Abstract
Human metapneumovirus (hMPV) is a recently described member of the Paramyxoviridae family/Pneumovirinae subfamily and shares many common features with respiratory syncytial virus (RSV), another member of the same subfamily. hMPV causes respiratory tract illnesses that, similar to human RSV, occur predominantly during the winter months and have symptoms that range from mild to severe cough, bronchiolitis, and pneumonia. Like RSV, the hMPV virus can be subdivided into two genetic subgroups, A and B. With RSV, a single monoclonal antibody directed at the fusion (F) protein can prevent severe lower respiratory tract RSV infection. Because of the high level of sequence conservation of the F protein across all the hMPV subgroups, this protein is likely to be the preferred antigenic target for the generation of cross-subgroup neutralizing antibodies. Here we describe the generation of a panel of neutralizing monoclonal antibodies that bind to the hMPV F protein. A subset of these antibodies has the ability to neutralize prototypic strains of both the A and B hMPV subgroups in vitro. Two of these antibodies exhibited high-affinity binding to the F protein and were shown to protect hamsters against infection with hMPV. The data suggest that a monoclonal antibody could be used prophylactically to prevent lower respiratory tract disease caused by hMPV.
Respiratory viruses account for a large proportion of upper and lower respiratory tract illness in humans. In the past few decades, many etiological agents of respiratory tract illness have been identified. Of these, respiratory syncytial virus (RSV) is the single most important cause of respiratory infections during infancy and early childhood (29). However, only 60% of clinically attended respiratory infections of infants and children are of a known etiology (21). Recently, van den Hoogen et al. (26) discovered and described human metapneumovirus (hMPV) and revealed that it may account for a portion of these previously unclassified infections. Prospective and retrospective studies suggest that hMPV infections account for between 3% and 15% of respiratory tract infections (5, 6, 8). hMPV has been found to be associated with respiratory tract illness that ranges from mild respiratory problems to severe cough, bronchiolitis, and pneumonia, a pattern similar to that seen for RSV (6, 19). Additionally, the seasonality of the infection is similar to that of RSV, peaking in the winter months (20, 30). Recent studies on lung transplant recipients, bone marrow recipients, and preterm infants have shown that hMPV can be isolated from patients with respiratory infections in this immunocompromised population, suggesting that some of the risk factors associated with RSV infection may be similarly associated with hMPV infection (8, 10, 14, 18, 25, 29).
hMPV is an RNA virus in the Pneumovirinae subfamily of the Paramyxoviridae family (26). hMPV shares a similar genetic structure with RSV but lacks the nonstructural genes NS1 and NS2 (27). Both viruses code for similar surface proteins that are defined as the surface glycoprotein (G) and the fusion (F) protein. Based upon differences between the amino acid sequences of the G and F proteins, both RSV and hMPV have been subdivided into A and B groups. However, in hMPV, there is a further bifurcation of A and B subgroups into A1, A2, B1, and B2 groupings (4, 28). For both viruses, the sequences of the G proteins display a wide variance between subgroups; with hMPV, the G protein has only 30% identity between the A and B subgroups. For both RSV and hMPV, the F protein is more conserved; across the known hMPV isolates, the F protein amino acid sequence is 94% conserved (3, 4, 28). Despite the similarities in structure of the viruses, the F proteins of hMPV and RSV share only a 33% amino acid sequence identity, and antisera generated against either RSV or hMPV do not neutralize across the Pneumoviridae group (32). Recently, it has been shown that a potent neutralizing response could be evoked in animals using virally vectored hMPV F protein; importantly, this neutralizing response could protect against challenge with heterologous virus (22, 24).
The presence of serum antibodies to hMPV in archival samples indicates that the human population has been exposed to this virus since at least 50 years ago (26). Currently, it is estimated that, by the age of 5, every person in the world has been exposed to this virus and has generated a serum antibody response to it (26, 30). Recent studies (15) that focused on the presence of antibodies that were specifically directed against the F protein of hMPV showed that there was a similar trend toward 90% seropositivity by age 5. These studies also demonstrated that there is significant anti-F protein reactivity against both the A and B subgroups of hMPV.
Our previous work with RSV established the effectiveness of prophylaxis both in animals and in at-risk populations with either polyclonal or monoclonal antibodies directed against RSV (1, 9, 11, 23). Moreover, palivizumab, a potent monoclonal antibody that shows high capacity for neutralization of RSV, protects against serious lower respiratory disease in at-risk populations (9, 23). In the work reported here, we extend this concept to hMPV and have generated high-affinity monoclonal antibodies specific to the F protein of hMPV that neutralize in both in vitro and in vivo models of viral infection.
MATERIALS AND METHODS
Cells and virus.
Vero, WI-38, and LLC-MK2 cells that were used for the propagation of hMPV and parainfluenza virus 3 (PIV3)-derived viruses were maintained in Eagle's minimal essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS). Adenovirus vectors were propagated in HEK-293 cells grown in Dulbecco's modified Eagle medium plus 10% FBS. The mouse myeloma cell line, NS0, was maintained in Dulbecco's modified Eagle medium plus 20% FBS; myeloma fusion cell lines were maintained in Excell 610 (JRH Biosciences, Lenexa, KS) plus 10% FBS. Titers of viral stocks were determined by 50% tissue culture infectious dose (TCID50) measurement (13) on Vero cells. Viral infection was determined by reactivity with antibodies directed against hMPV, as described in subsequent sections.
For the propagation of hMPV, semiconfluent cell monolayers were infected at a multiplicity of infection of 0.1 TCID50/cell in EMEM plus 2.5 μg/ml trypsin without FBS; at 5 to 9 days postinfection, the virions were harvested by freeze-thaw disruption of the cells. Viral samples were stabilized by the addition of 10× SPG (2.18 M sucrose, 0.038 M KH2PO4, 0.054 M l-glutamate) and stored at −80°C. The prototype hMPV strains studied were A1 NL100, A2 NL1700, B1 NL199, and B2 NL194, which have a 98% amino acid identity among the A and B groups and a 94% amino acid identity between the A and B groups.
PIV3-vectored hMPV F protein virus (b/hPIV3/hMPV F) has been reported previously and was propagated in Vero cells as described previously (24). The viral concentration of PIV3 constructs was estimated by determining PFU per milliliter of viral stock on Vero cells. Adenovirus constructs expressing the F protein sequences from strains NL100 and NL199 were produced using the AdEasy adenoviral system with the transfer vector pShuttle-CMV (AdEasy; Stratagene, La Jolla, CA). The resultant adenovirus was propagated in HEK-293 cells according to the manufacturer's instructions. Viral titers for adenovirus were determined using a TCID50 assay with cytopathic effect as the readout.
