Abstract
Chloroviruses are large, double-stranded-DNA, plaque-forming viruses that infect certain eukaryotic chlorella-like green algae. The prototype of the genus is Paramecium bursaria chlorella virus 1 (PBCV-1). Chlorovirus genomes contain various amounts of methylated nucleotides due to virus-encoded DNA methyltransferases (MTases); about 25% of the MTases are associated with companion DNA site-specific (restriction) endonucleases (REases). These enzymes constitute virally encoded restriction-modification (R/M) systems. Although several of the chlorovirus R/M systems are characterized, their biological functions are unknown. The PBCV-1 proteome reveals that two virus-encoded REases, but not their companion MTases, are virion associated, suggesting that viral REases might help degrade the host DNA early in infection. To test this hypothesis, host chromosomal DNA from PBCV-1-infected cells was examined by pulsed-field gel electrophoresis. Initiation of host chromosomal DNA degradation occurred within 5 min postinfection (p.i.). The DNA degradation was insensitive to protein synthesis inhibitors or UV inactivation of virus particles, consistent with the agent being a small protein associated with the virion. Nuclease activities, including those of the two predicted REases and an uncharacterized general nuclease(s), were detected in disrupted PBCV-1 particles. The general nuclease(s) degraded both host and viral DNAs in vitro, although the viral DNA was not degraded in vivo, suggesting differential intracellular trafficking of the virion-associated nucleases. Infection with chloroviruses lacking an R/M system(s) resulted in either delayed host chromosomal DNA degradation or no detectable host chromatin changes. These immediate-early events associated with chlorovirus infections may facilitate rapid switching of the host transcriptional apparatus to viral transcription, which begins within 5 to 10 min p.i.
Chloroviruses are large, icosahedral, plaque-forming, genetically diverse but morphologically similar, linear, double-stranded-DNA (dsDNA) viruses (315 to 380 kb) that belong to the family Phycodnaviridae and the genus Chlorovirus (8, 22). The prototype chlorovirus, Paramecium bursaria chlorella virus 1 (PBCV-1), has a 331-kb genome that contains 366 protein-encoding genes and a polycistronic tRNA gene encoding 11 tRNAs. The chloroviruses infect certain freshwater, unicellular, chlorella-like green algae (e.g., Chlorella strain NC64A or Chlorella strain Pbi), which normally exist as endosymbionts in the protozoan Paramecium bursaria. Viruses that infect Chlorella NC64A (NC64A viruses) are serologically different from viruses that infect Chlorella Pbi (Pbi viruses). NC64A viruses neither infect nor attach to Chlorella Pbi, and vice versa (16).
One distinctive feature of the chloroviruses is that their genomes contain methylated nucleotides: 5-methylcytosine (m5C) levels range from 0.1% to 47% of the total cytosines, and N6-methyladenine (m6A) levels vary from 0% to 37% of the total adenines (25, 26). The discovery that m5C and m6A occur in specific nucleotide sequences led to the finding that the viruses often carry multiple DNA methyltransferases (MTases) and that about 25% of the DNA MTases have companion DNA site-specific (restriction) endonucleases (REases), collectively referred to as restriction-modification (R/M) systems. REases recognize and cleave specific nucleotide sequences, while their companion DNA MTases protect DNA from cleavage by methylating one of the bases in the same nucleotide sequence. The number of virus-encoded MTases and REases varies from virus to virus.
PBCV-1 carries two R/M systems. The CviAI R/M system consists of the REase R.CviAI (cleaves /GATC but not GmATC sites) and the adenine MTase M.CviAI (methylates GATC sequences) (30, 31). The other system, named CviAII, consists of R.CviAII (cleaves C/ATG but not CmATG sites) and the adenine MTase M.CviAII (methylates CATG sequences) (34). In addition to these two R/M systems, PBCV-l carries three MTases that lack companion REases (22).
Two more chlorovirus genomes have been sequenced recently. The NC64A virus NY-2A contains 2 REase- and 18 MTase-encoding genes, and the methylation site has been established for 7 of the MTases (4, 35). The NY-2A genome is heavily methylated, with 45% m5C and 37% m6A (14). In contrast, the Pbi virus MT325 encodes only one putative MTase and no recognizable REases (L. A. Fitzgerald, M. V. Graves, H. Ogata, and J. L. Van Etten, unpublished data).
The biological function(s) of the chlorovirus-encoded R/M systems is unknown. The following two functions have been proposed for the R/M enzymes: (i) the REases help to degrade host DNA to provide deoxynucleotides for reincorporation into virus DNA, and methylation of nascent virus DNAs by the cognate MTases protects the virus DNA from self-digestion; and (ii) the REases prevent infection of the host by a second DNA virus. Several observations are consistent with the first hypothesis. (a) PBCV-1 infection results in degradation of host nuclear and chloroplast DNAs, beginning at 1 to 2 h postinfection (p.i.) (24). (In the previous study, nuclear DNA degradation was monitored by its isopycnic position in CsCl equilibrium density gradients.) (b) Degradation of the host nuclear and chloroplast DNAs in vivo coincides with the appearance of PBCV-1 REase synthesis (30). (c) Initiation of PBCV-1 DNA synthesis (at 60 to 90 min p.i.) concurs with the appearance of MTase activity (24). (d) In vitro, host DNA, but not virus DNA, is cleaved by the two PBCV-1-carried REases.
The virus IL-3A carries the CviJI R/M system. R.CviJI cleaves RG/CY but not RGmCY sequences, and M.CviJI methylates RGCY sequences (32). The isolation of three IL-3A deletion mutants lacking functional genes encoding M.CviJI and R.CviJI allowed us to test the host DNA degradation hypothesis (3). Unexpectedly, beginning after 1 h p.i., host nuclear and chloroplast DNAs were degraded at the same rates when infected with either wild-type IL-3A or the three mutants. We concluded from these experiments that R.CviJI activity is not essential for degradation of the host DNAs. However, the integrity of the chromosomal DNA was monitored by CsCl equilibrium centrifugation, which is not a very sensitive method.
To test the other hypothesis, i.e., that REases exclude infection of a host cell by a second virus, chlorella cells were dually inoculated with different viruses, and plaques arising from infective centers were distinguished by immunoblotting (5). These experiments established that the chloroviruses, like certain bacteriophages, exclude one another; however, this exclusion is independent of the REase activities of the viruses.
Thus, the biological function of the chlorovirus-encoded R/M systems remains unknown. This report revisits the issue because of the discovery that the two PBCV-1-encoded REases, but not their companion MTases, are packaged into PBCV-1 virions. We focused on changes that occur in the host chromosomal DNA in the first few minutes after infection (0 to 30 min p.i.). Furthermore, the integrity of the DNA was monitored by pulsed-field gel electrophoresis (PFGE) instead of CsCl centrifugation because PFGE can detect changes produced by a few cleavages.
MATERIALS AND METHODS
Strains and culture conditions.
