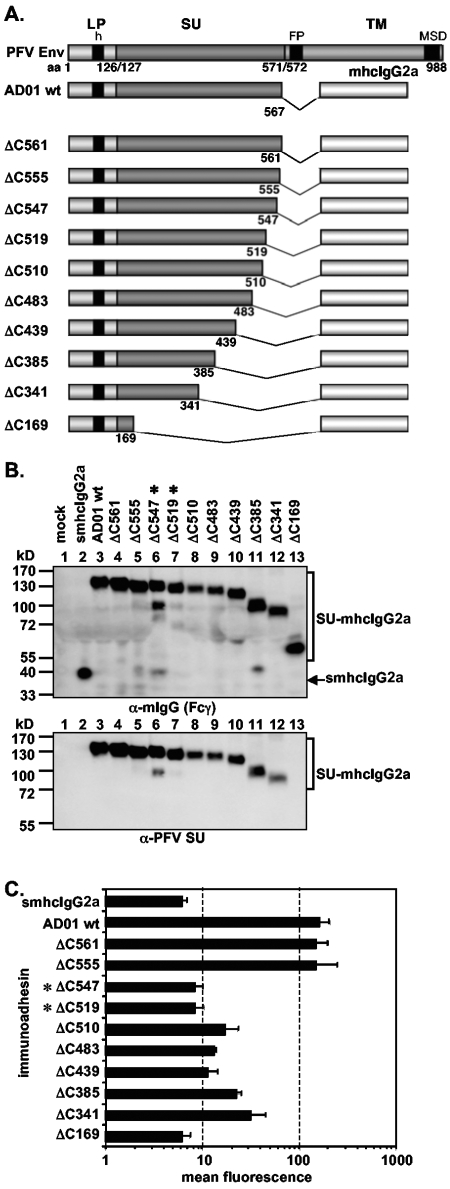
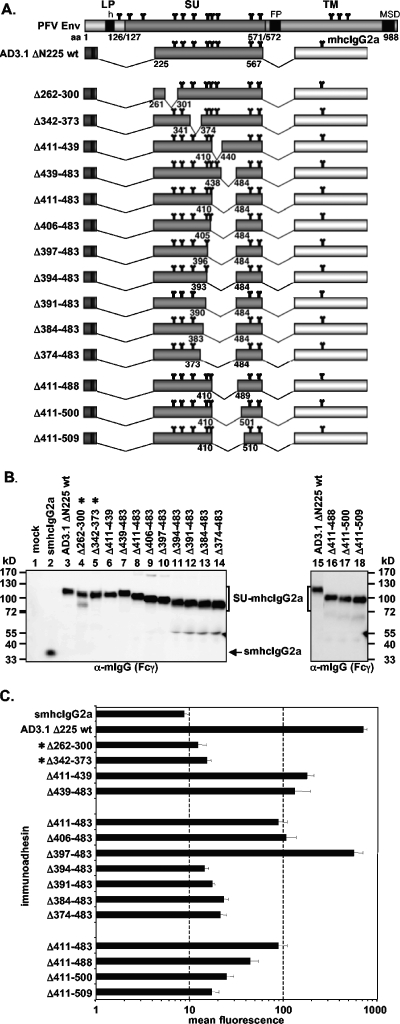
The following constructs with N-terminal truncations were derived from pAD3.1: pAD3.1ΔN170, pAD3.1ΔN225, pAD3.1ΔN233, pAD3.1ΔN262, pAD3.1ΔN301, pAD3.1ΔN342, pAD3.1ΔN386, and pAD3.1ΔN533 (see Fig. 3A). Based on pAD01, the following constructs with C-terminal truncations were generated: pAD01ΔC561, pAD01ΔC555, pAD01ΔC547, pAD01ΔC519, pAD01ΔC510, pAD01ΔC483, pAD01ΔC439, pAD01ΔC385, pAD01ΔC341, and pAD01ΔC169 (see Fig. 4A). The constructs with various internal deletions in PFV SU, pAD3.1Δ262-300, pAD3.1Δ342-373, pAD3.1Δ411-439, pAD3.1Δ439-483, pAD3.1Δ411-483, pAD3.1Δ406-483, pAD3.1Δ397-483, pAD3.1Δ394-483, pAD3.1Δ391-483, pAD3.1Δ384-483, pAD3.1Δ374-483, pAD3.1Δ411-488, pAD3.1Δ411-500, and pAD3.1Δ411-509 were based on pAD3.1ΔN225 (see Fig. 7A). In addition, several point mutants based on construct pAD3.1 or pAD01 were generated. Based on pAD3.1, the following cysteine point mutants were generated: pAD3.1ΔC5 (C228S), pAD3.1ΔC6 (C235S), pAD3.1ΔC7 (C256S), pAD3.1ΔC8 (C318S), pAD3.1ΔC9 (C381S), pAD3.1ΔC10 (C404S), pAD3.1ΔC11 (C418S), pAD3.1ΔC12 (C435S), pAD3.1ΔC13 (C449S), pAD3.1ΔC14 (C457S), pAD3.1ΔC15 (C486S), pAD3.1ΔC16 (C506S), and pAD3.1ΔC17 (C565S) (see Fig. 6A). In constructs pAD01ΔN8 (N391Q), pAD01ΔN8.1 (S393V), and pAD01ΔN8.2 (T392V), the N-glycosylation site consensus motif N-X-S/T of the eighth potential N-glycosylation site of PFV Env at position aa 391 to 393 was mutated (see Fig. 5A). In the case of pAD01ΔN8 and pAD01ΔN8.1, the mutations prevent N-glycosylation at this site, whereas in pAD01ΔN8.2, N-glycosylation is not affected (28). These expression constructs were generated by transferring the mutations from the full-length Env context of the PFV Env gp130Env expression construct pczHFVenvEM109 (ΔN8), pczHFVenvEM131 (ΔN8.1), or pczHFVenvEM151 (ΔN8.2), described previously (28), by subcloning a KspAI/BamHI fragment into the pAD01 immunoadhesin backbone. Furthermore, two additional point mutants, pAD01ΔN8.3 (Y394N) and pAD01ΔN8.4 (S396N), based on pAD01ΔN8, were generated which have the natural eighth N-glycosylation site inactivated but a new N-glycosylation consensus motif introduce downstream of the original position. These N-glycosylation site mutants were generated by recombinant PCR techniques (15).