Production of hybridoma cell lines.
Armenian hamsters (Cytogen Research and Development, Inc., Boston, MA) and BALB/c mice (Jackson Laboratory, Bar Harbor, ME) were immunized using a combination of some or all of the following: intranasal infection with hMPV at 106 TCID50 per animal of either NL1700, NL100, or NL199; intranasal infection with 106 PFU b/hPIV3/hMPV FNL100; intraperitoneal injection with adenovirus-vectored hMPV FNL100 or hMPV FNL199 at a dose of 9 × 107 TCID50; purified soluble hMPV F protein derived from NL100 and NL199 sequences injected intraperitoneally with either GERBU MM adjuvant (CC Biotech, Valley Center, CA) or in an adjuvant-free solution. Four days after the final immunization, splenic lymphocytes were isolated and fused to NS0 cells using polyethylene glycol as described previously (7). Fusions were plated either in semisolid medium (ClonaCell; Stem Cell Technologies, Vancouver, BC) or in liquid medium in 96-well plates. Hybridoma supernatants that produced hMPV-specific antibodies were identified by enzyme-linked immunosorbent assay (ELISA) on hMPV-infected cells.
Identification and sequencing of monoclonal antibodies (MAbs).
RNA was isolated from hybridoma cells expressing the antibodies of interest using the RNeasy system (QIAGEN, Germantown, MD). The complementarity-determining region (CDR) sequences were amplified by PCR using commercially available probes (EMD Biosciences, La Jolla, CA) and were cloned into topoisomerase-bound TA overhang plasmid vectors (Invitrogen, Carlsbad, CA). Multiple clones of the CDR-containing plasmid vectors were isolated and sequenced using BigDye Terminator v3 (ABI, Foster City, CA) reactions and run on either an ABI 3100 or ABI 3730 sequencer to derive a consensus sequence of the hypervariable regions.
MAb purification.
Hamster monoclonal antibodies were purified on MEP Hypercel (Pall Corp., East Hills, NY) columns using 50 mM citrate at pH 4.0 to elute the MAb; eluates were immediately neutralized with a 1:10 volume of 1 M Tris-HCl, pH 8.0. Mouse monoclonal antibodies were purified on protein A-Sepharose; mouse immunoglobulin G1 (IgG1) was loaded in hybridoma medium containing 50 mM Tris, pH 8.5, and 1 M Na2SO4 while all other mouse subtypes were loaded directly from hybridoma medium. The protein A columns were eluted with 0.1 M glycine, pH 2.8, and the eluates were neutralized immediately with a 1:10 volume of 1 M Tris-HCl, pH 8.0.
hMPV F protein construct generation.
Full-length and truncates of F protein that lacked the transmembrane domain were made using plasmids containing full-length sequences of the fusion protein from isolates NL100 and NL199, respectively, as the template for PCRs. To obtain a soluble histidine-tagged form of the hMPV F protein, the following oligonucleotides were used to generate clones: from the NL100 sequence, 5′-AACCAAAAGCTTCACCATGTCTTGGAAAGTGGTGATC-3′and 5′-TTAATTGAATTCTTAGTGATGGTGATGGTGATGGCCAGTGTTTCCTTTCTCTGC-3′; from the NL199 sequence, 5′-TTCCTTAAGCTTCACCATGTCTTGGAAAGTGATGATCATC-3′and 5 ′-TTAATTGGATCCTTAGTGATGGTGATGGTGATGACCAGTGTTTCCTTTTTCTGCACT-3′. The PCR products were cleaved using the restriction endonucleases EcoRI and HindIII for the NL100 sequence and BamHI and HindIII for the NL199 sequence and then ligated to the vector pcDNA3.1(+) cleaved with the same endonucleases. HEK-293 cells were transiently transfected with the pcDNA clones, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) to introduce the DNA into the cells. To make stable hMPV F protein-expressing constructs, the same plasmid source of DNA was used to generate PCR products; however, alternate primers were used for both the NL/1/00 and NL/1/99 sequences: 5′-AATCAACGGTCCGCCACCATGTCTTGGAAAGTG-3′and 5′-TTAATTGAATTCTTAGTGATGGTGATGGTGATGGCCAGTGTTTCCTTTCTCTGC-3′. The PCR products were cleaved with RsrII and EcoRI and ligated to the pEE15.1 (Lonza, Allendale, NJ) vector cleaved with the same restriction endonucleases. Stable NS0 cell lines were made as described by Bebbington et al. (2). Full-length F protein constructs were made using the following oligonucleotides: for NL100, 5′-AACCAAAAGCTTCACCATGTCTTGGAAAGTGGTGATC-3′and 5′-AATTAAGGATCCTAATTATGTGGTATGAAGCCATT-3′; for NL199, 5′-TTCCTTAAGCTTCACCATGTCTTGGAAAGTGATGATCATC-3′and 5′-AATTAAGGATCCTAATTATGTGGTATGAAACCGCC. PCR products were cleaved with BamHI and HindIII endonucleases and were ligated to pcDNA3.1(+) cleaved with the same endonucleases. These vectors were used as the source of DNA for the construction of the adenovirus transfer vector pShuttle-CMV. The full-length F protein-containing fragments were obtained by cleavage of the pcDNA3.1 clones with the restriction endonucleases HindIII and EcoRV and were ligated to the pShuttle-CMV vector cleaved with the same endonucleases.
hMPV F protein purification.
Histidine-tagged soluble F protein was initially purified by Ni-nitrilotriacetic acid (QIAGEN, Germantown, MD) chromatography, which yielded protein that was 60% pure, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Subsequently, after isolation of the F protein-specific monoclonal antibody (MAb 1017), the F protein was purified by affinity chromatography on MAb 1017 coupled to cyanogen bromide-activated Sepharose and eluted with 0.1 M glycine, pH 2.8; the eluate was neutralized with a 1:10 volume of 1 M Tris-HCl, pH 8.0, and was dialyzed into phosphate-buffered saline (PBS). Affinity-purified F protein was >90% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
ELISAs.
