Abstract
The Fv1 protein is an endogenous factor in mice that confers resistance to infection by certain classes of murine leukemia virus, a phenomenon referred to as restriction. The mechanism of restriction is not understood, and the low endogenous level of Fv1 in cells has prevented any biochemical or biophysical analysis of the protein. We have now purified recombinant Fv1n protein from a baculovirus system and demonstrate that Fv1 exists in a multimeric form. Furthermore, we have mapped the position of two domains within the protein using limited proteolysis. Biophysical characterization of the N-terminal domain reveals that it comprises a highly helical and extended dimeric structure. Based on these biochemical and biophysical data, we propose a model for the arrangement of domains in Fv1 and suggest that dimerization of the N-terminal domain is necessary for Fv1 function to allow the protein to interact with multiple capsid protomers in retroviral cores.
Mammalian cells have developed innate intracellular mechanisms to inhibit infection by retroviruses, and several factors responsible for this inhibition have now been identified. Among these are factors that interfere with accumulation of viral mRNA in the cytoplasm (12, 14), enzymes that cause hypermutation of nascent retroviral reverse transcripts (4, 15, 25, 52), and proteins that block infection before integration can occur (2, 46). Trim5α and Fv1 are restriction factors that fall into this final category.
Fv1 is present in mice as two major alleles Fv1n and Fv1b, where Fv1n confers resistance to B-tropic murine leukemia virus (MLV) and Fv1b renders mice resistant to infection by N-tropic MLV (16). Trim5 is a member of a large family of Trim proteins present in mammals (18, 36). In humans and primates, the splice variant Trim5α confers species specific resistance to infection by a variety of retroviruses (17, 33, 46). It has been demonstrated that both Fv1 and Trim5α block retroviral infection at a stage after viral entry and before proviral integration into the host genome. However, Trim5α acts before reverse transcription, whereas restriction by Fv1 occurs after (19, 35, 46). Nevertheless, the target for both Fv1 and Trim5α restriction activity appears to be retroviral capsid protein in the context of a preintegration complex (7-9, 20, 41).
The Trim family of proteins are characterized by the presence of a tripartite RBCC motif comprised of a RING domain, a B-box, and a coiled coil. Trim5α contains this tripartite motif at its N terminus together with a carboxyl-terminal PRYSPRY domain responsible for the specificity of the restriction activity (28, 32, 37, 47, 50). Inspection of the Fv1 protein sequence reveals a degree of homology with the Gag region of the human endogenous retrovirus L or murine endogenous retrovirus L family of endogenous retroviruses (1, 2). Contained within this region is a sequence corresponding to the major homology region common to all retroviral capsids (3). The protein also contains a region of predicted coiled coil, but no other recognized sequence motifs or domains are apparent.
The differential specificity of Fv1n and Fv1b restriction is determined by three sequence elements located in the C-terminal region of the protein (3, 5), and genetic studies have indicated that a central region of the protein is dispensable for restriction activity (3). Structural data would greatly enhance mechanistic studies of these restriction factors, but to date, no biochemical or biophysical analysis of Fv1 has been reported due mainly to the low level of endogenous expression and the insolubility of the protein in Escherichia coli. We present the first purification of a retroviral restriction factor along with an analysis of the domain organization and the oligomeric state of the molecule. Based on these data, together with recent structural studies on the target for Fv1, the MLV capsid protein (27), we propose a model for restriction where dimeric Fv1 molecules act through association with an array of capsid molecules present in the viral reverse transcription complex (RTC).
MATERIALS AND METHODS
Details of constructs and baculoviruses.
An Fv1n baculovirus expression construct was prepared using the BD BacPAK baculovirus expression system from Clontech. Briefly, the coding sequence of Fv1n (2) and an additional sequence encoding an N-terminal hexahistidine tag was inserted into a pBacPAK9 transfer vector and cotransfected along with linearized BD BacPAK6 baculovius DNA into Spodoptera frugiperda (Sf9) cells. The cells were incubated at 27°C in SF-900 II serum-free insect cell culture medium for 3 days, and recombinant viruses expressing Fv1 were then isolated from individual plaques and amplified using the BacPAK rapid titer kit. For protein expression, 3 liters of Trichoplusia ni 5 (Tn5) cells (density of 5 × 105 cells/ml) were infected at a multiplicity of infection of 3, and the culture was grown for 72 h postinfection. The cells were harvested by centrifugation, and Fv1n was purified from the clarified insect cell extract using Ni affinity chromatography.
The DNA sequence coding for Fv1 residues 20 to 200 was amplified by PCR using a plasmid DNA template. The PCR product was inserted into a pET22b expression vector (Novagen) between the NdeI and XhoI restriction sites. The N-terminal primer contained sequences encoding MPHHHHHH, and the C-terminal primer contained a stop codon after the last coding triplet to produce an N-terminal hexahistidine fusion with the sequence MP at the N terminus. The nucleotide sequence of the expression clone was verified by automated DNA sequencing, and the protein was expressed in E. coli BL21(DE3) by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to a mid-log culture. Fv1(20-200)-ht was then purified from clarified crude cell extracts using a combination of anion exchange, immobilized metal ion affinity, and gel filtration chromatography. The purity and monodispersity of preparations were monitored by electrospray ionization mass spectrometry (MS), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), photon correlation spectroscopy, and analytical size-exclusion chromatography. Protein concentration was determined from the absorbance at 280 nm using a molar extinction coefficient derived by summing the contributions from tyrosine and tryptophan residues [Fv1(20-200)-ht, 26,700−1 cm−1].
Peptide analysis and proteolysis.