The host chlorella cells were grown on MBBM medium (Chlorella NC64A) or on FES medium (Chlorella Pbi) with continuous light and shaking (200 rpm) at 25°C, with an initial cell concentration at 106 cells/ml (16, 23). Cultures were grown in 500-ml flasks, and growth was monitored by direct cell counts with a hemocytometer.
Chlorella virus purification.
Chlorella cells (1.5 × 107 to 2 × 107 cells/ml) were inoculated with filter-sterilized (0.45-μm filter pore size) virus at a multiplicity of infection (MOI) of ∼0.001 and incubated for 2 to 3 days at 25°C with continuous light and shaking. Cell lysates were centrifuged at 4,000 × g for 5 min at 4°C; Triton (to 1%) was added to the supernatant and kept at room temperature with constant mixing for 1 h. The sample was then centrifuged at 4°C for 50 min at 53,000 × g to pellet the virus. The virus pellet was resuspended in a small volume of Tris buffer (TB; 50 mM Tris-HCl, pH 7.8) and incubated with proteinase K (0.02 mg/ml) for 1 h at 45°C. The virus suspension was then layered onto 10 to 40% linear sucrose density gradients equilibrated with TB and centrifuged in a swinging bucket rotor at 4°C for 20 min at 72,000 × g. The virus band was removed from the gradient with a sterile needle, diluted with TB, and centrifuged for 3 h at 80,000 × g. The virus in the pellet was resuspended and subjected to a second sucrose density gradient centrifugation and sedimentation process to ensure viral purity. Virus concentrations were determined on a UV spectrophotometer as A260 values and by plaque assay (PFU/ml). One A260 unit of PBCV-1 routinely yields 1.5 × 1010 to 2.5 × 1010 PFU/ml. Isolated and purified virus was stored at 4°C.
Identification of proteins in virions.
Virion proteins were solubilized and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (21). The gel was stained with SyproRuby (Molecular Probes), and the revealed bands were excised. The proteins in the gel slices were hydrolyzed by in-gel trypsin digestion (20). The resulting peptide fragments were eluted from the gel pieces and then analyzed using a Q-TOF Ultima tandem mass spectrometer (Micromass/Walters) with electrospray ionization. The instrument was calibrated using the fragment ion masses of doubly protonated Glu-fibrinopeptide. The tandem mass spectrometry results were processed using Masslynx software (Micromass) to produce peak lists for database searching. MASCOT (Matrix Science) was used as the search engine. Results were searched against the PBCV-1 nonredundant database.
PFGE.
PFGE studies were carried out according to the procedure of McCluskey et al. (9), with slight modifications. Chlorella cells were harvested from 4-day-old cultures (1.2 × 107 to 2.0 × 107 cells/ml) by centrifugation at 5,000 × g for 5 min and then resuspended in growth medium at a concentration of 8.6 × 107 cells/ml. Typically, chloroviruses were added at an MOI of 10 PFU/cell. Infected chlorella cells (25 ml) were collected in centrifuge tubes (kept on ice), immediately mixed with formaldehyde to bring the concentration to 4% (vol/vol), and placed on ice. After centrifugation at 5,000 × g for 5 min, the samples were washed three times by resuspending them in MBBM amended with 50 mM EDTA and, following centrifugation, resuspending them in 0.5 ml of SB (25 mM Tris-HCl, pH 7.5, 1 M sorbitol, 25 mM EDTA, pH 8.0). The resuspended fixed cells were mixed with an equal volume of 2% low-melting-point agarose (Bio-Rad) in SB at 45°C, poured into plug molds (Bio-Rad, Hercules, CA), and placed at 4°C for 15 min to solidify. Agarose blocks were incubated in approximately 2 ml of 1-mg/ml proteinase K in DB (250 mM EDTA, pH 9.5, 1% N-lauroylsarcosine) for 24 h. After digestion, samples were washed two times for 30 min with DB and cut into small pieces that fit into gel wells. Samples were sealed with 1% low-melting-point agarose at 45°C in electrophoresis buffer. Chromosomal DNAs were separated in a CHEF-DR II (Bio-Rad) unit in a 1% agarose gel. Electrophoresis conditions and running buffer were selected to resolve the target chromosome sizes (1). The exact conditions are described in the figure legends. Hansenula wingei chromosomes (1.05 to 3.13 Mb) (Bio-Rad) and Saccharomyces cerevisiae chromosome PFGE markers (225 to 1,900 kb) (New England BioLabs, Beverly, MA) were used as DNA size markers. Gels were stained with 0.5 μg/ml ethidium bromide for 30 min and destained in water for 2 h, and digital images were made with a ChemiDoc EQ system (Bio-Rad).
Cell-free virion-associated nuclease activity.
Chlorella and virus DNAs were prepared in agarose plugs similar to those used for PFGE. Chlorella NC64A cells (2.2 × 109 cells/ml) and PBCV-1 virions (2.2 × 1010 PFU/ml) were embedded, either separately or together, in 1% agarose. Agarose plugs were treated with proteinase K (1 mg/ml) for 24 h. The plugs were cut into small pieces to fit the gel wells and washed four times (at least 30 min for each wash) with 2 ml of Tris-EDTA buffer. Reactions with the REases MboI (10 U) and CviAII (10 U) were performed according to the manufacturer's instructions (New England BioLabs). After overnight incubation at 37°C, the reactions were stopped with DB.
For virion-associated, cell-free nuclease activities, virus preparations (250 μl) at a concentration of 2 × 1011 PFU/ml were sonicated (Tekmar Ultrasonic processor; 100-W model) for 15 s (three pulses of 5 s each) on ice and then mixed with buffer (50 mM NaCl, 50 mM Tris-HCl, 1 mM dithiothreitol, 25 μg/ml bovine serum albumin). Agarose plugs with isolated PBCV-l DNA were incubated with the sonicated virus in buffer solution (50 mM NaCl, 50 mM Tris-HCl, 1 mM dithiothreitol, 25 μg/ml bovine serum albumin) overnight at 37°C; the reaction was stopped with DB. Electrophoresis conditions and running buffers were selected to resolve DNA sizes (1). The exact conditions used are described in the figure legends.
In-gel nuclease activity assay.
