Abstract
Nuclear domains called ND10 or PML nuclear bodies contain interferon (IFN)-upregulated proteins like PML and Sp100. Paradoxically, herpes simplex virus 1 (HSV-1) begins its transcriptional cascade at aggregates of ND10-associated proteins, which in turn are destroyed by the HSV-1 immediate-early protein ICP0. While PML is essential in the formation of ND10, the function of Sp100 in the cells' defense against viral infection is unknown. In this study we investigated the potential antiviral effect of IFN-β-induced Sp100. We found that IFN-β treatment leads to a differential accumulation of four Sp100 isoforms in different cell lines. Using an HEK293 cell line derivative, 293-S, producing no detectable amounts of Sp100 even after IFN exposure, we analyzed individual Sp100 isoforms for their effect on HSV-1 infection. Sp100 isoforms B, C, and HMG, but not Sp100A, suppressed ICP0 and ICP4 early after infection. Isoforms B, C, and HMG suppressed expression from the ICP0 promoter in transient transfection, whereas Sp100A enhanced expression. Moreover, Sp100A localized in ND10, whereas the repressive isoforms were either dispersed within the nucleus or, at unphysiologically higher expression levels, formed new aggregates. The repressive activity was dependent on an intact SAND domain, since Sp100B bearing a W655Q mutation in the SAND domain lost this repressive activity and accumulated in ND10. Using RNA interference to knock down the repressive Sp100 isoforms B, C, and HMG, we find that they are an essential part of the IFN-β-mediated suppression of ICP0 expression. These data suggest that repression by the Sp100 isoforms B, C, and HMG takes place outside of ND10 and raise the possibility that viral genomes at Sp100A accumulations are more likely to start their transcription program because of a more permissive local environment.
Virus-infected cells respond to infection by inducing antiviral programs that limit viral replication. This response is characterized by the induction of cellular signaling pathways that lead to the transcription and production of type I interferons (IFNs). These newly synthesized IFNs are secreted from the host cell and act in autocrine and paracrine modes to activate a global antiviral response in the infected cell and the surrounding tissue (53). The IFN-induced antiviral state is characterized by the transcriptional activation of hundreds of cellular genes involved in blocking virus replication. To be productively successful, viruses must first overcome the innate antiviral response of the cell (reviewed in reference 33). In vivo, the antiviral effects of IFN are mediated by the immune system and by intracellular antiviral pathways. The latter mechanisms underline the direct effects of IFNs on virus replication, which have been studied primarily in cultured cells (reviewed in reference 50), where the immune system does not contribute to an antiviral effect of IFN. However, viruses have evolved to block the IFN-induced antiviral response and allow viral persistence in the infected host. In fact, many viruses encode proteins that specifically target the production of IFNs (28, 50, 53).
IFNs mediate their effect by binding to cell receptors that activate members of the JAK kinase family of proteins. Activated JAK kinases phosphorylate the signal transducers and activators of the transcription (STAT) family of transcription factors, which activate transcription of hundreds of IFN-stimulated genes (ISGs). Products of these genes are primary effectors of the IFN response. The promyelocytic leukemia protein (PML), which is accumulated at specific nuclear domains (called ND10) and acts as the matrix of the domains (24), is among the set of IFN effector proteins, as are the ND10-associated proteins Daxx, Sp100, and Sp140 (2, 8, 15-17, 19). The PML promoter contains functional IFN-α-stimulated response elements and IFN-γ activation sites (52). The Sp100 gene also has an IFN-α-stimulated response element and IFN-γ activation site (8, 30). To date, more than 30 proteins have been found to colocalize with PML in ND10, either transiently or constitutively (43, 47). Additionally, both PML and Sp100 are modified by small ubiquitin-related modifier 1 (SUMO-1) (3, 26, 27, 41, 54), which is essential in the recruitment of Daxx (24).
Herpes simplex virus 1 (HSV-1) is a common human pathogen in which lytic infection starts a cascade of gene expression that initiates with the induction of immediate-early (IE) genes ICP0, ICP4, ICP22, ICP27, and ICP47 by the virion protein VP16, shortly after the release of the viral genome into the nucleus (1, 6, 21, 22). Expression continues with early genes and is followed by DNA replication, and this program is dependent on the expression of IE genes (21, 22). Viral IE gene expression is decreased in cells pretreated with IFNs (37, 44, 45, 56). One of the IFN-upregulated proteins, the Sp100B isoform, has recently been shown to inhibit viral promoters (57). In cell culture, HSV-1 is relatively resistant to the effects of IFNs, in part by counteracting an IFN-induced block to virus transcription (20, 36, 38, 39). ICP0 is an essential component of IFN resistance (20, 39) and is sufficient to inhibit activation of IFN-responsive genes (9). However, in the absence of ICP0, HSV-1 can still inhibit expression of ISGs and as such can replicate, but only at high multiplicities of infection. This suggests that more than one viral gene product inhibits the innate immune response (36, 39) and that ICP0 enhances expression of those viral genes.
Newly synthesized ICP0 accumulates initially in ND10 and promotes dispersion of this domain (12, 34) and degradation of ND10 components, including PML and especially sumoylated isoforms of these proteins (11) and Sp100 (7, 46), although there is no evidence that ICP0 interacts directly with any components of ND10. In cells in which ND10 are not disrupted (e.g., cells infected with an ICP0 mutant or under conditions where PML is overexpressed), viral DNA localizes adjacent to ND10-associated protein complexes prior to replication, and replication occurs in close association with PML aggregates (13, 35, 51). This association of viral genomes with ND10-associated proteins requires ICP4 and ICP27 and is likely mediated by the interaction of these proteins with Daxx (55).
As a step toward identifying the molecular mechanisms underlying the function of ND10-associated proteins in viral regulation, we dissected the nuclear distribution of Sp100 isoforms derived from the same IFN-upregulated transcript and determined whether any of the isoforms are part of an IFN-induced antiviral effect. We found that IFN-β treatment led to the accumulation of transcripts of all Sp100 isoforms in HEp-2 cells, whereas HEK293 (human embryonic kidney) cells produced mostly the Sp100A isoform. Moreover, we derived cell line 293-S, in which Sp100 proteins are not detectable, from HEK293 cells. Individually expressed Sp100B, -C, and -HMG isoforms in 293-S or HEp-2 cells preferentially accumulated at sites different from ND10, whereas Sp100A specifically accumulated at ND10. Apparently, only the isoforms not segregated to ND10 suppress HSV-1 IE viral protein expression. We also show that Sp100 isoforms B, C, and HMG, but not A, suppress ICP0 promoter activity and that the suppressive effect of the Sp100B isoform depends on an intact SAND (Sp100, AIRE-1, NucP41/75, and DEAF-1) domain, since mutation in that domain relaxed the suppression and shifted the nuclear distribution of this protein toward accumulation at ND10. Moreover, knockdown of Sp100B expression in HEp-2 cells with short hairpin RNA (shRNA) allowed increased expression of HSV-1 IE viral proteins even after IFN-β treatment. These results identify Sp100 isoforms B, C, and HMG as part of the downstream IFN repressive pathway that can inhibit HSV-1 gene expression and predict a different role for Sp100A in viral regulation.