An ELISA was developed to detect anti-hMPV antibodies in hybridoma supernatants or animal sera using hMPV-infected WI-38 cell monolayers. Cell monolayers in 96-well plates were infected with hMPV at a multiplicity of infection of 1.0 and were incubated subsequently for 3 to 5 days postinfection. The supernatants were removed, and the cells were desiccated at 37°C and stored at 4°C until use. For ELISA, the plates were blocked with PBS containing 0.1% (vol/vol) Tween 20 and 0.5% (wt/vol) bovine serum albumin (BSA). This and all subsequent steps were performed at room temperature. Diluted serum samples or hybridoma supernatants were incubated on the plates for 1 h, and the plates were then washed with PBS-Tween. Horseradish peroxidase (HRP)-conjugated anti-mouse or anti-Armenian hamster biotinylated antibody (Jackson ImmunoReasearch, West Grove, PA) was added, and the plates were incubated for an additional hour and then washed. For the hamster samples, streptavidin-HRP (Amersham Biosciences, Piscataway, NJ) was added and incubated for 1 h. Plates were developed with SureBlue TMB substrate (KPL, Gaithersburg, MD). End point titers of serum samples were defined as the last dilution that achieved a minimal twofold increase in absorbance over the control absorbance.
Competition ELISA experiments were performed using biotinylated MAb 242, MAb 338, MAb 659, MAb 757, MAb 836, MAb 1017, and MAb 1025. Antibodies were biotinylated using either biotin-XX-SSE (Invitrogen, Carlsbad, CA) or biotin-XX-SE (Vector Laboratories, Burlingame, CA) according to the manufacturers' instructions. A standard binding curve for each of the biotinylated antibodies was generated on hMPV-infected WI-38 cells using streptavidin-HRP as the detection reagent. The concentration of the biotinylated antibodies that gave a half maximal signal on the standard curve was used in the competition ELISA. The competing monoclonal antibodies were used at concentrations ranging from 50 to 0.03 μg/ml. Unlabeled competitive MAb that gave a >50% reduction in signal at a concentration less than or equal to 100 times the biotinylated antibody concentration were scored as competing.
To determine the serum concentration of injected antibodies, capture ELISAs were performed as follows. Soluble hMPV F protein (50 ng/well) from NL100 was coated onto Nunc Maxisorp (Nalge Nunc, Rochester, NY) microtiter plates overnight at 4°C in PBS buffer (Pierce, Rockford, IL). The following day, the plates were blocked using 1% casein in PBS. Serum samples were diluted into PBS and applied to the plate. A standard curve generated using matched antibody in the same concentration of normal hamster serum was used to calculate the serum concentration of antibody. Anti-mouse HRP conjugate was used for detection with SureBlue TMB reagent.
Neutralization assays.
Serial twofold dilutions of sera, hybridoma supernatants, or purified antibodies were incubated with 50 to 1,000 TCID50 of virus at 37°C for 1 h. After incubation, the virus-antibody mixtures were added to monolayers of Vero cells in 96-well plates; the plates were then centrifuged at 2,000 × g for 15 min at 25°C. The medium was removed from the cells, and the cells were washed in fresh medium without FBS and finally overlaid with EMEM without FBS and supplemented with trypsin at 2.5 μg/ml. The cells were grown for 5 to 7 days at 37°C, after which the medium was removed, and the cells were then fixed by the addition of 80% acetone at 4°C for 20 min. After this, the plates were air dried. Prior to development, the plates were blocked with 1% (wt/vol) casein and then probed with either polyclonal sera obtained from animals immunized with virus or biotinylated MAb 1017. A streptavidin-horseradish peroxidase conjugate was used to detect the biotinylated antibody. Alternatively, an anti-species-specific secondary antibody conjugated to horseradish peroxidase was used for detection of polyclonal sera. The plates were developed using SureBlue reagent. For hybridoma supernatants or polyclonal sera, the neutralization titer was defined as the last dilution that gave an absorbance that was less than twofold over the uninfected control cell absorbance. IC50s (50% inhibitory concentrations) for purified MAb were determined using GraphPad Prism software with curve fitting for a nonlinear sigmoid dose response.
Biacore analysis.
Kinetic analysis was performed to determine the binding constants for antibodies MAb 338 and MAb 234 to immobilized soluble hMPV F protein. Soluble FNL100 and soluble FNL199 proteins were immobilized on CM5 sensor chips (Biacore, Uppsala, Sweden) using an amine coupling kit as described previously (12) at an immobilization density between 80 and 300 resonance units (RU). Excess reactive esters were quenched with 70 μl of a 1 M ethanolamine hydrochloride, pH 8.5, solution. The surfaces were connected to a BiaCore 3000 in series. Two hundred fifty microliters of each MAb solution was injected at concentrations ranging from either 0.39 nM to 400 nM or from 3.13 nM to 100 nM at a flow rate of 75 μl/min, and 15 min of dissociation data was collected. Between injections, the surfaces were regenerated with a 1-min pulse of 1 M NaCl-50 mM NaOH. Data were analyzed using the BIAevaluation software supplied by Biacore, Inc.
In vivo assessment of protection.
Six- to eight-week-old Golden Syrian hamsters (6 to 7 animals/group) were injected intramuscularly with various concentrations of purified monoclonal antibody or bovine serum albumin in a volume of 100 μl the day prior to challenge. The following day, the animals were anesthetized with isoflurane and bled, and 100 to 200 μl of virus (1 × 107 TCID50/ml) was instilled intranasally. At 4 days postinfection, the animals were euthanized by CO2 asphyxiation, and the lungs were removed and homogenized in Hank's balanced salt solution using a Dounce homogenizer. Nasal turbinates were isolated and ground using a mortar and pestle in Hank's balanced salt solution. TCID50 determinations from lung and nasal turbinate homogenates were performed as follows: homogenates and sequential 10-fold dilutions of the homogenates were applied to washed LLC-MK2 cells and incubated for 1 h at room temperature. The supernatants were removed, and cells were overlaid with Opti-MEM (Invitrogen, Carlsbad, CA) medium containing 5 μg/ml of porcine-derived trypsin (Biowhittaker, Walkersville, MD). The cells were incubated at 37°C for 6 to 7 days. The medium was removed, and the cells were fixed using 80% methanol. Plates were blocked in 5% nonfat dried milk for 30 min. Polyclonal sera raised from animals infected with hMPV were used to stain the cells. Species-specific secondary antibody conjugated to HRP was used for detection using the 4CN peroxidase substrate (KPL, Gaithersburg, MD). Infection was assessed by visual inspection of individual wells and presented as log10 TCID50/gram tissue as previously described (19). Statistical analysis of the data was carried out using the GraphPad Prism software.