To sequence the N terminus of baculovirus-produced Fv1n and accompanying degradation products, the proteins were separated by electrophoresis on a 14% SDS-polyacrylamide gel and transferred onto a Problott membrane by electroblotting at 50 V for 40 min {transfer buffer, 10 mM CAPS [3-(cyclohexylamino)propanesulfonic acid (CAPS); pH 11], 10% methanol}. Bands were visualized by staining the filter with Coomassie brilliant blue and then excised. N-terminal sequencing was carried out using automated Edman degradation (Microchemical facility, Babraham Institute, Cambridge, United Kingdom).
Limited proteolytic digests were carried out as described previously (42). Briefly, samples were dialyzed overnight against digestion buffer, 100 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl), 20 mM Tris-HCl, pH 7.8. Digests were carried out using either trypsin or chymotrypsin at a protein concentration of 0.4 mg/ml and an enzyme-to-substrate ratio of 1:250 (wt/wt). Products of the digests were then separated on 14% SDS-polyacrylamide gels and visualized by staining with colloidal blue (Novex). Bands were excised from the gel, the stain was extracted, and the samples were reduced and alkylated, followed by digestion with trypsin. Peptides produced from tryptic hydrolysis were then extracted and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) (MS).
Analytical size-exclusion chromatography.
A sample of Ni affinity-purified Fv1n derived from baculovirus-infected cells was fractionated by size exclusion on a Superdex 200 (10/30) column calibrated with molecular weight markers. The column was equilibrated in 20 mM Tris-HCl, 150 mM NaCl at a flow rate of 0.4 ml/min, and the absorbance of the eluent was recorded at 210, 220, and 280 nm. The column was mounted on a high-performance liquid chromatography system (Jasco) controlled by the Borwin software package, enabling retention times to be accurately measured. Fractions were collected every minute, and a sample from each was separated by SDS-PAGE and probed by Western blotting using an anti-Fv1n antibody. The retention time of a sample of purified Fv1(20-200) was also determined using the same column and solution conditions.
Sedimentation equilibrium.
Sedimentation equilibrium experiments were performed in a Beckman Optima XLA analytical ultracentrifuge using six-channel centerpieces in an An-60 Ti rotor. Prior to centrifugation, samples were dialyzed exhaustively against the buffer blank, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM TCEP. After centrifugation for 18 h, cells were scanned radially at 2-h intervals (λ = 280 nm) until no further change in the absorbance profile was observed. The rotor speed was then increased and the procedure repeated. Data were collected at 10,000 and 15,000 rpm together with a scan at 42,000 rpm to determine the degree of absorbance offset. Data were collected on nine samples of Fv1(20-200) where the concentration varied from 3 to 30 μM. Initially, the program SEDPHAT (38, 48) was used to determine weight-averaged molecular masses by nonlinear fitting of individual equilibrium profiles (A versus r) to a single-species ideal solution model. Inspection of these data revealed that the molecular masses showed no concentration dependency, so global fitting incorporating the data from multiple speeds and sample concentrations was applied to extract the final molecular mass.
Sedimentation velocity.
Sedimentation velocity experiments were performed in a Beckman Optima XLA analytical ultracentrifuge using conventional double sector quartz cells in an An-60 Ti rotor. The rotor speed was 42,000 rpm, and the temperature was maintained at 293 K. Protein partial specific volumes and the solvent density were calculated from tabulated values (22). Prior to centrifugation, protein samples were dialyzed exhaustively against the buffer blank, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM TCEP. Experiments were carried out at protein concentrations varying from 0.7 to 0.9 mg/ml, and radial scans, λ = 280 nm, were collected every 300 s. The data recorded from moving boundaries were analyzed in terms of a single-species model or to the continuous size distribution functions, C(S) and C(M), using the program SEDFIT (39, 40).
CD spectroscopy.
Far UV circular dichroism (CD) spectra (260 to 190 nm) were recorded using a Jasco J715 spectropolarimeter purged with nitrogen gas. Spectra were recorded in 0.1-cm cells at 20°C in 20 mM NaH2PO4/Na2HPO4, pH 7.6 at a protein concentration of 150 μg/ml. Quantitative analysis of the spectra for protein structure secondary structure content was carried out using the CONTIN software package (34) implemented in the CDPro program suite (43). For denaturation studies, the CD at 222 nm was recorded at increasing concentrations of urea (0 to 8 M). Typically, the protein was allowed to equilibrate after each addition of urea until a steady CD signal was obtained. Data were then recorded in scan mode until equivalent spectral signal-to-noise ratio was obtained for each addition. Unfolding data were fit as a two-state transition, and ΔGH20 and m were then derived by linear extrapolation (31).
RESULTS
Production of Fv1 in baculovirus and proteolytic mapping of domains.
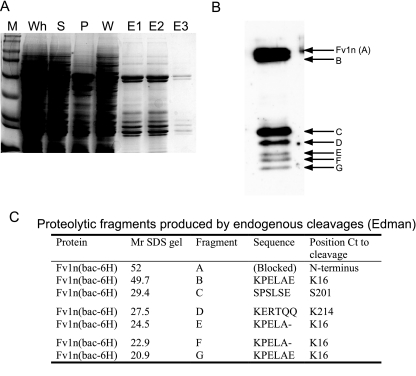
Expression and subsequent purification of full-length Fv1 in E. coli is hampered by the insolubility of the protein. To overcome the problem, Fv1n was overexpressed as an N-terminal histidine tag fusion from a recombinant baculovirus. Using this system, Fv1n was detected in the soluble fraction of virus-infected insect cells, and the protein could be purified using nickel chelate affinity chromatography exploiting the N-terminal histidine tag (Fig. 1a). However, intact Fv1n was always accompanied by several smaller-molecular-weight species, indicating a substantial amount of degradation had occurred, and after affinity chromatography, many of the fragments copurified with the full-length protein.