MATERIALS AND METHODS
Expression plasmids.
The pmRFP-Sp100A mammalian expression vector was obtained from Peter Hemmerich (Institute of Molecular Biotechnology, Germany), pSG5-Sp100B, pSG5-Sp100HMG, and pICP0-luc were obtained from J. Taylor (Medical College of Wisconsin), and pSG5-Sp100C was obtained from Ann Dejean (Institut Pasteur, France). pmRFP-Sp100B, pmRFP-Sp100C, and pmRFP-Sp100HMG were created by replacing the XbalI fragment from pmRFP-Sp100A with XbalI fragments from pSG5-Sp100B, pSG5-Sp100C, and pSG5-Sp100HMG, respectively. As a control, plasmid pmRFP-NLS (pmRT1) was constructed, encoding monomeric red fluorescent protein (mRFP) with a nuclear localization signal (NLS) from simian virus 40 large T antigen (amino acid sequence, PKKKRKV) by subcloning a BamHI-XbalI fragment from pET (42) into the C-terminal end of the mRFP open reading frame between the BamHI and XbalI sites on pmRFP-Sp100A. All constructs were verified by sequencing.
Site-specific mutagenesis of Sp100.
Site-directed mutagenesis was performed on pmRFP-Sp100B or pSG5-Sp100B using the QuikChange site-directed mutagenesis protocol (Stratagene, La Jolla, CA) with the following oligonucleotides (nucleotide changes are shown in boldface): Sp100W655Q-For, 5′-GGAGCATCCAAGAACCAGAAGCTAAGTATACGC-3′; Sp100W655Q-Rev, 5′-GCGTATACTTAGCTTCTGGTTCTTGGATGCTCC-3′. These were used to create plasmids pmRFP-Sp100B-W655Q and pSG5HA-Sp100B-W655Q.
All constructs were verified by sequencing. Transient transfections were carried out using the SuperFect reagent (QIAGEN, Valencia, CA) according to the manufacturer's recommendations.
Cells and viruses.
HEp-2 carcinoma, HEK293, and African green monkey kidney (Vero) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% antibiotics. All cells were grown at 37°C in a humidified 5% CO2 atmosphere. For immunohistochemical staining, cells were grown on round coverslips in 24-well plates (Corning Glass Inc., NY) until they were approximately 80% confluent before fixation. HSV-1 strain 17+ was obtained from R. D. Everett. Titers of all virus stocks were determined in Vero cells.
Cell infection.
In all experiments, replicate cell cultures grown in six-well plates were either mock infected or exposed to virus at the PFU/cell ratio as indicated in Results. Cells were exposed to virus for 1 h at 37°C in DMEM with 1% FCS. The inoculum was replaced with DMEM supplemented with 10% FCS and 1% antibiotics, and cultures were incubated for 3 h at 37°C.
Sp100 knockdown.
For lentiviral vector-based knockdown, clones TRCN000019224 (targeting the sequence of nucleotides 1759 to 1780: 5′-GCCAACACTAGACCTTTGAA-3′, designated as Sp100si-2) and TRCN000019226 (targeting the sequence of nucleotides 1871 to 1891: 5′-GCACTCATATAAGGAGCGA-3′, designated as Sp100si-3) that expressed the RNAi Consortium human shRNA from the pLKO.1 vector were purchased from Open BioSystems (Huntsville, AL). These shRNAs will recognize targeting sequences common in mRNAs of Sp100 isoforms B, C, and HMG. Lentiviral vectors were cotransfected with packaging plasmids pCMV-VSVG and pCMV-deltaR8.2 into 293T cells on a 6-cm-diameter dish by standard calcium phosphate precipitation. Viral supernatants were collected from the transfected 293T cells 24, 36, and 48 h posttransfection, clarified by centrifugation, filtered, and used for infection of 5 × 104 target HEp-2 cells grown on six-well plates in the presence of 4 μg/ml of polybrene (Sigma, St Louis, MO). Stably transduced cell populations were selected in DMEM medium with 0.5 μg/ml puromycin for 3 days.
Indirect immunofluorescence (IF).
Two days after plating on round glass coverslips, cells were fixed at room temperature for 15 min with freshly prepared 1% paraformaldehyde in phosphate-buffered saline (PBS) and treated as described previously (23). Cells were analyzed using a Leica confocal laser scanning microscope. Leica image enhancement software was used in balancing signal strength, and fourfold scanning was used to separate signal from noise. Because of the variability among cells in any given culture, the most prevalent cells were photographed and are presented as small groups of cells at high magnification. ND10 were visualized using the following antibodies: monoclonal antibody (MAb) PG-M3, which recognizes PML (Santa Cruz Biotechnology, Santa Cruz, CA) (1:500 dilution); rabbit antibody AB1380 against Sp100 (Chemicon International, Temecula, CA) (1:1,000 dilution); anti-ICP0 MAb 11060, which has been described previously (10) (1:5 dilution); and anti-ICP4 MAb (American Type Culture Collection, Rockville, MD) (1:2 dilution).
Preparation of cellular extracts.
Total cellular protein extract was prepared from 105 HEK293 cells per six-well plate, treated as indicated above, and washed with PBS, lysed in radioimmunoprecipitation assay buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.5% sodium dodecyl sulfate [SDS]) supplemented with protease inhibitor cocktail (PIC) (1 mM EDTA, 100 μg/ml phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin) at room temperature for 5 min. Extraction was carried out with rotation at 4°C for 20 min, and extracts were passed through a 22-gauge needle five times and left for an additional 10 min at 4°C. Extracts were clarified by centrifugation at 12,000 × g for 20 min at 4°C. For fractionation, cells were scraped, rinsed with PBS, resuspended in hypotonic lysis buffer (20 mM HEPES, 10 mM KCl, 1 mM EDTA, 0.1 mM sodium orthovanadate, 1 mM dithiothreitol, 0.2% NP-40, 10% glycerol, PIC), lysed for 5 min on ice, and centrifuged at 13,000 rpm for 10 s to separate the cytoplasmic (supernatant) and nuclear (pellet) fractions. The cytoplasmic fraction was transferred into a new tube. The pelleted nuclei were gently rinsed with hypotonic buffer and pelleted again as described above. The nuclear fractions were resuspended in hypertonic lysis buffer (20 mM HEPES, 10 mM KCl, 1 mM EDTA, 0.1 mM sodium orthovanadate, 1 mM dithiothreitol, 0.2% NP-40, 20% glycerol, 500 mM NaCl, PIC), lysed for 30 min on ice, and centrifuged at 13,000 rpm for 5 min to remove the insoluble fraction. Protein concentration was quantified by the Bradford method (Bio-Rad). The final protein concentrations in the nuclear extracts ranged from 1 to 5 mg/ml.