RESULTS
We set out to generate a panel of monoclonal antibodies that was both specific for hMPV F protein and which, at low concentration, neutralized hMPV. Numerous immunization strategies were employed to elicit robust F protein-specific immune responses in both Armenian hamsters and BALB/c mice. In all cases, the first immunization was an intranasal infection with a wild-type hMPV virus to prime the animals; this was followed by immunizations employing either recombinant adenovirus or recombinant bovine parainfluenza virus expressing hMPV F protein. In some cases, subsequent immunization with soluble recombinant hMPV F protein was carried out (Table 1). In this manner, we were able to generate high-titer anti-F protein-specific responses in the animals. In addition, to generate antibody responses that were reactive to both subgroups of hMPV, we employed immunizations containing both prototype A (NL100) and prototype B (NL199) F protein sequences. As shown in Table 1, this immunization strategy resulted in the production of high-titered antibodies in both mice and hamsters that neutralized one or both types of hMPV.
TABLE 1.
Immunizations producing serum titers and monoclonal antibodies
| Animal | Immunization route and immunogena | End point infected cell titer for strain (subgroup) determined byb:
|
MAb obtained | |||
|---|---|---|---|---|---|---|
| ELISA
|
Microneutralization
|
|||||
| NL100 (A1) | NL199 (B1) | NL100 (A1) | NL199 (B1) | |||
| Hamster | i.n. NL1700 | 1:781,250 | 1:781,250 | 1:1,250 | <1:50 | 1017 |
| i.n./i.p. PIV3/FNL100 | 757 | |||||
| Hamster | i.n. NL199 | 1:25,600 | 1:25,600 | 1:100 | 1:800 | 836 |
| i.n. NL100 | 659 | |||||
| i.p. adeno FNL100/FNL199 | 967 | |||||
| i.p. soluble FNL100/FNL199 | ||||||
| Mouse | i.n. NL199 | 1:312,500 | 1:312,500 | 1:100 | 1:200 | 338 |
| i.n. PIV3/FNL100 | 234 | |||||
| i.p. adeno FNL100/FNL199 | 224 | |||||
| i.p. soluble FNL100/FNL199 | ||||||
| Mouse | i.n. NL199 | 1:312,500 | 1:312,500 | 1:50 | 1:1,600 | 710 |
| i.n. NL100 | 344 | |||||
| i.p. adeno FNL100/FNL199 | 628 | |||||
| i.p. soluble FNL100/FNL199 | ||||||
Immunization routes were intranasal (i.n.) and intraperitoneal (i.p.).
End point titers were determined as described in Materials and Methods.
Following the immunizations, the spleens of the mice and hamsters were fused with myeloma cells to generate hybridoma cells, and the hybridoma supernatants were screened for reactivity toward cells infected with the hMPV (NL100) or uninfected cells. A minimal fivefold differential in absorbance between infected and uninfected cells was used as the criterion to select antibodies for the next stage of analysis. Hybridoma supernatants which were reactive with infected cells were expanded and tested as unfractionated supernatants in viral neutralization assays. The hybridoma supernatants varied greatly in the quantity of antibody they contained but were tested for hMPV neutralization without concentration adjustment. Therefore, this screening method selected for hybridomas that either produced high levels of antibody or produced antibody at low concentrations but with high neutralization activity. Hybridoma supernatants that exhibited neutralization activity at dilutions greater than 1:2 against at least one hMPV type were cloned by limited dilution and were then expanded to generate antibody for purification and further analysis. Not all hybridomas that had neutralizing activity in their supernatants could be cloned by limited dilution. Table 2 shows the IC50 titers of all of the antibodies that could be isolated by limited dilution and that produced sufficient antibody to assess their potency. In four cases, the same monoclonal antibodies were isolated from multiple cell lines, as determined by reverse transcription-PCR and sequencing of the heavy and light chains. A single isolate of each sequence was carried forward for full evaluation of neutralization potency. A wide range of neutralization potencies was seen, and many of the isolated antibodies did not neutralize all of the four prototype viruses tested. For the top three antibodies, MAb 338, MAb 234, and MAb 628, the neutralization capacity against all 4 prototype stains was lower than 2 μg/ml IC50, demonstrating a strong broadly neutralizing capacity. MAb 1017 showed neutralization across all 4 prototypes but was 25- to 100-fold less potent than the top 3 antibodies described above. Some antibodies showed enhanced neutralization (10- to 100-fold lower IC50) against B type viruses, as seen for MAb 242, MAb 757, MAb 710, and MAb 344. MAb 1025 and MAb 967 are more potent at neutralizing A type viruses. However, they differ in the following ways: MAb 1025 has essentially no neutralizing capacity against both B1 and B2 prototypes, whereas MAb 967 has the capacity to neutralize the B2 prototype but not the B1 prototype. Two antibodies, MAb 659 and MAb 836, show a difference in neutralizing capacity that does not split along the A and B subgroups. These antibodies neutralize the A2 and B1 subgroups better than the A1 and B2 subgroups, suggesting that amino acid changes that are not subgroup specific are playing a role in the binding of these antibodies to their epitopes.
TABLE 2.
IC50 determination of purified monoclonalsa
| MAbb | IC50 (μg/ml) for strain (subgroup):
|
|||
|---|---|---|---|---|
| NL100 (A1) | NL1700 (A2) | NL199 (B1) | NL194 (B2) | |
| 338 | 0.38 | 0.8 | 0.03 | 0.17 |
| 234 | 0.59 | 1.2 | 0.01 | 0.18 |
| 628 | 0.39 | 1.7 | 0.15 | 0.15 |
| 242 | 10.4 | 23.6 | 0.2 | 1.0 |
| 757 | 11.9 | 10.4 | 1.4 | 5.1 |
| 659 | 17.0 | 2.0 | 2.4 | 19.0 |
| 836 | 36.2 | 7.20 | 3.0 | 39.6 |
| 1017 | 16.7 | 38.4 | 12.8 | 29.6 |
| 967 | 0.15 | 0.15 | >100 | 4.8 |
| 710 | 33.8 | >100 | 2.4 | 16.4 |
| 344 | >100 | >100 | 0.8 | 24.0 |
| 1025 | 1.80 | 48.6 | >100 | >100 |
IC50 determinations were calculated as described in Materials and Methods.
MAbs in boldface type were isolated from more than one clone.