FIG. 3.
Analysis of different N-terminal truncation mutants. (A) Schematic outline of the PFV Env domain structure and the N-terminal PFV Env LP-SU immunoadhesin deletion mutants. For abbreviations, see legend to Fig. 2. (B) Western blot analysis of 293T supernatants (50 μl) containing different PFV Env immunoadhesins or purified immunoadhesins (marked by asterisks) and controls, as indicated, using polyclonal anti-mouse IgG-Fcγ or monoclonal anti-PFV SU-specific antibodies. The identities of the individual proteins are given on the right. (C) Mean fluorescences and corresponding standard deviations (n = 3) of different immunoadhesins and controls on HT1080 target cells. Staining was done using 100 ng immunoadhesin or control in a volume of 450 μl.
FIG. 4.
Analysis of different C-terminal truncation mutants. (A) Schematic outline of the PFV Env domain structure and the C-terminal PFV Env LP-SU immunoadhesin deletion mutants. For abbreviations, see legend to Fig. 2. (B) Western blot analysis of 293T supernatant (50 μl) containing different PFV Env immunoadhesins or purified immunoadhesins (marked by asterisks) and controls, as indicated, using polyclonal anti-mouse IgG-Fcγ or monoclonal anti-PFV SU-specific antibodies. The identities of the individual proteins are given on the right. (C) Mean fluorescences and corresponding standard deviations (n = 3) of different immunoadhesins and controls on HT1080 target cells. Staining was done using 100 ng immunoadhesin or control in a volume of 350 μl.
FIG. 7.
Analysis of different internal deletion mutants. (A) Schematic outline of the PFV Env domain structure and the N-terminal PFV Env LP-SU immunoadhesin deletion mutants. For abbreviations, see legend to Fig. 2. (B) Western blot analysis of 293T supernatant (50 μl) containing different PFV Env immunoadhesins or purified immunoadhesins (marked by asterisks) and controls, as indicated, using polyclonal anti-mouse IgG-Fcγ-specific antibodies. The identities of the individual proteins are given on the right. (C) Mean fluorescences and corresponding standard deviations (n = 3) of different immunoadhesins and controls on HT1080 target cells. Staining was done using 100 ng immunoadhesin or control in a volume of 300 μl.
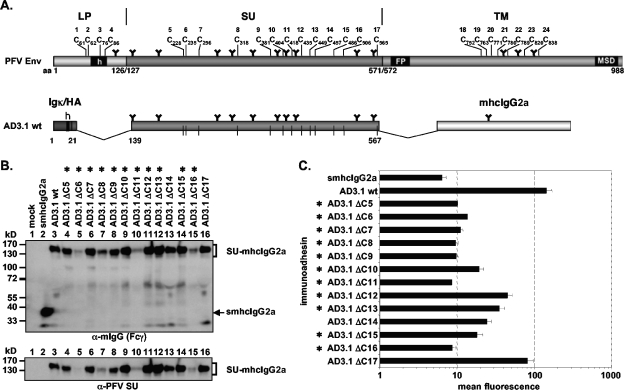
FIG. 6.
Analysis of PFV SU cysteine point mutants. (A) Schematic outline of the PFV Env domain organization and the PFV Env SU immunoadhesin AD3.1 with annotated positions of the individual cysteine residues. For abbreviations, see legend to Fig. 2. (B) Western blot analysis of 293T supernatant (50 μl) containing different PFV Env immunoadhesins or purified immunoadhesins (marked by asterisks) and controls, as indicated, using polyclonal anti-mouse IgG-Fcγ or monoclonal anti-PFV SU-specific antibodies. The identities of the individual proteins are given on the right. (C) Mean fluorescences and corresponding standard deviations (n = 1 to 3) of different immunoadhesins and controls on HT1080 target cells. Due to the poor secretion, some mutants could be analyzed for binding activity only once (ΔC6, ΔC11) or twice (ΔC5, ΔC8, ΔC16). Staining was done using 100 ng immunoadhesin or control in a volume of 435 μl.
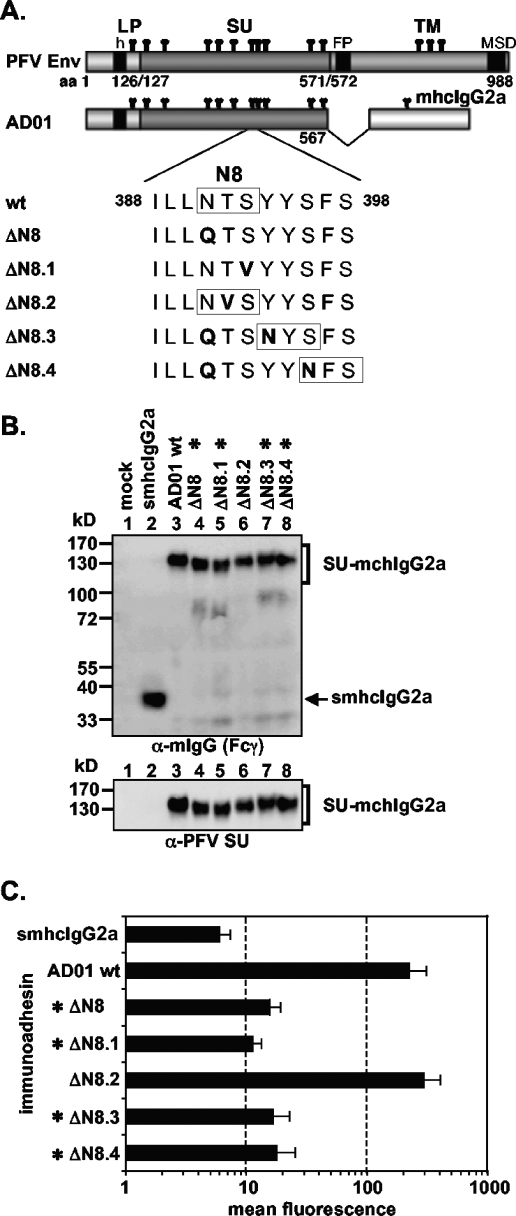
FIG. 5.