Mammalian cell transfection and reporter assays.
DNA for transfection was prepared by CsCl gradient centrifugation. HEp-2 cells HEK293 or 293-S cells were cultured in DMEM plus 10% FCS and transfected with plasmids using the SuperFect reagent (QIAGEN) according to the manufacturer's protocol. For luciferase reporter gene assays, 1 × 105 cells were seeded in each well of a 12-well plate and transfected with total of 2 μg of plasmid DNA, which routinely included 0.1 μg of pTKmin-β-gal to monitor transfection efficiency. In transfections using increasing doses of effector plasmids, the total amount of plasmid containing simian virus 40 or cytomegalovirus enhancer was maintained by addition of the empty expression vector pSG5. Cells were harvested 24 to 48 h posttransfection, washed with PBS, and lysed in 100 μl of hypotonic lysis buffer. The cytoplasmic fraction was used to measure protein content (1 μl), β-galactosidase activity (2 μl), and luciferase activity (20 μl). The nuclear fraction was used to check expression of effector proteins. Three independent transfections were carried out, and mean fold repression was calculated relative to the basal luciferase activity obtained from cells transfected with pSG5. Because the minimal HSV-1 thymidine kinase promoter in the pTKmin-β-gal plasmid was repressed approximately twofold by all Sp100 isoforms except Sp100A, normalization of luciferase activity to β-galactosidase activity was not appropriate in many experiments. Absolute luciferase values and fold changes of activation or repression differed among similar experiments performed on different days. Thus, all data within a panel were derived from transfections performed on the same day and assayed as a set.
Western blotting.
Total cellular protein extract was prepared from cells by lysis as described above, except that the radioimmunoprecipitation assay buffer contained 2% SDS. Protein concentration was quantitated by the Bradford method (Bio-Rad Laboratories, Hercules, CA). Total cellular protein (30 μg) was fractionated by SDS-4 to 12% gradient PAGE and electrotransferred to a nylon membrane by standard techniques. Immunoblotting was performed using 5% nonfat dry milk in PBS containing 0.1% Tween-20 as the blocking agent. Filters were blocked overnight at 4°C, incubated with primary antibody for 1 h at room temperature, washed three times for 10 min per wash in PBS containing 0.1% Tween-20, and incubated for 1 h with goat anti-mouse or anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase (GIBCO/BRL, Rockville, MD) according to the manufacturer's recommendations. Filters were washed again as described above, and reactivity was detected using an enhanced chemiluminescence kit (Amersham Bioscience Corp, Piscataway, NJ) according to the manufacturer's recommendations. The following antibodies were applied: rabbit anti-Sp100 antibody AB1380 (1:30,000 dilution), MAb anti-ICP0 (1: 50 dilution), MAb anti-ICP4 (1:25 dilution), and MAb antitubulin (1:10,000 dilution) (Sigma, St Louis, MO).
RT-PCR.
Cells grown in 100-mm-diameter dishes were treated with 500 to 1,000 U/ml of IFN-β for 18 h, and total cellular RNA was prepared using the Trizol reagent (Life Technologies, Rockville, MD). RNA samples were treated with DNase I at 37°C for 30 min. Reverse transcription was performed for 60 min in a 30-μl volume with 5 μg of total RNA with a reverse transcription (RT)-PCR system kit (Amersham Bioscience Corp) according to the manufacturer's protocol. The following primers were used: Sp100 All (consensus primer for all Sp100 isoforms) sense (5′-GCACACAGCCACGATTTG) and antisense (5′-CAGGTTAAATGTCTTCTC), Sp100A sense (5′-TGGGAACTCCTTTTTGCATT) and antisense (5′-CAAACGACAATGATGTCAACC), Sp100B sense (5′-AGGAAGCGATTCAAACAAGGA) and antisense (5′-AGACGAGACATTGGCAGAAG), Sp100C sense (5′-AAGCCAATCAGGTCATCAGG) and antisense (5′-ATGTCCTGCACAAACCCTTC), Sp100HMG sense (5′-TAGCCCTGTCCTGGTGGTAT) and antisense (5′-TGTCAACAAAACAGCTGCAA), PKR sense (5′-ATGATGGAAAGCGAACAAGGA) and antisense (5′-GTTTCAAAAGCAGTGTCACATACATG), ISG54 sense (5′-CTGAAGAGTGCAGCTGCCTG) and antisense (5′-CACTTTAACCGTGTCCACCC), ISG15 sense (5′-GACCTGACGGTGAAGATGC) and antisense (5′-GACCTGACGGTGAAGATGC), and GAPDH sense (5′-ACCACAGTCCATGCCATCAC) and antisense (5′-TCCACCACCCTGTTGCTGTA). To estimate relative transcript levels, PCRs were performed on serially diluted samples of RT products. PCR products were run on 2% agarose gels.
RESULTS
IFN-β treatment preferentially induces the Sp100A isoform in HEp-2 cells.
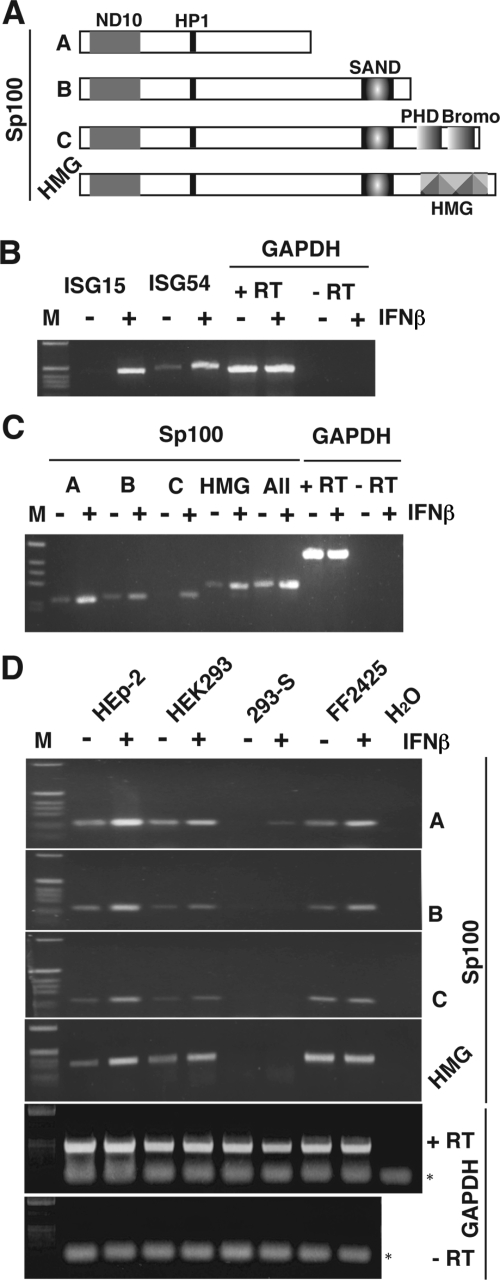
Cells treated with IFN show highly upregulated Sp100 proteins with many characteristic bands, which might represent different isoforms of Sp100, shown schematically in Fig. 1A, and/or sumoylated or otherwise modified versions of these isoforms. To identify which Sp100 species they represent, we first investigated IFN-stimulated transcription in HEp-2 cells. RT-PCR analysis of total RNA isolated from HEp-2 cells treated with 500 U/ml of IFN-β revealed strong induction of ISG54 and ISG15 transcripts (Fig. 1B), both of which have previously been characterized as genes upregulated by IFN treatment. Thus, the HEp-2 cell line responds to IFN-β. To examine the effect of IFN-β on transcript abundance of the different Sp100 isoforms, we designed primer sets that specifically recognize Sp100 isoforms A, B, C, and HMG. Use of these primers on the same RNA as described above led to identification of isoforms A and HMG as major species in untreated cells, whereas all isoforms were upregulated after IFN-β treatment (Fig. 1C). RT-PCR with different numbers of cycles used for amplification, followed by semiquantitative analysis, showed that HEp-2 cells produced at least 10-fold more Sp100A than Sp100B or Sp100C (data not shown).