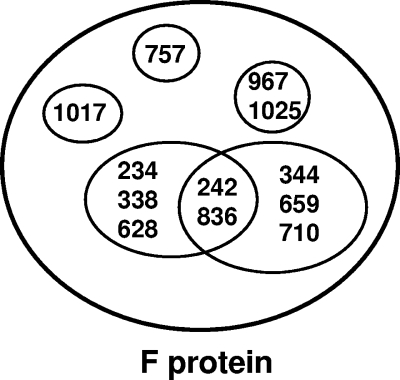
To determine the number of antigenic sites that these antibodies recognize, competition ELISA matrices were set up. Seven of the 12 isolates were biotinylated, and the ability of unlabeled monoclonal antibodies to compete for their binding to hMPV-infected cells was determined. The results of these experiments are shown in Table 3. In all, there are 6 distinct patterns of competition. Two of the antibodies (MAb 1017 and MAb 757) are only competed by matched antibody. MAb 967 and MAb 1025 compete for the same or overlapping sites. The remainder of the monoclonal antibodies have a more complex order of competition. These data suggest the epitope map shown in Fig. 1. This map is not intended to indicate actual sites on the F protein but rather to illustrate the overlapping nature of the epitopes.
TABLE 3.
Competition ELISA using biotinylated MAba
| MAb | Competition with biotinylated MAb
|
||||||
|---|---|---|---|---|---|---|---|
| 242 | 338 | 659 | 757 | 836 | 1017 | 1025 | |
| 234 | − | + | − | − | − | − | − |
| 242 | + | − | + | − | + | − | − |
| 338 | + | + | − | − | + | − | − |
| 344 | + | − | + | − | + | − | − |
| 628 | + | + | − | − | + | − | − |
| 659 | − | − | + | − | − | − | − |
| 710 | + | − | + | − | + | − | − |
| 757 | − | − | − | + | − | − | − |
| 836 | + | − | + | − | + | − | − |
| 967 | − | − | − | − | − | − | + |
| 1017 | − | − | − | − | − | + | − |
| 1025 | − | − | − | − | − | − | + |
+, competition (as described in Materials and Methods); −, no competition.
FIG. 1.
Depiction of the epitopes recognized by the hMPV F protein-specific monoclonal antibodies. Each circle represents an individual epitope on hMPV F, with the MAb binding to that epitope shown inside the circle. MAb numbers inside the intersection of circles are those monoclonal antibodies that have recognition sites comprised of a portion of two epitopes.
Of all the hMPV F protein-specific monoclonal antibodies that were selected, antibodies 234 and 338 had the most potent virus-neutralizing activity across all four hMPV subgroups (Table 2). To determine their binding characteristics, Biacore analysis using immobilized soluble hMPV NL100 or NL199 F proteins (sFNL100 and sFNL199, respectively) was performed. The kon, koff, and Kd values derived from the Biacore analysis are shown in Table 4; antibodies 234 and 338 showed comparable on and off rates and nanomolar affinities for the F protein of both hMPV types.
TABLE 4.
Biacore determinations of binding constantsa
| MAb | Antigen | Immobilized RU | kon (105/Ms) | koff(10−4/s) | Kd (10−9 M) |
|---|---|---|---|---|---|
| 234 | sFNL199 | 80 | 1.92 | 4.63 | 2.41 |
| 234 | sFNL100 | 134 | 7.83 | 3.52 | 4.49 |
| 338 | sFNL199 | 80 | 1.92 | 2.72 | 1.42 |
| 338 | sFNL100 | 134 | 1.58 | 2.76 | 1.74 |
Binding constants were determined and calculated as described in Materials and Methods using the Biaevaluation software.
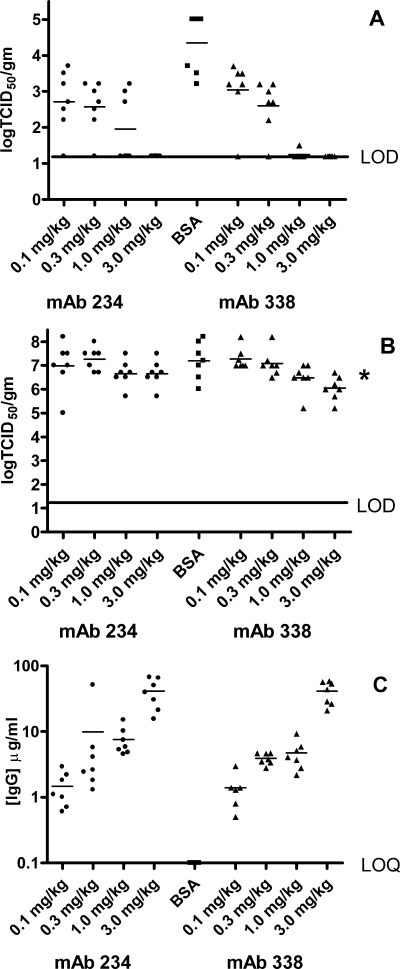
To examine further the ability of the antibodies to neutralize hMPV, MAbs 234 and 338 were tested in vivo in a prophylactic viral infection model using Golden Syrian hamsters as described previously (17). The MAbs were administered to the hamsters (7 animals/group) by intramuscular injection, 24 h prior to intranasal challenge with hMPV NL100 at a dose of 1 × 106 to 2 × 106 TCID50. Control animals received BSA instead of antibody. Animals were euthanized 4 days postchallenge, and the quantities of hMPV in the lungs and nasal turbinates of the animals were measured. Animals that had received either MAb 234 or 338 at doses greater than or equal to 3 mg/kg showed no detectable levels of virus in their lungs. To determine the minimum effective dose of the antibodies, a dose titration of each antibody between 0.1 and 3.0 mg/kg is shown in Fig. 2. At 3.0 mg/kg, both MAbs 338 and 234 gave rise to a minimum 3-log reduction (relative to the BSA control) in lung virus titer (Fig. 2, panel A). At 1 mg/kg, MAb 338 still caused a minimum 3-log reduction in lung virus titer, whereas MAb 234 caused an average 2-log reduction. Doses of 0.3 and 0.1 mg/kg resulted in higher levels of virus in the lungs; however, for these dose groups, the reductions in lung virus titers relative to the control were still statistically significant. Thus, antibodies 338 and 234 were able to decrease the viral burden in the lungs of animals at doses as low as 0.1 mg/kg. Prevention of viral replication in the upper airways was much less marked and only seen with higher doses of antibody. The log10 TCID50/gram of hMPV recovered from nasal turbinates of animals receiving intramuscular injection with MAbs 338 and 234 at doses between 3 and 0.1 mg/kg is shown in Fig. 2B. A reduction in viral titers relative to the controls was observed at doses of either antibody of 3.0 and 1.0 mg/kg but was statistically significant only for the 3-mg/kg dose of MAb 338.
FIG. 2.
