Abstract
The influence of the transcriptional enhancer of the pea plastocyanin gene (PetE) on the acetylation of histones was examined with chromatin immunoprecipitation (ChIP) experiments using antibodies that recognize acetylated or nonacetylated histones H3 and H4. In transgenic tobacco plants containing the pea PetE promoter fused to uidA, both acetylated and nonacetylated histones H3 and H4 were present on the integrated transgene. Linking the PetE enhancer to the transgene resulted in increased β-glucuronidase activity and increased amounts of acetylated histones H3 and H4 present on the promoter, suggesting that the enhancer may increase transcription by mediating the acetylation of histones. Trichostatin A and sodium butyrate, which are potent inhibitors of histone deacetylases (HDAs), activated expression from the PetE promoter by fourfold, with a concomitant increase in the acetylation states of histones H3 and H4, as determined by ChIP, indicating that the acetylation of histones has a direct positive effect on transcription. The HDA inhibitors did not increase expression from the PetE promoter when it was linked to the enhancer, consistent with preexisting hyperacetylated histones on the transgene. Mapping of histone acetylation states along the reporter gene indicated that the histones H3 and H4 associated with the promoter and the 5′ region of uidA were hyperacetylated in the presence of the PetE enhancer. The PetE enhancer bound to isolated tobacco nuclear matrices in vitro and was associated with the nuclear matrix in nuclei isolated from transgenic tobacco plants. These results suggest that the pea PetE enhancer activates transcription by associating with the nuclear matrix, mediating the acetylation of histones on the promoter and the nearby coding region and resulting in an altered chromatin structure.
INTRODUCTION
DNA in eukaryotic nuclei is packaged into nucleosomes, which are composed of two turns of DNA wound around a histone octamer complex containing two molecules each of histones H2A, H2B, H3, and H4 (Wolffe and Hayes, 1999). The Lys residues in the N-terminal tails of histones H3 and H4 can be acetylated by histone acetyltransferases (HATs) (reviewed by Strahl and Allis, 2000). The acetylated histones H3 and H4 can increase transcription by two postulated mechanisms: (1) acetylation of the Lys residues neutralizes the positive charges of histone proteins, weakens histone-DNA contacts, and facilitates the displacement of nucleosomes from promoters of genes, thereby allowing the entry of transcription factors (Fernández et al., 2001; Eberharter and Becker, 2002); (2) the acetylated histones stabilize the assembly of transcriptional complexes on promoters through protein factors that contain bromodomain modules that interact specifically with acetylated histone tails (Strahl and Allis, 2000; Hassan et al., 2001). Recent evidence has indicated that two classes of cis-acting elements, transcriptional enhancers and matrix-attachment regions (MARs), may regulate transcription through the acetylation of histones (Agalioti et al., 2000; Fernández et al., 2001).
The first class of these cis-acting elements, the transcriptional enhancers, contains DNA sequences that augment gene expression from homologous or heterologous promoters in a position- or orientation-independent manner (Blackwood and Kadonaga, 1998; Struhl, 2001). Various enhancer binding proteins interact with HATs, which acetylate the N-terminal tails of histone H3 and H4 proteins present on enhancers and/or promoters (Brown et al., 2000). Acetylation of histones plays critical roles in the functions of mammalian transcriptional enhancers, including the human interferon-β gene (Agalioti et al., 2000), the bovine β-casein gene BCE-1 (Myers et al., 1998), and the human immunodeficiency virus-1 (HIV-1) enhancers (Sheridan et al., 1997). Treatment with histone deacetylase (HDA) inhibitors, such as trichostatin A or sodium butyrate, resulted in the hyperacetylation of histones and the activation of the BCE-1 and HIV-1 enhancers, suggesting that histone acetylation is necessary for enhancer-mediated transcriptional activation (Sheridan et al., 1997; Myers et al., 1998). In the human interferon-β gene, the enhanceosome complex assembled on the enhancer recruits the GCN5 complex, which acetylates the nucleosomes present on the enhancer and the nearby promoter (Thanos and Maniatis, 1992; Agalioti et al., 2000). The acetylated histones are necessary for the subsequent recruitment of the TATA-box protein and the RNA polymerase II holoenzyme, which initiate transcription (Agalioti et al., 2000).
The second class of cis elements, the MARs, is defined operationally as DNA sequences that associate specifically with the residual fibrillar proteinaceous framework that remains in eukaryotic nuclei after DNA, RNA, and soluble nuclear proteins are removed (Mirkovitch et al., 1984; Cockerill and Garrard, 1986). MARs may be responsible for organizing DNA into chromatin loops by attaching to the nuclear matrix (Mirkovitch et al., 1984; Gasser and Laemmli, 1986). MARs also can function as boundary elements that insulate the expression of reporter genes from the effects of neighboring chromatin, thus reducing transformant-to-transformant variation in gene expression (Allen et al., 1993, 1996; Mlynárová et al., 1994, 1995) and conferring copy number–dependent expression (Blasquez et al., 1989; Stief et al., 1989). In addition, MARs often colocalize with transcriptional enhancers and can augment the activity of flanking enhancers through the acetylation of histones (Forrester et al., 1994; Aronow et al., 1995; Fernández et al., 2001). For example, the MARs that flank the murine immunoglobulin μ enhancer can generate an extended domain of acetylated histones, thereby activating expression from promoters located as far as 1 kb away (Forrester et al., 1999; Fernández et al., 2001). The MAR of the human topoisomerase-I gene binds to the nuclear matrix protein SAF-A, which interacts with the HAT p300 to result in the acetylation of histones and the subsequent activation of transcription (Martens et al., 2002).
The PetE gene encodes the plastocyanin protein, which transfers electrons from the cytochrome f protein in the cytochrome bf complex to the P700 reaction center of photosystem I in the light reactions of photosynthesis. In pea, PetE is expressed only in photosynthetic tissues, and its transcription is activated by light (Last and Gray, 1989; Chua et al., 2001). A 268-bp sequence in the PetE promoter (−444 to −177, with +1 as the translation start site) functions as a general transcriptional enhancer by activating transcription in a promoter-, position-, orientation-, and tissue-independent manner; the sequence upregulates expression from the minimal Cauliflower mosaic virus 35S, patatin, and PetE promoters when present in both orientations upstream or downstream of reporter genes in tobacco leaves and roots or potato tubers (Sandhu et al., 1998). Experimental evidence suggests that the pea PetE enhancer increases transcription by inducing changes in chromatin structure and chromatin accessibility to transcription factors. First, the enhancer interacts with HMG-I/Y and HMG-1 proteins (Pwee et al., 1994; Webster et al., 1997), which are architectural chromosomal proteins that regulate transcription by modulating DNA conformation (Grosschedl et al., 1994; Grasser, 1998). Second, the accessibility of the pea PetE enhancer to nuclease digestion increases when the gene is expressed, suggesting that it adopts a more open chromatin structure for interactions with transcription factors (Chua et al., 2001). Finally, histones H3 and H4 present on the enhancer are acetylated when the transcription of pea PetE is activated by light (Chua et al., 2001).
Many plant nuclear genes also possess cis-acting elements that stimulate the expression of reporter genes only when integrated into chromatin; therefore, they are likely to activate transcription through chromatin structure. Examples include MAR sequences from the bean phaseolin gene (van der Geest et al., 1994), the soybean Gmhsp17.6-L gene (Schöffl et al., 1993), and the tobacco RB7 gene (Allen et al., 1996). However, little is known about the mechanism by which these plant sequences activate transcription. Here, we examine the acetylation states of histones present on various regions of reporter genes using chromatin immunoprecipitation (ChIP) and show that the pea PetE enhancer mediates the acetylation of histones and increases transcription. Moreover, the PetE enhancer associates with the nuclear matrix, suggesting that MARs may function as entry sites for HATs during transcriptional activation.
