Abstract
The Epstein-Barr virus (EBV) is an oncogenic human herpesvirus. EBV latent membrane protein 1 (LMP-1) is a viral oncogene that manifests its oncogenic phenotype through activation of cellular signaling pathways involved in cell growth, survival, differentiation, and transformation. Lytic LMP-1 (lyLMP-1) is a related EBV gene without oncogenic properties. The lyLMP-1 gene is found in 60% of the EBV strains circulating in nature, but it is not found in EBV strains associated with nasopharyngeal carcinoma. We recently demonstrated that lyLMP-1 down-regulates the half-life of LMP-1 in epithelial cells. Therefore in this study, we tested the hypothesis that lyLMP-1 concomitantly down-regulates LMP-1 oncogenic activity. The results demonstrated that lyLMP-1 inhibits LMP-1-mediated intracellular signaling activation, epithelial cell growth and survival, and fibroblast cell transformation in a dose-dependent manner. Lytic LMP-1 manifested this effect through the promotion of LMP-1 degradation and a reduction in the expressed quantity of LMP-1. Thus, lyLMP-1 functions as a posttranslational negative regulator of LMP-1 oncogenesis. These results support a model of EBV-associated epithelial oncogenesis in which lyLMP-1 may act in vivo to reduce the risk of LMP-1-mediated transformation and is therefore subjected to negative selection in nasopharyngeal carcinoma pathogenesis.
Epstein-Barr virus (EBV) is an oncogenic human herpesvirus associated with a broad spectrum of benign and malignant diseases, including infectious mononucleosis, oral hairy leukoplakia, African Burkitt's lymphoma, Hodgkin's disease lymphoma, lymphoproliferative disorders of immunocompromised hosts, and nasopharyngeal carcinoma. The EBV latent membrane protein 1 (LMP-1) is a viral oncogene (30) that is believed to be important in the pathogenesis of many EBV-associated diseases, including nasopharyngeal carcinoma (25). In the B958 EBV strain, LMP-1 is a 63-kDa protein of 386 amino acids encoded by the BNLF1 gene. LMP-1 localizes to cellular membranes and functions as a constitutively active tumor necrosis factor receptor homologue that propagates intracellular signaling, including the NF-κB, cJun N-terminal protein kinase/AP-1, and Janus kinase/STAT pathways (5, 12, 15, 27). Through these signaling pathways, LMP-1 generates a myriad of effects on host cell growth, differentiation, and apoptosis, including growth promotion and survival in epithelial cells (4, 11, 16, 23, 32) and transformation of rodent fibroblasts (3, 30). Although the mechanisms by which LMP-1 influences cell biology have been intensively studied, little is known about the mechanisms that regulate LMP-1 oncogenic activity.
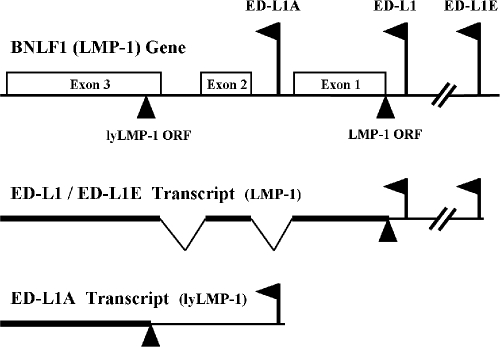
Some EBV strains also encode an amino-terminally truncated form of LMP-1 called lytic LMP-1 (lyLMP-1). Transcription of lyLMP-1 is driven by the ED-L1A promoter that is located in the first intron of the LMP-1 gene and that is present in all EBV strains (6, 17, 29) (Fig. 1). However, the presence or absence of the lyLMP-1 open reading frame (ORF) is determined by the sequence at LMP-1 gene codon number 129 in the third exon: ATG (methionine) serves as the lyLMP-1 initiator, whereas ATT (isoleucine) prohibits translation of lyLMP-1 from the ED-L1A transcript (6). Molecular epidemiologic studies demonstrate that the lyLMP-1 ORF is present in 60% of EBV strains that circulate in nature (6) but consistently absent from the LMP-1 genes found in nasopharyngeal carcinoma (6, 14, 19), despite apparent transcriptional activity of the ED-L1A promoter in nasopharyngeal carcinoma (20). These observations suggest that selective pressures act in favor of LMP-1 expression, but against lyLMP-1 expression, in the pathogenesis of nasopharyngeal carcinoma.
FIG. 1.
LMP-1 promoters and gene expression. The ED-L1 and ED-L1E promoters are located upstream of the LMP-1 ORF. Transcripts from either of these two promoters express complete LMP-1. The ED-L1A promoter is located in the first intron of the LMP-1 gene. Transcripts from the ED-L1A promoter express lyLMP-1 only when LMP-1 gene codon number 129 encodes ATG (methionine) and creates the lyLMP-1 ORF.
The biological function of lyLMP-1 is largely unknown. Unlike LMP-1, lyLMP-1 neither activates signaling nor transforms cells, and lyLMP-1 may even negatively regulate some LMP-1 signaling activity (7, 31). Recently, we demonstrated that expression of lyLMP-1 significantly down-regulates the half-life of LMP-1 in a dose-dependent manner in epithelial cells (24). In this study, we tested the hypothesis that lyLMP-1 possesses the ability to down-regulate a broad range of LMP-1 signaling activities and the associated oncogenic cell phenotype induced by LMP-1 signaling. We also present evidence suggesting that lyLMP-1 down-regulates LMP-1 oncogenic activity by promoting LMP-1 degradation and thereby reducing the quantity of expressed LMP-1. Finally, we discuss the clinical implications of these data for the pathogenesis of nasopharyngeal carcinoma.
MATERIALS AND METHODS
LMP-1 and lyLMP-1 expression constructs.