In vivo protection against NL100 challenge. Golden Syrian hamsters were injected 24 h prior to intranasal challenge with NL100 with different does of MAb 234 (solid circles), MAb 338 (solid triangles), or with BSA (solid squares). Animals were bled prior to challenge to determine the levels of serum antibodies present at time of challenge. At 4 days postinfection, lungs (panel A) and nasal turbinates (panel B) were harvested, and virus titers were determined as described in Materials and Methods. The limit of detection (LOD) for the viral titers was 1.2 log/g tissue. For MAb quantification, animal serum samples were diluted 1:100 and 1:500 (panel C). The limit of the quantitation (LOQ) for this assay as performed was 0.1 μg MAb/ml serum. P values for MAb 234 were <0.0001, 0.0004, 0.0013, and 0.0042 for doses of 3 mg/kg, 1 mg/kg, 0.3 mg/kg, and 0.1 mg/kg, respectively. P values for MAb 338 were <0.0001, <0.0001, 0.0014, and 0.0145 for doses of 3.0 mg/kg, 1.0 mg/kg, 0.3 mg/kg, and 0.1 mg/kg, respectively. The nasal turbinate P value, indicated by an asterisk, was 0.008. IgG, immunoglobulin G.
Serum concentrations of the antibodies were determined 24 h following intramuscular administration (Fig. 2C). Serum samples were collected just prior to intranasal challenge with hMPV. As expected, higher doses of administered antibody resulted in higher concentrations of antibody measured in the serum of the animals. These data suggest that a serum concentration of MAb 338 between 5 and 10 μg/ml correlates with a minimum 3-log reduction in viral titer in the lungs of infected animals. MAb 234 appears to be slightly less potent at reducing virus in both the upper and lower airways, and a circulating concentration of >10 μg/ml of this antibody is required to decrease the lung viral titers to undetectable levels.
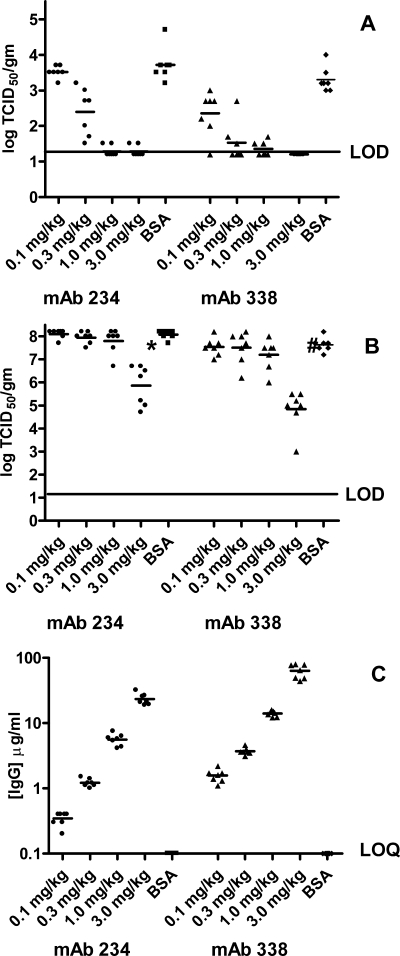
A similar set of experiments were performed using the B1 subgroup prototype virus. In separate experiments, Syrian hamsters received an intramuscular injection with either MAb 338 or MAb 234 24 h prior to nasal challenge with NL199. As in the previous experiment, control animals received BSA. The results of both experiments are plotted on the same axes in Fig. 3. The maximum level of hMPV NL199 recovered from the lungs of the control animals was lower than that obtained with NL100. Because of this, we were unable to observe a 3-log reduction with either MAb 234 or MAb 338 due to the limit of detection of the assay. Nevertheless, similar to what was seen with the NL100 virus, at doses of 3 mg/kg and 1 mg/kg, both antibodies reduced the lung viral titers by greater than 2 logs (Fig. 3A). Again, at a dose of 0.3 mg/kg, there was a statistically significant reduction in lung viral titer with both MAb 338 and MAb 234; however, at the 0.1-mg/kg dose, only MAb 338 showed a statistically significant reduction in lung viral titers. We noted a difference in the calculated serum concentrations of MAb 234 and MAb 338 in this experiment, with the concentration of MAb 338 being higher than that of MAb 234 at the same dose (Fig. 3C). However, taking these concentrations into consideration, there was a 2-log reduction in lung titers at a serum concentration of both MAbs similar to what was seen in the experiment with the NL100 virus.
FIG. 3.
In vivo protection against NL199 challenge. Golden Syrian hamsters were injected 24 h prior to intranasal challenge with NL199 with different doses of MAb 234 (solid circles), MAb 338 (solid triangles), or with BSA (solid squares). Animals were bled prior to challenge to determine the levels of serum antibodies present at time of challenge. At 4 days postinfection, lungs (panel A) and nasal turbinates (panel B) were harvested, and virus titers were determined as described in Materials and Methods. Limit of detection (LOD) for the viral titers was 1.2 log/g tissue. For MAb quantification, animal serum samples were diluted 1:100 and 1:500 (panel C). The limit of quantitation (LOQ) for this assay as performed was 0.1 μg MAb/ml serum. P values for MAb 234 were <0.0001, <0.0001, 0.0016, and 0.325 for doses of 3 mg/kg, 1 mg/kg, 0.3 mg/kg, and 0.1 mg/kg, respectively. P values for MAb 338 were <0.0001, <0.0001, <0.0001, and 0.0064 for doses of 3.0 mg/kg, 1.0 mg/kg, 0.3 mg/kg, and 0.1 mg/kg, respectively. Nasal turbinate P values of statistical significance were 0.0006 ( ) and <0.0001 (#). IgG, immunoglobulin G.
) and <0.0001 (#). IgG, immunoglobulin G.
The protection of the upper airway against the NL199 virus was more marked than that which was observed with the NL100 virus. Administration of MAb 338 and MAb 234 at the 3-mg/kg dose resulted in statistically significant decreases in the viral titers obtained from the nasal turbinates of mice challenged with NL199 (Fig. 3B); MAb 234 caused a 2-log reduction and MAb 338 gave a 2.5-log reduction. These data suggest that the antibodies present in the upper airways were more effective against the B type virus than the A type virus. This difference could possibly be correlated to the differences seen in vitro in IC50s for these antibodies (Table 2). The IC50s for MAb 338 are 10-fold lower with the B1 virus than with the A1 virus, and the values for MAb 234 differ by 50-fold when comparing A1 neutralization and B1 neutralization.