Analysis of different N-glycosylation site 8 point mutants. (A) Schematic outline of the PFV Env domain structure and the PFV Env LP-SU immunoadhesin AD01 and point mutants thereof. The amino acid sequence flanking N-glycosylation site 8 of the wild-type and different AD01 point mutants are listed below. N-glycosylation site consensus sequences (N-X-S/T) are boxed. For abbreviations, see legend to Fig. 2. (B) Western blot analysis of 293T supernatant (50 μl) containing different PFV Env immunoadhesins or purified immunoadhesins (marked by asterisks) and controls, as indicated, using polyclonal anti-mouse IgG-Fcγ or monoclonal anti-PFV SU-specific antibodies. The identities of the individual proteins are given on the right. (C) Mean fluorescences and corresponding standard deviations (n = 3) of different immunoadhesins and controls on HT1080 target cells. Staining was done using 100 ng immunoadhesin or control in a volume of 430 μl.
Generation of cell culture supernatants containing immunoadhesin glycoproteins.
293T cells were transfected with the immunoadhesin expression constructs by the calcium-phosphate coprecipitation method as described previously (23). Forty-eight hours posttransfection, cell-free supernatants containing the individual secreted PFV Env immunoadhesins were harvested after pelleting cellular debris by centrifugation for 5 min at 1,700 × g.
Purification of immunoadhesin proteins from cell culture supernatant.
To concentrate certain immunoadhesins showing reduced secretion rates, recombinant proteins were purified from the cell culture supernatant of transiently transfected 293T cells using the Proteus protein A spin column kit (ProChem) according to the instructions of the manufacturer.
Antisera and Western blot expression analysis and quantification of secreted proteins.
Western blot expression analysis of cell culture supernatant was performed essentially as described previously (25), using polyclonal antisera specific for murine IgG (mIgG) Fcγ (Jackson Immuno Research) or the PFV Env LP (25) or a hybridoma supernatant specific for the SU subunit of PFV Env (clone P3E10) (10). The chemiluminescence signal was digitally recorded using a LAS-3000 imager (Fujifilm) and quantified using the Image Gauge software package (Fujifilm). The concentrations of secreted proteins in individual cell culture supernatants or purified immunoadhesin preparations were determined using serial dilutions of a commercially available IgG2a preparation with a defined concentration (Dianova) as the standard in the individual immunoblots. An example is shown in Fig. 2B.
Analysis of receptor binding capacity.
The flow cytometric assay to quantitatively determine and compare the capability of individual immunoadhesin preparations to specifically bind to HT1080 target cells is schematically depicted in Fig. 1A. Defined amounts of the individual immunoadhesins as indicated, harvested either as plain cell culture supernatants or purified immunoadhesins diluted in cell culture medium, were incubated with 1 × 105 HT1080 cells in a total volume of 300 to 1,270 μl on ice for 1 h. Notably, the same amounts of different immunoadhesins or control proteins and identical total volumes were used within the individual assays. Subsequently, the cells were washed once with cold fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline, 1% fetal calf serum) to remove excess recombinant proteins and incubated for an additional hour on ice with a secondary phycoerythrin-conjugated polyclonal donkey anti-mouse IgG (heavy plus light) antiserum (1:200; Jackson ImmunoResearch) in a total volume of 200 μl. Following a second wash in cold FACS buffer, the pelleted samples were resuspended in 200 μl FACS buffer, stored on ice, and analyzed by flow cytometry using a FACSCalibur (Becton Dickinson). The mean fluorescence of 10,000 events per sample was subsequently determined using the Cell Quest software package (Becton Dickinson). An example of the different types of histogram profiles is given in Fig. 1B.
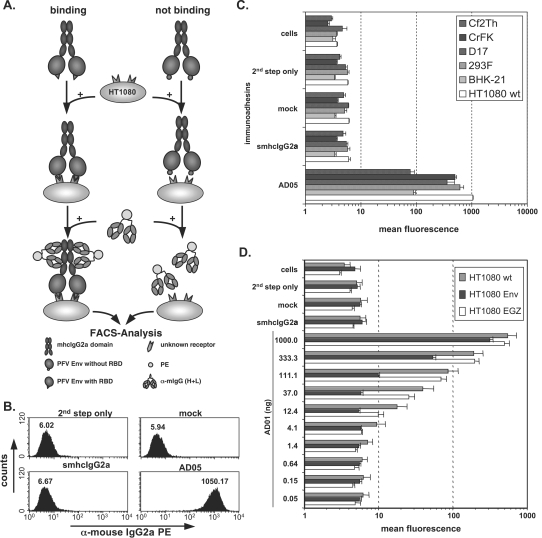
FIG. 1.