Three different expression vectors were constructed in the pSG5 plasmid (Stratagene, La Jolla, Calif.) encoding the simian virus 40 promoter, as previously described (24) and briefly characterized as follows. (i) Clone B958WT encodes the LMP-1 ORF from the B958 EBV strain. Clone B958WT encodes ATG (methionine) at codon 129 and intrinsically expresses lyLMP-1 in addition to LMP-1. (ii) Clone K is identical to clone B958WT in LMP-1 ORF sequence with the exception of a single nucleotide mutation at codon 129 (ATG to ATT, methionine to isoleucine) that ablates the lyLMP-1 ORF. Clone K expresses only LMP-1 and is incapable of expressing lyLMP-1. (iii) Clone lyLMP-1 encodes the lyLMP-1 ORF derived from the B958 EBV strain and expresses only lyLMP-1.
Two additional fluorescent fusion protein expression vectors were constructed for LMP-1 and lyLMP-1. The LMP-1 coding sequence of clone K was subcloned into the phrGFPII-1 plasmid (Stratagene, La Jolla, Calif.). This clone, called LMP-1(K)-GFP, expresses the humanized recombinant green fluorescent protein (GFP; variant II) of Renilla reniformis fused to the carboxy terminus of LMP-1. The lyLMP-1 coding sequence of clone lyLMP-1 was subcloned into the pDsRed-Monomer-Hyg-N1 plasmid (BD Biosciences-Clontech, Mountain View, Calif.). This clone, called lyLMP-1-DsRed, expresses the monomeric mutant of the Discosoma spp. red fluorescent protein fused to the carboxy terminus of lyLMP-1.
Cell lines, transient transfection, and protein expression.
RHEK-1 is a nontumorigenic immortalized normal human keratinocyte cell line (26). BALB/3T3 (clone A31; American Type Culture Collection, Manassas, Va.) is a rodent fibroblast cell line (1). Both cell lines have previously been utilized to study the oncogenic and transforming properties of LMP-1 (3, 8, 16, 30, 32). Both cell lines were maintained in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% fetal calf serum and 2 millimolar glutamine and incubated at 37°C with 5% CO2. LMP-1 and lyLMP-1 plasmid expression constructs were introduced into both cell lines by electroporation at 260 V and 975 μF using a Gene Pulser II electroporation system (Bio-Rad Laboratories, Hercules, Calif.). Expression of LMP-1 and lyLMP-1 was detected by Western blotting using the S12 monoclonal antibody (BD Biosciences-Pharmingen, San Diego, Calif.). Western blot band intensity was measured by spot densitometry (AlphaImager system; Alpha Innotech Corporation, San Leandro, Calif.) as the mean pixel value enclosed in the band after background correction, and protein expression data were recorded as numeric values of densitometry units. Expression of LMP-1 and lyLMP-1 fluorescent fusion proteins was detected and analyzed by flow cytometry (FACSort flow cytometer with CellQuest software; Becton-Dickinson, Franklin Lakes, N.J.), and protein expression data were recorded as numeric values of fluorescence intensity units.
LMP-1 signaling activation reporter assays.
A Mercury pathway profiling system (BD Biosciences-Clontech, Mountain View, Calif.) and a Dual-Luciferase reporter assay system (Promega, Madison, Wis.) were used for LMP-1 signaling activation studies. RHEK-1 cells (2 × 105) were transiently cotransfected with (i) 1 μg of a reporter plasmid encoding the firefly luciferase gene driven by a specific cis-acting DNA enhancer element responsive to either NF-κB, AP-1, STAT1 homodimer, STAT1/STAT2 heterodimer, or STAT3 homodimer; (ii) 0.5 μg of an internal control plasmid encoding the Renilla luciferase gene; and (iii) 4 to 40 μg of clones B958WT, K, and/or lyLMP-1. After 6 h of incubation in complete medium, cells were fed with fresh medium containing 5% fetal calf serum (serum starvation). At 24 h after transfection, cells were washed with phosphate-buffered saline and lysed in reporter lysis buffer. Firefly and Renilla luciferase activities were measured in 20 μl of cleared lysate with a TD-20/20 luminometer (Promega, Madison, Wis.). Renilla luciferase values were used to normalize the firefly luciferase results as a control for transfection efficiency between experiments. All experiments were performed in triplicate, and mean values and standard deviations were calculated. The t test was used to evaluate differences between means, and P values less than 0.050 were regarded as statistically significant.
LMP-1 cell growth and survival assay.
RHEK-1 cells (2 × 105) were harvested in the exponential growth phase, transiently transfected with 4 to 40 μg of clones B958WT, K, and/or lyLMP-1, and transferred into a culture dish with fresh media containing either 10%, 1%, or 0.1% fetal calf serum. The cells were fed with fresh growth media twice weekly. At set time points over a 14-day period, the cells were trypsinized, and the viable cells (trypan blue exclusion method) were counted using a hemacytometer. Cell density did not reach confluence for any of the cultures over the 14-day period of the experiment. All experiments were performed in duplicate or triplicate, and mean values and standard deviations were calculated. Different mean comparator values with nonoverlapping standard deviations were regarded as statistically significant.
LMP-1 cell transformation assay.
Transformation of BALB/3T3 cells by LMP-1 was assayed by anchorage-independent colony formation in soft agar. The culture base layer was prepared as DMEM with 0.5% low-melting-temperature agarose. BALB/3T3 cells were transiently transfected with 4 to 40 μg of clones B958WT, K, and/or lyLMP-1. The culture feed layer was prepared by resuspending 2 × 105 transfected cells per milliliter in DMEM with 0.25% low-melting-temperature agarose and then pouring it onto the previously prepared base layer. The agarose was set by placing the culture plates at 4°C for 20 min before incubation. Cells were fed with fresh growth media twice weekly until colonies grew to a suitable size for observation (2 to 3 weeks). Colonies larger than 100 μm in diameter were considered to be evidence of cell transformation and were counted by light microscopy. All experiments were performed in triplicate, and mean values and standard deviations were calculated. The t test was used to evaluate differences between means, and P values less than 0.050 were regarded as statistically significant.