RESULTS
Hyperacetylated Histones H3 and H4 Are Present on the PetE Promoter When Linked to the PetE Enhancer
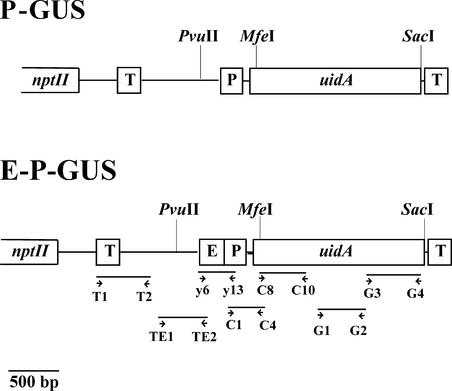
The pea PetE gene possesses a promoter of 124 bp (−176 to −53, with +1 as the translation start site) and an A/T-rich enhancer sequence (−444 to −177) immediately upstream of the promoter. In a previous promoter-deletion analysis, removal of the PetE enhancer sequence from the PetE promoter resulted in a 40-fold reduction in reporter gene expression in stable transgenic tobacco plants, suggesting that the sequence functions as a positive regulatory sequence (Pwee and Gray, 1993). Further investigations indicated that the sequence is a general transcriptional enhancer that activated reporter expression when present upstream or downstream of different heterologous promoters in either orientation in tobacco leaves and roots or potato tissues (Sandhu et al., 1998). To investigate whether the PetE enhancer may increase transcription through histone acetylation, we performed ChIP using second-generation transgenic tobacco plants from Pwee and Gray (1993). These plants contained the uidA gene fused to the −176 to +4 (P-GUS [β-glucuronidase]) or −444 to +4 (E-P-GUS) region of the pea PetE promoter (Figure 1). Three independently transformed lines containing P-GUS or E-P-GUS were examined with ChIP.
Figure 1.
Scheme of Transgenes Containing the Pea PetE Promoter.
The locations of primers are indicated by arrowheads. E, pea PetE enhancer sequence (−444 to −177 of pea PetE); MfeI, PvuII, and SacI, restriction sites used for the experiments shown in Figure 9; nptII, neomycin phosphotransferase selectable marker gene; P, PetE promoter (−176 to +4 of pea PetE); T, nos 3′ region; uidA, β-glucuronidase reporter gene.
Nuclei were prepared from shoots of tobacco seedlings (four-leaf stage) and treated immediately with formaldehyde to cross-link nuclear proteins to their cognate-DNA binding sites. The chromatin then was sheared to an average length of 300 to 700 bp by sonication and immunoprecipitated with antibodies specific for acetylated or nonacetylated Lys residues in the N-terminal tails of histones H3 and H4. The immunocomplexes were harvested with protein A–Sepharose beads and heated at 65°C for 5 h to release DNA cross-linked to the immunoprecipitated histone proteins. The DNA was analyzed by semiquantitative PCR for the presence of specific sequences with the primers shown in Figure 1. To assess nonspecific binding, the immunoprecipitation reaction was also performed in the absence of antibody or with nonimmune rabbit serum. Our initial attempts at ChIP with tobacco nuclei yielded high levels of nonspecific background in the PCR, which was eliminated by preclearing the chromatin solution with protein A–Sepharose beads before immunoprecipitation with antibodies (see Methods). The ChIP samples were analyzed by PCR in parallel with DNA purified from a fraction of sonicated nuclei (total DNA) to allow quantification relative to input chromatin. The coding region of the actin gene Tac9 was used as an internal control for the immunoprecipitation reactions.
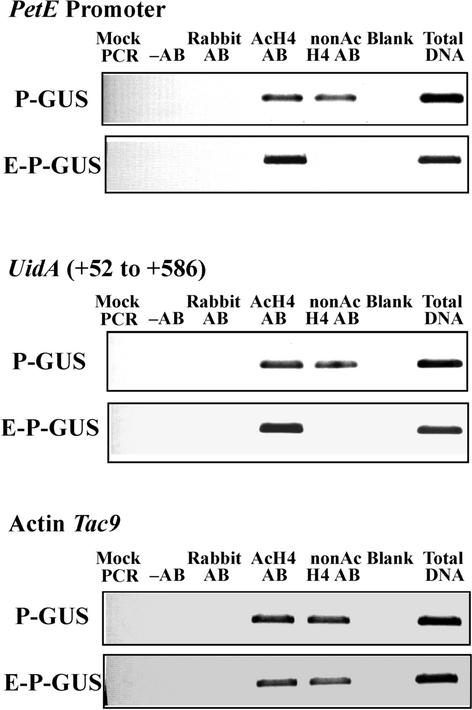
In ChIP assays performed with tobacco seedlings containing E-P-GUS, the PetE promoter and the 5′ region of the uidA coding sequence (+52 to +586, with +1 as the translation start site) were enriched in the immunoprecipitates with anti-acetylated H4 antibody but depleted in the precipitates with anti-nonacetylated H4 antibody (Figure 2). This finding suggests that the Lys residues in the tails of histone H4 proteins in the nucleosomes present on the PetE promoter and the +52 to +586 region of uidA were hyperacetylated in E-P-GUS plants. However, in plants containing P-GUS, the PetE promoter and the +52 to +586 region of uidA were detected in both anti-acetylated and anti-nonacetylated H4 immunoprecipitates, indicating that these sequences were associated with a mixture of acetylated and nonacetylated histone H4 proteins (Figure 2). This finding suggests that the PetE promoter and the 5′ uidA coding regions are associated with hyperacetylated histones when linked to the PetE enhancer. The histone H4 acetylation patterns of the coding region of the actin gene Tac9 were similar in plants containing E-P-GUS or P-GUS (Figure 2).
Figure 2.
Acetylation Patterns of Histone H4.
Nuclei isolated from tobacco seedlings containing P-GUS or E-P-GUS were cross-linked with formaldehyde and immunoprecipitated with antibody against acetylated histone H4 (AcH4 AB) or nonacetylated histone H4 (nonAc H4 AB). The amounts of PetE promoter sequence (analyzed with primer pairs C1 and C4) and the +52 to +586 region of uidA (analyzed with primer pairs C8 and C10) in the immunoprecipitates were quantified with PCR. Immunoprecipitation reactions were also performed with no antibody (−AB) or with nonimmune rabbit antiserum (Rabbit AB). PCR was performed with no template (Mock PCR) or with DNA isolated from a fraction of chromatin extract before immunoprecipitation (Total DNA). Three independently transformed lines containing P-GUS or E-P-GUS were analyzed. ChIP was performed in triplicate for each line, and similar results were obtained.
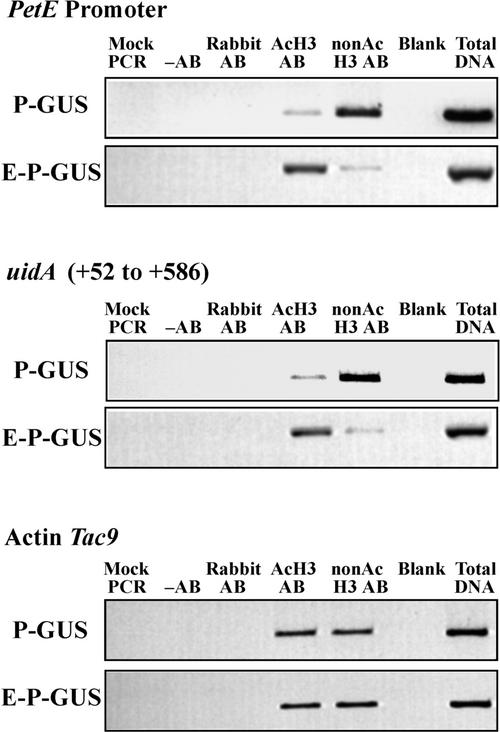
ChIP assays also were performed with antibodies against acetylated or nonacetylated H3 to examine histone H3 acetylation patterns (Figure 3). In E-P-GUS plants, the promoter and the +52 to +586 region of uidA were associated with histone H3 possessing mainly acetylated Lys residues, although a low level of nonacetylated Lys residues also was detected (Figure 3). However, in plants containing P-GUS, the PetE promoter and the +52 to +586 region of uidA were more enriched in the anti-nonacetylated histone H3 antibody immunoprecipitates than in the anti-acetylated H3 antibody immunoprecipitates (Figure 3). This finding suggests that the histone H3 present on the PetE promoter and the +52 to +586 region of uidA is acetylated in the presence of the PetE enhancer. The acetylation patterns of histone H3 present on Tac9 were similar in plants containing E-P-GUS or P-GUS (Figure 3). Because the specific GUS activities of leaf extracts from E-P-GUS plants were ∼30-fold higher than those of extracts from P-GUS plants (see Figure 5 legend), the ChIP results suggest that the PetE enhancer may increase transcription from the PetE promoter through the acetylation of histones H3 and H4.
Figure 3.
Acetylation Patterns of Histone H3.