Polyacrylamide gels were prepared as described elsewhere (18), with some modifications (2, 17). Generally, 5% polyacrylamide stacking gels and either 12.5% or 15.0% resolving gels (1.5-mm thickness) were amended with 150 μg/ml calf thymus or virus DNAs prior to polymerization (SDS-DNA-PAGE). Virus preparations (1.5 mg/ml) were heated at 60°C for 30 min and then centrifuged at 15,000 × g for 10 min at 4°C. This step removed most of the major capsid protein (12). After centrifugation, virus pellets were resuspended in TB, mixed with 2× sample buffer (2% SDS, 10% glycerol, 0.0625 M Tris-HCl, pH 6.8, bromophenol blue [trace]), heated for 10 min at 90°C, and centrifuged for 5 min at 15,000 × g. Twenty-microliter samples were loaded into each gel well. Electrophoresis was conducted in Tris-glycine (0.025 mM Tris base, 0.19 mM glycine, pH 8.3) and 0.1% SDS buffer at 100 V until bromophenol blue eluted. Following electrophoresis, the gel was washed twice for 15 min with 100 ml of wash buffer (25% 2-butanol, 40 mM Tris-HCl, pH 7.6, 2 mM MgCl2) and then for 1 h with nuclease incubation buffer (40 mM Tris-HCl, pH 7.6, 2 mM MgCl2, and 0.02% sodium azide). After one buffer change, the gel was incubated overnight at 37°C. Nuclease buffer was replaced with fresh buffer supplemented with 2 mM CaCl2 and incubated for 8 h. The gel was stained with 0.5 μg/ml ethidium bromide for 10 min and periodically visualized with a ChemiDoc EQ system (Bio-Rad). In-gel DNase activity was indicated by a loss of fluorescence.
Restriction endonuclease extraction, purification, and assay.
Virus preparations (15 mg/ml) were mixed with an equal volume of 2× Tris-acetate buffer with potassium acetate (40 mM Tris-acetate, pH 8.5, 2 mM EDTA, 20 mM 2-mercaptoethanol, 1 M potassium acetate, 40 μg/ml phenylmethylsulfonyl fluoride), and the extracts were clarified by centrifugation for 30 min at 15,000 × g at 4°C. The supernatant fraction (S-15) was diluted sixfold and applied to a heparin-Sepharose column. Fractions were eluted with a 0.1 to 0.8 M potassium acetate gradient, pH 7.6, in Tris-acetate buffer (20 mM Tris-acetate, pH 8.5, 1 mM EDTA, 2 mM 2-mercaptoethanol), and these fractions were dialyzed against Tris-acetate buffer (20 mM Tris-acetate, pH 8.5, 1 mM EDTA, 2 mM 2-mercaptoethanol) without potassium acetate (34). Active fractions were assayed at 23°C by adding 36 μl of eluted and dialyzed fractions in Tris-acetate buffer with 10 mM magnesium acetate to 4 μl of DNA (250 μg/μl).
Copurifying R.CviAI and R.CviAII activities were distinguished from each other by using different PCR-generated DNA amplicons (∼2 kb) from the PBCV-1 genome as substrates (amplicon B was from nucleotides 12844 to 14953, and amplicon C was from nucleotides 130538 to 132820). The DNA amplicon B has cleavage sites for both PBCV-1-encoded DNA REases. R.CviAI generates two fragments (1,920 and 189 bp), and R.CviAII makes five fragments (1,046, 591, 252, 184, and 34 bp). Only R.CviAII cleaves DNA amplicon C (five fragments, of 715, 625, 392, 383, 97, and 71 bp). Commercially manufactured REases CviAII and MboI (New England Biolabs) were used as positive controls. DNA was electrophoresed in a 1% agarose gel in 0.5× TBE buffer (80 mM Tris-borate, 2 mM EDTA), stained with 0.5 μg/ml ethidium bromide, and visualized with a ChemiDoc EQ system (Bio-Rad).
RESULTS
PBCV-1 virion-associated restriction endonucleases.
Experiments to identify all of the PBCV-1-encoded proteins that are packaged in the virion (D. D. Dunigan, R. Cerny, L. C. Lane, B. J. Kronschnabel, and J. L. Van Etten, unpublished data) indicated that the two PBCV-1-encoded REases, R.CviAI and R.CviAII, but not their corresponding MTases, are packaged in the virions. The virion-associated peptide fragments covered ∼6% of the REase R.CviAI (Mowse score, 49) and ∼7% of R.CviAII (Mowse score, 68). One concern was that the two REases might be contaminating proteins on the exteriors of the virions. However, treatment of the virions with proteinase K during their purification did not remove the REase proteins. (Note that protease K has no effect on PBCV-1 infectivity [J. L. Van Etten, unpublished results].) This finding suggested that the two PBCV-1-encoded REases might have a physiological role early in virus infection.
Host nuclear DNA is degraded rapidly after PBCV-1 infection.
The discovery that PBCV-1 packages its two REases in the virion prompted a reinvestigation of host chromosomal integrity following virus infection. However, unlike previous studies, PFGE was used to monitor intact chromosomal DNAs, and the samples were processed within min of infection. Infection experiments were stopped rapidly by mixing infected chlorella cells with an equal volume of ice-cold 8% formaldehyde to prevent chromosomal changes during sample processing.
Previous reports indicated that Chlorella NC64A has a genome size of 39 to 45 Mb and that the DNA is distributed into 13 chromosomes that vary from 1.1 to 6.5 Mb (7, 9). In the current study, the PFGE conditions were optimized to allow a single gel to monitor DNAs ranging from 50 kb to slightly above 3.2 Mb (1). These PFGE conditions separated Chlorella NC64A chromosomal DNAs into seven bands (Fig. 1A). Six bands represent seven chlorella chromosomes that range from 1.1 to 3.2 Mb; the 2.8-Mb band probably contains two chromosomes. The intense ∼3.3-Mb band near the top of the gel represents compressed chromosomal DNA, which is beyond the resolution limit of the applied switch interval and presumably consists of seven chromosomes, including three 3.5-Mb, one 4.2-Mb, and two 6.5-Mb chromosomes (9).
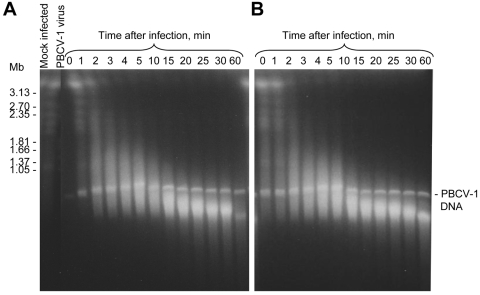
FIG. 1.
Kinetics of DNA degradation of Chlorella NC64A cells upon infection with PBCV-1 virus. (A) No CHX; (B) 5 μg/ml CHX. Times (min p.i.) are listed in the figures. Electrophoresis conditions were as follows: 100 V with pulses of 250 to 900 s for 60 h at 14°C in 1× Tris-acetate-EDTA (TAE) buffer.
PBCV-1 infection at an MOI of 10 caused rapid degradation of the host chromosomal DNAs (Fig. 1A). Most chromosomal DNAs were degraded to an average size of 150 to 200 kb by 10 min p.i., and this pattern remained fairly constant until at least 30 min p.i. (Fig. 1A). Additional DNA degradation occurred after 60 min p.i. (data not shown). The 330-kb PBCV-1 genomic DNA was present in all the virus-infected cells and was essentially unchanged during this 60-min experiment (Fig. 1A). This DNA was from viruses that either infected the host cells or simply attached to the cells without injecting DNA. To verify that the 330-kb PBCV-1 DNA was not from free virions that were coincidently pelleted by centrifugation, virions were mixed with nonhost Chlorella Pbi cells, and the samples were processed by the same procedure. As expected, no 330-kb DNA band existed in these samples (data not shown).