RESULTS
Lytic LMP-1 inhibits LMP-1 activation of responsive promoters.
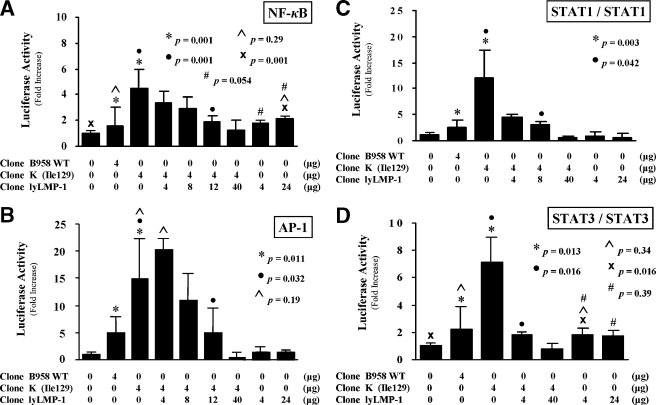
LMP-1 constitutively activates multiple signaling pathways (5, 12, 15, 27). In this study, LMP-1-transfected RHEK-1 epithelial cells were analyzed for their ability to induce luciferase expression from responsive promoters. LMP-1 expressed from clone K stimulated NF-κB-, AP-1-, STAT1 homodimer-, and STAT3 homodimer-responsive promoters to a significantly greater degree than did LMP-1 expressed from clone B958WT (Fig. 2A to D). Consistent with previous data on STAT activation by LMP-1 (12, 15), the STAT1/STAT2 heterodimer-responsive promoter was not activated by LMP-1 expressed from either clone (data not shown). These results suggest that the absence of lyLMP-1 enhances the ability of LMP-1 to activate cellular signaling pathways.
FIG. 2.
Effect of lyLMP-1 on LMP-1-mediated signaling activity. Luciferase reporter assays in standardized quantities of transfected RHEK-1 epithelial cells were used to study LMP-1 and lyLMP-1 activation of specific promoters responsive to (A) NF-κB, (B) AP-1, (C) STAT-1 homodimer, and (D) STAT3 homodimer. The results are presented as bar graphs representing the mean (with standard deviation) n-fold increase in luciferase activity relative to nontransfected cells. P values are shown for comparative bars as indicated by matching symbols. The quantities of the LMP-1 and lyLMP-1 expression plasmids transfected or cotransfected for each experiment are shown below each bar of the graph. Taken together, these results indicate that clone K activates LMP-1-mediated signaling activity better than clone B958WT in epithelial cells and that lyLMP-1 specifically down-regulates clone K LMP-1-mediated signaling activity in a dose-dependent manner.
Cotransfection of clone lyLMP-1 with clone K resulted in a significant dose-dependent inhibition of LMP-1-mediated activation of each of four responsive promoters, restoring a low-signaling phenotype similar to that of clone B958WT (Fig. 2A to D). Cotransfection of clone lyLMP-1 with clone K at a plasmid quantity ratio of 10:1 suppressed LMP-1 signaling activation to levels even below those of clone B958WT and similar to those of the nontransfected cells (Fig. 2A to D). Transfection of clone lyLMP-1 alone demonstrated low-level but statistically significant activation of the NF-κB- and STAT3 homodimer-responsive promoters but not the AP-1- and STAT1 homodimer-responsive promoters (Fig. 2A to D). This activation was similar to that of clone B958WT, was not dose responsive, and was interpreted to be nonspecific and biologically insignificant. These results indicate that lyLMP-1 inhibits LMP-1-mediated NF-κB, AP-1, and STAT signaling activity in epithelial cells in a dose-dependent manner.
Lytic LMP-1 inhibits LMP-1-mediated cell growth and survival.
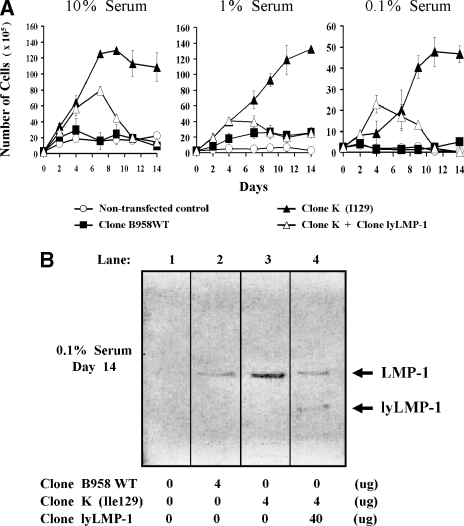
LMP-1 promotes epithelial cell growth and survival (4, 11, 16, 23, 32). In this study, LMP-1-transfected RHEK-1 epithelial cells were analyzed for growth and survival in culture media with different serum concentrations. Cells transfected with clone K continued in logarithmic growth phase and survived for longer periods of time than did cells transfected with clone B958WT and nontransfected cells (Fig. 3A). The effect of clone K LMP-1 was apparent at all three serum concentrations but was especially evident at the lowest serum concentration (0.1%). These results suggest that the absence of lyLMP-1 enhances the ability of LMP-1 to promote cell growth and survival.
FIG. 3.