Nuclei isolated from tobacco plants containing P-GUS or E-P-GUS were cross-linked with formaldehyde and immunoprecipitated with antibody against acetylated histone H3 (AcH3 AB) or nonacetylated histone H3 (nonAc H3 AB). The amounts of PetE promoter sequence (analyzed with primer pairs C1 and C4) and the +52 to +586 region of uidA (analyzed with primer pairs C8 and C10) in the immunoprecipitates were quantified with PCR. Immunoprecipitation reactions were also performed with no antibody (−AB) or with nonimmune rabbit antiserum (Rabbit AB). PCR was performed with no template (Mock PCR) or with DNA isolated from a fraction of chromatin extract before immunoprecipitation (Total DNA). Three independently transformed lines containing P-GUS or E-P-GUS were analyzed. ChIP was performed in triplicate for each line, and similar results were obtained.
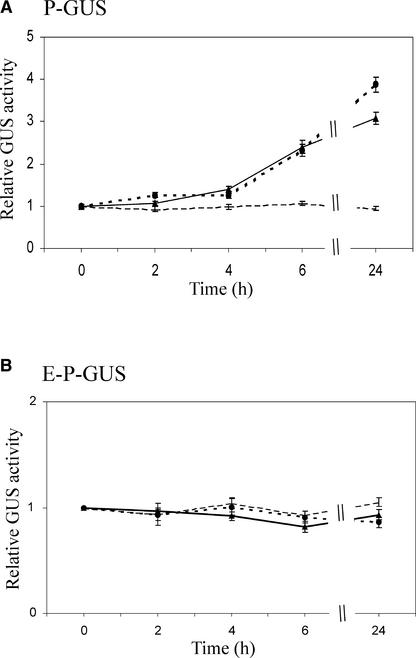
Figure 5.
Effects of Trichostatin A and Sodium Butyrate on Reporter Gene Expression.
Tobacco seedlings containing P-GUS (A) or E-P-GUS (B) were transferred to Murashige and Skoog (1962) medium containing no inhibitors (dashed line), 10 μM trichostatin (circles), or 10 mM sodium butyrate (triangles) for 0, 2, 4, 6, or 24 h. Specific GUS activities in tobacco leaves were determined and expressed as a fraction of GUS activities in nontreated control seedlings. The specific GUS activities presented are averages of six replicates from each of the three individually transformed lines containing P-GUS or E-P-GUS. The GUS activities in nontreated seedlings from the three lines containing P-GUS were 177 ± 6.6, 489 ± 36, and 646 ± 19 pmol methylumbelliferone·min−1·μg−1 protein, and those for the three lines containing E-P-GUS were 1094 ± 75, 11,900 ± 370, and 24,550 ± 339 pmol methylumbelliferone·min−1·μg−1 protein. Error bars represent standard errors of the mean.
Treatment with Histone Deacetylase Inhibitors Results in Histone Hyperacetylation and Increased Gene Expression
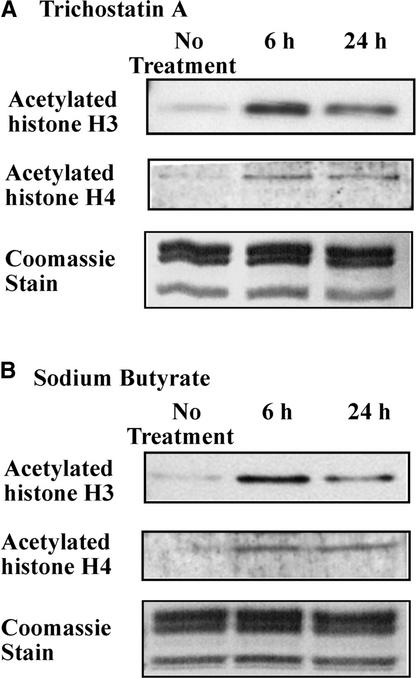
To determine whether the acetylation of histones H3 and H4 is necessary for enhancer-mediated gene activation or is a consequence of transcription, transgenic tobacco plants containing P-GUS were treated with trichostatin A (Yoshida et al., 1990) or sodium butyrate (Kruh, 1982), which are inhibitors of HDA, to increase the acetylation of the histones present on the PetE promoter. Trichostatin A at 1 to 100 μM, or sodium butyrate at 5 to 50 mM, caused an accumulation of acetylated histones H3 and H4 in mammalian cells (Kruh, 1982; Yoshida et al., 1990; Myers et al., 1998). In plants, Brassica napus seedlings treated with 10 μM trichostatin A or 10 mM sodium butyrate for 2 weeks also possessed increased levels of acetylated histone proteins (Chen and Pikaard, 1997). However, prolonged treatment with sodium butyrate for >6 h has been reported to cause chromatin modifications unrelated to histone acetylation, such as the inhibition of histone H1 and H2A phosphorylation and changes in the abundance of histone H1 variants in mammalian cells (Boffa et al., 1981; Annunziato et al., 1988). Furthermore, in alfalfa cell suspensions, the levels of acetylated histones declined 6 h after the addition of trichostatin A, reaching histone acetylation states similar to those of nontreated cells by 12 to 20 h (Waterborg and Kapros, 2002). When we treated tobacco seedlings with 10 μM trichostatin A or 10 mM sodium butyrate for 2 weeks, the plants appeared stunted and possessed fewer roots and there was no significant increase in the levels of acetylated bulk histone proteins (data not shown), whereas treatment for 6 or 24 h resulted in increased acetylation states of bulk histones H3 and H4, as determined by immunoblot analysis with antibodies specific for acetylated histone H3 or H4 protein (Figure 4).
Figure 4.
Effects of Trichostatin A and Sodium Butyrate on the Acetylation States of Bulk Histone Proteins.
Tobacco seedlings were treated with 10 μM trichostatin A (A) or 10 mM sodium butyrate (B) for 6 or 24 h. Nuclei were isolated, solubilized in SDS-PAGE loading buffer, and analyzed on SDS-PAGE gels. Immunoblot analysis was performed with antibodies against acetylated histone H3 or H4. Coomassie blue staining of protein preparations shows similar loading of histone proteins.
The effects of HDA inhibitors on transgene expression in 2-week-old plants containing P-GUS or E-P-GUS were investigated in liquid Murashige and Skoog (1962) medium containing no inhibitor, 10 μM trichostatin A, or 10 mM sodium butyrate for 2 to 24 h (Figure 5). Both trichostatin A and sodium butyrate activated the expression of P-GUS by threefold to fourfold after treatment for 6 or 24 h (Figure 5A). However, the inhibitors had no detectable effects on GUS activity in plants containing E-P-GUS (Figure 5B), in which the PetE promoter was already associated with hyperacetylated histones H3 and H4, as suggested by earlier ChIP results (Figures 2 and 3). Therefore, activated expression from the PetE promoter in the P-GUS reporter gene is correlated with the acetylation of bulk histone proteins by HDA inhibitors after 6 h of treatment.
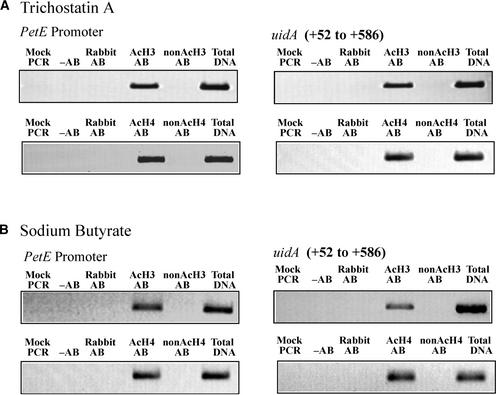
To confirm that the HDA inhibitors had increased the acetylation states of histones on P-GUS at the single-gene level, ChIP was performed on P-GUS tobacco seedlings treated with trichostatin A for 6 h. The PetE promoter and the +52 to +586 region of uidA were enriched in the immunoprecipitates with anti-acetylated H3 or H4 antibody but depleted in the immunoprecipitates with anti-nonacetylated H3 or H4 antibody (Figure 6A). This finding suggests that these sequences in P-GUS plants are associated with hyperacetylated histone H3 and H4 after treatment with trichostatin A. Similarly, after treatment with sodium butyrate for 6 h, the PetE promoter and the +52 to +586 region of uidA were no longer present in the anti-nonacetylated H3 or H4 antibody immunoprecipitates, suggesting that the histone H3 and H4 proteins associated with these sequences were hyperacetylated (Figure 6B). After HDA inhibitor treatment, the histone acetylation patterns on the PetE promoter in P-GUS were similar to the patterns on the promoter sequence in E-P-GUS, in which the promoter was linked to the PetE enhancer (Figures 2 and 6). This finding suggests that the acetylation of histones H3 and H4 on the PetE promoter, either by linking to the enhancer sequence or by treatment with HDA inhibitors, can increase transcription. However, the HDA inhibitors increased the activity of GUS to a lesser extent than the enhancer (Figure 5 legend), possibly as a result of a depletion of HATs or other transcriptional activators in the vicinity of the PetE promoter in the tobacco cells. The HDA inhibitors induce the acetylation of histones present on the promoters of other tobacco genes, as determined by microarray analyses of promoter regions immunoprecipitated in ChIP experiments (Y.L. Chua, E. Mott, A.P.C. Brown, and J.C. Gray, unpublished results), and the acetylated histones are likely to compete for similar transcription factors.