Cell binding assay. (A) Schematic outline of the cell-based, flow cytometric immunoadhesin binding assay. (B) Representative examples of FACS histogram profiles from HT1080 cells incubated with different controls or the AD05 immunoadhesin supernatant. Staining was done in a volume of 830 μl using 50 ng immunoadhesin. (C) Graphic representation of the mean fluorescence and standard deviation (n = 3) of different target cell lines, as indicated, stained with the AD05 immunoadhesin or the corresponding control stains. (D) Mean fluorescence values and the corresponding standard deviations (n = 3) for serial dilutions of the AD01 immunoadhesin or different controls on HT1080 cells stably expressing wild-type PFV Env and EGZ (HT1080 Env) or expressing only EGZ (HT1080 EGZ) or of unmodified cells (HT1080 wt). For the smhcIgG2a control, 1,000 ng recombinant protein was used. Staining of all samples was done in a total volume of 1,270 μl. Abbreviations: cells, unstained cells; 2nd step only, incubation with FACS buffer and 2nd step reagent only; mock, supernatant from 293T cells transfected with the empty pczCFG5IEGZ retroviral vector; smhcIgG2a, supernatant from 293T cells transfected with psmhcIgG2a secreting only the IgG2a domains; PE, phycoerythrin; α, anti.
RESULTS
Establishment of a cell-based quantitative FV Env receptor-binding assay.
One hallmark of FVs is their very broad host range; to date, no cell line or cell type has been identified that is not permissive for FV Env-mediated entry (16, 17, 31). Primarily for this reason, the cellular receptor molecules for FVs have eluded identification. A recombinant immunoadhesin, containing the chimpanzee FV isolate (SFVcpz) LP-SU domains linked to a human IgG1 heavy chain Fc fragment and produced by baculovirus infection of insect cells has been shown to bind specifically to permissive cells (14). However, neither a further delineation of an FV Env RBD nor the contribution of glycosylation to receptor binding, which is different in insect cells, has been assessed until now. In the absence of knowledge of the nature of the FV cellular receptor(s), we established a flow cytometric assay based on this initial report by Herchenröder et al. (14). This assay allowed the rapid and quantitative characterization of various immunoadhesins containing different subdomains of PFV Env linked to the Fc region of mouse IgG2a and produced in mammalian cells with respect to their specific target cell-binding capacity and permitted delineation of an RBD for the PFV glycoprotein (Fig. 1). A schematic outline of the cell-based assay is shown in Fig. 1A, and the details are described in Materials and Methods. In brief, the different recombinant immunoadhesins were produced by transient transfection in 293T cells and harvested directly as cell culture supernatants from transfected cells or concentrated by protein A affinity chromatography. Equal amounts of recombinant immunoadhesins or controls were incubated with target cells to allow for specific binding. Following a wash step to remove unbound, recombinant immunoadhesins, the cells were incubated with a fluorochrome-labeled, anti-mouse antiserum recognizing the Fc region of any immunoadhesin molecules bound to the surface of target cells. After a final wash step, target cells stained with immunoadhesin and fluorochrome-labeled anti-mouse antibodies were identified and quantified by flow cytometry. A typical example of the histogram profiles of positive (AD05), negative (smhcIgG2a), and control (mock, second step only) HT1080 target cell samples is shown in Fig. 1B. Specific binding, up to about 170-fold above background, was detectable by flow cytometric analysis of an immunoadhesin comprising the complete extracellular domains of PFV Env (aa 1 to 936) fused to the mouse IgG2a Fc region (AD05) and incubated with various target cell lines of different species (Fig. 1C). In contrast, no specific binding was observed by incubation with an identical amount of an immunoadhesin lacking PFV sequences (smhcIgG2a), similar volumes of control supernatant (mock), or cells incubated with the second-step reagent only (second step only) (Fig. 1C). In addition, the signal intensity, measured as mean fluorescence of the phycoerythrin staining, of the AD01 immunoadhesin, containing aa 1 to 567 of PFV Env, was dose dependent on unmodified (HT1080 wild type [wt]) or EGZ-expressing HT1080 (HT1080 EGZ) cell populations over a concentration range from approximately 5 to 1,000 ng immunoadhesin per 1 × 105 target cells (Fig. 1D). In contrast, similar to what has been described previously for an SFVcpz immunoadhesin (14), a dose-dependent reduction of signal intensity was observed on a HT1080 cell population stably expressing the wild-type PFV Env protein (HT1080 Env) (Fig. 1D), which has been described previously to result in superinfection resistance against MLV FV Env vector pseudotypes (4). Furthermore, binding of PFV Env immunoadhesins to target cells was competitively inhibited by coincubation with PFV vector particles harboring PFV Env but not by mutant PFV particles lacking any glycoprotein or MLV particles pseudotyped with either amphotropic MLV Env or vesicular stomatitis virus glycoprotein G (data not shown). Taken together, these data demonstrate the feasibility of using this assay to detect differences in the capacity of various PFV Env immunoadhesins to bind specifically to target cells to characterize the PFV Env RBD.
The PFV Env SU domain is sufficient to mediate specific cell binding.
Unlike other retroviral glycoproteins, three mature PFV Env glycoprotein processing products are particle-associated LP, SU, and TM. To determine which glycoprotein domains are essential for interaction with the putative cellular receptor molecules, we analyzed various PFV Env immunoadhesins that differed in their composition of PFV Env domains (Fig. 2A) for differences in cell-binding capacity. Western blot analysis of transiently transfected 293T supernatants using a mIgG Fcγ-specific antiserum or a PFV SU-specific monoclonal antibody showed the efficient secretion of all immunoadhesins (Fig. 2B, lanes 2 to 8). Immunoadhesins AD05 and AD08, lacking only the PFV Env membrane-spanning domain and cytoplasmic domain of the TM subunit, were processed into SU and TM IgG2a Fc subunits, although some protein representing an intermediate with unprocessed SU/TM subunits was present in the supernatant preparations (Fig. 2B, lanes 3 and 4). Immunoadhesins AD01, AD02, AD05, and AD08 harbor the complete or most of the PFV Env LP domain. Whereas in lysates of cells transfected with these constructs the precursor proteins and LP cleavage products were readily detected using a LP-specific antiserum, no LP-specific protein bands were detectable in the corresponding supernatant samples (data not shown). This indicated that all immunoadhesins secreted into the cell culture supernatant were properly processed at the LP/SU cleavage site and no LP cleavage products remained associated with the immunoadhesins. Testing of the different immunoadhesins in the cell-binding assay, using similar amounts of recombinant proteins, revealed specific binding for all constructs (Fig. 2C). The AD05 and AD08 immunoadhesins displayed a two- to fourfold-higher mean fluorescence intensity then all other proteins, including those lacking either the complete TM domain sequences (AD01, AD02) or lacking both the TM and LP domains (AD3.1, AD04). Thus, the RBD resides predominantly within the SU subunit of PFV Env.