FIG. 1.
Induction of Sp100 isoforms in response to IFN-β treatment. (A) Structural and functional domains of Sp100 isoforms. (B) Induction of ISG15 and ISG54 mRNA was assessed by semiquantitative RT-PCR analysis. HEp-2 cells were treated with 500 U/ml of IFN-β for 18 h before total cellular RNA was extracted. (C) Levels of Sp100 isoforms were analyzed by semiquantitative RT-PCR from the same samples. (D) Induction of Sp100 isoforms in different cell lines. Levels of GAPDH mRNA were used to control an equal amount of mRNAs. To control DNA contamination we used RNA samples without RT (-RT). Asterisks indicate positions of GAPDH primers. M, molecular size marker.
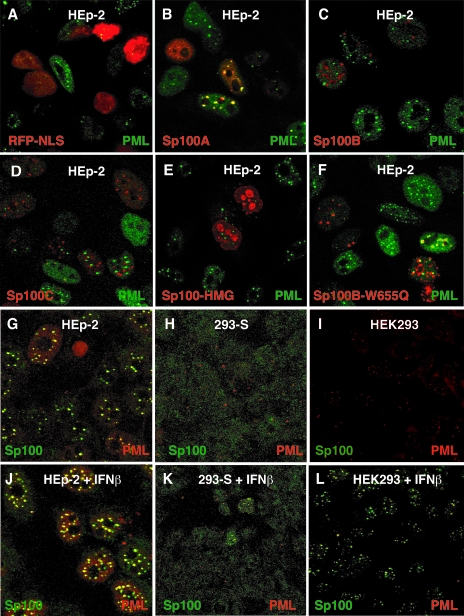
We then analyzed the intracellular localization of these isoforms tagged with mRFP since isoform specific antibodies are not available. HEp-2 cells on coverslips were transfected with RFP-tagged Sp100 isoforms, fixed, and stained 18 h later with MAb against endogenous PML (Fig. 2). RFP-NLS alone (vector control) distributed diffusely throughout the nucleus (Fig. 2A). The RFP-tagged Sp100A isoform preferentially colocalized with endogenous PML (Fig. 2B). Isoforms B, C, and HMG were found throughout the nucleus and, if expressed at higher concentrations, in new aggregates. The cause of the aggregate formation remains unknown. The aggregated isoforms colocalized only occasionally with endogenous PML as previously reported (48) or appeared juxtaposed to it (Fig. 2C, D, and E). The same experiment using hemagglutinin (HA)-tagged Sp100 revealed an equivalent distribution (data not shown), ruling out a potential influence of the RFP tag on Sp100 distribution.
FIG. 2.
Nuclear distribution of Sp100 protein. (A to F) HEp-2 cells grown on coverslips were transfected with plasmid constructs expressing mRFP-tagged NLS (A), Sp100A (B), Sp100B (C), Sp100C (D), Sp100HMG (E), or Sp100B-W655Q (F) isoforms and analyzed by IF assay with MAb PG-M3 (green) against endogenous PML. (G to L) HEp-2, HEK293, and 293-S cells grown on coverslips were treated with 1,000 U/ml of IFN-β for 18 h (J, K, and L) or left untreated (G, H, and I) and double-labeled with polyclonal Ab 1380 against endogenous Sp100 (green) and MAb PG-M3 against endogenous PML (red). Colocalization appears as a yellow signal. Note that we artificially increased the background on panels H and K to detect the lowest amount of Sp100.
The 293-S cell line is deficient in Sp100 protein expression.
The differential distribution of Sp100 isoforms raised the possibility of functional differences among the Sp100 isoforms with respect to the antiviral effect of IFN. Testing this possibility requires a cell line that does not express endogenous Sp100, such as the embryocarcinoma cell line NT2/6 (42). Unfortunately, this cell line proved very difficult to transfect with plasmid DNA. We screened several human cell lines by indirect IF assay with polyclonal rabbit antiserum against Sp100 for cell lines without Sp100. Double-staining for PML and Sp100 in HEp-2 and HEK293 cells visualized the normal extensive colocalization of the two proteins in multiple foci (Fig. 2G and I), even though HEK293 cells express much less PML and Sp100 than HEp-2 cells. During this search we identified one laboratory clone of the HEK293 cell line, designated 293-S, which was negative for Sp100. This clone also showed fewer PML foci and none labeled with Sp100 (Fig. 2H), but we do not know what additional changes this cell line might have. Even after treatment with IFN-β, fewer than 5% of these cells revealed the presence of Sp100 (Fig. 2K shows two such cells among the negative ones). Equivalent treatment of HEp-2 and HEK293 cells leads to pronounced accumulation of PML and Sp100 proteins in ND10 (Fig. 2J and L). Similar results were obtained with cells treated with IFN-α or IFN-γ (data not shown), although those exposed to IFN-β consistently gave the strongest signal.