To determine if the host DNA degradation caused by PBCV-1 virus infection was MOI sensitive, infection experiments were conducted with various MOIs (0.1 to 10). The results indicated that a single PFU per cell was necessary and sufficient to initiate host DNA degradation (data not shown).
Protein synthesis is not required for host chromosomal DNA degradation.
Host chromosomal DNA degradation should occur in the absence of protein synthesis if cleavage is mediated by REases packaged in the PBCV-1 virion. This prediction was tested by adding 5 μg/ml of cycloheximide (CHX) to the chlorella cells 10 min before infecting them with PBCV-1 (note that 5 μg/ml of CHX inhibits cytoplasmic protein synthesis in 10 min [24]). The CHX treatment had no effect on virus-mediated chromosomal DNA cleavage (Fig. 1B), indicating that de novo protein synthesis was not required for host DNA degradation.
PBCV-1 virions were also UV irradiated prior to infection to determine if an infectious plaque-forming virus was required for host DNA degradation. PBCV-1 was exposed to sufficient UV to reduce its infectivity by several orders of magnitude (Fig. 2A). This experiment, which was carried out in the dark to prevent DNA repair by the host's light-dependent photolyase, revealed that host chromosomal DNAs were cleaved to an average length of 150 to 200 kb with approximately the same kinetics as untreated PBCV-1 (Fig. 2B).
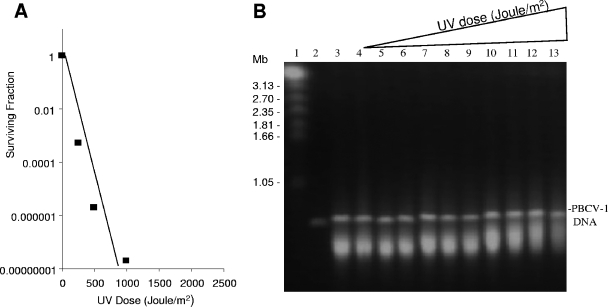
FIG. 2.
Effect of UV treatment on PBCV-1 virus. (A) Effect of UV radiation dose on PBCV-1 virus replication. (B) UV radiation dose and ability of PBCV-1 virus to cause Chlorella NC64A chromatin degradation. Lanes: 1, mock-infected NC64A cells at 30 min p.i.; 2, PBCV-1 virus DNA; 3, NC64A cells infected with PBCV-1 virus at 30 min p.i.; 4 to 13, NC64A cells at 30 min p.i. with UV-treated PBCV-1 virus receiving the following doses (Joule/m2): 4, 0; 5, 0.5 × 103; 6, 1.2 × 103; 7, 2.5 × 103; 8, 4 × 103; 9, 5 × 103; 10, 7.5 × 103; 11, 10 × 103; 12, 15 × 103; and 13, 20 × 103. Electrophoresis conditions were as follows: 100 V with pulses of 250 to 900 s for 60 h at 14°C in 1× TAE buffer.
These results establish that the UV inactivation target size for DNA degradation is much smaller than the target size for virus replication, which is presumed to be the viral genome. This relatively small UV-sensitive target is consistent with the size of a protein. Prior exposure of PBCV-1 to very high dosages of UV (2 × 104 J/m2) resulted in slightly less chromosomal degradation (Fig. 2B, lane 13), demonstrating that the virion has a UV-sensitive target, albeit a small one relative to the target size required for infectivity.
Nuclease activity is associated with PBCV-1 virions.
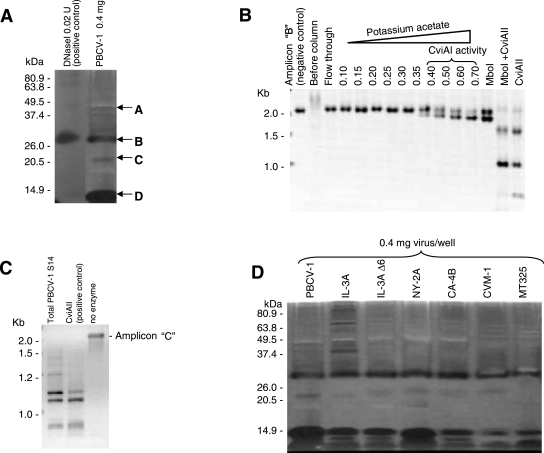
Although proteomic analyses established that the R.CviAI and R.CviAII proteins are associated with the PBCV-1 virion, these results do not prove that the two proteins are enzymatically active. To evaluate REase activity, an in-gel activity assay was developed to detect virion-associated nucleases after separation by SDS-PAGE. This assay depends on the abilities of many nucleases to renature and to regain enzymatic activity after removal of SDS from the gels (17). Calf thymus DNA was used as a substrate by adding it to the SDS-acrylamide mixture prior to polymerization. Following electrophoresis and removal of the SDS, enzyme activities were visualized by ethidium bromide staining. Dark bands in a fluorescent background of unhydrolyzed DNA indicate DNase activity. SDS-DNA-PAGE of PBCV-1 virion proteins revealed several bands with nuclease activity (Fig. 3A). Band A migrates at ∼40 kDa, corresponding to the size of R.CviAII, and band C migrates at ∼21 kDa, corresponding to the size of R.CviAI. The more prominent 29-kDa (band B) and 14-kDa (band D) activities have not been assigned to a known nuclease(s). The regions with nuclease activity were excised, digested with trypsin, and analyzed by tandem mass spectrometry for protein identification. This experiment detected R.CviAI in the 21-kDa protein band (band C) (Mowse score of 70 with 7% coverage) and R.CviAII in the 40-kDa protein band (band A) (Mowse score of 79 with 7% coverage).
FIG. 3.
Nuclease activities of virion-associated proteins. (A and D) SDS-DNA-PAGE gel assay; (B and C) REase activities of virion-associated proteins. (A) SDS-DNA-PAGE gel assay of PBCV-1 virion-associated proteins. (B) Heparin-Sepharose column chromatography of enzyme extracts prepared from PBCV-1 virions eluted with potassium acetate. The fractions were assayed on DNA amplicon B. (C) R.CviAII activity of disrupted PBCV-1 supernatant fraction (S-15) assayed on DNA amplicon C. DNA fragments were electrophoresed in a 1% agarose gel in 0.5× TBE buffer at 5.5 V/cm. (D) SDS-DNA-PAGE assay of virion-associated proteins from different chlorella viruses.
To determine if the virion-associated nucleases had REase activities, PBCV-1 virions were suspended in a high-salt buffer and disrupted by sonication, extracts were clarified by centrifugation and applied to a heparin-Sepharose column, and protein was eluted with a potassium acetate gradient. The eluted fractions were dialyzed and assayed using two ∼2-kb DNA amplicons with known sequences. DNA amplicon B has cleavage sites for both virus-encoded REases (Fig. 3B). R.CviAI and MboI (which cleave at identical sites and have identical methylation sensitivities) cleaved amplicon B into two fragments, and R.CviAII produced five fragments. DNA amplicon C only has R.CviAII restriction sites. R.CviAI-like activity eluted from the column starting at 0.4 M potassium acetate, with a peak at 0.7 M (Fig. 3B). The enzyme activity produced a DNA restriction pattern for DNA amplicon B that was identical to that obtained with MboI.