Effect of lyLMP-1 on LMP-1-mediated cell growth and survival. A cell growth and survival assay using culture media with different serum concentrations was used to study the effect of LMP-1 in transfected RHEK-1 epithelial cells. (A) The results are presented as the mean (with standard deviation) number of cells present in culture at each time point over a 14-day period. Note the different scale on the vertical axis of the 0.1% serum graph, illustrating that the magnitude of the clone K effect is relatively greater here than for the higher serum concentrations. Four micrograms each of clone B958WT plasmid and clone K plasmid were used for transfection. Forty micrograms of clone lyLMP-1 plasmid was cotransfected with four micrograms of clone K plasmid (ratio = 10:1) to ensure expression levels of lyLMP-1 that would achieve a detectable inhibitory effect on LMP-1. (B) A Western blot illustrates that both LMP-1 and lyLMP-1 were expressed in the transfected cells for the duration of the 14-day experiment. All available cells at the 14-day time point for each of the four transfection cultures were harvested, lysed, and loaded into the designated lane. Taken together, these results indicate that clone K confers a greater growth and survival advantage to epithelial cells than does clone B958WT at all three serum concentrations and that lyLMP-1 completely reverses the growth and survival advantage of clone K.
The growth and survival advantage of clone K LMP-1 was completely reversed by cotransfection of clone lyLMP-1 at a 10:1 lyLMP-1-to-LMP-1 plasmid quantity ratio at all three serum concentrations (Fig. 3A). Interestingly, this inhibitory effect became evident only after 4 to 7 days, reaching its maximum by 11 to 14 days (Fig. 3A). Western blotting demonstrated both LMP-1 and lyLMP-1 expression at 14 days (Fig. 3B), indicating that the reversal of the cell growth and survival effect was not due to a loss of transient LMP-1 expression. It was shown by the luciferase promoter studies that LMP-1 signaling activation is significantly inhibited by a 10:1 lyLMP-1-to-LMP-1 plasmid quantity ratio within 24 h of cotransfection (Fig. 2A to D). Thus, the delayed onset of the inhibitory effect of lyLMP-1 on cell growth and survival may represent the time to reverse the oncogenic cell phenotype induced by LMP-1 signaling that may have occurred briefly after transfection and before lyLMP-1 could exert its down-regulatory effect on LMP-1. These results indicate that lyLMP-1 inhibits the ability of LMP-1 to promote epithelial cell growth and survival.
Lytic LMP-1 inhibits LMP-1-mediated cell transformation.
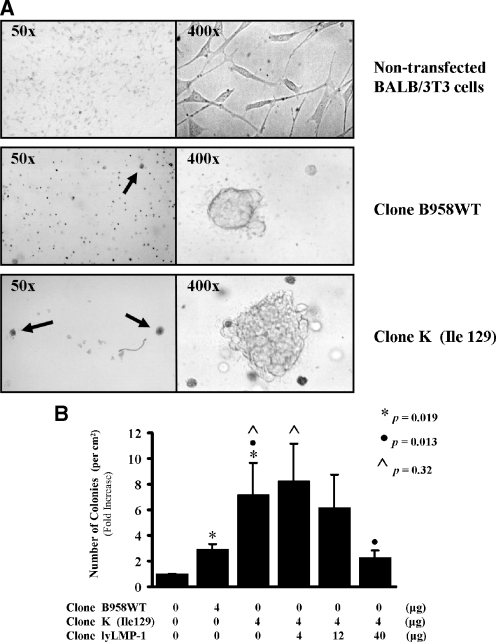
LMP-1 is recognized as an oncogene for its ability to transform rodent fibroblast cells, as defined by colony formation and anchorage-independent growth in soft agar (3, 30). In this study, LMP-1-transfected BALB/3T3 rodent fibroblast cells were analyzed for induction of the transformed phenotype. Cells transfected with clone K formed colonies in soft agar in greater numbers and of greater size than did cells transfected with clone B958WT and nontransfected cells (Fig. 4). These results suggest that the absence of lyLMP-1 enhances the ability of LMP-1 to transform BALB/3T3 cells. Cotransfection of clone lyLMP-1 with clone K significantly reduced colony formation in a dose-dependent manner, restoring a less-transforming phenotype similar to that of clone B958WT (Fig. 4B). These results indicate that lyLMP-1 inhibits LMP-1-mediated transformation of rodent fibroblast cells.
FIG. 4.
Effect of lyLMP-1 on LMP-1-mediated cell transformation. BALB/3T3 rodent fibroblast cells were used to study the transformation efficiency of LMP-1, as defined by colony formation and anchorage-independent growth in soft agar. (A) Colony formation and growth is illustrated with representative photomicrographs for cultures of nontransfected cells and for cells transfected with clone B958WT or clone K. The 50× magnification photomicrographs illustrate that colony formation (indicated by an arrow) was commonly detected in LMP-1-transfected cells but rarely detected in nontransfected cells. The 400× magnification photomicrographs illustrate that colonies of the clone K-transfected cells were generally larger in size than colonies of clone B958WT-transfected cells. The nontransfected cells retained their diffuse single-layer, spindle-shaped morphology. (B) Colony count results are presented as bar graphs representing the mean (with standard deviation) n-fold increase in the number of colonies per unit area relative to nontransfected cells. P values are shown for comparative bars as indicated by matching symbols. The quantities of the LMP-1 and lyLMP-1 expression plasmids transfected or cotransfected for each experiment are shown below each bar of the graph. Taken together, these results indicate that clone K transforms rodent fibroblast cells more efficiently than does clone B958WT and that lyLMP-1 inhibits LMP-1-mediated transformation in a dose-dependent manner.
Lytic LMP-1 reduces the quantity of expressed LMP-1.
The inhibition of LMP-1 oncogenic activity by lyLMP-1 occurred in a parallel manner across several different functional assays, suggesting that lyLMP-1 acts upon LMP-1 at a proximal level rather than upon each LMP-1 effector pathway individually. The known ability of lyLMP-1 to down-regulate LMP-1 half-life and promote LMP-1 degradation (24) would constitute such a proximal effect and would predict that lyLMP-1 down-regulates LMP-1 oncogenic activity by reducing the quantity of expressed LMP-1.