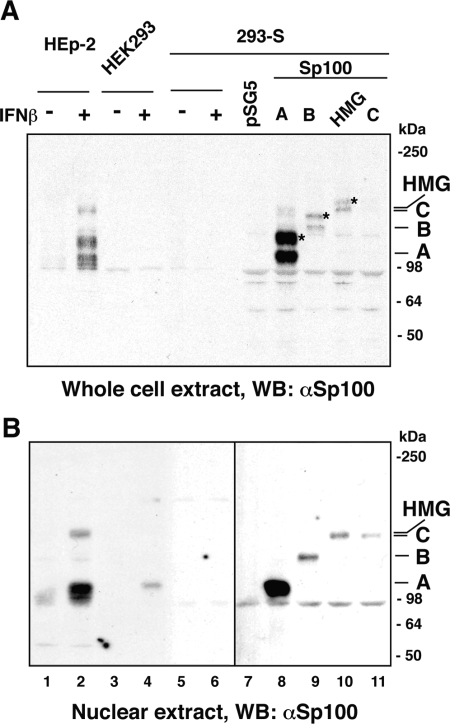
Sp100 is a very-low-abundance protein, and Western blot analysis of total cell extracts of HEp-2 cells showed no specific band. Only after IFN-β treatment were many bands revealed, because the SUMO modification was preserved. Untreated and treated HEK293 and 293-S cells had no detectable Sp100 proteins (Fig. 3A, lanes 3 to 6). We tried to enrich Sp100 by isolating the nuclear fraction. After such enrichment a faint band is revealed in untreated HEp-2 cells (Fig. 3B, lane 1), but the SUMO modification bands, which were present in the whole cell extracts (Fig. 3A), were lost in the IFN-β-treated sample (Fig. 3B, lane 2). In HEK293 nuclei Sp100 could be demonstrated only after induction by IFN-β (lane 4), in contrast to the high-level expression of this protein in the whole extract and the nuclear fraction of IFN-β-treated HEp-2 cells (Fig. 3, lanes 2). Under none of the conditions used was Sp100 demonstrated in the 293-S clone (Fig. 3B [lanes 1 to 6 were overexposed to show the absence]). We also tested whether any transcripts were present. RT-PCR analysis of total RNA from HEK293 cells (Fig. 1D) indicated a moderate increase in all Sp100 isoforms after IFN treatment. Similar analysis of 293-S cells revealed no transcripts for the Sp100B, -C, or -HMG isoforms, and only a very slight induction of Sp100A, which probably corresponds to the Sp100 detected by IF assay in a small number of these cells after IFN-β treatment. Human fibroblasts (FF2425 cells) also increased transcripts of Sp100A and -B, but little increase was seen in Sp100C and -HMG after IFN treatment. Interestingly, in this cell line mRNA for Sp100HMG is the dominant Sp100 isoform. This indicates that cell type-dependent differential splicing may take place.
FIG. 3.
Accumulation of Sp100 isoforms in HEp-2, HEK293, and 293-S cells. Whole-cell extracts (A) or nuclear fractions (B) of replicate HEp-2, HEK293, and 293-S cell cultures were stimulated with 500 U/ml of IFN-β for 18 h or left untreated as a control. 293-S cells were transfected with 2 μg of pSG5 vector DNA or constructs expressing Sp100A (A), Sp100B (B), Sp100HMG (HMG), or Sp100C (C). Note that sumoylation of Sp100 proteins, indicated by asterisks, is preserved in whole-cell extracts but not in nuclear fractions. WB, Western blot.
We wanted to determine whether the different levels of mRNA correspond to different amounts of expressed protein. To identify the Sp100 isoforms recognized by the common epitope in the N terminus we compared them with individually expressed Sp100 isoforms A, B, C, and HMG in 293-S cells. This Western blot analysis indicated that Sp100A is the predominant species induced by IFN-β in HEp-2 cells (compare Fig. 3A and B, lanes 2 and 8). Substantially less Sp100B was produced in HEp-2 cells (compare double band from the transfected Sp100B in lane 9 with barely recognizable amounts in Fig. 3A, lane 2, and the single band from the nuclear fraction; also compare Fig. 3B, lanes 9 and 2). These results are consistent with the results from RT-PCR analysis. Due to the resolution limit of the Western blot, analysis of Sp100C or Sp100HMG is more problematic as it was not possible to determine which isoform was the slowest migrating (Fig. 3B, compare lanes 2, 10, and 11). Based on the intensity of this band (lane 2), both these isoforms might be represented. Except for Sp100A none of the other isotypes were recognized in the HEK293 cells (Fig. 3B, lane 4) by Western blotting. Thus, Sp100A appears to be the major isoform induced by IFN-β in HEK293 cells. Neither Sp100 isoform was recognized in 293-S cells, indicating that the 293-S clone has little or no Sp100 and that Sp100 cannot be significantly upregulated by IFN-β in this clone. In addition, the results show that because of Sp100 aggregation at ND10, the IF assay is more sensitive than Western blotting. Since the 293-S cell line shows no Sp100 isotype by Western blot, none by immunofluorescent assay and only very little Sp100A by RT-PCR, this cell line was used to test the effects of the individual Sp100 isoforms on HSV-1 infection as no interference with endogenous Sp100 was to be expected.
The 293-S cell line is deficient in the repression of HSV-1 IE proteins.
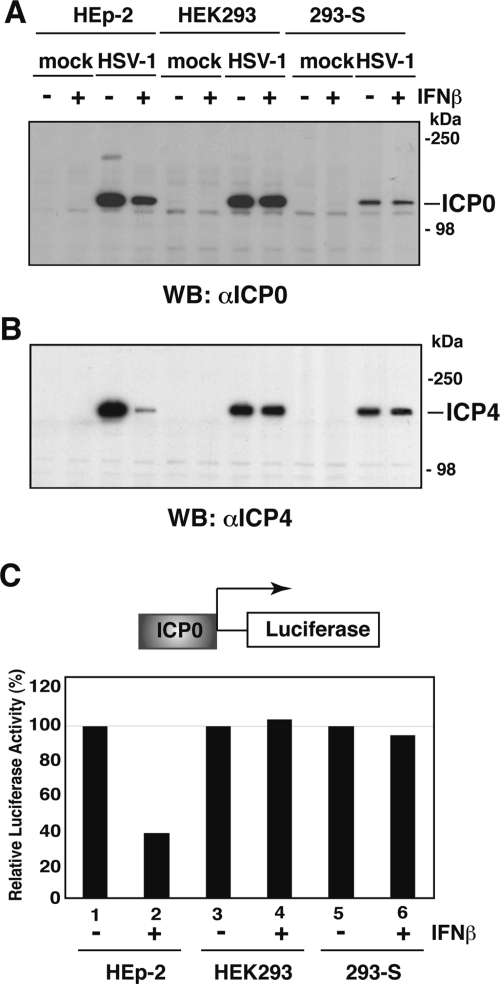
It has been shown previously that IFN can suppress the expression of ICP0 of HSV-1 (29, 37, 44, 45, 55). To determine whether Sp100 participates in this suppression, IFN-β (1,000 U/ml)-pretreated or untreated HEp-2, HEK293, and 293-S cells were infected with 0.5 PFU/cell of HSV-1, collected at 3 h postinfection, and examined by Western blot analysis using anti-ICP0 antibodies. The results indicate the expected suppression of ICP0 expression by IFN in HEp-2 cells, but no suppression was found in HEK293 or 293-S cells (Fig. 4A). Similar results were obtained for ICP4 (Fig. 4B). These results, together with the differential expression of Sp100 in these cell lines, suggest that Sp100 isoforms may have distinctive effects on the expression of HSV-1 IE proteins.
FIG. 4.