Figure 6.
Effects of HDA Inhibitors on the Acetylation States of Histones Present on Transgenes.
Tobacco seedlings containing P-GUS were treated with 10 μM trichostatin A (A) or 10 mM sodium butyrate (B) for 6 h. Nuclei were isolated from tobacco plants, cross-linked with formaldehyde, and immunoprecipitated with antibodies against acetylated histone H3 (AcH3 AB), nonacetylated histone H3 (nonAcH3 AB), acetylated histone H4 (AcH4 AB), or nonacetylated histone H4 (nonAcH4 AB). The amounts of PetE promoter sequence (analyzed with primer pairs C1 and C4) and the +52 to +586 region of uidA (analyzed with primer pairs C8 and C10) in the immunoprecipitates were quantified with PCR. Immunoprecipitation reactions also were performed with no antibody (−AB) or with nonimmune rabbit antiserum (Rabbit AB). PCR was performed with no template (Mock PCR) or with DNA isolated from a fraction of chromatin extract before immunoprecipitation (Total DNA). Similar results were obtained for duplicates of ChIP reactions performed on each of the three lines of P-GUS treated with trichostatin A or sodium butyrate.
The PetE Enhancer Mediates the Acetylation of Histones Present on the Promoter and the 5′ End of uidA
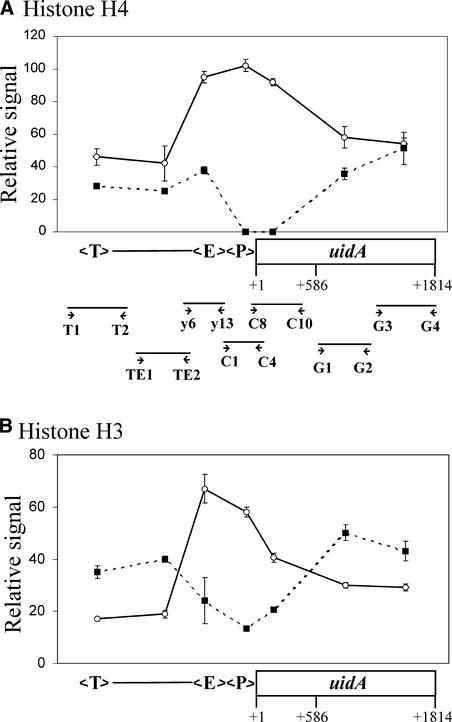
To investigate whether the PetE enhancer increased histone acetylation states beyond the promoter, we examined the histone acetylation patterns of various regions of the reporter gene with ChIP (Figure 7). The amount of sequence present in the ChIP samples was quantified and expressed as a percentage of the total input DNA.
Figure 7.
Mapping of Histone Acetylation States along Transgenes.
The acetylation states of histone H4 (A) or histone H3 (B) on various regions of the E-P-GUS transgene were analyzed. ChIP was performed in triplicate for each of the three individually transformed lines containing E-P-GUS. The primers used for PCR are indicated by arrowheads. The amounts of PCR products obtained in the immunoprecipitates are expressed as percentages of total DNA. ChIP was performed using anti-acetylated histone H4 antibody (open circles) or anti-nonacetylated histone H4 antibody (closed squares) (A) or using anti-acetylated histone H3 antibody (open circles) or anti-nonacetylated histone H3 antibody (closed squares) (B). Error bars represent standard errors of the mean. E, pea PetE enhancer sequence (−444 to −177 of pea PetE); P, PetE promoter (−176 to +4 of pea PetE); T, nos 3′ region.
The sequences between the nos 3′ region and the PetE enhancer possessed comparable amounts of acetylated and nonacetylated histone H4 (Figure 7A). The amounts of acetylated histone H4 associated with the enhancer, the promoter, and the 5′ end of the uidA coding region (+52 to +586, with +1 as the translation start site) were nearly threefold higher than the amounts associated with the sequences upstream of the PetE enhancer and the +587 to +1814 region of uidA. The amounts of nonacetylated histone H4 present on the PetE enhancer and the sequences upstream, and on the +587 to +1814 region of uidA, were similar, whereas no nonacetylated histone H4 was present on the promoter and the +52 to +586 region of uidA. This finding suggests that histone H4 in nucleosomes present on the enhancer, the promoter, and the +52 to +586 region of uidA are hyperacetylated relative to the other regions of the transgene.
For histone H3, the amounts of acetylated forms present on the enhancer, the promoter, and the +52 to +586 region of uidA were twofold to threefold higher than those in the rest of the transgene (Figure 7B). Correspondingly, the amounts of nonacetylated histone H3 on these regions were approximately twofold lower than those on the other regions of the reporter gene. These ChIP results suggest that the histones H3 and H4 present on the PetE enhancer, the PetE promoter, and the 5′ end of uidA were hyperacetylated in transgenes containing the enhancer sequence. Histone acetylation mediated by the PetE enhancer also appeared to be directional, extending into the 5′ end of the coding region of uidA but not into the upstream nos 3′ region.
The Pea PetE Enhancer Associates with the Nuclear Matrix
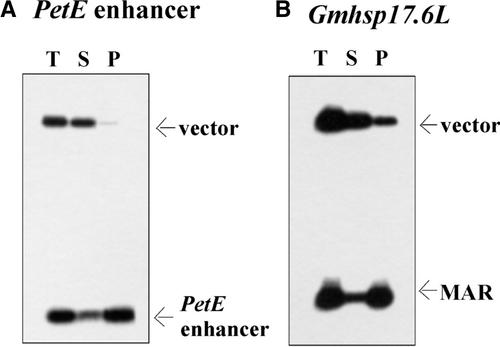
In addition to transcriptional enhancers, some MARs regulate gene expression through the acetylation of histones, as indicated by the murine immunoglobulin μ gene MAR (Fernández et al., 2001) and the human topoisomerase-I gene MAR (Martens et al., 2002). To determine whether the pea PetE enhancer is associated with the nuclear matrix, matrix binding experiments were performed in vitro. Nuclear matrices were prepared from isolated tobacco leaf nuclei by extraction of histones with 20 mM lithium diiodosalicylate (LIS) followed by digestion with EcoRI and HindIII to remove loop DNA (Hatton and Gray, 1999). Figure 8A shows the preferential association of the 268-bp PetE enhancer with the nuclear matrix, whereas the pUBS vector fragment remained unbound. A control experiment with the soybean Gmhsp17.6-L MAR (Schöffl et al., 1993) revealed a similar association with the nuclear matrix preparation (Figure 8B). This finding indicates that the 268-bp DNA fragment containing the pea PetE enhancer has the ability to bind to the nuclear matrix in vitro.
Figure 8.
Interactions of the PetE Enhancer Region with Nuclear Matrices in Binding Assays in Vitro.
The pea PetE enhancer and a non-MAR pUBS vector fragment (A) or the soybean Gmhsp17.6-L MAR and the pUC18 vector (B) were labeled with α-32P-dATP and incubated with tobacco matrices. The nuclear matrices were collected by centrifugation, and DNA was isolated from the supernatant (S) and pellet (P) fractions, analyzed by agarose gel electrophoresis, and exposed to x-ray films. T represents a total DNA sample taken before centrifugation.