FIG. 1.
Analysis of the expression, purification, and in vivo proteolysis of Fv1n in recombinant baculovirus-infected insect cells. (A) Coomassie brilliant blue-stained SDS gel of nickel-chelate affinity purification. M, molecular weight markers; Wh, whole-cell extract; S, soluble fraction; P, pellet fraction; W, wash; E1 to E3, fractions from imidazole elution. (B) Western blot showing purified Fv1n (A) and copurifying fragments (B to G). (C) Table of the N-terminal sequences determined by automated Edman degradation. Fragments and Fv1n are labeled as in panel B. Relative molecular weights (Mr) are in thousands. Ct, C-terminal.
To identify the N-terminal sequence of these smaller fragments and confirm the correct sequence of the full-length protein, each band was analyzed by automated Edman degradation (Fig. 1b and c). No sequence could be obtained from the intact protein, suggesting that the protein was N-terminally blocked. However, sequence was obtained from band B, migrating just below full-length protein and several of the copurifying fragments that migrate between 20 to 30 kDa. The N terminus of band B corresponds to a cleavage between F15 and K16. This is also the N terminus of the 3 shortest fragments, E, F, and G, migrating at 24.5, 22.9, and 20.9 kDa. The two remaining products, C and D, are the result of cleavages after A200 and S213, respectively. It is likely that these fragments are the result of proteolytic cleavages in unstructured regions and at domain boundaries in the protein. Bearing this in mind, the pattern of cleavages broadly defines two, approximately 20-kDa, protease-resistant domains within Fv1; one contained within the N-terminal region of the protein and the other located toward the C terminus.
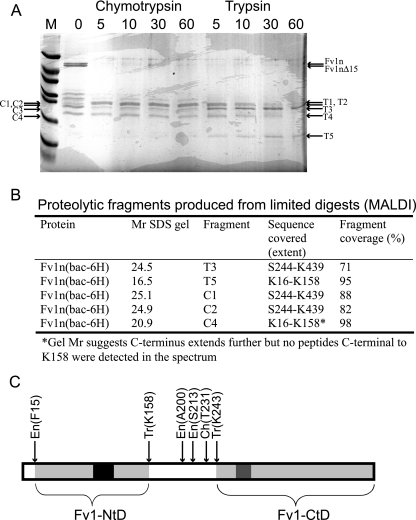
To define more precisely the amino and carboxyl termini of these protease-resistant domains, the mixture of Fv1n together with the N- and C-terminal fragments was digested with trypsin or chymotrypsin under limiting conditions. After this treatment, there was no intact Fv1n, but several protease-resistant fragments were present, migrating between 15 and 25 kDa. These bands were excised, and the peptides resulting from a total tryptic hydrolysis of each product were analyzed using MALDI-TOF (MS). The results of these experiments are presented in Fig. 2. After 60 min of limited tryptic digestion, two distinct products remain, T3 and T5 (Fig. 2a). Similarly, after 60 min of limited chymotryptic digestion, three products remain, C1 and C2, which are barely resolved, and C4. Analysis of the tryptic fingerprint produced from these bands reveals that T3, C1, and C2 correspond to a C-terminal protease-resistant domain in Fv1n (Fv1n-CtD) and that T5 and C4 result from a protease-resistant domain at the N terminus (Fv1n-NtD). The limits of the domain boundaries were determined by looking for the most N-terminal and C-terminal tryptic peptides that were present in the MALDI-TOF spectra. The spectrum resulting from tryptic hydrolysis of T3 contained an ion at an m/z of 2,691.47 (calculated m/z, 2,691.49), corresponding to the tryptic peptide D414-K439. This locates the C-terminal boundary of Fv1n-CtD only a single residue from the C terminus of the entire protein. The most N-terminal peptide detected in the digest of T3 was S244-K252, located C-terminal to the endogenous cleavages at A200 and S213. Similar results were obtained from the tryptic hydrolysis of C1 and C2. The tryptic peptides S244-K252 and D414-K439 were also detected in these spectra. However, several other ions were present that had peptide masses consistent with multiple chymotryptic cleavages between Q226 and T231, each with a constant C terminus located at K243. Inspection of the MALDI-TOF spectra produced from tryptic hydrolysis of T5 and C4 allowed the identification of the domain boundaries of Fv1n-NtD. The most C-terminal tryptic peptides present in the T5 digest were L144-R157 and L144-K158, and the most N-terminal was K16-R38, corresponding to the N terminus defined by the endogenous cleavages and a C terminus further N-terminal to the A200 and S213 cleavages. Analysis of C4 gave similar results. K16-R38 was the most N-terminal peptide, and L144-K158 was the most C-terminal. However, as the electrophoretic mobility of C4 is less than that of T5, the C terminus of C4 is likely to be located closer to the endogenous cleavage sites at A200 and S213. The absence of tryptic peptides from this region in the C4 spectrum is probably due to the nature of the distribution of basic residues between V159 and S213. The peptides produced from hydrolysis are either too small for the m/z range or within range but contain no basic residues.
FIG. 2.
MALDI-TOF analysis of limited tryptic and chymotryptic digestion of purified Fv1n. (A) SDS gel analysis of time course of limited digestion. The times of digestion in minutes with either chymotrypsin or trypsin are shown above each lane, and the fragments that were extracted and analyzed by MALDI are indicated. (B) Table of sequences determined by MALDI-TOF, fragments are labeled as in panel A. Relative molecular weights (Mr) are in thousands. (C) Schematic representation of Fv1 showing the endogenous (En), tryptic (Tr), and chymotryptic (Ch) sites of proteolytic cleavage. The stable domains resistant to proteolytic degradation are highlighted in light gray. The black region highlights the region of sequence strongly predicted to be coiled coil by the “coils” program (24), and the dark gray region is the proposed Fv1 major homology region sequence (2).