Characterization of N- and C-terminal PFV Env RBD boundaries.
Having shown that, similar to other retroviral glycoproteins, the RBD of PFV Env is contained within the SU subunit, we next sought to determine the boundaries of a minimal, continuous PFV Env RBD. Therefore, various N- and C-terminal truncation mutants of the AD3.1 and AD01 immunoadhesins were generated (Fig. 3A and Fig. 4A), and recombinant proteins were analyzed with respect to their cell-binding capacity. All truncation constructs were expressed; however, some were secreted only poorly (data not shown) and had to be purified and concentrated by protein A affinity chromatography prior to analysis (Fig. 3B and Fig. 4B). Western blot analysis of the truncation mutants using the PFV SU-specific monoclonal antibody P3E10 indicated that its epitope resides between amino acids 170 and 225 within SU (Fig. 3B and Fig. 4B, lower panels). Analysis of the cell-binding capacity of the individual immunoadhesins by the flow cytometric assay revealed a loss of high-affinity binding when N-terminal deletion extended beyond aa 225 (Fig. 3C) and C-terminal deletions extended further than aa 555 (Fig. 4C). Interestingly, several immunoadhesins with some of the greatest truncations (e.g., AD01 ΔC341 or AD3.1 ΔN386) displayed an intermediate staining intensity clearly above background (4- to 5-fold) but also significantly below (5- to 10-fold) the respective parental immunoadhesins (Fig. 3C and Fig. 4C). In contrast, immunoadhesin AD3.1 ΔN225 showed a three- to fourfold-stronger signal than the parental protein AD3.1 (Fig. 3C). Purification and concentration of immunoadhesins by protein A affinity chromatography itself did not influence the binding activities of the individual immunoadhesins (data not shown). Taken together, these data indicated that the minimal continuous PFV Env RBD comprises aa 225 to 555.
N-glycosylation site 8 is important for receptor interaction.
Previous characterization of PFV Env N-glycosylation suggested that, of all SU N-glycosylation sites, only site 8 (N391) potentially contributed to Env function (28). Therefore, different AD01 immunoadhesin variants with various mutations at or C-terminal to N-glycosylation site 8 were generated (Fig. 5A). In general, mutant immunoadhesins preventing N-glycosylation at the original N-glycosylation site 8 (AD01 ΔN8, ΔN8.1, ΔN8.3, ΔN8.4) were secreted less efficiently than either the parental immunoadhesin (AD01 wt) or a mutant protein not affecting glycosylation at the original site (AD01 ΔN8.2) (data not shown). As a consequence of poor secretion, these immunoadhesins had to be concentrated by protein A affinity chromatography. Mutation of the invariant residues N391 (AD01 ΔN8) and S393 (AD01 ΔN8.1), but not the variant residue T392 (AD01 ΔN8.2), within the N-X-S/T N-glycosylation site 8 consensus sequence led to secretion of mutant immunoadhesins with a higher electrophoretic mobility than the parental immunoadhesin (AD01 wt), consistent with the absence of a polysaccharide chain at this position (Fig. 5B, lanes 3 to 6). Immunoadhesins with the original N-glycosylation site 8 inactivated and a new N-glycosylation site consensus sequence introduced further downstream (AD01 ΔN8.3, ΔN8.4) showed an electrophoretic mobility similar to the parental immunoadhesin, consistent with attachment of oligosaccharide chains at the newly introduced positions (Fig. 5B, lanes 3, 7, 8). Analysis of the cell-binding capacity of the different immunoadhesin point mutants revealed the requirement of oligosaccharide attachment at the natural N-glycosylation site 8 for specific binding activity (Fig. 5C). All mutants lacking N-glycosylation at the original site in PFV SU (AD01 ΔN8, ΔN8.1, ΔN8.3, ΔN8.4) displayed signal intensities within a threefold range above background, even when that critical N-glycosylation site was shifted a few amino acids C-terminal to the original site (AD01 ΔN8.3, ΔN8.4) (Fig. 5C). In contrast, an immunoadhesin harboring a mutation in the eighth N-glycosylation site consensus sequence not affecting carbohydrate attachment at this position (AD01 ΔN8.2) (Fig. 5C) or an immunoadhesin with inactivated N-glycosylation site 12 (data not shown) had a binding activity similar to the parental AD01 immunoadhesin. Thus, the addition of oligosaccharide chains at the original N-glycosylation site 8 is important for high-affinity binding.
Importance of evolutionarily conserved PFV Env SU subunit cysteine residues for RBD formation.