DISCUSSION
It has been established that immunization with a viral vector containing hMPV F protein can induce a potent neutralizing immune response that can protect against hMPV challenge (22, 24). These earlier studies demonstrate that an immune response, specific to the F protein alone, can neutralize virus in vivo in the absence of cellular or humoral immunity to other hMPV-specific antigens. However, neither of these papers demonstrated that the protection was solely due to the presence of neutralizing antibody. A recent paper has described the isolation of neutralizing monoclonal antibodies to hMPV that were obtained by immunization with hMPV-infected cells (16). These antibodies were reactive with the F protein of hMPV and neutralized virus in a PCR-based assay. However, purified antibody was not used in these experiments, so the potency of the MAb neutralization was not determined. In addition, this study did not look for in vivo protection. Thus, the studies so far have not shown that MAbs to F protein alone can protect animals from virus challenge.
In this report, we show that neutralizing monoclonal antibodies can be obtained from animals immunized with human metapneumovirus F protein and that these antibodies can protect cells from infection in vitro and protect animals from infection in vivo. We found that only a small number of antibodies cross-neutralized all 4 hMPV prototypic subgroups, even though the conservation of F protein sequence (94%) might have suggested that the majority of antibodies would be pan-neutralizing. Many of the antibodies that were isolated were not able to neutralize at least one of the 4 virus types with comparable potency, which suggests that the neutralizing epitopes may be in the regions of highest variability, presumably as a result of selective pressures.
A comparison of the differential abilities of the antibodies to neutralize the viral subgroups and the ability of the MAbs to cross-compete with each other for binding to the F protein, led us to identify 6 epitopes. Of these, we found 3 distinct nonoverlapping epitopes that are recognized by MAb 1017, MAb 757, and the MAb 967/MAb 1025 pair. The remaining 3 epitopes have antibodies that recognize one of two independent epitopes or recognize the overlap between these two epitopes (Fig. 1). We are generating monoclonal antibody-resistant mutants of the hMPV virus that will allow the determination of the precise location on the F protein sequence to which the MAbs bind.
Although we isolated only 3 MAbs that neutralized all 4 subgroup prototypes at an IC50 of <5 μg/ml, two of these antibodies, MAb 234 and MAb 338, have characteristics that make them appealing for further study. Both of these antibodies have properties that are similar to the properties of palivizumab, which is currently used for the prophylaxis of RSV infection in at risk infants (11, 31). A comparison of the neutralization and binding properties of the hMPV antibodies with the neutralization and binding properties of palivizumab to its RSV target are shown in Table 5. Both hMPV-specific antibodies, MAb 234 and MAb 338, show high-affinity binding to soluble F protein from both an A group and a B group sequence. Both MAb 234 and MAb 338 have kon rates of 2 × 105 to 8 × 105 M−1 s−1 against both types (A and B) of soluble F protein. These kon rates are comparable to the kon rate of palivizumab for soluble RSV F protein (1.2 × 105 M−1 s−1) (31). The koff rates of MAb 234 and MAb 338 are comparable to the koff of palivizumab against RSV F protein (7 × 10−4 s−1). The Kd values of the two anti-hMPV MAbs and palivizumab are less than 10 nM.
TABLE 5.
Comparison of anti-hMPV and anti-RSV monoclonals
| Antibodya | In vitro data
|
In vivo 2-log reduction data
|
|||||
|---|---|---|---|---|---|---|---|
| Virus type | kon(105 M−1 s−1) | koff(10−4 M−1) | Kd (nM) | IC50 neutralization (μg/ml) | Dose (mg/kg) | Concn of IgG in serum (μg/ml) | |
| MAb 234 | A | 7.83 | 3.52 | 4.49 | 1.2-0.6 | 1.0 | 8 |
| B | 1.92 | 4.63 | 2.41 | 0.2-0.01 | 1.0 | 6 | |
| MAb 338 | A | 1.58 | 2.76 | 1.74 | 0.8-0.4 | 1.0 | 5 |
| B | 1.92 | 2.72 | 1.42 | 0.2-0.03 | 0.3 | 4 | |
| Palivizumab | A | 1.27 | 4.3 | 3.39 | 0.453 | 2.5 | ∼30 |
| B | NDb | ND | ND | 0.06 | 2.5 | ∼30 | |
The in vitro neutralization capacity of MAb 234 and MAb 338 against the A subgroup viruses (IC50 between 1.2 and 0.4 μg/ml) was comparable to the IC50 of palivizumab against its A group virus (0.5 μg/ml). The neutralization seen with MAb 234 and MAb 338 against the B subgroup viruses is somewhat more potent, with an IC50 between 0.2 and 0.01 μg/ml for the neutralization of the B2 and B1 subgroups, respectively. The increased potency against B group isolates has also been seen with palivizumab. Remarkably, the in vivo potency of the hMPV monoclonal antibodies was comparable to that of palivizumab in reducing the viral load in the lungs of rodents. Further testing of the ability of MAb 234 and MAb 338 to neutralize a broader range of viral isolates and prevent virus-induced pathology is under way. Thus, humanization and optimization of MAbs 338 and 234 may result in viable clinical candidates with the ability to prevent lower respiratory tract disease that results from hMPV infection. Ultimately, this could extend our ability to protect those individuals at greatest risk from serious lung infections.