This potential lyLMP-1 effect on LMP-1 quantity was initially investigated by Western blot and quantitative densitometry of lyLMP-1- and LMP-1-transfected RHEK-1 epithelial cells. It was confirmed that lyLMP-1 is intrinsically expressed from clone B958WT but not from clone K (Fig. 5). Cotransfection of clone lyLMP-1 with clone K at a plasmid quantity ratio of 1:1 demonstrated lyLMP-1 expression without appreciable change in the expressed LMP-1 quantity (Fig. 5). However, an lyLMP-1-to-LMP-1 plasmid quantity ratio of 10:1 reduced the total expressed LMP-1 quantity by over fourfold (Fig. 5). The specificity of this lyLMP-1 effect was shown by cotransfecting an EBNA-2-expressing plasmid with LMP-1 at a 10:1 plasmid quantity ratio, which resulted in an insignificant change in the expressed LMP-1 quantity (Fig. 5). While these Western blot results generally supported the hypothesis that lyLMP-1 reduces expressed LMP-1 quantity, a few inconsistencies were noted at the lower lyLMP-1-to-LMP-1 plasmid quantity ratios between the down-regulatory effects in the signal activation and transformation assays and the relative quantities of protein expression detected by Western blotting.
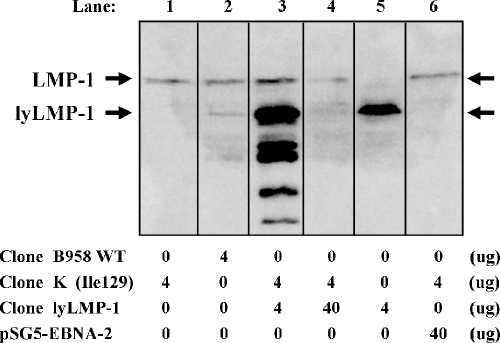
FIG. 5.
Effect of lyLMP-1 on expressed LMP-1 quantity detected by Western blotting. Western blotting illustrates the quantities of LMP-1 and lyLMP-1 expressed at 24 h after RHEK-1 cell transfection with the quantities of the plasmids indicated below each lane. Equal numbers of cells were transfected, lysed (including both adherent and nonadherent cells), and loaded for each lane. Densitometry quantitations of the LMP-1 and lyLMP-1 bands were performed. The total expressed LMP-1 quantity was reduced by more than fourfold when clone lyLMP-1 was cotransfected with clone K at a 10:1 plasmid quantity ratio (lane 4 LMP-1 = 2.2 versus lane 1 LMP-1 = 9.7). Cotransfection of a pSG5-based control clone expressing the Epstein-Barr virus nuclear antigen-2 gene (pSG5-EBNA-2) with clone K at a 10:1 plasmid quantity ratio did not have a significant effect on LMP-1 quantity (lane 6 LMP-1 = 8.6 versus lane 1 LMP-1 = 9.7), indicating that nonspecific effects cannot explain this reduction in LMP-1 quantity. The greater than threefold reduction in lyLMP-1 quantity despite a 10-fold increase in transfected plasmid quantity (lane 4 lyLMP-1 = 12.9 versus lane 5 lyLMP-1 = 41.4) suggests that the interaction between LMP-1 and lyLMP-1 may accelerate the degradation of lyLMP-1 as well. Taken together, these results generally support the hypothesis that lyLMP-1 coexpression with LMP-1 reduces the expressed quantity of LMP-1 in RHEK-1 epithelial cells.
Under transient cotransfection conditions, expressed protein quantities are determined not only by the quantity of each plasmid transfected and its relative transfection efficiency but also by the relative rates of synthesis and degradation for each protein. It is especially difficult to determine the stoichiometry of a protein-protein interaction when one of the coexpressed proteins acts to promote the degradation of the other, as lyLMP-1 does to LMP-1 (24) and as LMP-1 may do to lyLMP-1 (Fig. 5). Given the inherent variability in plasmid cotransfection efficiencies, the reciprocal effects that lyLMP-1 and LMP-1 exert upon each other, and the relative insensitivity of Western blot densitometry to distinguish between small changes in protein quantity, we sought a more sensitive and accurate method to quantify changes in LMP-1 expression in these transient cotransfection experiments.
The question of an lyLMP-1 effect on LMP-1 quantity was further investigated by transient transfection and expression of red and green fluorescent fusion proteins of lyLMP-1 and LMP-1 in RHEK-1 epithelial cells, with quantitative detection of protein expression by two-color flow cytometry. It has previously been demonstrated that green fluorescent protein fusions to the carboxy terminus of LMP-1 do not influence LMP-1 stability, signaling, proliferation, transformation, or tumorigenicity (28). Consequently, the phenotypes of the red and green fluorescent fusion proteins of lyLMP-1 and LMP-1 are expected to be identical to those of clones lyLMP-1 and K, respectively. Unlike Western blotting, which detects only total protein expression across an entire cell population, this flow cytometry approach has the ability to quantify variations in lyLMP-1 and LMP-1 expression at the individual cell level.