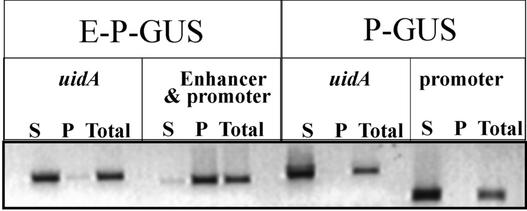
To determine whether endogenous DNA containing the PetE enhancer sequence is associated with the nuclear matrix in isolated nuclei, nuclei were prepared from transgenic tobacco seedlings containing the E-P-GUS construct at the four-leaf stage. The nuclei were treated with LIS and incubated with PvuII, MfeI, and SacI for 4 h to separate the PetE enhancer, the uidA coding region, and the nos 3′ region into different DNA fragments (Figure 1). The nuclei matrices were collected by centrifugation, and DNA was isolated from the pellet and the supernatant fractions by phenol/chloroform extraction. Sequences associated with the nuclear matrix should be pelleted, whereas loop non-MAR regions should be recovered in the supernatant (Slatter et al., 1991). The presence of the PetE enhancer and the uidA coding region in these fractions was investigated by semiquantitative PCR. In nuclei prepared from the E-P-GUS seedlings, the PetE enhancer and promoter regions, amplified with primers y6 and y13 (Figure 1), were enriched in the pelleted matrix fraction, whereas the uidA coding region, amplified with primers G1 and G2, was enriched in the released loop fraction (Figure 9). In nuclei prepared from the P-GUS seedlings, both the PetE promoter, amplified with primers c2 (overlapping with C1) and y13, and the uidA coding region were enriched in the supernatant fraction (Figure 9), suggesting that the PetE promoter and uidA were not bound to the nuclear matrix. These results suggest that the PetE enhancer is associated with the nuclear matrix in isolated nuclei.
Figure 9.
Association of the PetE Enhancer Region with Nuclear Matrices in Isolated Nuclei.
Nuclei were isolated from E-P-GUS or P-GUS seedlings, extracted with LIS, and digested with PvuII, MfeI, and SacI to separate the enhancer, the uidA coding region, and the nos 3′ region into individual fragments. The nuclear matrices were collected by centrifugation, and DNA was isolated from the pellet (P) and supernatant (S) fractions and analyzed by semiquantitative PCR. Total represents a sample of the digestion mixture taken before centrifugation. The enhancer and promoter regions of PetE were amplified with primer pairs y6 and y13, the uidA coding region was amplified with primer pairs G1 and G2, and the PetE promoter region was amplified with primer pairs c2 and y13.
DISCUSSION
The Pea PetE Enhancer Induces the Acetylation of Histones
The results presented here indicate that the PetE enhancer stimulated the expression of uidA from the PetE promoter and increased the acetylation states of histones H3 and H4 present on the promoter and the +1 to +586 region of uidA, suggesting that the enhancer region is likely to promote transcription by mediating the acetylation of histones. The pea PetE enhancer appears to be similar to the human interferon-β gene enhancer (Agalioti et al., 2000), the bovine β-casein gene BCE-1 enhancer (Myers et al., 1998), the human HIV-1 enhancer (Sheridan et al., 1997), and the murine immunoglobulin μ MAR (Fernández et al., 2001), which regulate gene expression by mechanisms that involve the hyperacetylation of histones present on promoter sequences (Agalioti et al., 2000; Fernández et al., 2001). Acetylated histones on promoters may increase transcription by facilitating the recruitment of transcription activators that recognize acetylated histone tails (Strahl and Allis, 2000; Hassan et al., 2001). Alternatively, the acetylation of histones can weaken histone–DNA interactions and facilitate the displacement of nucleosomes for the entry of transcriptional machinery (Eberharter and Becker, 2002).
In the absence of the PetE enhancer region, the PetE promoter became associated with hyperacetylated histones H3 and H4 after treatment with HDA inhibitors, and expression from the promoter was increased by almost fourfold. This finding strongly supports the notion that histone acetylation promotes transcription in plants. Moreover, because the tobacco plants were treated for only 6 h, increased GUS activity was likely to be a direct consequence of histone acetylation and not a result of secondary effects of the HDA inhibitors (Boffa et al., 1981; Annunziato et al., 1988). Recent evidence has also indicated that histone acetylation is associated closely with gene expression in plants. Treatment of Brassica napus seedlings with HDA inhibitors led to the accumulation of acetylated histones, stimulating the expression of silent rRNA genes (Chen and Pikaard, 1997). When the Arabidopsis homologs of yeast HD2 and RPD3 histone deacetylases were targeted to promoters of reporter genes, the expression of reporter genes was repressed, suggesting that reducing histone acetylation states decreases transcription (Wu et al., 2000a, 2000b). Furthermore, in transgenic Arabidopsis plants expressing the antisense mRNA sequence of ATHD1 (Arabidopsis HD1 histone deacetylase gene), silenced genes were expressed as a result of the accumulation of acetylated histones (Tian and Chen, 2001). The Arabidopsis PCAT2 possesses HAT activities and upregulates reporter gene expression (Bordoli et al., 2001). In pea, activated PetE expression is correlated with increased acetylation states of histones H3 and H4 present on enhancer, promoter, and coding regions of the gene (Chua et al., 2001). Moreover, in an Arabidopsis mutant containing a knockout in the histone deacetylase HDA6 gene, reporter genes fused to auxin-responsive promoters displayed auxin-independent and increased auxin-inducible expression, suggesting a link between histone acetylation and gene expression (Murfett et al., 2001). In Arabidopsis, the transcriptional activation of cold-regulated genes containing the CRT/DRE regulatory element is likely to involve the recruitment of the HAT GCN5 to promoters (Stockinger et al., 2001).
The Pea PetE Enhancer Colocalizes with a MAR
The 268-bp fragment containing the PetE enhancer was shown to associate with the nuclear matrix in vitro using two different assays. This finding indicates that the enhancer and MAR are colocalized within a short region of A/T-rich DNA adjacent to the minimal PetE promoter. The enhancer activity has been shown to be caused by the A/T content of the region (Sandhu et al., 1998). Multiple copies of a random 26-bp sequence consisting solely of dA and dT nucleotides were able to enhance the expression of the GUS reporter gene from the minimal PetE promoter, as were various shorter regions of the 268-bp fragment (Sandhu et al., 1998). A high A/T content is a general feature of MARs (Käs et al., 1989), and MAR binding to the nuclear matrix in vitro is highly sensitive to distamycin, which binds to the narrow minor groove of A/T-rich DNA (Hatton and Gray, 1999). These features suggest a close relationship between enhancer and MAR activities. The colocalization of enhancer and MAR sequences has been shown previously for various genes, including the murine immunoglobulin μ (Forrester et al., 1999; Fernández et al., 2001), the bean phaseolin (van der Geest et al., 1994), the soybean Gmhsp17.6-L (Schöffl et al., 1993), and the tobacco RB7 (Allen et al., 1996) genes.
The localization of a MAR adjacent to the PetE promoter indicates that the chromosomal loop containing the pea PetE gene is likely to be 9 kb. The 3′ MAR of pea PetE resides within a 1.4-kb fragment located 8 kb downstream of the coding region (Slatter et al., 1991).
Enhancer and MAR sequences have been proposed to mediate histone acetylation by recruiting HATs (Sheridan et al., 1997; Agalioti et al., 2000). Previous studies have demonstrated that the pea PetE enhancer interacts with HMG-I/Y proteins (Pwee et al., 1994; Webster et al., 1997, 2000), which are MAR binding proteins in native chromosomes (Saitoh and Laemmli, 1994). It seems likely that the pea PetE enhancer mediates histone acetylation by first interacting with HMG-I/Y proteins and associating with the nuclear matrix, bringing the minimal promoter in close proximity to the nuclear matrix, which is enriched in protein factors involved in transcription (Razin and Gromova, 1995). The HMG-I/Y proteins consequently facilitate the recruitment of specific transcription activators and HATs through protein–protein interactions, resulting in the acetylation of histones present on the nearby promoter region. In the human interferon-β gene, HMG-I/Y proteins also facilitate the assembly of an enhanceosome complex that contains the transcription factors NF-κB, IRFs, and ATF-2/c-Jun on the enhancer (Thanos and Maniatis, 1992). The enhanceosome complex next recruits the GCN5 complex, which acetylates the nucleosomes present on the enhancer and the nearby promoter for activated transcription (Agalioti et al., 2000). Therefore, HMG-I/Y proteins are likely to play critical roles in MAR-mediated histone acetylation (Agalioti et al., 2000). Consistent with this possibility, the tobacco Rb7 MAR stimulates reporter gene expression to a greater extent in suspension culture cells (40-fold) than in stably transformed tobacco plants (2-fold), possibly as a result of the greater amounts of HMG-I/Y proteins present in actively dividing suspension culture cells (Ülker et al., 1999). The identification of protein complexes assembled on the pea PetE enhancer during gene activation will provide useful insights into how enhancer and MAR sequences may direct the recruitment of HATs to increase expression.