Taken together, these data allow us to divide Fv1 into three regions. At the N terminus, the protein contains a trypsin-resistant domain, Fv1-NtD, that spans from K16-R157. The central region of Fv1 (residues 157 to 243) encompasses a protease-sensitive region of the molecule, and at the C terminus, another trypsin-resistant fragment, residues S244-L440, defines the Fv1 C-terminal domain, Fv1n-Ctd. The position of Fv1 protease-resistant domains together with the location of the cleavage sites is summarized in Fig. 2C.
Self-association of Fv1n.
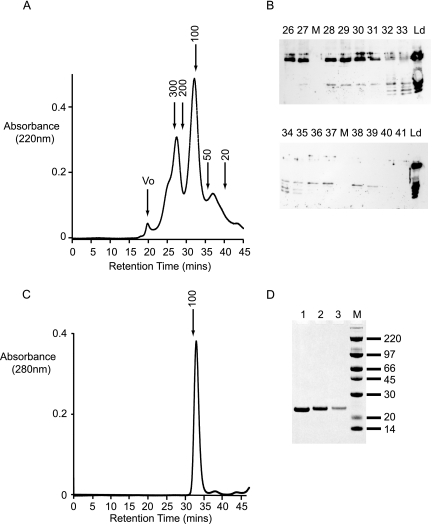
The presence of such a large number of copurifying fragments not containing the His6 affinity tag is a strong indicator that Fv1n is able to self-associate. To test this hypothesis, the mixture was fractionated using a Superdex (10/30) size-exclusion column. The resulting chromatogram is presented in Fig. 3a. The size-exclusion column clearly resolves the sample into four peaks, a small peak eluting at the void volume of the column, followed by three others with apparent molecular masses of 270 kDa, 110 kDa, and 40 kDa. Each fraction was analyzed by Western blotting using anti-Fv1n antibodies (Fig. 3b). The peak in the void volume contains no Fv1-derived material. Intact Fv1n and Fv1nΔ15 co-elute in the 270-kDa peak, accompanied by a 25-kDa Fv1n-CtD fragment in later fractions. The 100-kDa peak consists of a mixture of both Fv1n-NtD and Fv1n-CtD fragments, while the 40-kDa peak appears to be the Fv1n-CtD alone. The molecular mass of an Fv1n monomer is 50 kDa, so an apparent solution molecular mass of 270 kDa is highly indicative of self-association between Fv1n monomers. Furthermore, co-elution of the Fv1n-CtD in these fractions is also indicative of an associative interaction between Fv1n proteins. The 110-kDa peak contains both Fv1n-NtD and Fv1n-CtD fragments. As each individual domain has a molecular mass of around 20 kDa, this suggests that even the isolated domains retain some of the self-association properties of the intact protein. However, it is not clear if fragments within this peak comprise homomeric Fv1nNtD-Fv1nNtD and Fv1nCtD-Fv1nCtd complexes, heteromeric Fv1nNtD-Fv1nCtD complexes, or perhaps, a mixture of all three. The final peak that elutes with an apparent molecular mass of 40 kDa contains only Fv1n-CtD and presumably consists of monomeric Fv1n-CtD or a homomeric assembly distinct from the material in the 110-kDa peak.
FIG. 3.
Fractionation of Fv1n and copurifying fragments by size-exclusion chromatography. (A) Chromatogram produced by fractionation of nickel-chelate affinity-purified Fv1n on a Superdex 200 (10/30) column. The column was calibrated with standards, and the expected molecular masses (kDa) based on this calibration together with the column void volume (Vo) are indicated. (B) Western blot analysis using anti-Fv1 antibodies of fractions eluted from the size-exclusion column. The lane number corresponds to the fraction number in panel A, Ld is the material loaded onto the column, and the lanes marked M contained molecular weight markers. (C) Elution profile of purified Fv1(20-200) applied to a Superdex 200 (10/30) column. (D) SDS-PAGE of the peak fractions from panel C, the gel is stained with Coomassie brilliant blue, and the molecular masses (kDa) of the size markers are indicated.
Sedimentation analysis.
Although full-length Fv1 is highly insoluble in bacteria, using the information gained by mapping domain boundaries, we were able to express soluble fragments of Fv1n in E. coli, and this allowed us to investigate Fv1 self-association properties more closely. Residues 20 to 200 of Fv1n, Fv1(20-200), were expressed in E. coli as an N-terminal His tag fusion, and the protein was purified to carry out in vitro biophysical studies. The N-terminal domain was chosen, as it contains a predicted coiled-coil motif (residues 88 to 115), and it seemed likely that this region of the protein might be responsible for the self-association properties of Fv1n. Analysis of Fv1(20-200) using size-exclusion chromatography (Fig. 3c and d) revealed that this domain eluted at a similar volume from the Superdex (10/30) column as the NtD fragments purified from the baculovirus-infected cells. Comparison with molecular mass standards provided an estimate of the apparent molecular mass of 91 kDa, consistent with the idea that baculovirus-produced Fv1-NtD fragments are homo-oligomers and that the same homomeric association is present in Fv1(20-200).