Similar to other retrovirus genera, the cysteine residues involved in disulfide bonds in the extracellular domains of FVs from different species are highly conserved (data not shown). The characterization of the minimal continuous RBD in PFV SU described above suggested that all cysteine residues except the most C-terminal (C17, C565) were essential for proper formation of the RBD (Fig. 3 and 4). We therefore analyzed the importance of individual cysteine residues for formation of the PFV Env RBD and binding to target cells by generation and analysis of PFV Env SU immunoadhesins bearing individual cysteine-to-serine exchanges (Fig. 6A). All mutants except one (ΔC11) were expressed intracellularly at levels comparable to those of the wild type (data not shown). However, most of the mutants were secreted poorly and therefore had to be purified and concentrated by protein A affinity chromatography prior to further analysis (Fig. 6B). Most mutants (ΔC5-9, ΔC11, ΔC16) displayed cell-binding activities within a twofold range of the mock control. Several mutants (ΔC10, ΔC12-15) retained some residual binding activity (within three- to sevenfold of background), and only one (ΔC17) displayed binding activity reaching 50% of that of the wild type. Taken together, these data indicated that any mutation of cysteines in the N-terminal part of the PFV SU is detrimental for RBD formation, whereas only some cysteine residues in the C-terminal part are completely essential.
Identification of a bipartite PFV Env RBD.
The analysis of N- and C-terminally truncated immunoadhesins as well as the cysteine point mutants suggested the contribution of domains located distantly between aa 225 and 555 to receptor binding. To characterize these subdomains in further detail, we generated additional immunoadhesins with internal deletions based on the AD3.1 ΔN225 immunoadhesin (Fig. 7A). All internal deletion constructs except two (AD3.1 Δ262-300, Δ342-373) were secreted efficiently (Fig. 7B) and could be analyzed without prior purification and concentration by protein A affinity chromatography. The results of the analysis of cell-binding capacity of the individual mutant immunoadhesins are summarized in Fig. 7C. First, internal deletions from aa 262 to 300 (Δ262-300) or from aa 342 to 373 (Δ342-373) resulted in a loss of binding activity. In contrast, internal deletions from aa 411 to 439 (Δ411-439), aa 439 to 483 (Δ439-483) or a combination of both from aa 411 to 483 (Δ411-483) only led to a four- to eightfold reduction in binding activities compared to the parental AD3.1 ΔN225 immunoadhesin (Fig. 7C). Next, based on the Δ411-483 internal deletion immunoadhesin, we further extended this internal deletion N- and C-terminally (Fig. 7A). Cell-binding activity analysis of these immunoadhesins revealed an N-terminal boundary for well-tolerated deletions at aa 397 (Fig. 7C). All immunoadhesins with deletions extending upstream of aa 397 (Δ394-483, Δ391-483, Δ384-483, Δ374-483) resulted in mean fluorescence signals within a threefold range of background (Fig. 7C). Interestingly, a deletion spanning aa 397 to 483 resulted in binding activities similar to the parental AD3.1 ΔN225 immunoadhesin (Fig. 7C). Extending the Δ411-483 deletion at the C terminus up to aa 509 gradually reduced the binding activity to background levels (Fig. 7C). Thus, only a deletion of aa 397 to 483 within a minimal continuous PFV Env RBD immunoadhesin was tolerated without significant reduction in binding activity.
DISCUSSION
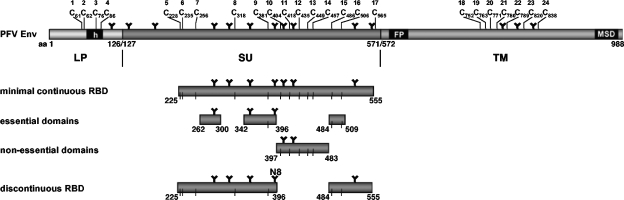
FVs are a special type of retroviruses with many unique features in their replication strategy distinguishing them from all other retroviruses (reviewed in reference 38). Due to some of these unique properties and their broad host range, FVs have gained increased interest in recent years as potential vector systems for gene therapeutic approaches (reviewed in reference 32). FV vector systems seem to be particularly well suited for an efficient gene transfer in cells of the hematopoietic lineage (18, 19, 22). However, to date, the cellular receptor molecules enabling FV glycoprotein-mediated entry have not been characterized and little structural and functional information on the extracellular Env domains involved in receptor recognition and entry into host cells is available. Since the FV glycoprotein undergoes a highly unusual biosynthesis (10, 11) and all three proteolytic cleavage products, LP, SU, and TM, are viral particle-associated (25, 40), it was not known which glycoprotein domains contribute to target cell binding. Therefore, in this study, we characterized the putative RBD of the PFV Env glycoprotein by flow cytometric analysis of chimeric PFV Env IgG Fc immunoadhesin protein binding to target cells. Our results, summarized schematically in Fig. 8, demonstrate that the PFV Env RBD is located in the SU subunit and that the LP and TM domains are dispensable for host cell binding, similar to other retroviral glycoproteins. The data define a 331-aa minimal continuous RBD, spanning aa 225 to 555 in the gp130Env precursor protein. With respect to the localization of the RBD within the processed SU subunit, the PFV Env RBD resembles human immunodeficiency virus type 1 (HIV-1) Env, which has an RBD contained within the C-terminal region of the SU subunit, rather than the MLV RBDs, which are located in the N-terminal domain of SU (2, 6, 13, 20, 34). As described in this report for PFV Env, small deletions at the C terminus of the HIV-1 gp120 SU subunit abolish the cell-binding capacity of secreted HIV-1 gp120 proteins (20).
FIG. 8.
Overview of the localization of PFV Env RBDs. Schematic outline of the PFV Env domain structure, the minimal continuous RBD, and essential or nonessential domains within, as characterized in this study.
Interestingly, deletion of the N-terminal 99 aa of the PFV SU subunit in the context of recombinant immunoadhesins increased the mean fluorescence signal intensity in the cell-binding assay significantly. This might suggest that these N-terminal sequences are unfavorable for optimal folding of the RBD or that they might mask the RBD in the context of a PFV Env SU-IgG Fc fusion protein. Perhaps, in the context of the complete PFV Env glycoprotein complex, these SU domain sequences are normally involved in interactions with the membrane-spanning LP and/or TM subunits. Consistent with this possibility, the minimal continuous PFV Env RBD protein immunoadhesin AD3.1 ΔN225 and the AD05 immunoadhesin containing, in addition to the complete SU subunit, the extracellular domains of TM had similar signal intensities.