R.CviAII activity is unstable and is inhibited by potassium or sodium acetate concentrations above 50 mM (34). Attempts to isolate R.CviAII activity by heparin-Sepharose chromatography using potassium acetate were unsuccessful. Therefore, R.CviAII activity was assayed with soluble extracts from disrupted virions. The extracts produced a DNA restriction pattern identical to that obtained with commercial CviAII with amplicon C as the substrate (Fig. 3C).
Two control experiments were conducted to (i) eliminate the possibility that contaminant exogenous REases copurify with PBCV-1 virions and (ii) demonstrate that the R.CviAI and R.CviAII REases are susceptible to proteinase K. In the first experiment, the REase HaeIII (150 U/ml) was added to a PBCV-1 virus preparation, and the preparation was subjected to the standard virus purification procedure, including proteinase K treatment (0.02 mg/ml at 45°C for 1 h). No HaeIII activity was detected in sonicated virus extracts from this sample, indicating that proteinase K treatment of the virus removes HaeIII not packaged in the virion. In contrast, HaeIII activity was detected in a non-proteinase K-treated virus sample (data not shown). In the second experiment, a PBCV-1 virus preparation was disrupted and treated with proteinase K (0.02 mg/ml at 45°C for 1 h) before electrophoresis in an SDS-DNA-PAGE gel to determine if the virion-free R.CviAI and R.CviAII REases were susceptible to proteinase K. Control experiments included disrupted virions without proteinase K treatment and intact virions treated with proteinase K. This experiment established that R.CviAI and R.CviAII were resistant to proteinase K treatment when they were in the virion but that the two REases were sensitive to proteinase K when the virions were disrupted (data not shown).
Additional Chlorella NC64A-infecting viruses lead to host DNA degradation.
The preceding results establish that the two PBCV-1-encoded REases are packaged in the virion and suggest that these enzymes are responsible for cleaving host chromosomal DNAs immediately after infection. An ideal experiment would be to construct PBCV-1 mutants that lack one or both REase-encoding genes and then determine if infection leads to host chromosomal DNA degradation. These experiments cannot be conducted, however, because procedures are not currently available for molecular manipulation of the PBCV-1 genome. As an alternative, we examined infections by other chloroviruses that have different R/M systems (Table 1). Some of these viruses infect Chlorella NC64A, while two infect Chlorella Pbi.
TABLE 1.
DNA restriction-modification phenotypes of tested chloroviruses
| Virus strain | Start of host DNA degradation (min p.i.) | Level of methylation (%)a
|
Identified REase gene(s)a | Identified MTase genes | Length of virus replication cycle (h) | |
|---|---|---|---|---|---|---|
| m5C | m6A | |||||
| Chlorella NC64A viruses | ||||||
| PBCV-1 | 2-5 | 1.9 | 1.5 | R.CviAI, R.CviAII | M.CviAI, M.CviAII, and three cytosine MTases | 6-8 |
| IL-3A | 15 | 9.7 | ND | R.CviJ | M.CviJI and one putative MTasea | 8-9 |
| IL-3A Δ6 | 25-30 | 1.6b | ND | 8-9 | ||
| NY-2A | 2-5 | 45 | 37 | R.CviQI plus Nt.CviQII | 18 MTasesc | 15-18 |
| CA-4B | 20-25 | 0.1 | ND | Unknown | Unknown | 6-8 |
| Chlorella Pbi viruses | ||||||
| MT-325 | No apparent degradation | Unknown | Unknown | Unknown | One cytosine MTased | 6-8 |
| CVM-1 | 2-5 | 41.9 | 10.1 | Unknown | Unknown | 6-8 |
As noted in the introduction, the virus IL-3A carries the CviJI R/M system and IL-3A deletion mutants exist that lack both the M.CviJI and R.CviJI genes (3). If the function of the virus-encoded REases is to degrade host DNA, and assuming that R.CviJI is packaged in the IL-3A virion, host chromosomal DNA degradation should begin earlier in cells infected with wild-type IL-3A than in cells infected with IL-3A deletion mutant 6 (Δ6). Wild-type IL-3A infection led to host chromosomal DNA degradation beginning at about 15 min p.i. (Table 1). As predicted, there was a delay of ∼15 min in the initiation of chromosomal DNA degradation in cells infected with virus IL-3A Δ6 compared to that in cells infected with wild-type virus IL-3A (Table 1). However, by 60 min p.i., most of the host chromosomal DNA was degraded to ∼200 kb in cells infected with either of the viruses. It should be noted that although IL-3A Δ6 lacks the CviJI R/M system, its genome has 1.6% m5C, indicating that the virus encodes a second m5C MTase. This second MTase could have a companion REase; however, attempts to detect REase activity in IL-3A Δ6-infected cells were unsuccessful (3).
We recently sequenced the genome of another NC64A virus, NY-2A. NY-2A encodes 18 putative MTases and at least two functional site-specific endonucleases (4, 35). One of the endonucleases, named R.CviQI, cleaves G/TAC sequences but not G/TmAC sequences (33). The other nuclease is a nicking endonuclease, named Nt.CviQII; it cleaves one strand of dsDNA at R/AG sites (4, 35). Proteomic analyses of the NY-2A virion established that Nt.CviQII is packaged in the particle without its corresponding MTase (data not shown). NY-2A infection of Chlorella NC64A resulted in host chromosomal DNA degradation at a rate similar to that for PBCV-1 infection (Table 1), even though the NY-2A life cycle is two to three times longer than that of PBCV-1 (27).
Host chromosomal DNA integrity was also monitored after infection with the NC64A virus CA-4B; CA-4B has a replication time similar to that of PBCV-1 (Van Etten, unpublished results). The CA-4B genome contains the lowest level of methylated nucleotides among the chloroviruses. It has no detectable m6A, and only 0.12% of the cytosines are methylated (28), suggesting that CA-4B may lack a complete R/M system. Attempts to detect REase activity in chlorella cells infected with CA-4B were unsuccessful (Y. Xia and J. L. Van Etten, unpublished results). Like infection with the IL-3A Δ6 deletion mutant, infection with CA-4B was predicted to cause a slower change in host chromosomal DNA structure. Indeed, CA-4B infection resulted in slower host DNA degradation than PBCV-1 infection, initiating degradation at approximately 20 min p.i. (Table 1).
Pbi virus infections.