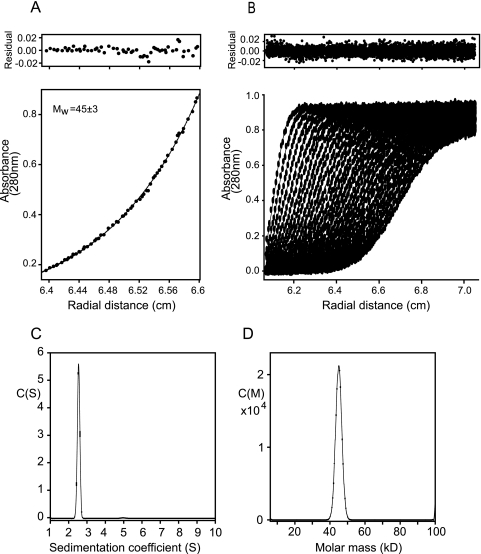
To determine the oligomeric state of Fv1(20-200), the solution molecular mass of the protein was measured using sedimentation equilibrium. Data were recorded for nine samples at two rotor speeds at concentrations ranging from 3 to 30 μM. A typical equilibrium distribution and curve of best fit are presented in Fig. 4a. Global fitting of the whole data set gave a molecular mass of 45 kDa, equivalent to the molecular mass of a dimer (monomer Mr, 21,482) (Table 1). Analysis of the data set gave no indication of any other species, either monomer or higher aggregate, and furthermore, no concentration dependency of the molecular mass was observed. Taken together, these data demonstrate that, at micromolar concentrations, the Fv1NtD forms a stable dimeric structure. Sedimentation velocity experiments were employed to examine the molecular shape of Fv1(20-200). Moving boundary data were collected from samples in the range of 0.7 to 0.9 mg/ml. The datasets were analyzed using finite element solutions to the Lamm equation for a single-species model and the continuous size distribution functions C(S) and C(M) (Fig. 4b to d). No apparent concentration dependency of the sedimentation coefficient was observed, consistent with the constant weight-averaged molecular mass determined by sedimentation equilibrium. The C(S) function that best fits the data shows a tight distribution with a maximum at 2.5 Sapp, indicating the sample likely contains only a single species. Similarly, the C(M) function contains a single peak centered at 45 kDa. Analyzing the data using a single-component model gives a good fit and, after conversion to standard conditions (20°C in water), gives an S20,w of 2.64 and a D20,w of 5.44 × 10−7 cm2 sec−1. These hydrodynamic data are summarized in Table 1 together with frictional ratios calculated from S20,w and D20,w. The value for the frictional ratio (f/fo) derived from these data is 1.7, and upon accounting for hydration of the molecule, the frictional ratio due to shape alone (f/fh) is 1.47. These values are rather higher than those observed for compact globular proteins and indicate that the dimer forms a highly extended structure.
FIG. 4.
Sedimentation analysis of Fv1(20-200). (A) Concentration distribution at sedimentation equilibrium for 16 μM Fv1(20-200) at 15,000 rpm. The curve is the best fit to the data, and the top panel shows the residuals to this fit. Mw, is the weight-averaged molecular weight derived from global fitting of all the data in thousands. (B) Analysis of Fv1(20-200) by sedimentation velocity. The data were produced from a sedimentation velocity experiment at 42,000 rpm, scanned every 300 s. The points are the measured data, and the lines are the best fit to the data using a C(S) size distribution function and f/fo of 1.7. Residuals from the fit are shown above. (C) The C(S) function corresponding to the best fit in panel A. (D) C(M) function corresponding to the best fit to the data in panel B. The fit quality and residuals are comparable to the C(S) function.
TABLE 1.
Hydrodynamic data for Fv1(20-200)
| Parameter | Value |
|---|---|
| Mr | 21,482 |
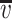 (ml g−1)a (ml g−1)a
|
0.725 |
| Ms (SE), avg (kDa)b | 45 ± 3 |
| Mw (SE), global (kDa)c | 45.2 |
| S20,w | 2.64 ± 0.03 |
| D20,w (10−7 cm2 s−1) | 5.4 ± 0.16 |
| Mw (SV), avg (kDa)d | 43 ± 1.5 |
| f/fo | 1.70 ± 0.03 |
| f/fhe | 1.47 |
Protein partial specific volume.
Weight-averaged molecular mass derived by averaging the masses obtained from all nine sedimentation equilibrium (SE) experiments analyzed individually.
Weight-averaged molecular mass derived by global fitting of all nine sedimentation equilibrium experiments.
Weight-averaged molecular mass derived by averaging the masses obtained from all sedimentation velocity (SV) experiments.
Value for the frictional ratio due to shape alone. Assuming a hydration of 40% (wt/wt), the frictional ratio due to hydration, fh/fo, is 1.156 where (f/fh) = (f/fo)/(fh/fo).
CD spectroscopy.
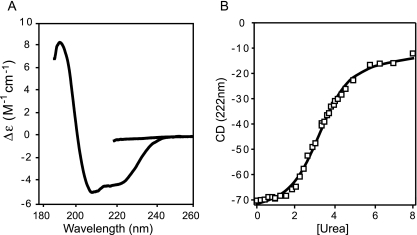
The high value of the frictional ratio observed for the Fv1-NtD dimer could be a result of the molecule containing a large amount of disordered residues. Alternatively, extensive coiled-coil interactions are likely to create a large degree of asymmetry in the molecule, a situation that would also result in a high frictional ratio. To test these two possibilities, CD spectroscopy was used to analyze the secondary structure content of Fv1(20-200). The Far UV CD spectrum, 190 to 250 nm, is shown in Fig. 5a. The spectrum is representative of a structured and predominantly α-helical protein characterized by a large negative Δɛ at 222 nm. A quantitative analysis of the spectrum (Table 2) suggests that around 43% of the residues are in an α-helical conformation, supporting the idea that the frictional ratio observed in the sedimentation experiments is a result of extended helical structure rather than disorder. The stability of the Fv1NtD dimer was examined by following the effects of urea denaturation on the far UV CD spectrum of Fv1(20-200). The intensity of the spectrum in 8 M urea (Fig. 5a) is largely diminished, indicating the loss of protein secondary structure. The denaturation transition induced by urea, monitored by CD at 222 nm, of Fv1(20-200) is presented in Fig. 5b. From these data, the free energy of unfolding in water ΔGH20, a measure of protein stability, and m, a measure of the change in solvent exposure between the native and denatured states, were then derived by linear extrapolation (31) (Table 2). The ΔGH20 for Fv1(20-200) derived in this manner is 2.6 ± 0.12 kcal · mol−1, and the slope of the extrapolation (m) is 0.81 ± 0.04 kcal · mol−1 M−1 (Table 2). These values are lower than values published for globular proteins with densely packed hydrophobic cores (30). Nevertheless, they do indicate that the Fv1NtD dimer is a folded stable structure.