REFERENCES
- 1.Atkins, J. T., P. Karimi, B. H. Morris, G. McDavid, and S. Shim. 2000. Prophylaxis for respiratory syncytial virus with respiratory syncytial virus-immunoglobulin intravenous among preterm infants of thirty-two weeks gestation and less: reduction in incidence, severity of illness and cost. Pediatr. Infect. Dis. 19:138-143. [DOI] [PubMed] [Google Scholar]
- 2.Bebbington, C. R., G. Renner, S. Thomson, D. King, D. Abrams, and G. T. Yarranton. 1992. High level expression of a recombinant antibody from myeloma cells using a glutamine synthetase gene as an amplifiable selectable marker. Bio/Technology 10:169-175. [DOI] [PubMed] [Google Scholar]
- 3.Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., I. Mackay, T. P. Sloots, S. Madhi, F. Freymuth, D. Wolf, Y. Shemer-Avni, H. Ludewick, G. C. Gray, and E. LeBlanc. 2004. Global genetic diversity of human metapneumovirus fusion gene. Emerg. Infect. Dis. 10:1154-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chano, F., C. Rousseau, C. Lafarriere, M. Couillard, and H. Charest. 2005. Epidemiological survey of human metapneumovirus infection in a large pediatric tertiary care center. J. Clin. Microbiol. 43:552-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowe, J. E. 2004. Human metapneumovirus as a major cause of human respiratory tract disease. Pediatr. Infect. Dis. J. 23:S215-S221. [DOI] [PubMed] [Google Scholar]
- 7.de St Groth, F. S., and D. Scheidegger. 1980. Production of monoclonal antibodies: strategy and tactics. J. Immunol. Methods 35:1-21. [DOI] [PubMed] [Google Scholar]
- 8.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 9.The IMpact-RSV Study Group. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduced hospitalization from respiratory syncytial virus infection in high risk infants. Pediatrics 102:531-537. [PubMed] [Google Scholar]
- 10.Ison, M. G., and F. G. Hayden. 2002. Viral infections in immunocompromised patients: what's new with respiratory viruses? Curr. Opin. Infect. Dis. 15:355-367. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, S., C. Oliver, G. A. Prince, V. G. Hemming, D. S. Pfarr, S. C. Wang, M. Dormitzer, J. O'Grady, S. Koenig, J. K. Tamura, R. Woods, G. Bansal, D. Couchenour, E. Tsao, W. C. Hall, and J. F. Young. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215-1224. [DOI] [PubMed] [Google Scholar]
- 12.Johnsson, B., S. Lofas, and G. Lindquist. 1991. Immobilization of proteins to a carboxymethyldextran-modified gold surface for bispecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 198:268-277. [DOI] [PubMed] [Google Scholar]
- 13.Karber, G. 1931. 50% end-point calculation. Arch. Exp. Pathol. Pharmakol. 162:480-483. [Google Scholar]
- 14.Larcher, C., C. Geltner, H. Fischer, D. Nachbaur, L. C. Muller, and H. P. Huemer. 2005. Human metapneumovirus infection in lung transplant recipients: clinical presentation and epidemiology. J. Heart Lung Transplant. 24:1891-1901. [DOI] [PubMed] [Google Scholar]
- 15.Leung, J., F. Esper, C. Weibel, and J. S. Kahn. 2005. Seroepidemiology of human metapneumovirus (hMPV) on the basis of a novel enzyme-linked immunosorbent assay utilizing hMPV fusion protein expressed in recombinant vesicular stomatitis virus. J. Clin. Microbiol. 43:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, X., R. Endo, T. Ebihara, N. Ishiguro, H. Ishiko, and H. Kikuta. 2005. Production and characterization of neutralizing monoclonal antibodies against human metapneumovirus F protein. Hybridoma 24:201-205. [DOI] [PubMed] [Google Scholar]
- 17.MacPhail, M., J. H. Schickli, R. S. Tang, J. Kaur, C. Robinson, R. A. Fouchier, A. D. Osterhaus, R. R. Spaete, and A. A. Halle. 2004. Identification of small-animal and primate models for evaluation of vaccine candidates for human metapneumovirus (hMPV) and implications for hMPV vaccine design. J. Gen. Virol. 85:1655-1663. [DOI] [PubMed] [Google Scholar]
- 18.Martino, R., R. P. Porras, N. Rabella, J. V. Williams, E. Ramila, N. Margall, R. Labeaga, J. E. Crowe, Jr., P. Coll, and J. Sierra. 2005. Prospective study of the incidence, clinical features, and outcome of symptomatic upper and lower respiratory tract infections by respiratory viruses in adult recipients of hematopoietic stem cell transplants for hematologic malignancies. Biol. Blood Marrow Transplant. 11:781-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mejías, A., S. Chávez-Bueno, and O. Ramillo. 2004. Human metapneumovirus: a not so new virus. Pediatr. Infect. Dis. J. 23:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Robinson, J. L., B. E. Lee, N. Bastein, and Y. Li. 2005. Seasonality and clinical features of human metapneumovirus infection in children in northern Alberta. J. Med. Virol. 76:98-105. [DOI] [PubMed] [Google Scholar]
- 21.Sinaniotis, C. A. 2004. Viral pneumoniae in children: incidence and aetiology. Paediatr. Respir. Rev. 5:S197-S200. [DOI] [PubMed] [Google Scholar]
- 22.Skiadopoulos, M. H., S. Biacchesi, U. J. Buchholz, J. M. Riggs, S. R. Surman, E. Amaro-Carambot, J. M. McAuliffe, W. R. Elkins, M. St. Claire, P. L. Collins, and B. R. Murphy. 2004. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J. Virol. 78:6927-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorrentino, M., T. Powers, et al. 2000. Effectiveness of palivizumab: evaluation of outcomes from the 1998 to 1999 respiratory syncytial virus season. Pediatr. Infect. Dis. J. 19:1068-1071. [DOI] [PubMed] [Google Scholar]
- 24.Tang, R. S., J. H. Schickli, M. MacPhail, F. Fernandes, L. Bicha, J. Spaete, R. A. Fouchier, A. D. Osterhaus, R. Spaete, and A. A. Haller. 2003. Effects of human metapneumovirus and respiratory syncytial virus antigen insertion in two 3′ proximal genome positions of bovine/human parainfluenza virus type 3 on virus replication and immunogenicity. J. Virol. 77:10819-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulloa-Gutierrez, R., P. Skippen, A. Synnes, M. Seear, N. Bastein, Y. Li, and J. C. Forbes. 2004. Life-threatening human metapneumovirus pneumonia requiring extracorporeal membrane oxygenation in a preterm infant. Pediatrics 114:517-519. [DOI] [PubMed] [Google Scholar]
- 26.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 28.van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo-Neto, R. L. de Swart, A. D. Osterhaus, and R. A. Fouchier. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10:658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welliver, R. C. 2003. Review of epidemiology and clinical risk factors for severe respiratory syncytial virus (RSV) infection. J. Pediatr. 143:S112-S117. [DOI] [PubMed] [Google Scholar]
- 30.Williams, J. V., P. A. Harris, S. J. Tollefson, L. L. Halburnt-Rush, J. Pingsterhaus,K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, H., D. S. Pfarr, Y. Tang, L. L. An, N. K. Patel, J. D. Watkins, W. D. Huse, P. A. Kiener, and J. F. Young. 2005. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J. Mol. Biol. 350:126-144. [DOI] [PubMed] [Google Scholar]
- 32.Wyde, P. R., S. N. Chetty, A. M. Jewell, G. Boivin, and P. A. Piedra. 2003. Comparison of the inhibition of human metapneumovirus and respiratory syncytial virus by ribavirin and immune serum globulin in vitro. Antivir. Res. 60:51-59. [DOI] [PubMed] [Google Scholar]