Trichostatin A and sodium butyrate increased expression from the minimal promoter but failed to increase GUS activity when the PetE promoter was linked to the enhancer, supporting the observation that the histones present on constructs containing the enhancer already were hyperacetylated. Treatment with deacetylase inhibitors cannot further increase the acetylation states of histones present on the promoter to increase expression. By contrast, trichostatin A and sodium butyrate were reported to stimulate the expression of reporter genes containing the interferon-β MAR and the potato ST-LS1 MAR to a greater extent than non-MAR control genes (Klehr et al., 1991; Schlake et al., 1994). These MARs can stabilize the open chromatin conformations induced by acetylated histones, because MARs have a propensity to assume single-stranded structures, thereby increasing the accessibility of reporter genes to transcription factors (Böde et al., 1992; Schlake et al., 1994). These MARs are likely to regulate gene expression through a mechanism different from that used by the pea PetE enhancer and the immunoglobulin μ MAR, which actively induce histone hyperacetylation on promoters to increase transcription. The MAR of the human topoisomerase-I gene also mediates histone acetylation by binding to the nuclear matrix protein SAF-A, which in turn interacts with the HAT p300 to result in the acetylation of histones (Martens et al., 2002). Based on the observed effects on gene expression after HDA inhibitor treatment, MARs appear to regulate transcription through at least two mechanisms: by functioning as entry sites for HATs and increasing transcription through the acetylation of histones, or by maintaining regulatory elements in an open chromatin structure for transcription factors.
PetE Enhancer–Mediated Histone Acetylation Is Directional
Mapping the acetylation states of histones along the transgene indicated that histone hyperacetylation occurred over a region of <1 kb containing the PetE enhancer (268 bp), the PetE promoter (176 bp), and the 5′ end of uidA (586 bp) (Figure 7). In the native PetE gene in pea, activated expression also is associated with the acetylation of histones present on a 1-kb region containing the enhancer (268 bp), the promoter (176 bp), and the PetE coding region (504 bp) (Chua et al., 2001). These results suggest that the PetE enhancer is likely to activate the expression of transgenes in tobacco through a mechanism similar to that used by the native gene in pea and that it selectively induces the acetylation of histones present on the promoter and the nearby coding regions. Furthermore, in the E-P-GUS transgene and the native pea PetE (Chua et al., 2001), histones present downstream but not upstream of the enhancer region are acetylated. It appears unlikely that the PetE enhancer provides information for the acetylation of histones present only in downstream sequences, because the enhancer is able to upregulate transgene expression to similar extents when fused in either orientation to homologous and heterologous promoters (Sandhu et al., 1998). These results suggest that the HATs recruited by the HMG-I/Y proteins may recognize sequences in the downstream promoter region or interact with proteins that bind to the promoter. This is likely to place the HATs on the promoter side of the protein complex assembled on the enhancer, resulting in the acetylation of histones present on the promoter and the nearby coding sequences. The histones on the coding regions also may be acetylated by protein components in the RNA polymerase machinery. For example, the human basal transcription factor TFIID and the Elp3 subunit of the yeast elongating RNA polymerase II holoenzyme possess HAT activities (Kornberg and Lorch, 1999; Wittschieben et al., 1999).
METHODS
Treatment of Plants with Histone Deacetylase Inhibitors
Tobacco seeds (Nicotiana tabacum) collected from self-fertilized first-generation transgenic plants generated by Pwee and Gray (1993) were surface-sterilized and germinated on Murashige and Skoog (1962) (MS) agar (0.46% MS salts, 3% sucrose, and 0.8% agar, pH 5.9) containing kanamycin at 50 μg/mL under continuous light (PAR of 150 μmol·m−2·s−1). Three individually transformed lines expressing high, middle, and low levels of uidA were selected for analysis. For inhibitor studies, seedlings at the four-leaf stage were placed in MS solution (0.46% MS salts and 3% sucrose, pH 5.9) or MS solution containing 10 mM sodium butyrate (Sigma) or 10 μM trichostatin A (Sigma) for various periods of time. Stocks of 3 M sodium butyrate (in water) and 10 mM trichostatin A (in methanol) were stored at −20°C before use.
β-Glucuronidase Assays
Quantitative β-glucuronidase (GUS) assays were performed using methylumbelliferyl-β-d-glucuronide as a substrate, according to Jefferson et al. (1987). Approximately 0.5 cm2 of the second leaf from the base was collected for analysis. All measurements of GUS activity were linear with time and with protein concentration. Protein in leaf extracts was determined using Bradford protein assay reagent (Bio-Rad Laboratories), and the values were used to calculate specific GUS activities (pmol methylumbelliferone·min−1·μg−1 protein).
Preparation of Nuclear Proteins and Protein Gel Blot Analysis
Nuclei were isolated from 10 g of tobacco leaf tissues based on Hatton and Gray (1999) with modifications. Tissues were homogenized with a Polytron homogenizer fitted with a PTA20SM head (Kinematica, Lucerne, Switzerland) in 100 mL of cold nuclear extraction buffer (0.44 M sucrose, 25 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 2.5% Ficoll 400, 5% dextran, 0.5% Triton X-100, 2 mM spermine, 2.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin), filtered through four layers of muslin cloth, and centrifuged at 3000g for 10 min. The nuclei were washed twice in 100 mL of nuclear extraction buffer without spermine and once with 10 mL of nuclei suspension buffer (NSB; 50% glycerol, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 2.5 mM DTT, 0.5 mM PMSF, 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin). All steps were performed at 4°C.
The nuclei pellet was suspended in 100 μL of 5× SDS-PAGE loading buffer (0.2 M Tris-HCl, pH 6.8, 25% glycerol, 25% SDS, and 12.5% 2-mercaptoethanol) and heated at 95°C for 10 min. The samples were centrifuged at 11,300g for 5 min, and the supernatant was collected. The supernatant (10 μL) was analyzed on 12% polyacrylamide gels using the buffer systems established by Laemmli (1970). The proteins were electroblotted onto nitrocellulose membranes with 0.45-μm pore sizes (Schleicher & Schuell) using an Atto Horizblot semidry electroblotting apparatus (Genetics Research Instrumentation, Braintree, Essex, UK) at 100 V for 1 h in blotting buffer (50 mM Tris, 380 mM Gly, and 20% methanol). The membrane was rinsed with Tris-buffered saline (TBS; 10 mM Tris-HCl, pH 7.5, and 150 mM NaCl) and blocked with 25 mL of blocking solution (2.5% skim milk powder, 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.2% Tween 20) for 1 h. The membrane was then incubated in 50 mL of incubation buffer (50 mL of blocking buffer mixed with 100 mL of TBS) containing 1 μg/mL anti-acetylated histone H4 antibody (Upstate Biotechnology, Lake Placid, NY) or 10 to 500 ng/mL anti-acetylated histone H3 antibody (Upstate Biotechnology) overnight at 4°C. After overnight incubation, the membrane was washed twice with TBST (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.2% Tween 20), once with TBS, and once with TBST. The membrane was then incubated in 50 mL of incubation buffer containing biotinylated donkey anti-rabbit IgG (Amersham International) at a dilution of 1:1500 for 1 h, washed twice with TBST, once with TBS, and once with TBST, and then incubated in 50 mL of incubation buffer containing streptavidin-biotinylated horseradish peroxidase complex (Amersham International) at a dilution of 1:1500 for 1 h. After washing twice with TBST and TBS, the bound horseradish peroxidase was detected with the Amersham ECL (enhanced chemiluminescence) protein gel blot detection system, and the membrane was exposed to Hyperfilm ECL film (Amersham International).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation experiments were performed according to Chua et al. (2001) with slight modifications. Nuclei were isolated from 10 g of tobacco leaf tissues as described previously and suspended in 1 mL of NSB for cross-linking with formaldehyde. After sonication, the supernatant was collected and mixed with 10 mL of immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1 mM PMSF, 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin). To reduce nonspecific background, the chromatin solution was precleared with 10 μL of 0.66 μg/mL sonicated bacteriophage λ DNA and 100 μL of protein A–Sepharose beads (50% slurry in TE [10 mM Tris-HCI, pH 8.0, 1 mM EDTA, and Na2] and 0.1% BSA). The mixture was incubated for 1 h at 4°C on a rotation wheel. The agarose beads were removed by centrifugation. The supernatant was divided into 1-mL aliquots and combined with the following amounts of rabbit antisera: 10 μL of anti-acetylated H4 (Upstate Biotechnology), 10 μL of anti-nonacetylated H4 (Serotec, Oxford, UK), 10 μL of anti-acetylated H3 (Upstate Biotechnology), or 30 μL of anti-nonacetylated H3 (Upstate Biotechnology). These antibodies were generated with the peptide sequences 5′-GRG[K*]GG[K*]GLG[K*]GGA[K*]RH-3′ for histone H4 and 5′-[K*]STGG[K*]APR[K*]-3′ for histone H3, where [K*] was acetylated, and recognized any of the acetylated Lys residues in the tails of histone H3 or H4 protein. The solutions were incubated overnight at 4°C on a rotation wheel.