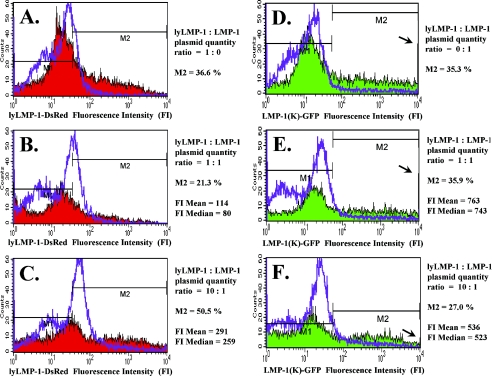
Transfection of lyLMP-1 or LMP-1 alone yielded similar transfection efficiency rates (Fig. 6A and D) (M2 = 36.6% and 35.3%, respectively). Notably, there was extremely wide cell-to-cell variation in the quantity of each protein expressed, especially for LMP-1 (Fig. 6D) (range from 5 × 101 to greater than 104 fluorescent intensity units). Compared to transfection of the same plasmid quantity of each clone alone, cotransfection of lyLMP-1 with LMP-1 at a 1:1 plasmid quantity ratio resulted in a reduced transfection efficiency specifically for clone lyLMP-1 (Fig. 6A and B) (M2 from 36.6% to 21.3%) and generally lower levels of expression for both lyLMP-1 (Fig. 6A and B) and LMP-1 (Fig. 6D and E). Compared to a 1:1 plasmid quantity ratio, cotransfection of lyLMP-1 with LMP-1 at a 10:1 ratio resulted in an increase in total lyLMP-1 expression (Fig. 6B and C and Table 1) and a decrease in total LMP-1 expression. Specifically, the decreased LMP-1 expression was evidenced by (i) a decrease in the number of cells expressing high levels of LMP-1 (Fig. 6E and F, arrows), (ii) a decrease in the number of cells expressing any LMP-1 (Fig. 6E and F) (M2 from 35.9% to 27.0%), (iii) a decrease in the number of cells expressing only LMP-1 (Table 1) (from 25.7% to 3.1%), and (iv) a decrease in both the mean and median values for expressed LMP-1 fluorescence intensity (Fig. 6E and F) (mean from 763 to 536, and median from 743 to 523), reflecting a reduction in the total amount of LMP-1 expressed among all transfected cells. Compared to a 1:1 plasmid quantity ratio, cotransfection of lyLMP-1 with LMP-1 at a 10:1 plasmid quantity ratio also resulted in an increase in the number of cells coexpressing both lyLMP-1 and LMP-1 (Table 1) (from 10.2% to 23.9%). Thus, the results of these flow cytometry analyses greatly extend the original Western blot densitometry results and strongly support the hypothesis that lyLMP-1 down-regulates LMP-1 oncogenic activity by promoting LMP-1 degradation and reducing the quantity of expressed LMP-1 in epithelial cells.
FIG.6.
Effect of lyLMP-1 on expressed LMP-1 quantity detected by flow cytometry. Flow cytometry results of RHEK-1 cells harvested 24 h after transfection with clone lyLMP-1-DsRed and/or clone LMP-1(K)-GFP at the indicated plasmid quantity ratio (0 = 0 μg; 1 = 4 μg; 10 = 40 μg) are shown as histogram plots of the number of cells (counts; vertical axis) versus the quantitative level of lyLMP-1 or LMP-1 fluorescent fusion protein expression as measured in log-scale fluorescence intensity units (horizontal axis). Red and green shaded areas represent protein expression data for the adherent (viable) cell population. Purple lines represent protein expression data for the nonadherent (nonviable) cell population and demonstrate that comparatively little lyLMP-1 or LMP-1 is expressed in these cells. M2, mean, and median fluorescence values were derived only from the protein expression data illustrated by the shaded areas (adherent, viable cells). (A to C) Red fluorescence data representing lyLMP-1 expression from clone lyLMP-1-DsRed. (D to F) Green fluorescence data representing LMP-1 expression from clone LMP-1(K)-GFP. The arrows draw attention to tall bars representing the presence of a number of cells expressing greater than 104 fluorescence intensity units (off scale), indicating very high levels of LMP-1 expression in those cells. The change in height in the bars between panel E and panel F indicates a decrease in the number of cells expressing very high levels of LMP-1 with the 10:1 compared to the 1:1 plasmid quantity ratio. Taken together, these flow cytometry results strongly support the hypothesis that lyLMP-1 coexpression with LMP-1 reduces the expressed quantity of LMP-1 in RHEK-1 epithelial cells.
TABLE 1.
Expression of lyLMP-1 and LMP-1 fluorescent fusion proteins detected by flow cytometry in adherent RHEK-1 cells at 24 h after cotransfection
| lyLMP-1:LMP-1a | Fluorescent protein expression (% of cells)
|
|||
|---|---|---|---|---|
| Double negative, no expression | Single positive, lyLMP-1 (red) | Single positive, LMP-1 (green) | Double positive, lyLMP-1 (red) and LMP-1 (green) | |
| 1:1 | 59.1 | 5.0 | 25.7 | 10.2 |
| 10:1 | 55.1 | 18.0 | 3.1 | 23.9 |
Cotransfected plasmid quantity ratio (red:green).
DISCUSSION
The results of this study strongly support our hypothesis that lyLMP-1 down-regulates LMP-1 oncogenic activity in human epithelial and rodent fibroblast cells. We observed statistically highly significant inhibitory effects of lyLMP-1 on LMP-1 using standard assays that are widely accepted as in vitro measures of LMP-1 oncogenic activity, including (i) LMP-1 signaling through the NF-κB, cJun N-terminal protein kinase/AP-1, and Janus kinase/STAT pathways, (ii) LMP-1 promotion of cell growth and survival, and (iii) LMP-1 transformation as measured by the induction of anchorage-independent cell growth and colony formation in soft agar. In each of these functional assays, a single nucleotide mutation and ablation of intrinsic lyLMP-1 expression greatly enhanced the oncogenic properties of LMP-1, while the less-oncogenic LMP-1 phenotype was restored to the mutated LMP-1 gene by ectopic coexpression of lyLMP-1. Thus, lyLMP-1 appears to function as a posttranslational negative regulator of LMP-1 oncogenesis.