FIG. 5.
CD spectroscopy. (A) Far UV CD spectra (250 to 190 nm) of Fv1(20-200) The spectra were recorded in 8 M urea (upper curve) and in CD buffer (lower curve). (B) Urea-induced denaturation of Fv1(20-200) monitored by the reduction of CD intensity at 222 nm.
TABLE 2.
CD spectroscopy for Fv1(20-200)
| Parameter | Fv1(A20-A200) result |
|---|---|
| No. of residues | 188 |
| Δɛ222 | −4.19 |
| % of α-helical residuesa | 43.3 |
| No. of helical residues | 81 ± 6 |
| ΔGH20 (kcal · mol−1) | 2.6 ± 0.12 |
| m (kcal · mol−1 M−1) | 0.81 |
Calculated from fitting the CD spectrum with Contin.
DISCUSSION
Domain organization of Fv1.
We have overexpressed and purified Fv1n using a recombinant baculovirus in an insect cell system. In contrast to expression in prokaryotic E. coli systems, Fv1n was produced in a soluble form but was accompanied by several degradation products that copurified with the intact protein. Analysis of these endogenous cleavage sites together with further cleavage sites produced from in vitro-limited proteolysis has allowed us to map the location of structural domains within the protein and show that Fv1n is organized into two approximately 20-kDa structured domains separated by a central, protease-sensitive linker region.
During the analysis of the proteolysis data, it became apparent that a large number of endogenously produced fragments, including a species migrating at ∼49 kDa (Fv1nΔ15), just smaller than the 52-kDa full-length protein, did not contain the N-terminal His tag used in the affinity purification. Furthermore, some of the smaller fragments were variants of the N-terminal domain of Fv1n, and others appeared to be derived from the C-terminal domain of Fv1n. The conclusion drawn from these data was that Fv1 molecules must be associated with each other for a His-tagged molecule to carry a nontagged molecule through the purification. Further size-exclusion chromatography data also supported the idea that Fv1n monomers are able to self-associate and that the individual N- and C-terminal domains retain some of the self-association properties of the whole protein.
To investigate the association properties of the molecule in more detail, a construct encompassing the N-terminal domain of Fv1, Fv1(20-200), was overexpressed in E. coli and examined using a variety of in vitro biophysical techniques. Measurements of the optical and hydrodynamic properties of this molecule produced data that are consistent with idea that the Fv1-NtD forms a stable dimeric and elongated α-helical structure. Interestingly, only residues 88 to 115 within this region of Fv1 are predicted to form a coiled-coil structure. However, our data show that the fraction of residues in an α-helical conformation within Fv1(20-200) is greater than can be accounted for by the number of helical residues within this predicted coiled-coil region alone. Furthermore, the value of the frictional ratio (f/fo = 1.70 ± 0.03) indicates that the molecule contains a larger degree of asymmetry than can be accounted for by just the extended structure within the predicted coiled coil. Taken together, our data suggest that the dimeric Fv1-NtD contains extensive extended helical structure outside the predicted coiled-coil region.
Taking into account the dimerization properties of this N-terminal domain along with the copurification of Fv1 fragments, it is interesting to reexamine the proteolysis data in terms of the possible arrangement and oligomerization of the amino- and carboxyl-terminal domains. The simplest explanation of the proteolysis data is that a protease-sensitive linker connects the N- and C-terminal domains and that the domains behave independently like “beads on a string.” However, noting that our data not only provide evidence of Fv1n self-association through the N-terminal dimerization domain but also demonstrate that Fv1 C-terminal domains retain the capacity to associate with intact Fv1, it is possible that the C-terminal domains could mediate further higher-order self-association of Fv1 dimers.
Comparison between Fv1 and Trim5α.
It is interesting to compare the arrangement of domains in Fv1 to that within Trim5α. Our proteolysis data provide evidence that Fv1 contains two structural regions. An N-terminal dimerization domain separated by a central unstructured region from a protease-resistant C-terminal domain. Previous genetic and virological studies have demonstrated that residues within this C-terminal domain direct the specificity of restriction of N- or B-tropic MLV (3) and that this specificity is targeted toward the viral capsid (7, 20, 29, 41, 44, 45). Similarly, the PRYSPRY domain at the C-terminal region of Trim5α is also responsible for the capsid specificity of restriction (28, 32, 37, 47, 50). In Trim5α, the N-terminal region contains the tripartite RBCC motif, and the coiled-coil part of the motif has been shown to be necessary for the multimerization properties of Trim5α (26, 32). Residues 88 to 115 of the Fv1 dimerization domain are also predicted to form a coiled coil, and although the coil is not predicted to be as extensive (40 residues in Fv1, 100 residues in Trim5α), our data now suggest that, in Fv1, the extended helical structure is likely to persist outside residues 88 to 115. This observation supports the idea that the topological arrangement of domains within these two different classes of restriction factor is actually rather similar. Both consist of an N-terminal coiled-coil domain connected to a C-terminal specificity domain, suggesting that some features of the mechanism of Fv1 and Trim5α restriction may be common, such as the necessity for a multivalent capsid-specific interaction provided by the combination of SPRY/Fv1-CtD and coiled-coil domains. However, N-terminal to the coiled coil, Trim5α contains additional RING and B-box motifs that are not present in Fv1 but are required for Trim5α restriction activity (32, 46), suggesting that, although the mode of restriction factor-capsid interaction may be common, the subsequent mechanism of Trim5α and Fv1 restriction is potentially different. Another important difference between Fv1 and Trim5α appears to be the stoichiometry of homo-oligomerization. Recently, it has been proposed that Trim5α associates into a trimeric structure through an interaction mediated by its coiled-coil domain (26). Our data clearly demonstrate that the N-terminal domain of Fv1 containing the coiled-coil region associates to form a dimeric structure. This presents an interesting difference between the two classes of restriction factor where a Trim5α-capsid interaction would be mediated through the interaction of three SPRY domains with the core/RTC, but the Fv1-core/RTC interaction is mediated by the interaction of only two Fv1-CtDs.