Effect of IFN-β treatment on expression of IE ICP0. (A and B) Replicate HEp-2, HEK293, and 293-S cell cultures were stimulated with 1,000 U/ml of IFN-β for 18 h or left untreated as a control. Half of each replicate was mock infected or inoculated with 0.5 PFU of HSV-1 (strain 17+) per cell as described in Materials and Methods. After 3 h of infection, cells were harvested, and total cell extract and the nuclear fractions (30 μg/lane) were electrophoretically separated on denaturing polyacrylamide gels, transferred to nitrocellulose membranes, and probed with antibodies to ICP0 (A) or ICP4 (B). WB, Western blot. (C) HEp-2, HEK293, and 293-S cells were transiently cotransfected with ICP0 promoter-luciferase and stimulated 24 h later with 1,000 U/ml of IFN-β for 18 h or left untreated. Cells were harvested and fractionated on nuclear and cytoplasmic fractions. Firefly luciferase activity was measured in the cytoplasmic fractions and normalized to protein concentration. Responsiveness to IFN-β was calculated as the ratio of normalized luciferase activity of treated cells to that of untreated cells.
To determine whether differential induction of Sp100 might participate in IE protein suppression at the transcriptional level, HEp-2, HEK293, and 293-S cells were transiently transfected with reporter plasmids driven by an ICP0 promoter and treated 24 h later with 1,000 U/ml IFN-β for 18 h or left untreated. Measurements of luciferase activity indicated the expected suppression of the ICP0 promoter in HEp-2 cells after IFN-β treatment (Fig. 4C, lanes 1 and 2). However, IFN-β had no effect on the ICP0 promoter-driven luciferase activity in HEK293 cells (Fig. 4C, lanes 3 and 4) or in 293-S cells (lanes 5 and 6). Thus, the cell lines in which IFN does not influence the expression of viral proteins after infection or from viral promoters after transfection can be used to determine the downstream effect of IFN-induced proteins like Sp100. Since these cell lines have very low or no expression of these IFN-upregulated isotypes, they can be used to evaluate their individual contribution to the IFN-based suppression of viral promoters like the ICP0 promoter at the level of transcription.
Sp100B, -C, and -HGM, but not Sp100A, isoforms suppress the expression of HSV-1 IE proteins.
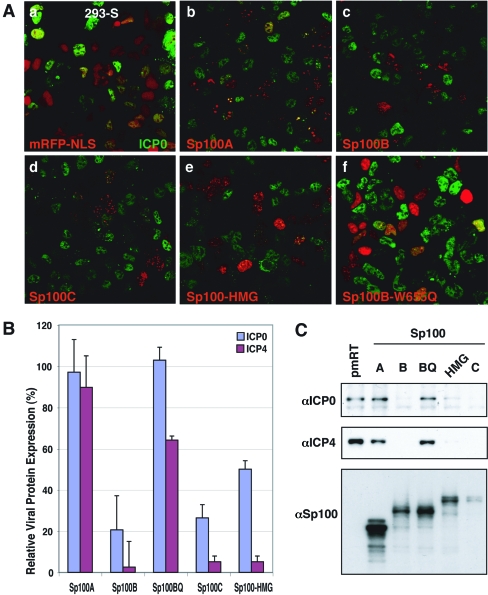
To test whether the Sp100 isoforms can differentially inhibit expression of HSV-1 IE proteins, we transiently expressed each of these proteins in 293-S cells and assessed the effects on ICP0 and ICP4 expression after HSV-1 infection. Transient transfection was used because despite repeated attempts, 293-S cell lines stably expressing Sp100B or Sp100C could not be obtained. Cells transfected with these isoforms underwent only a few cell cycles before losing the Sp100 portion of the fusion protein, and only cells with truncated Sp100 survived after selection on antibiotics (data not shown). Apparently a low level of expression and/or the combination of these IFN-upregulated isoforms is necessary for long-term cell survival. On the other hand, HEK293 and 293-S cells were very efficiently transfected at rates of ∼40 to 60% (data not shown). Input virus had been titrated to result in infection of approximately 90% of cells as assayed by ICP0 production using IF. The normalized number of cells producing IE proteins in vector-transfected versus Sp100 isoform-transfected cells was determined using IF at 3 h postinfection in cells transfected 18 h previously. Although ICP0 was segregated with Sp100A (Fig. 5A, panel a), the number of cells producing ICP0 in vector-transfected cells was very similar to that in Sp100A-transfected cells (Fig. 5A, panel b). These sites also contained PML (data not shown). However, when 293-S cells were transfected with B, C, or HMG isoforms, most cells showing the red marker produced no ICP0, whereas the untransfected cells did produce ICP0 (Fig. 5A, panels c to e).
FIG. 5.
Effect of Sp100 isoforms on expression of IE HSV-1 proteins. (A) 293-S cells grown on coverslips were transfected with plasmid constructs expressing mRFP-tagged NLS (a), Sp100A (b), Sp100B (c), Sp100C (d), Sp100-HMG (e), or Sp100B-W655Q (f). Eighteen hours later, cells were inoculated with HSV-1 (strain 17+) as described in Materials and Methods. After 3 h of infection, cells were fixed and analyzed by IF assay with rabbit anti-ICP0 antibodies (green). (B) 293-S cells grown on coverslips were transfected with plasmid constructs expressing mRFP-tagged NLS (pmRT), Sp100A, Sp100B, Sp100C, Sp100-HMG, or Sp100B-W655Q (Sp100BQ). Eighteen hours later, cells were inoculated with HSV-1 (strain 17+) and analyzed by counting cells labeled for ICP0 or ICP4 in cells expressing the respective Sp100 isotype. At 3 h postinfection approximately 1,000 cells expressing red fusion protein were analyzed for the presence of ICP0 or ICP4. Bars indicate the numbers of those Sp100-expressing cells that exhibited ICP0 or ICP4 staining relative to the vector control (pmRT). (C) Western blot analysis of the nuclear fractions (30 μg/lane) from the above experiment were electrophoretically separated, transferred to nitrocellulose membranes, and probed with antibodies to ICP0 (top), ICP4 (middle), or Sp100 (bottom).
To quantitate this observation we counted transfected cells in five independent fields for expression of ICP0 and ICP4. Data presented in Fig. 5B show that B, C, and HMG isoforms very effectively suppressed expression of ICP0 and ICP4 proteins, but the Sp100A isoform did not, in full agreement with the results of the Western blot analysis of samples probed with antibodies against ICP0 or ICP4 (Fig. 5C). These data suggest that Sp100 can regulate expression of HSV-1 IE proteins and that the Sp100B, -C and -HMG isoforms repress IE proteins and may block virus production.