The results of this study also lend strong support to the hypothesis that lyLMP-1 exerts its down-regulatory effect on LMP-1 by promoting LMP-1 degradation and thereby reducing the quantity of LMP-1 available to generate oncogenic signals. In this study, we demonstrated a dose-dependent reduction in LMP-1 oncogenic activity by lyLMP-1 that mirrors the previously demonstrated dose-dependent reduction in LMP-1 half-life by lyLMP-1 (24). More importantly, we demonstrated a reduction in expressed LMP-1 quantity by lyLMP-1. Our results are consistent with previous studies noting possible correlations between LMP-1 quantity and phenotype (3, 9, 10, 13, 16, 18, 30, 31) and between LMP-1 turnover rate and signal activation (21, 22, 28). However, our demonstration that lyLMP-1 induces parallel quantitative changes in both LMP-1 expression and activity greatly expands upon the previous evidence and supports a strong correlation between expressed LMP-1 quantity and the oncogenic LMP-1 phenotype.
The results of this study have important technical implications for future studies of the lyLMP-1 interaction with LMP-1. The flow cytometry analyses of the transient cotransfections clearly illustrate the dual phenomena of variable cotransfection efficiency and reciprocal protein stability effects. It is important to understand that increasing the cotransfected plasmid quantity ratio from 1:1 to 10:1 did not necessarily increase the quantity of lyLMP-1 expressed in each individual cell, but it did significantly increase the number of cells in which lyLMP-1 was coexpressed with LMP-1. These cells coexpressing both lyLMP-1 and LMP-1 are the only cells in which the interaction between lyLMP-1 and LMP-1 could potentially occur. At the lower plasmid quantity ratio, the ability of Western blot densitometry or a functional assay to detect an lyLMP-1 effect on LMP-1 quantity or oncogenic activity is confounded by the overwhelming excess of cells expressing noninteracting lyLMP-1 or LMP-1 alone, an unavoidable anomaly of this experimental system. Increasing the plasmid quantity ratio increases the prevalence of coexpressing cells, which serves primarily to increase the signal-to-noise ratio of the Western blot and functional assays and thereby to permit the prodegradation or down-regulatory effect of the lyLMP-1 interaction with LMP-1 to be somewhat more readily detectable. LMP-1 degradation is known to occur through ubiquitination and proteasome cleavage (2, 28), but the actual molecular mechanism by which lyLMP-1 accelerates the process of LMP-1 degradation remains to be determined. Future studies to determine the intracellular stoichiometry and the molecular mechanism of the lyLMP-1 interaction with LMP-1 will depend upon the ability to obtain a pure population of cells that stably coexpress both proteins.
The clinical significance of this study is the identification of lyLMP-1 as a virally encoded regulator of LMP-1 oncogenic activity with the potential to function in vivo as an EBV tumor suppressor mechanism. EBV-associated oncogenesis is a complex process with multiple contributing factors, including host susceptibilities, environmental influences, and viral determinants such as EBV strain variation. The EBV oncogene LMP-1 is believed to play a critical role in the cell transformation process that leads to malignancy, especially in nasopharyngeal carcinoma (25). The discovery that lyLMP-1 down-regulates LMP-1 oncogenic activity in epithelial cells is especially interesting in light of recent studies suggesting strong negative selection pressure in nasopharyngeal carcinoma against EBV strains encoding the lyLMP-1 ORF (6, 14, 19).
Drawing upon currently available data, it is now possible to envision a model in which the EBV strain-dependent level of LMP-1 oncogenic activity is a viral determinant of nasopharyngeal carcinoma pathogenesis. In the presence of other nasopharyngeal carcinoma risk factors, infection of nasopharyngeal epithelial cells with an EBV strain that expresses LMP-1 alone would contribute a given quantity of transformation risk, while infection with an EBV strain that coexpresses lyLMP-1 with LMP-1 would contribute a relatively reduced quantity of transformation risk by virtue of the down-regulatory effect of lyLMP-1 on expressed LMP-1 quantity and oncogenic activity. Unlike the artificial situation of transient cotransfection of cultured cells in vitro, lyLMP-1-encoding EBV strains in vivo would presumably possess the ability to coexpress lyLMP-1 in every cell in which LMP-1 is expressed, thereby reducing the risk of oncogenic transformation at the level of each individual cell. Testing this hypothesized protective effect of lyLMP-1 in nasopharyngeal carcinoma pathogenesis will require molecular epidemiologic studies of at-risk populations in areas where nasopharyngeal carcinoma is endemic.
Acknowledgments
We thank Johng S. Rhim for providing the RHEK-1 cells, Paul D. Ling for providing the pSG5-EBNA-2 expression vector, and Jean A. Niles for assistance with the flow cytometry.
This work was supported in part by a USPHS research grant NIH R01-DE12323 to Dennis M. Walling, a grant from the John Sealy Memorial Endowment Fund for Biomedical Research to Dennis M. Walling, and a USPHS postdoctoral training grant NIH T32-AI07536 to Jyotsna Pandya.