Matrix-Binding Assays
Nuclei were isolated from 10 g of tobacco leaves as described previously, and nuclear matrices were prepared according to Hatton and Gray (1999) with no modifications and suspended in 200 μL of binding buffer (20 mM Tris-HCl, pH 7.5, 7 mM MgCl2, 100 mM KCl, 2.5 mM DTT, 1 mM PMSF, 100 μg/mL BSA, 15 μg/mL digitonin, and 0.2 μg/mL leupeptin). Plasmids containing the Gmhsp17.6L MAR in pUC18 (Schöffl et al., 1993) and the PetE enhancer in pUBS were digested with EcoRI and HindIII, respectively, and labeled with α-32P-dATP by end-filling with Klenow fragment as described by Hatton and Gray (1999). The DNA probes (2 ng) were incubated with 20 μL of tobacco nuclear matrix preparations according to Hatton and Gray (1999). Nuclear matrices were collected by centrifugation, and both the supernatant and pellet fractions were treated with proteinase K at 1 mg/mL in 1% SDS at 65°C for 1 h. DNA was extracted from the pellet and supernatant fractions by phenol/chloroform extraction and ethanol precipitation. The DNA was separated on a 1% agarose gel, which was fixed in 7% trichloroacetic acid for 30 min, dried, and autoradiographed.
The association of endogenous DNA sequences with the matrix in isolated nuclei was examined according to the method of Slatter et al. (1991) with slight modifications. Nuclei were isolated from 10 g of tobacco leaf tissues as described previously and suspended in 10 mL of NSB. A 500-μL aliquot of nuclei was washed three times with buffer A (15 mM Tris-HCl, pH 8.0, 20 mM KCl, 60 mM NaCl, 0.5 mM EDTA, 0.5 mM spermine, 0.125 mM spermidine, 1% [v/v] thiodiglycol, and 0.1% digitonin) to solubilize membranes, and the nuclear matrices were stabilized with copper sulfate and extracted with 20 mM lithium diiodosalicylate to remove histone proteins. After six washes with buffer D (20 mM Tris-HCl, pH 8.0, 70 mM NaCl, 20 mM KCl, 10 mM MgCl2, 0.125 mM spermidine, 0.05 mM spermine, and 0.1% digitonin), the matrices were suspended in 500 μL of buffer D, and 200 units of PvuII, MfeI, and SacI (New England Biolabs, Beverly, MA) were added. Digestion was performed at 37°C on a rotating wheel. After 2 h, 200 units more of each enzyme was added, and the digestion mixture was incubated at 37°C for another 2 h. A 200-μL aliquot was removed as a total DNA sample. The digestion mixture was centrifuged at 2400g for 10 min, and the pellet and supernatant fractions were collected. The pellet was washed twice in 1 mL of buffer D, and the supernatant fractions were pooled. The pellet was then suspended in 500 μL of buffer D. The suspended pellet, supernatant, and total DNA fractions were treated with proteinase K at 1 mg/mL in the presence of 0.1% SDS at 30°C overnight. DNA was isolated by phenol/chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen. The DNA pellets were then suspended in 20 μL of TE.
Quantitative PCR
The amounts of DNA present in the samples from chromatin immunoprecipitation and matrix association assays were determined by quantitative PCR. The primers used were as follows: T1, 5′-GATATCTTGCTGCGTTCGGATA-3′; T2, 5′-ATAGCAGCACCGTAATCAGTAGC-3′; TE1, 5′-AAGTCGGTGACGGTGATAATTC-3′; TE2, 5′-CTTGCTCTATGAAATAATCAATGAAG-3′; y6, 5′-AGCTTAGTTAATCATGTTAAACAAC-3′; y13, 5′-AATCGAAAGAGAGACACACAGAAGG-3′; C1, 5′-CACATATTCTTCCACACATCTTAGCC-3′; C2, 5′-CACATCTACATTATCTAAATCACATATTC-3′; C4, 5′-TTCGCGCTGATACCAGACGTTGCC-3′; C8, 5′-CTGTGGGCATTCAGTCTGGATCGCG-3′; C10, 5′-CTTGCGCGACATGCG-TCACCACGG-3′; G1, 5′-CGGGTGAAGGTTATCTCTATGAAC-3′; G2, 5′-GGCAATACTCCACATCACCAC-3′; G3 5′-CACTTACAGGCGATTAAAGAGCTG-3′; and G4 5′-GAAGATCCCTTTCTTGTTACCG-3′.
The PCR conditions used were as described by Chua et al. (2001). The PCR fragments were resolved by electrophoresis on a 2% agarose gel, stained with ethidium bromide, and quantified with an Alpha Imager 1200 (Alpha Innotech, San Leandro, CA). The enhancer and minimal promoter sequences used in this study correspond to nucleotides 369 to 637 and 638 to 806, respectively, of the pea PetE gene (accession number X16082).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The accession number for the pea PetE gene is X16082.
Acknowledgments
We thank Tristan Dyer, Julian Hibberd, Nicola Ramsay, and Tai Wai Yeo for valuable discussions. L.A.W. was supported by a research studentship from the Science and Engineering Research Council (UK). This work was supported by research grants from the Biotechnology and Biological Sciences Research Council (UK).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011825.