Self-association of restriction factors.
The presence of a stable dimeric structure within Fv1 leads to the question of what the functional significance of restriction factor self-association might be. The importance of restriction factor multimerization is underlined by the dominant-negative effect of Trim5γ, a splice variant of Trim5α that lacks the SPRY domain. Expression of Trim5γ in cells that also express Trim5α results in abolition of the anti-human immunodeficiency virus type 1 restriction activity (46). Furthermore, expression of Trim5α deletion mutants that contain the coiled-coil domain and also have the capacity to multimerize but have no SPRY and or RING/B-box strongly suppresses HeLa cell N-tropic MLV restriction (32).
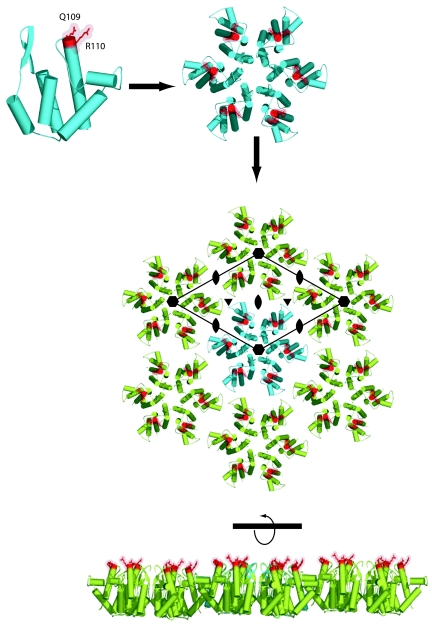
At present, there is mounting evidence that the target for Fv1 and Trim5α activity is the capsid protein in the context of an uncoated viral core or RTC (8, 9). The exterior surface of retroviral cores consists of an interlocking network of hexagonal discs comprised from six capsid monomers (6, 11, 23, 27), and in both uncoated cores and RTCs, there is still likely to be a large degree of this long-range multimeric organization present. A result of this arrangement of hexagonal discs is that the residues that are responsible for viral tropism in MLV and direct the specificity of restriction by Fv1 and Trim5α (21, 44, 49) are located on the outer surface of these discs (27). Furthermore, a consequence of lattice formation is that these residues are related to each other by local 2-, 3-, and 6-fold symmetry axes and so form a defined and regular array of restriction factor interaction sites, illustrated in Fig. 6.
FIG. 6.
Arrangement on a capsid lattice of residues responsible for viral tropism and for the specificity of restriction. (Top) A ribbon representation of the structure of the N-terminal domain (NtD) of the N-tropic MLV capsid. The residues that are strong determinants of viral tropism Q109 and R110 are highlighted in red. (Middle) Assembly of six capsid NtDs into the capsid hexamer observed in the crystal structure (27). The residues involved in tropism are highlighted as in panel A. (Bottom) A layer of hexameric molecules, taken from the crystal lattice that results from the packing of NtD hexameric disks. The residues involved in the specificity of MLV restriction form an array of restriction factor binding sites on the outer surface of the lattice. The 6-fold rotation axes (
) together with the local interhexamer 2-fold ( ) and 3-fold (▾) rotation axes where multimeric restriction factors may target are indicated.
) and 3-fold (▾) rotation axes where multimeric restriction factors may target are indicated.
When the restriction factor-capsid interaction is examined with the idea of a binding site array in mind, it is easy to see why a multivalent interaction would significantly enhance the potency of restriction, enabling the effects of a single restriction factor to be felt by many capsid monomers. Furthermore, as the lattice contains local 3-fold rotation axes as well as 2-fold axes, it is also easy to reconcile the differences in the apparent stoichiometry of the Fv1-capsid and Trim5α-capsid interactions. The common factor is that, regardless of the oligomerization state, it is the capacity of the restriction factor to interact with multiple capsid monomers spanning across hexameric disks that is likely to be important to implement the restriction effect. Moreover, the existence of extended helical and/or coiled-coil structures in these molecules not only provides the oligomerization required to generate multivalent interactions but also has the potential to provide the correct spacing of recognition domains to facilitate interactions with the binding site array.
Our data also provide evidence for further Fv1 self-association mediated through the C-terminal domain, and although it has been demonstrated that the PRYSPRY domain of Trim5α and the CtD of Fv1 contain the residues key to specificity of restriction, a recent publication has suggested that SPRY domains themselves have the capacity to associate weakly (13). One possibility is that the CtD of Fv1 and the PRYSPRY of Trim5α are not only responsible for the specificity of restriction but have the capacity to recruit further multimeric molecules to a targeted RTC through CtD-mediated interactions.