REFERENCES
- 1.Aaronson, S. A., and G. J. Todaro. 1968. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J. Cell. Physiol. 72:141-148. [DOI] [PubMed] [Google Scholar]
- 2.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275:23491-23499. [DOI] [PubMed] [Google Scholar]
- 3.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 4.Dawson, C. W., A. G. Eliopoulos, S. M. Blake, R. Barker, and L. S. Young. 2000. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology 272:204-217. [DOI] [PubMed] [Google Scholar]
- 5.Eliopoulos, A. G., S. M. Blake, J. E. Floettmann, M. Rowe, and L. S. Young. 1999. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J. Virol. 73:1023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson, K. D., C. Berger, W. F. Coffin III, E. Schiff, D. M. Walling, and J. M. Martin. 2003. Unexpected absence of the Epstein-Barr virus (EBV) lyLMP-1 open reading frame in tumor virus isolates: lack of correlation between Met129 status and EBV strain identity. J. Virol. 77:4415-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson, K. D., and J. M. Martin. 2000. The late lytic LMP-1 protein of Epstein-Barr virus can negatively regulate LMP-1 signaling. J. Virol. 74:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahraeus, R., L. Rymo, J. S. Rhim, and G. Klein. 1990. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature 345:447-449. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, N., B. Kopper, N. Graf, J. R. Schlehofer, F. A. Grasser, and N. Mueller-Lantzsch. 1999. Functional analysis of different LMP1 proteins isolated from Epstein-Barr virus-positive carriers. Virus Res. 60:41-54. [DOI] [PubMed] [Google Scholar]
- 10.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 11.Fries, K. L., W. E. Miller, and N. Raab-Traub. 1996. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 70:8653-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt, W., B. Sugden, and V. R. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry, S., C. Sacaze, L. Berrajah, H. Karray, M. Drira, A. Hammami, J. Icart, and B. Mariame. 2001. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int. J. Cancer 91:698-704. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi, M., E. Kieff, and K. M. Izumi. 2002. The Epstein-Barr virus latent membrane protein 1 putative Janus kinase 3 (JAK3) binding domain does not mediate JAK3 association or activation in B-lymphoma or lymphoblastoid cell lines. J. Virol. 76:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, L. F., F. Chen, X. Zheng, I. Ernberg, S. L. Cao, B. Christensson, G. Klein, and G. Winberg. 1993. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene 8:1575-1583. [PubMed] [Google Scholar]
- 17.Hudson, G. S., P. J. Farrell, and B. G. Barrell. 1985. Two related but differentially expressed potential membrane proteins encoded by the EcoRI Dhet region of Epstein-Barr virus B95-8. J. Virol. 53:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, S. N., Y. S. Chang, and S. T. Liu. 1996. Effect of a 10-amino acid deletion on the oncogenic activity of latent membrane protein 1 of Epstein-Barr virus. Oncogene 12:2129-2135. [PubMed] [Google Scholar]
- 19.Lin, J. C., J. M. Cherng, H. J. Lin, C. W. Tsang, Y. X. Liu, and S. P. Lee. 2004. Amino acid changes in functional domains of latent membrane protein 1 of Epstein-Barr virus in nasopharyngeal carcinoma of southern China and Taiwan: prevalence of an HLA A2-restricted ‘epitope-loss variant. ’ J. Gen. Virol. 85:2023-2034. [DOI] [PubMed] [Google Scholar]
- 20.Martel-Renoir, D., V. Grunewald, R. Touitou, G. Schwaab, and I. Joab. 1995. Qualitative analysis of the expression of Epstein-Barr virus lytic genes in nasopharyngeal carcinoma biopsies. J. Gen. Virol. 76:1401-1408. [DOI] [PubMed] [Google Scholar]
- 21.Martin, J., and B. Sugden. 1991. The latent membrane protein oncoprotein resembles growth factor receptors in the properties of its turnover. Cell Growth Differ. 2:653-600. [PubMed] [Google Scholar]
- 22.Martin, J., and B. Sugden. 1991. Transformation by the oncogenic latent membrane protein correlates with its rapid turnover, membrane localization, and cytoskeletal association. J. Virol. 65:3246-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson, L. J., P. Hopwood, I. Johannessen, J. R. Salisbury, J. Codd, D. Thorley-Lawson, and D. H. Crawford. 1997. Epstein-Barr virus latent membrane protein does not inhibit differentiation and induces tumorigenicity of human epithelial cells. Oncogene 15:275-283. [DOI] [PubMed] [Google Scholar]
- 24.Pandya, J., and D. M. Walling. 2004. Epstein-Barr virus latent membrane protein 1 (LMP-1) half-life in epithelial cells is down-regulated by lytic LMP-1. J. Virol. 78:8404-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pathmanathan, R., U. Prasad, R. Sadler, K. Flynn, and N. Raab-Traub. 1995. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N. Engl. J. Med. 333:693-698. [DOI] [PubMed] [Google Scholar]
- 26.Rhim, J. S., J. Gilbert, P. Arnstein, F. M. Price, K. K. Sanford, and S. Aronson. 1985. Neoplastic transformation of human epidermal keratinocytes by AD12-SV40 and Kirsten sarcoma viruses. Science 227:1250-1252. [DOI] [PubMed] [Google Scholar]
- 27.Sylla, B. S., S. C. Hung, D. M. Davidson, E. Hatzivassiliou, N. L. Malinin, D. Wallach, T. D. Gilmore, E. Kieff, and G. Mosialos. 1998. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc. Natl. Acad. Sci. USA 95:10106-10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tellam, J., G. Connolly, N. Webb, J. Duraiswamy, and R. Khanna. 2003. Proteasomal targeting of a viral oncogene abrogates oncogenic phenotype and enhances immunogenicity. Blood 102:4535-4540. [DOI] [PubMed] [Google Scholar]
- 29.Torii, T., K. Konishi, J. Sample, and K. Takada. 1998. The truncated form of the Epstein-Barr virus LMP-1 is dispensable or complimentable by the full-length form in virus infection and replication. Virology 251:273-278. [DOI] [PubMed] [Google Scholar]
- 30.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 31.Wang, D., D. Liebowitz, and E. Kieff. 1988. The truncated form of the Epstein-Barr virus latent-infection membrane protein expressed in virus replication does not transform rodent fibroblasts. J. Virol. 62:2337-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, X., F. Yuan, L. Hu, F. Chen, G. Klein, and B. Christensson. 1994. Effect of beta-lymphocyte- and NPC-derived EBV-LMP1 gene expression on in vitro growth and differentiation of human epithelial cells. Int. J. Cancer 57:747-753. [DOI] [PubMed] [Google Scholar]