Previous mutational analysis of PFV N-glycosylation revealed a functional role for N-glycosylation of N-glycan attachment site 8 in PFV SU. Whereas mutation of the other nine N-glycosylation sites in SU showed no phenotypic consequences on PFV Env function, the site 8 mutant did not support PFV particle export, most probably due to an intracellular transport defect of the mutant glycoprotein (28). In this study, all immunoadhesins with mutations at the N-glycosylation site 8 consensus sequence that abolished carbohydrate addition at the original site were secreted less efficiently. On the one hand, this might be an indication for structural abnormalities of these mutant proteins, but on the other hand, this may be only a consequence of slower folding or impaired intracellular transport. A similar reduction in the release of a secreted mutant of the Friend MLV Env RBD, having the two N-glycosylation sites inactivated, has been observed (3). This MLV Env RBD N-glycosylation mutant was still able to interact with the receptor, as measured by an interference assay, although the receptor interaction of the mutant protein seemed to be weaker than that of the wild type. However, this mutant was not analyzed in a cell-binding assay. In contrast to the MLV Env RBD N-glycosylation mutant, receptor interaction of all PFV immunoadhesin mutants lacking the original N-glycosylation site 8, measured as cell-binding activities, were reduced to nearly background levels and could not be restored by introduction of new N-glycosylation sites just a few amino acids downstream of the original site. The importance of N-glycosylation site 8 for cell-binding activity was further supported by the analysis of immunoadhesin mutants with internal deletions in the minimal continuous PFV Env RBD. This analysis showed that the minimal continuous PFV Env RBD allowed internal deletions in the C-terminal but not the N-terminal or central regions. In addition, it revealed that optimal cell binding activity was only achieved for a deletion mutant retaining N-glycosylation site 8 and at least three flanking C-terminal amino acids. Thus, these data indicate that glycosylation at the evolutionarily conserved N-glycosylation site 8 in PFV SU either is important for the overall folding of the PFV Env RBD or, alternatively, might be directly involved in the interaction with the receptor. Similar to other retrovirus genera, the cysteine residues of glycoproteins from different FV species are evolutionarily highly conserved (data not shown). However, in contrast to the MLV Env (26, 27) and HIV-1 Env (21), the intrachain disulfide bond pattern of the FV glycoprotein has not yet been characterized. The mutation of cysteine residues in the RBD of MLV Env in the context of the full-length protein causes partial to complete loss of function, including receptor binding (29). Our analysis of the cell-binding activities of various PFV Env SU subunit cysteine mutants in the context of a PFV Env LP-SU immunoadhesin correlates very well with results using the N- and C-terminal SU truncation mutants or the minimal PFV Env RBD internal deletion constructs. Analogous to these, individual mutation of the first five cysteine residues in PFV SU (C5 to C9) located in the N-terminal half of SU abolished the binding activity, whereas six of eight cysteine mutants in the C-terminal half of SU retained residual binding activity. This suggests that the domain spanning aa 397 to 483 is nonessential for correct folding of the PFV Env RBD and direct interaction with the cellular receptor. However, the successive reduction in cell-binding activity upon C-terminal extension of the internal deletion and the lack of binding activity of the C16 mutant indicates that the C-terminal region spanning aa 484 to 555 is required for correct folding of the PFV Env RBD and/or interaction with the receptor molecules.
Taken together, our results suggest that the PFV Env RBD is formed by a bipartite sequence motif in the SU subunit. Furthermore, the identification of immunoadhesins with point mutations in N-glycosylation site 8 and lacking any detectable cell-binding activity should provide valuable tools for the characterization of as yet unknown FV receptor molecules.
Acknowledgments
We thank Axel Rethwilm, Ottmar Herchenröder, and Welkin Johnson for critically reading the manuscript.
This work was supported by grants from the DFG (Li621/3-1, Li621/4-1) and the BMBF (01ZZ0102) to D.L.