The steps in viral infection that occur between viral uncoating and provirus integration are poorly understood. However, it is clear that the absolute stability of the viral core is important, as mutations that both increase and decrease core stability result in a loss of viral infectivity (10). In our model, a single restriction factor is able to affect many capsid monomers within either intact cores or partially disassembled RTCs. This means that, even at low stoichiometric ratios, restriction factors would be expected to strongly influence the core stability, possibly explaining why retroviral restriction factors are efficient even though they are expressed at very low levels in host cells (51; unpublished data). Targeting of the RTC by restriction factors might be expected to upset the finely balanced process of core disassembly. On one hand, premature disruption of the core may prevent completion of reverse transcription. Conversely, inhibition of core disassembly could result in retardation of the core in the cytoplasm or a block to nuclear entry. Taking all of these possibilities into account, unraveling the molecular details of restriction factor-RTC interactions provides an exciting challenge.
Acknowledgments
We thank Barry Ely for assistance with baculovirus expression and the MRC, United Kingdom, for support.
REFERENCES
- 1.Benit, L., N. De Parseval, J. F. Casella, I. Callebaut, A. Cordonnier, and T. Heidmann. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 71:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, K. N., M. Bock, G. Towers, and J. P. Stoye. 2001. Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J. Virol. 75:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 5.Bock, M., K. N. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs, J. A., T. Wilk, R. Welker, H. G. Krausslich, and S. D. Fuller. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DesGroseillers, L., and P. Jolicoeur. 1983. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J. Virol. 48:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodding, M. P., M. Bock, M. W. Yap, and J. P. Stoye. 2005. Capsid processing requirements for abrogation of Fv1 and Ref1 restriction. J. Virol. 79:10571-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forshey, B. M., J. Shi, and C. Aiken. 2005. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in owl monkey cells. J. Virol. 79:869-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganser, B. K., A. Cheng, W. I. Sundquist, and M. Yeager. 2003. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 22:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703-1706. [DOI] [PubMed] [Google Scholar]
- 13.Grutter, C., C. Briand, G. Capitani, P. R. Mittl, S. Papin, J. Tschopp, and M. G. Grutter. 2006. Structure of the PRYSPRY-domain: implications for autoinflammatory diseases. FEBS Lett. 580:99-106. [DOI] [PubMed] [Google Scholar]
- 14.Guo, X., J. W. Carroll, M. R. Macdonald, S. P. Goff, and G. Gao. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78:12781-12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 16.Hartley, J. W., W. P. Rowe, and R. J. Huebner. 1970. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J. Virol. 5:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 19.Jolicoeur, P. 1979. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol. 86:67-122. [DOI] [PubMed] [Google Scholar]
- 20.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300-305. [DOI] [PubMed] [Google Scholar]
- 21.Lassaux, A., M. Sitbon, and J. L. Battini. 2005. Residues in the murine leukemia virus capsid that differentially govern resistance to mouse Fv1 and human Ref1 restrictions. J. Virol. 79:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 23.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 24.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 25.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 26.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortuza, G. B., L. F. Haire, A. Stevens, S. J. Smerdon, J. P. Stoye, and I. A. Taylor. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431:481-485. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama, E. E., H. Miyoshi, Y. Nagai, and T. Shioda. 2005. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5α determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79:8870-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou, C. Y., L. R. Boone, C. K. Koh, R. W. Tennant, and W. K. Yang. 1983. Nucleotide sequences of Gag-Pol regions that determine the Fv-1 host range property of BALB/c N-tropic and B-tropic murine leukemia viruses. J. Virol. 48:779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pace, C. N. 1990. Conformational stability of globular proteins. Trends Biochem. Sci. 15:14-17. [DOI] [PubMed] [Google Scholar]
- 31.Pace, C. N. 1986. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 131:266-280. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Provencher, S. W., and J. Glockner. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33-37. [DOI] [PubMed] [Google Scholar]
- 35.Pryciak, P. M., and H. E. Varmus. 1992. Fv-1 restriction and its effects on murine leukemia virus integration in vivo and in vitro. J. Virol. 66:5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuck, P. 2003. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320:104-124. [DOI] [PubMed] [Google Scholar]
- 39.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuck, P., M. A. Perugini, N. R. Gonzales, G. J. Howlett, and D. Schubert. 2002. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82:1096-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebastian, S., and J. Luban. 2005. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedgwick, S. G., I. A. Taylor, A. C. Adam, A. Spanos, S. Howell, B. A. Morgan, M. K. Treiber, N. Kanuga, G. R. Banks, R. Foord, and S. J. Smerdon. 1998. Structural and functional architecture of the yeast cell-cycle transcription factor swi6. J. Mol. Biol. 281:763-775. [DOI] [PubMed] [Google Scholar]
- 43.Sreerama, N., S. Y. Venyaminov, and R. W. Woody. 2001. Analysis of protein circular dichroism spectra based on the tertiary structure classification. Anal. Biochem. 299:271-274. [DOI] [PubMed] [Google Scholar]
- 44.Stevens, A., M. Bock, S. Ellis, P. LeTissier, K. N. Bishop, M. W. Yap, W. Taylor, and J. P. Stoye. 2004. Retroviral capsid determinants of Fv1 NB and NR tropism. J. Virol. 78:9592-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoye, J. P. 1998. Fv1, the mouse retrovirus resistance gene. Rev. Sci. Tech. 17:269-277. [DOI] [PubMed] [Google Scholar]
- 46.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 47.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vistica, J., J. Dam, A. Balbo, E. Yikilmaz, R. A. Mariuzza, T. A. Rouault, and P. Schuck. 2004. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 326:234-256. [DOI] [PubMed] [Google Scholar]
- 49.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 51.Yap, M. W., and J. P. Stoye. 2003. Intracellular localisation of Fv1. Virology 307:76-89. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]