References
- Agalioti, T., Lomvardas, S., Parekh, B., Yie, J., Maniatis, T., and Thanos, D. (2000). Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103, 667–678. [DOI] [PubMed] [Google Scholar]
- Allen, G.C., Hall, G., Michalowski, S., Newman, W., Spiker, S., Weissinger, A.K., and Thompson, W.F. (1996). High-level transgene expression in plant cells: Effects of a strong scaffold attachment region from tobacco. Plant Cell 8, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, G.C., Hall, G.E., Childs, L.C., Weissinger, A.K., Spiker, S., and Thompson, W.F. (1993). Scaffold attachment regions increase reporter gene expression in stably transformed plant cells. Plant Cell 5, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato, A.T., Frado, L.L., Seale, R.L., and Woodcock, C.L. (1988). Treatment with sodium butyrate inhibits the complete condensation of interphase chromatin. Chromosoma 96, 132–138. [DOI] [PubMed] [Google Scholar]
- Aronow, B.J., Ebert, C.A., Valerius, M.T., Potter, S.S., Wiginton, D.A., Witte, D.P., and Hutton, J.J. (1995). Dissecting a locus control region: Facilitation of enhancer function by extended enhancer-flanking sequences. Mol. Cell. Biol. 15, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood, E.M., and Kadonaga, J.T. (1998). Going the distance: A current view of enhancer action. Science 281, 60–63. [DOI] [PubMed] [Google Scholar]
- Blasquez, V.C., Xu, M., Moses, S.C., and Garrard, W.T. (1989). Immunoglobulin-kappa-gene expression after stable integration. I. Role of the intronic MAR and enhancer in plasmacytoma cells. J. Biol. Chem. 264, 21183–21189. [PubMed] [Google Scholar]
- Böde, J., Kohwi, Y., Dickinson, L., Joh, T., Klehr, D., Mielke, C., and Kohwi-Shigematsu, T. (1992). Biological significance of unwinding capacity of nuclear matrix-associating DNAs. Science 255, 195–197. [DOI] [PubMed] [Google Scholar]
- Boffa, L.C., Gruss, R.J., and Allfrey, V.G. (1981). Manifold effects of sodium butyrate on nuclear function: Selective and reversible inhibition of phosphorylation of histones H1 and H2A and impaired methylation of lysine and arginine residues in nuclear protein fractions. J. Biol. Chem. 256, 9612–9621. [PubMed] [Google Scholar]
- Bordoli, L., Netsch, M., Lüthi, U., Lutz, W., and Eckner, R. (2001). Plant orthologs of p300/CBP: Conservation of a core domain in metazoan p300/CBP acetyltransferase related proteins. Nucleic Acids Res. 29, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.E., Lechner, T., Howe, L., and Workman, J.L. (2000). The many HATs of transcription coactivators. Trends Biochem. Sci. 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J., and Pikaard, C.S. (1997). Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 16, 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, Y.L., Brown, A.P.C., and Gray, J.C. (2001). Targeted histone acetylation and altered nuclease accessibility over short regions of the pea plastocyanin gene. Plant Cell 13, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill, P.N., and Garrard, W.T. (1986). Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44, 273–282. [DOI] [PubMed] [Google Scholar]
- Eberharter, A., and Becker, P.B. (2002). Histone acetylation: A switch between repressive and permissive chromatin. EMBO Rep. 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, L.A., Winkler, M., and Grosschedl, R. (2001). Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21, 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester, W.C., Fernandez, L.A., and Grosschedl, R. (1999). Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 13, 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester, W.C., Vangenderen, C., Jenuwein, T., and Grosschedl, R. (1994). Dependence of enhancer-mediated transcription of the immunoglobulin-μ gene on nuclear matrix attachment regions. Science 265, 1221–1225. [DOI] [PubMed] [Google Scholar]
- Gasser, S.M., and Laemmli, U.K. (1986). The organization of chromatin loops: Characterization of a scaffold attachment site. EMBO J. 5, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser, K.D. (1998). HMG1 and HU proteins: Architectural elements in plant chromatin. Trends Plant Sci. 3, 260–265. [Google Scholar]
- Grosschedl, R., Giese, K., and Pagel, J. (1994). HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10, 94–100. [DOI] [PubMed] [Google Scholar]
- Hassan, A.H., Neely, K.E., and Workman, J.L. (2001). Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nu-cleosomes. Cell 104, 817–827. [DOI] [PubMed] [Google Scholar]
- Hatton, D., and Gray, J.C. (1999). Two MAR DNA-binding proteins of the pea nuclear matrix identify a new class of DNA-binding proteins. Plant J. 18, 417–429. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs, E., Izaurralde, E., and Laemmli, U.K. (1989). Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin: The role of oligo(dA).oligo(dT) tracts. J. Mol. Biol. 20, 587–599. [DOI] [PubMed] [Google Scholar]
- Klehr, D., Maass, K., and Bode, J. (1991). Scaffold-attached regions from the human interferon β domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry 30, 1264–1270. [DOI] [PubMed] [Google Scholar]
- Kornberg, R.D., and Lorch, Y. (1999). Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294. [DOI] [PubMed] [Google Scholar]
- Kruh, J. (1982). Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol. Cell. Biochem. 42, 65–82. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Last, D.I., and Gray, J.C. (1989). Plastocyanin is encoded by a single-copy gene in the pea haploid genome. Plant Mol. Biol. 12, 655–666. [DOI] [PubMed] [Google Scholar]
- Martens, J.H., Verlaan, M., Kalkhoven, E., Dorsman, J.C., and Zantema, A. (2002). Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol. Cell. Biol. 22, 2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovitch, J., Mirault, M.-E., and Laemmli, U.K. (1984). Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell 39, 223–232. [DOI] [PubMed] [Google Scholar]
- Mlynárová, L., Jansen, R.C., Conner, A.J., Stiekema, W.J., and Nap, J.-P. (1995). The MAR-mediated reduction in position effect can be uncoupled from copy number-dependent expression in transgenic plants. Plant Cell 7, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynárová, L., Loonen, A., Heldens, J., Jansen, R.C., Keizer, P., Stiekema, W.J., and Nap, J.-P. (1994). Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix-associated region. Plant Cell 6, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Murfett, J., Wang, X.-J., Hagen, G., and Guilfoyle, T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13, 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, C.A., Schmidhauser, C., Mellentin-Michelotti, J., Fragoso, G., Roskelley, C.D., Casperson, G., Mossi, R., Pujuguet, P., Hager, G., and Bissell, M.J. (1998). Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation. Mol. Cell. Biol. 18, 2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pwee, K.-H., and Gray, J.C. (1993). The pea plastocyanin promoter directs cell-specific but not full light-regulated expression in transgenic tobacco plants. Plant J. 3, 437–449. [DOI] [PubMed] [Google Scholar]
- Pwee, K.-H., Webster, C.I., and Gray, J.C. (1994). HMG protein binding to an A/T-rich positive regulatory region of the pea plastocyanin gene promoter. Plant Mol. Biol. 26, 1907–1920. [DOI] [PubMed] [Google Scholar]
- Razin, S.V., and Gromova, I.I. (1995). The channels model of nuclear matrix structure. Bioessays 17, 443–450. [DOI] [PubMed] [Google Scholar]
- Saitoh, Y., and Laemmli, U.K. (1994). Metaphase chromosome structure: Bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76, 609–622. [DOI] [PubMed] [Google Scholar]
- Sandhu, J.S., Webster, C.I., and Gray, J.C. (1998). A/T-rich sequences act as quantitative enhancers of gene expression in transgenic tobacco and potato plants. Plant Mol. Biol. 37, 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake, T., Klehr-Wirth, D., Yoshida, M., Beppu, T., and Bode, J. (1994). Gene expression within a chromatin domain: The role of core histone hyperacetylation. Biochemistry 33, 4197–4206. [DOI] [PubMed] [Google Scholar]
- Schöffl, F., Schroder, G., Kliem, M., and Rieping, M. (1993). An SAR sequence containing 395 bp DNA fragment mediates enhanced gene-dosage-correlated expression of a chimaeric heat shock gene. Transgenic Res. 2, 93–100. [DOI] [PubMed] [Google Scholar]
- Sheridan, P.L., Mayall, T.P., Verdin, E., and Jones, K.A. (1997). Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 11, 3327–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatter, R.E., Dupree, P., and Gray, J.C. (1991). A scaffold-associated DNA region is located downstream of the pea plastocyanin gene. Plant Cell 3, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief, A., Winter, D.M., Strätling, W.H., and Sippel, A.E. (1989). A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature 341, 343–345. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J., Mao, Y., Regier, M.K., Triezenberg, S.J., and Thomashow, M.F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl, K. (2001). A paradigm for precision. Science 293, 1054–1055. [DOI] [PubMed] [Google Scholar]
- Thanos, D., and Maniatis, T. (1992). The high mobility group protein HMG I(Y) is required for NF-KB-dependent virus induction of the human IFN-β gene. Cell 71, 777–789. [DOI] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker, B., Allen, G.C., Thompson, W.F., Spiker, S., and Weissinger, A.K. (1999). A tobacco matrix attachment region reduces the loss of transgene expression in the progeny of transgenic tobacco plants. Plant J. 18, 253–263. [Google Scholar]
- van der Geest, A.H.M., Hall, G.E., Jr., Spiker, S., and Hall, T.C. (1994). The β-phaseolin gene is flanked by matrix attachment regions. Plant J. 6, 413–423. [Google Scholar]
- Waterborg, J.H., and Kapros, T.B. (2002). Kinetic analysis of histone acetylation turnover and trichostatin A induced hyper- and hypoacetylation in alfalfa. Biochem. Cell Biol. 80, 279–293. [DOI] [PubMed] [Google Scholar]
- Webster, C.I., Cooper, M.A., Packman, L.C., Williams, D.H., and Gray, J.C. (2000). Kinetic analysis of high-mobility-group proteins HMG-1 and HMG-I/Y binding to cholesterol-tagged DNA on a supported lipid monolayer. Nucleic Acids Res. 28, 1618–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, C.I., Packman, L.C., Pwee, K.H., and Gray, J.C. (1997). High mobility group proteins HMG-1 and HMG-I/Y bind to a positive regulatory region of the pea plastocyanin gene promoter. Plant J. 11, 703–715. [DOI] [PubMed] [Google Scholar]
- Wittschieben, B., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., Li, Y., Allis, C.D., Tempst, P., and Svejstrup, J.Q. (1999). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Wolffe, A.P., and Hayes, J.J. (1999). Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000. a). Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol. Biol. 44, 167–176. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000. b). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., Kijima, M., Akita, M., and Beppu, T. (1990). Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179. [PubMed] [Google Scholar]