The Chlorella Pbi karyotype differs from that of Chlorella NC64A (compare Fig. 1A and 4), even though the total genome sizes of the two species are similar. Chlorella Pbi chromosomal DNA partially separated into five bands, from 1.7 Mb to 3.5 Mb. Chloroviruses that infect Chlorella Pbi are morphologically and genetically similar to NC64A viruses (16). The Pbi virus DNAs, like those from the NC64A viruses, also contain m5C (14% to 43% of the total cytosines) and m6A (0 to 18% of the total adenines), indicating that they also carry R/M systems (13). The Pbi virus CVM-1 genome is highly methylated, with 42% m5C and 10% m6A (16). CVM-1 infection of Chlorella Pbi causes rapid changes in host chromosomal DNA structure (Table 1). Degradation begins within 2 to 5 min of CVM-1 infection, and by 10 min p.i., chromosomal DNAs are degraded to 150- to 300-kb fragments. Like the case with PBCV-1, inhibition of host protein synthesis with CHX had no effect on this DNA degradation (results not shown).
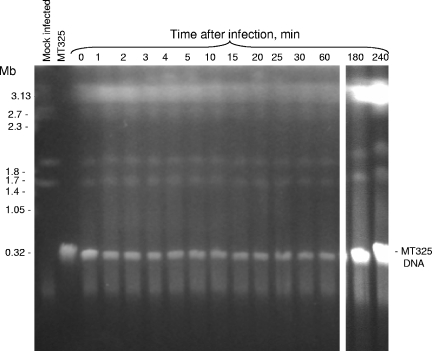
FIG. 4.
Kinetics of DNA degradation of Chlorella Pbi cells upon infection with MT325 virus. Electrophoresis conditions were as follows: 100 V with pulses of 250 to 900 s for 60 h at 14°C in 1× TAE buffer.
The Pbi virus MT325 genome has also been sequenced recently, and relevant to this study, MT325 only encodes one putative MTase and no recognizable REases (Fitzgerald et al., unpublished data). Unlike infections with the other viruses, MT325 infection of Chlorella Pbi did not cause detectable changes in the host chromosomal DNA (Fig. 4), i.e., the chromosomal integrity was unchanged through 4 h of infection. The 313-kb MT325 genome was present in all the infected cells. In fact, there was a large increase in viral DNA at 3 and 4 h p.i. due to virus replication (Fig. 4, last two lanes). Thus, the chlorovirus MT325 can infect and initiate DNA replication without degrading host chromosomal DNA.
Virions package additional nucleases.
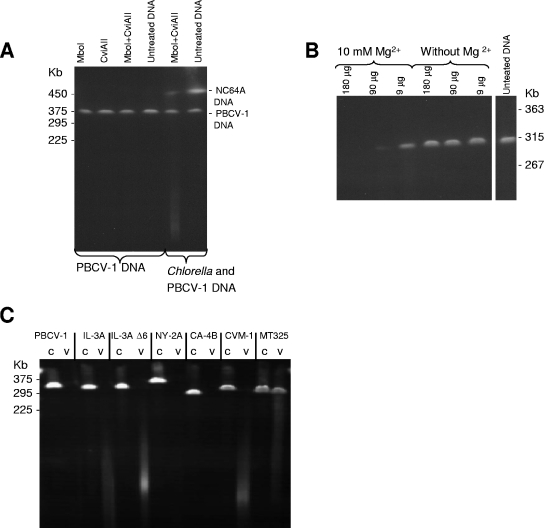
The results described above are consistent with the hypothesis that virion-associated, virus-encoded REases are involved in host chromosomal degradation. To further evaluate this hypothesis, virus and host chromosomal DNAs were incubated with extracts from disrupted virus particles in vitro. As a control, host and PBCV-1 DNAs were incubated with two commercial purified REases, i.e., MboI, an isoschizomer of R.CviAI (30), and R.CviAII. The experiment confirmed that PBCV-1 DNA was resistant to the two REases (Fig. 5A), while host chromosomal DNA was degraded (Fig. 5A). However, both PBCV-1 and host chromosomal DNAs were degraded with extracts from the PBCV-1 virions (Fig. 5B). The DNA degradation was apparently enzymatic because it required Mg2+ for activity (Fig. 5B). Although these results are consistent with the virion-packaged REases being responsible for host chromosomal DNA degradation, it indicates that PBCV-1 packages one or more general nucleases that can degrade both viral and chlorella chromosomal DNAs. The general nuclease(s) could be the 14- and/or 29-kDa band detected in the SDS-DNA-PAGE activity assay (Fig. 3A, bands B and D).
FIG. 5.
Digestion of viral and Chlorella DNAs with virion-associated nucleases. (A) PBCV-1 and Chlorella NC64A DNAs treated with MboI (an isoschizomer of R.CviAI), R.CviAII, and MboI plus R.CviAII. (B) PBCV-1 DNA in agarose plugs (1 μg/plug) treated with different concentrations of sonicated PBCV-1 virus extract. (C) Digestion of viral DNAs in agarose plugs (1 μg/plug) with sonicated virus extracts. c, untreated viral DNA (control); v, viral DNA treated with sonicated virus (150 μg/treatment) in the presence of 10 mM Mg2+. Electrophoresis conditions were as follows: 200 V with pulses of 5 to 30 s for 25 h at 14°C in 0.5× TBE buffer.
The same experiment was conducted with the other chloroviruses (Fig. 5C). In each case, the disrupted virions digested their own DNAs, indicating that a general nuclease(s) is packaged in all the virus particles. To further test this observation, the chloroviruses were assayed for activity by in-gel SDS-DNA-PAGE. Each of the viruses tested, including MT325, had general nucleases that migrated to similar positions in the gels (Fig. 3D).
DISCUSSION
The first chlorovirus-encoded R/M system was described about 20 years ago (30, 31). Since then, several virus-encoded REases and MTases have been characterized (13). However, the biological function(s) of these systems in the viral life cycle has remained a mystery. Bacterial R/M systems confer resistance to foreign DNAs and DNA viruses (e.g., see reference 29). In fact, the name “restriction” refers to their role in excluding foreign DNA. We assume that the virus-encoded enzymes serve an important function(s) because some viruses, such as NY-2A, encode as many as 18 MTases and at least two site-specific endonucleases (4, 35). Thus, at least 20 of NY-2A's predicted 400 protein-encoding genes (5% of the genes) are involved in R/M functions. However, other chloroviruses, such as MT325, survive with only one MTase and no apparent REase(s).
We presented two hypotheses on the function of the virus R/M systems in the introduction, namely, a host DNA degradation hypothesis and a virus exclusion hypothesis. Several results described in this report are consistent with the hypothesis that the REases are involved in degrading host chromosomal DNA, as follows. (i) Where known (i.e., PBCV-1 and NY-2A), the virus-encoded REases, but not their companion MTases, are packaged in the virions. (ii) Infection by many, but not all, chloroviruses results in a rapid change in the structure of the host chromosomal DNA, i.e., DNA degradation begins within min of infection. (iii) Host chromosomal DNA degradation occurs in the absence of de novo protein synthesis and with UV-treated viruses that are no longer able to produce viable progeny.