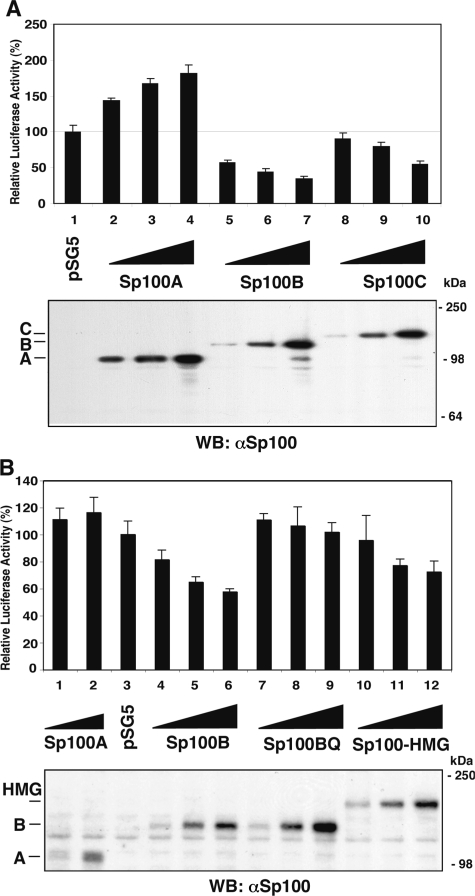
Sp100B, -C, and -HMG, but not Sp100A, isoforms repress the ICP0 promoter.
To determine whether Sp100 isoforms can suppress the ICP0 promoter, 293-S cells were transiently transfected with increasing amounts of expression plasmid encoding HA-tagged Sp100 isoforms together with a reporter plasmid driven by an ICP0 promoter. Luciferase activity was measured 48 h after transfection. The Sp100A isoform stimulated the ICP0 promoter (Fig. 6A, lanes 2 to 4) relative to the vector control (lane 1), whereas Sp100B significantly suppressed the ICP0 promoter in a dose-dependent manner (lanes 5 to 7), as did the C (lanes 8 to 10) and HMG (Fig. 6B, lanes 10 to 12) isoforms, though to a lesser extent. All isoforms were expressed to comparable levels (Fig. 6, bottom panels). Similar results were obtained with RFP-tagged Sp100 isoforms in 293-S cells and HEp-2 cells (data not shown). Thus, Sp100 isoforms can modulate expression from the ICP0 promoter and as such are involved in the regulation of HSV-1, at least at an early time point of viral infection.
FIG. 6.
Effect of Sp100 isoforms on luciferase expression from the ICP0 promoter. (A) 293-S cells were transiently cotransfected with luciferase reporter plasmid under the ICP0 promoter, minimum thymidine kinase promoter-β-galactosidase reporter, and increasing amounts (0.5, 1, and 2 μg) of effector plasmids expressing HA-tagged Sp100 isoforms A, B, and C. (B) 293-S cells were transiently cotransfected with luciferase reporter plasmid under the ICP0 promoter and increasing amounts (0.1, 0.3, and 1 μg) of effector plasmids expressing HA-tagged Sp100 isoforms A, B, BQ (the W655Q mutant of isoform B), and HMG. Cells were harvested 48 h later, and luciferase activity was determined from the cytoplasmic fraction and normalized to protein concentration (top). Results are presented as percentages of luciferase activity of the pSG5 vector. Mean values (± standard deviation) are from three independent experiments. Nuclear fractions (30 μg/lane) were electrophoretically separated on denaturing polyacrylamide gels, transferred to nitrocellulose membranes, and probed with antibodies to Sp100 to assure similar levels of expression. WB, Western blot.
The SAND domain of Sp100B is necessary for repression of the ICP0 promoter.
All repressive isoforms of Sp100 share the same N-terminal 684 amino acids, which includes the SAND domain. This domain has been implicated in DNA binding in other proteins (5). Because all isoforms also share the N-terminal 476 amino acids of Sp100A, which does not repress the ICP0 promoter, we analyzed the role of the SAND domain in repression of ICP0 promoter activity. We mutated the tryptophan at position 655 to glutamate (W655Q) in the SAND domain of Sp100B, since this mutation has been shown to strongly affect DNA binding of SAND domains (5). Measurement of luciferase activity 48 h after transfection of 293-S cells with various amounts of expression plasmid encoding the HA-tagged wild-type or W665Q mutant Sp100 isoform along with a reporter plasmid driven by the ICP0 promoter indicated that the mutation relieved the repression by Sp100B (Fig. 6B, compare lanes 4 to 6 with lanes 7 to 9). This mutation did not affect the stability of the Sp100B isoform (Fig. 6B, bottom panel).
The cellular localization of the mutated Sp100B isoform was analyzed in HEp-2 cells transfected with RFP-tagged Sp100 isoforms, fixed after 18 h, and stained with MAb against endogenous PML. This mutant preferentially colocalized with endogenous PML (Fig. 2F), indicating that the W655Q mutation in the SAND domain shifts the location of Sp100B to ND10 as for the Sp100A isoform. Moreover, mutated Sp100B no longer suppressed ICP0 and ICP4 expression after HSV-1 infection (Fig. 5A, panel f, and B and C), nor did it repress the ICP0 promoter in transient transfection experiments (Fig. 6B). These findings indicate the requirement for an intact SAND domain of Sp100B to form new domains in repressing the ICP0 promoter and in IE protein expression in the viral context.
Sp100 isoforms with SAND domains are necessary for IFN-β-mediated suppression of ICP0 expression.
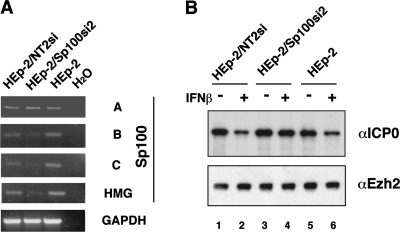
We have demonstrated that IFN-β suppresses expression of ICP0 upon HSV-1 infection of HEp-2 cells (Fig. 4A). To test whether the SAND domain-containing repressive Sp100 isoforms B, C, and HMG are required for IFN-β-mediated suppression of ICP0 expression, we utilized lentivirus-based vector pLKO expressing shRNA to suppress expression of chosen Sp100 isoforms. We selected lentiviral vectors that are able to suppress the expression of the repressive Sp100 isoforms, but not Sp100A. HEp-2 cells were transduced with lentiviral vector Sp100si-2 or Sp100si-3 expressing shRNA to produce cell populations in which Sp100 levels were suppressed (HEp-2/Sp100si-2 and HEp-2/Sp100si-3). Control cells containing the vector with shRNA against an unrelated protein, NT2, were produced in parallel. We confirmed that expression of Sp100 isoform B, C, and HMG, but not Sp100A, mRNA was reduced in HEp-2/Sp100si-2 cells in comparison with HEp-2 and HEp-2/NT2si cells (Fig. 7A). Results were similar with HEp-2/Sp100si-3 (data not shown).
FIG. 7.