REFERENCES
- 1.Bansal, A., K. L. Shaw, B. H. Edwards, P. A. Goepfert, and M. J. Mulligan. 2000. Characterization of the R572T point mutant of a putative cleavage site in human foamy virus Env. J. Virol. 74:2949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini, J. L., S. C. Kayman, A. Pinter, J. M. Heard, and O. Danos. 1994. Role of N-linked glycosylation in the activity of the Friend murine leukemia virus SU protein receptor-binding domain. Virology 202:496-499. [DOI] [PubMed] [Google Scholar]
- 4.Berg, A., T. Pietschmann, A. Rethwilm, and D. Lindemann. 2003. Determinants of foamy virus envelope glycoprotein mediated resistance to superinfection. Virology 314:243-252. [DOI] [PubMed] [Google Scholar]
- 5.Crandell, R. A., C. G. Fabricant, and W. A. Nelson-Rees. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176-185. [DOI] [PubMed] [Google Scholar]
- 6.Davey, R. A., C. A. Hamson, J. J. Healey, and J. M. Cunningham. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J. Virol. 71:8096-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimitrov, D. S. 2004. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinev, D., B. W. Jordan, B. Neufeld, J. D. Lee, D. Lindemann, U. R. Rapp, and S. Ludwig. 2001. Extracellular signal regulated kinase 5 (ERK5) is required for the differentiation of muscle cells. EMBO Rep. 2:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du Bridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duda, A., A. Stange, D. Lüftenegger, N. Stanke, D. Westphal, T. Pietschmann, S. W. Eastman, M. L. Linial, A. Rethwilm, and D. Lindemann.2004. Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity. J. Virol. 78:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiselhart, V., P. Bastone, T. Kempf, M. Schnolzer, and M. Löchelt. 2004. Furin-mediated cleavage of the feline foamy virus Env leader protein. J. Virol. 78:13573-13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiselhart, V., A. Schwantes, P. Bastone, M. Frech, and M. Löchelt. 2003. Features of the Env leader protein and the N-terminal Gag domain of feline foamy virus important for virus morphogenesis. Virology 310:235-244. [DOI] [PubMed] [Google Scholar]
- 13.Heard, J. M., and O. Danos. 1991. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J. Virol. 65:4026-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herchenröder, O., D. Moosmayer, M. Bock, T. Pietschmann, A. Rethwilm, P. D. Bieniasz, M. O. McClure, R. Weis, and J. Schneider. 1999. Specific binding of recombinant foamy virus envelope protein to host cells correlates with susceptibility to infection. Virology 255:228-236. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 16.Hill, C. L., P. D. Bieniasz, and M. O. McClure. 1999. Properties of human foamy virus relevant to its development as a vector for gene therapy. J. Gen. Virol. 80:2003-2009. [DOI] [PubMed] [Google Scholar]
- 17.Hooks, J. J., and C. J. Gibbs, Jr. 1975. The foamy viruses. Bacteriol. Rev. 39:169-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josephson, N. C., G. Trobridge, and D. W. Russell. 2004. Transduction of long-term and mobilized peripheral blood-derived NOD/SCID repopulating cells by foamy virus vectors. Hum. Gene Ther. 15:87-92. [DOI] [PubMed] [Google Scholar]
- 19.Josephson, N. C., G. Vassilopoulos, G. D. Trobridge, G. V. Priestley, B. L. Wood, T. Papayannopoulou, and D. W. Russell. 2002. Transduction of human NOD/SCID-repopulating cells with both lymphoid and myeloid potential by foamy virus vectors. Proc. Natl. Acad. Sci. USA 99:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 21.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 22.Leurs, C., M. Jansen, K. E. Pollok, M. Heinkelein, M. Schmidt, M. Wissler, D. Lindemann, C. Von Kalle, A. Rethwilm, D. A. Williams, and H. Hanenberg. 2003. Comparison of three retroviral vector systems for transduction of nonobese diabetic/severe combined immunodeficiency mice repopulating human CD34(+) cord blood cells. Hum. Gene Ther. 14:509-519. [DOI] [PubMed] [Google Scholar]
- 23.Lindemann, D., M. Bock, M. Schweizer, and A. Rethwilm. 1997. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J. Virol. 71:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindemann, D., and P. A. Goepfert. 2003. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 277:111-129. [DOI] [PubMed] [Google Scholar]
- 25.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linder, M., D. Linder, J. Hahnen, H. H. Schott, and S. Stirm. 1992. Localization of the intrachain disulfide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur. J. Biochem. 203:65-73. [DOI] [PubMed] [Google Scholar]
- 27.Linder, M., V. Wenzel, D. Linder, and S. Stirm. 1994. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J. Virol. 68:5133-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lüftenegger, D., M. Picard-Maureau, N. Stanke, A. Rethwilm, and D. Lindemann. 2005. Analysis and function of prototype foamy virus envelope N glycosylation. J. Virol. 79:7664-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macpherson, I., and M. Stoker. 1962. Polyoma transformation of hamster cell clones-an investigation of genetic factors affecting cell competence. Virology 16:147-151. [DOI] [PubMed] [Google Scholar]
- 31.McClure, M. O., and O. Erlwein. 1995. Foamy viruses-pathogenic or therapeutic potential? Rev. Med. Virol. 5:229-237. [Google Scholar]
- 32.Mergia, A., and M. Heinkelein. 2003. Foamy virus vectors. Curr. Top. Microbiol. Immunol. 277:131-159. [DOI] [PubMed] [Google Scholar]
- 33.Nelson-Rees, W. A., R. B. Owens, P. Arnstein, and A. J. Kniazeff. 1976. Source, alterations, characteristics and use of a new dog cell line (Cf2Th). In Vitro 12:665-669. [DOI] [PubMed] [Google Scholar]
- 34.Nygren, A., T. Bergman, T. Matthews, H. Jornvall, and H. Wigzell. 1988. 95- and 25-kDa fragments of the human immunodeficiency virus envelope glycoprotein gp120 bind to the CD4 receptor. Proc. Natl. Acad. Sci. USA 85:6543-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picard-Maureau, M., G. Jarmy, A. Berg, A. Rethwilm, and D. Lindemann. 2003. Foamy virus envelope glycoprotein-mediated entry involves a pH-dependent fusion process. J. Virol. 77:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 74:4474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasheed, S., W. A. Nelson-Rees, E. M. Toth, P. Arnstein, and M. B. Gardner. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027-1033. [DOI] [PubMed] [Google Scholar]
- 38.Rethwilm, A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 39.Riggs, J. L., R. M. McAllister, and E. H. Lennette. 1974. Immunofluorescent studies of RD-114 virus replication in cell culture. J. Gen. Virol. 25:21-29. [DOI] [PubMed] [Google Scholar]
- 40.Stanke, N., A. Stange, D. Lüftenegger, H. Zentgraf, and D. Lindemann. 2005. Ubiquitination of the prototype foamy virus envelope glycoprotein leader peptide regulates subviral particle release. J. Virol. 79:15074-15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilk, T., F. de Haas, A. Wagner, T. Rutten, S. Fuller, R. M. Flügel, and M. Löchelt. 2000. The intact retroviral Env glycoprotein of human foamy virus is a trimer. J. Virol. 74:2885-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilk, T., V. Geiselhart, M. Frech, S. D. Fuller, R. M. Flügel, and M. Löchelt. 2001. Specific interaction of a novel foamy virus Env leader protein with the N-terminal Gag domain. J. Virol. 75:7995-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]