We were surprised at how quickly host chromosomal DNA degradation occurred after infection with some of the viruses, e.g., beginning at 2 to 5 min p.i. for viruses PBCV-1, NY-2A, and CVM-1. Infection by the viruses CA-4B and IL-3A Δ6, which may not encode an REase(s), initiated DNA degradation beginning at 20 and 25 min p.i., respectively. Finally, infection with the virus MT325, which encodes one MTase and no recognizable REase, did not alter the integrity of the host's chromosomal DNA, even after MT325 DNA replication occurred (3 and 4 h p.i.) (Fig. 4).
The difference in the initiation of host chromosomal DNA degradation is not related to how quickly the viruses replicate. For example, NY-2A infection degrades host DNA at a rate comparable to that of PBCV-1, but the NY-2A virus replication cycle is about 2.5 times longer than that of PBCV-1 (27). Likewise, PBCV-1 and CA-4B replicate at about the same rate; however, CA-4B, which probably lacks an REase, takes ∼10 times longer to initiate host DNA degradation. Therefore, chlorovirus infections produce a variety of responses that are virus specific.
Another explanation that might account for the speed with which host DNA degradation begins is that virus infection triggers a preformed host signaling pathway that leads to DNA cleavage. In fact, infections with all the tested chlorella viruses produced rapid depolarization of the host membrane (6, 10). However, the onset of host DNA degradation depends on the virus. These different responses would appear to eliminate the signaling activation hypothesis because available evidence indicates that all the viruses initiate infection at about the same rate, i.e., depolarization occurs just after virus attachment and cell wall degradation (6). Furthermore, infection with MT325 does not result in chromosomal DNA degradation even by the time virus DNA is replicating (Fig. 4).
One issue to consider is how many REase molecules (REases typically function as homodimers) would need to be packaged in a virion to produce the ∼200-kb DNA degradation products from a 40-Mb genome in the time that this occurs. The DNA would need to be cleaved approximately 200 times if one assumes that the genome is a single chromosome. The number of required cleavages is smaller if one includes the 13 chromosomes in the calculations. The average cleavage rate for type II REases is 1 to 10 cleavages per min in vitro (11, 15). The rate may be higher in vivo due to the high concentration of DNA. If each PBCV-1 REase makes 10 cleavages per min, it would require ∼10 min to cleave 40 Mb of DNA to 200 kb. Assuming that it takes 2 min for the REases to reach the nucleus, this estimation agrees well with the actual PBCV-1-induced DNA degradation revealed by PFGE experiments (∼15 min) (Fig. 1). These calculations also suggest that packaging a minimum of two molecules of each REase in the PBCV-1 virion would be sufficient to achieve the observed level of chromatin degradation that occurs during PBCV-1 infection. However, the kcat values for these enzymes are unknown.
The virus disruption experiments revealed that all of the virions packaged as many as three general nucleases in addition to the REases (Fig. 3D). These nucleases, which are similar in size for all the viruses, digest both host and virus DNA in vitro (Fig. 5C). Their function during infection is unknown, but presumably the virus DNA and nucleases do not interact during infection; otherwise, the interaction would be lethal to the virus. These observations also suggest that the general nucleases are physically separate from the viral DNA in the virion. However, these general nucleases do not appear to be responsible for the initial events in host DNA degradation because they are also present in the MT325 virus, yet MT325 infection does not lead to host nuclear DNA degradation.
It is important that the experiments to detect nuclease activity in the disrupted virions by SDS-DNA-PAGE assay (e.g., see Fig. 3A and D) are not quantitative. That is, REases that recognize four nucleotides, such as R.CviAI and R.CviAII, statistically cleave DNA at about every 256 nucleotides; in contrast, a general nuclease presumably cleaves DNA into small oligonucleotides. Since the assay depends on diffusion of DNA degradation products from the gel, REase products will not diffuse as rapidly and consequently show up as bands with lower apparent activities.
There are at least two explanations, which are not mutually exclusive, for why a virus might package enzymes that aid in degrading its host's DNA. (i) The REases may help to degrade host DNA to allow recycling and reincorporation of deoxynucleotides into virus DNA. Chloroviruses have large dsDNA genomes, and a large concentration of deoxynucleotides is required for virus DNA synthesis. PBCV-1 infection increases the total DNA in the chlorella cell at least three- to fourfold by 4 h p.i. (24). Some of the deoxynucleotide intermediates for viral DNA synthesis may be recycled from chlorella DNA, although this would account for only about 20 to 25% of the required DNA intermediates. The burst size of the MT325 virus is similar to that of PBCV-1, so this virus compensates for the lack of host DNA degradation by some other mechanism. (ii) Host DNA degradation may contribute to the rapid inhibition of host-directed DNA and RNA syntheses that occurs in PBCV-1-infected chlorella cells (19, 24). Inhibiting host RNA synthesis is important because the chloroviruses do not carry a recognizable RNA polymerase, nor is RNA polymerase activity associated with the virion (22). Therefore, a host DNA-dependent RNA polymerase(s) has to shift from transcribing host DNA to transcribing virus DNA. This switching occurs quickly in PBCV-1 infection because early PBCV-1 transcripts can be detected within 5 to 10 min p.i. (e.g., see reference 36). However, a complicating factor in understanding the biological role of the virus-encoded REases is that some viruses apparently lack such genes and survive in nature without cleaving their host DNA during infection.
Acknowledgments
We thank Ron Cerny for help with the mass spectrometry experiments and Drake Stanger for critically reading the manuscript.
This investigation was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service award 1 T32 AIO60547 from the National Institute of Allergy and Infectious Diseases (I.A.), by NIH COBRE grant P20RR015635 (D.D.), and by NIH grant R01 GM32441 (J.V.E.).
REFERENCES
- 1.Birren, B., and E. Lai. 1993. Pulsed field gel electrophoresis. A practical guide. Academic Press, Inc., San Diego, Calif.
- 2.Blank, A., R. H. Sugiyama, and C. A. Dekker. 1982. Activity staining of nucleolytic enzymes after dodecyl sulfate-polyacrylamide gel electrophoresis: use of aqueous isopropanol to remove detergent from gels. Anal. Biochem. 120:267-275. [DOI] [PubMed] [Google Scholar]
- 3.Burbank, D. E., S. L. Shields, A. M. Schuster, and J. L. Van Etten. 1990. 5-Azacytidine resistant mutants of Chlorella virus IL-3A. Virology 176:311-315. [DOI] [PubMed] [Google Scholar]
- 4.Chan, S.-H., Z. Zhu, D. D. Dunigan, J. L. Van Etten, and S.-Y. Xu. 2006. Cloning of Nt.CviQII nicking endonuclease and its cognate methyltransferase: M.CviQII methylates AG sequences. Protein Expr. Purif. [Epub ahead of print.] [DOI] [PubMed]
- 5.Chase, T. E., J. A. Nelson, D. E. Burbank, and J. L. Van Etten. 1989. Mutual exclusion occurs in a chlorella-like green alga inoculated with two viruses. J. Gen. Virol. 70:1829-1836. [DOI] [PubMed] [Google Scholar]
- 6.Frohns, F., A. Kasmann, D. Kramer, B. Schafer, M. Mehmel, M. Kang, J. L. Van Etten, S. Gazzarrini, A. Moroni, and G. Thiel. 2006. Potassium ion channels of chlorella viruses cause rapid depolarization of host cells during infection. J. Virol. 80:2437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashiyama, T., and T. Yamada. 1991. Electrophoretic karyotyping and chromosomal gene mapping of Chlorella. Nucleic Acids Res. 19:6191-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang, M., D. D. Dunigan, and J. L. Van Etten. 2005. Chlorovirus: a genus of Phycodnaviridae that infects certain chlorella-like green algae. Mol. Plant Pathol. 6:213-224. [DOI] [PubMed] [Google Scholar]
- 9.McCluskey, K., M. V. Graves, D. M. Mills, and R. H. Meints. 1992. Replication of Chlorella virus PBCV-1 and host karyotype determination studied with pulsed-field gel electrophoresis. J. Phycol. 28:846-850. [Google Scholar]
- 10.Mehmel, M., M. Rothermel, T. Meckel, J. L. Van Etten, A. Moroni, and G. Thiel. 2003. Possible function for virus encoded K+ channel Kcv in the replication of chlorella virus PBCV-1. FEBS Lett. 552:7-11. [DOI] [PubMed] [Google Scholar]
- 11.Modrich, P., and R. J. Roberts. 1982. Type-II restriction and modification enzymes, p. 109-155. In S. M. Linn and R. J. Roberts (ed.), Nucleases. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Nandhagopal, N., A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing, DNA virus. Proc. Natl. Acad. Sci. USA 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson, M., D. E. Burbank, and J. L. Van Etten. 1998. Chlorella viruses encode multiple DNA methyltransferases. Biol. Chem. 379:423-428. [DOI] [PubMed] [Google Scholar]
- 14.Nelson, M., Y. Zhang, and J. L. Van Etten. 1993. DNA methyltransferases and DNA site-specific endonucleases encoded by chlorella viruses, p. 186-211. In J. P. Jost and H. P. Saluz (ed.), DNA methylation: molecular biology and biological significance. Birkhauser Verlag Publishers, Basel, Switzerland. [DOI] [PubMed]
- 15.Pingoud, A., J. Alves, and R. Geiger. 1993. Restriction enzymes, p. 107-200. In M. M. Burrell (ed.), Methods of molecular biology, vol. 16. Enzymes of molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 16.Reisser, W., D. E. Burbank, S. M. Meints, R. H. Meints, B. Becker, and J. L. Van Etten. 1988. A comparison of viruses infecting two different chlorella-like green algae. Virology 167:143-149. [DOI] [PubMed] [Google Scholar]
- 17.Rosenthal, A. L., and S. A. Lacks. 1977. Nuclease detection in SDS-polyacrylamide gel electrophoresis. Anal. Biochem. 80:76-90. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. A8.40-A8.49. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Schuster, A. M., D. E. Burbank, B. Meister, M. P. Skrdla, R. H. Meints, S. Hattman, D. Swinton, and J. L. Van Etten. 1986. Characterization of viruses infecting a eukaryotic chlorella-like green alga. Virology 150:170-177. [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 21.Skrdla, M. P., D. E. Burbank, Y. Xia, R. H. Meints, and J. L. Van Etten. 1984. Structural proteins and lipids in a virus, PBCV-1, which replicates in a Chlorella-like alga. Virology 135:308-315. [DOI] [PubMed] [Google Scholar]
- 22.Van Etten, J. L. 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37:153-195. [DOI] [PubMed] [Google Scholar]
- 23.Van Etten, J. L., D. E. Burbank, Y. Xia, and R. H. Meints. 1983. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology 126:117-125. [DOI] [PubMed] [Google Scholar]
- 24.Van Etten, J. L., D. E. Burbank, J. Joshi, and R. H. Meints. 1984. DNA synthesis in a chlorella-like alga following infection with the virus PBCV-1. Virology 134:443-449. [DOI] [PubMed] [Google Scholar]
- 25.Van Etten, J. L., C. H. Van Etten, J. K. Johnson, and D. E. Burbank. 1985. A survey for viruses from fresh water that infect a eukaryotic Chlorella-like green alga. Appl. Environ. Microbiol. 49:1326-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Etten, J. L., A. M. Schuster, L. Girton, D. E. Burbank, D. Swinton, and S. Hattman. 1985. DNA methylation of viruses infecting a eukaryotic chlorella-like green alga. Nucleic Acids Res. 13:3471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Etten, J. L., A. M. Schuster, and R. H. Meints. 1988. Viruses of eukaryotic chlorella-like algae, p. 411-428. In Y. Koltin and M. J. Leibowitz (ed.), Viruses of fungi and simple eukaryotes. Marcel Dekker, Inc., New York, N.Y.
- 28.Van Etten, J. L., L. C. Lane, and R. H. Meints. 1991. Unicellular plants also have large dsDNA viruses. Semin. Virol. 2:71-77. [Google Scholar]
- 29.Wilson, G. G., and N. E. Murray. 1991. Restriction and modification systems. Annu. Rev. Genet. 25:585-627. [DOI] [PubMed] [Google Scholar]
- 30.Xia, Y., D. E. Burbank, L. Uher, D. Rabussay, and J. L. Van Etten. 1986. Restriction endonuclease activity induced by PBCV-1 virus infection of a chlorella-like green alga. Mol. Cell. Biol. 6:1430-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia, Y., and J. L. Van Etten. 1986. DNA methyltransferase induced by PBCV-1 virus infection of a chlorella-like green alga. Mol. Cell. Biol. 6:1440-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia, Y., D. E. Burbank, L. Uher, D. Rabussay, and J. L. Van Etten. 1987. IL-3A virus infection of a chlorella-like green alga induces a DNA restriction endonuclease with novel sequence specificity. Nucleic Acids Res. 15:6075-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia, Y., K. E. Narva, and J. L. Van Etten. 1987. The cleavage site of the RsaI isoschizomer, CviII, is G/TAC. Nucleic Acids Res. 15:10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, Y., M. Nelson, J. W. Nietfeldt, D. E. Burbank, and J. L. Van Etten. 1992. Characterization of chlorella virus PBCV-1 CviAII restriction and modification system. Nucleic Acids Res. 20:5351-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y., M. Nelson, J. Nietfeldt, Y. Xia, D. E. Burbank, S. Ropp, and J. L. Van Etten. 1998. Chlorella virus NY-2A encodes at least twelve DNA endonuclease/methyltransferase genes. Virology 240:366-375. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Y., I. Calin-Jageman, J. R. Gurnon, T.-J. Choi, B. Adams, A. W. Nicholson, and J. L. Van Etten. 2003. Characterization of a chlorella virus PBCV-1 encoded ribonuclease III. Virology 317:73-83. [DOI] [PubMed] [Google Scholar]