Effect of depletion of Sp100 isoforms containing SAND domains on IFN-β-mediated suppression of ICP0 expression. (A) Knockdown of Sp100 mRNA. Total RNA from HEp-2, and stable cell lines based on lentiviral infected HEp-2 cells, was extracted and subjected to semiquantitative RT-PCR for Sp100 isoforms and GAPDH. (B) Replicates of HEp-2 and stable cell lines expressing Sp100si-2 or NT2si were stimulated with 1,000 U/ml of IFN-β or left untreated as a control. Eighteen hours later cells were inoculated with 0.5 PFU of HSV-1 (strain 17+) per cell as described in Materials and Methods. After 3 h of infection, cells were harvested, and the nuclear fractions (30 μg/lane) were electrophoretically separated, transferred to nitrocellulose membranes, and probed with antibodies to ICP0 (top). Ezh2 was used as a loading control (bottom).
Replicates of HEp-2, HEp-2/NT2si, and HEp-2/Sp100si-2 cells, untreated or treated with 1,000 U/ml of IFN-β for 18 h, were infected with 0.5 PFU/cell of HSV-1 and collected at 3 h postinfection, and nuclear extracts were examined by Western blot analysis using anti-ICP0 antibodies or antibodies against the nuclear protein EZH2 as a loading control. In the controls, i.e., HEp-2/NT2si and HEp-2 cells, ICP0 protein levels were suppressed after treatment with IFN-β as expected (Fig. 7, compare lanes 1 and 2 and 5 and 6, respectively). However, no significant reduction in ICP0 protein levels was found in the HEp-2/shSp100si-2 cells after treatment with IFN-β (Fig. 7B, lanes 3 and 4). Similar results were observed with HEp-2/Sp100si-3 (data not shown). These findings suggest that the suppression of the IFN-upregulated Sp100 isoforms with SAND domains allow expression of ICP0 proteins, thus identifying these isoforms as downstream IFN-regulated mediators of HSV repression.
DISCUSSION
IFNs were first recognized and defined by their antiviral properties. However, IFNs also have pleiotropic effects on many aspects of cell physiology, including cell growth, cellular differentiation, and both the adaptive and innate arms of the immune response (reviewed in references 31 and 53). In order to sustain infection of an animal host, a virus must counteract innate and adaptive antiviral mechanisms. Efficient viruses enter the host cell and suppress its ability to mount an antiviral response on many levels by impairing recognition of the virus, activation of STAT, and expression of ISGs. In this respect, some viruses are more efficient in specific cell types, depending on the level of expression of these antiviral genes. Three distinct pathways, the double-stranded RNA-dependent protein kinase R, the 2′-5′ oligodenylate synthetase/RNase L, and the Mx proteins, mediate the known IFN-inducible antiviral response. However, cells from mice deficient in all of these pathways retain some IFN-induced antiviral activity (58), suggesting that other IFN-inducible proteins play a role in the antiviral response.
Our present evidence supports the hypothesis that IFN-upregulatable Sp100 might be one of the proteins with antiviral properties, at least during early HSV-1 infection (see also reference 57). By analyzing the IFN-dependent variability of Sp100 mRNA in different cells, we show that untreated HEp-2 cells express predominantly the Sp100A isoform and IFN-β treatment leads to increased accumulation of all isoforms, but mostly Sp100A (Fig. 1 and 3). In human fibroblasts, Sp100HMG is the predominant isoform, whereas Sp100A is the second most abundant isoform and neither Sp100HMG nor Sp100C is induced after IFN-β treatment. The parental HEK293 cells express less Sp100 than HEp-2 cells with A and HMG as major isoforms, the levels of which did not significantly change after IFN-β treatment. The 293-S clone had no recognizable Sp100, and only after treatment with relatively high levels of IFN-β (1,000 U/ml) was Sp100 detected in not more than ∼5% of the cells by IF assay. As determined by RT-PCR (Fig. 2) the isoform in these few cells was Sp100A. The low inducibility in both 293 cell lines may be due to their adenovirus 5-transformed state and, as such, a compromised STAT/JNK pathway (32). These data indicate that regulation of splicing and expression of Sp100 isotypes can vary in different cell types and might define the viral progression during early parts of the replication cycle. The status of the 293-S cells also made it possible to test downstream effects of IFN by transiently increasing the expression of specific IFN-upregulated proteins, such as Sp100.
When studied by IF assays, the Sp100 isoforms show differences in intranuclear distribution: Sp100A preferentially accumulated at ND10, but B, C, and HMG isoforms are diffusely present throughout the nucleus, and at high expression levels they aggregate at new domains quite different from ND10 in appearance and composition. These results are similar to, although slightly different from, an early report on the distribution of Sp100HMG and Sp100C (48). Possible explanations are that Sp100 isoforms may have different posttranslational modifications in the cell lines we tested and/or that they might express different isoforms of PML, which are known to affect the composition of ND10 in different cell lines. Moreover, we succeeded only in establishing stable 293-S cell lines expressing Sp100A or mutated Sp100B (W665Q) proteins, but not Sp100B, -C or -HMG proteins. The cells transfected with the latter proteins did not survive selection. These data suggest different roles for Sp100A than for the B, C, and HMG isoforms and may have relevance for the activity of immediate-early transcription of viral genomes, depending on their location within the nucleus.
We confirmed early observations that IFN-β can suppress ICP0 expression upon HSV-1 infection (Fig. 4A) (29, 37, 44, 45, 55). Suppression of Sp100 isoforms B, C, and HMG with RNA interference relieved IFN-β repression of ICP0 expression after HSV-1 infection (Fig. 7). This suggests that the repressive effect of IFN-β on ICP0 expression depends on the upregulation of those Sp100 isoforms that have SAND domains. In addition, these Sp100 isoforms can repress both expression of IE proteins after HSV-1 infection in transient transfection (Fig. 4 and 5) and expression from the HSV-1 ICP0 promoter (Fig. 6). We don't think that repressive Sp100 isoforms alone are responsible for this effect; most likely additional proteins, probably also inducible with IFN and present at ND10, are working together with these Sp100 isoforms. Nevertheless, these repressive Sp100 isoforms are significant parts of an intrinsic antiviral defense. Interestingly, Sp100A is not required for IFN-mediated effect on ICP0, even though this major Sp100 isoform was induced with IFN-β. It is possible that the Sp100A isoform is involved in viral repression either at a different stage of the HSV-1 replication cycle or for different viruses. Further investigation is required to clarify these observations.
The SAND domain-containing Sp100 isoforms are not found in recognizable amounts at ND10. This suggests that they are present throughout the nucleus under physiological concentrations and that repression of viral/transfected DNA occurs outside of ND10 or ND10-like aggregates. Only the Sp100A isoform accumulated preferentially at ND10 and did not suppress ICP0 expression. Moreover, Sp100A slightly increased luciferase expression from the ICP0 promoter. This effect may be specific through interaction with some proteins necessary for transcriptional activation, such as CBP, or nonspecific through removal of proteins involved in repression, such as HP1. Both HP1 and CBP are transiently accumulated in ND10 (4, 49). We are currently investigating whether HP1 is required for Sp100-mediated repression.
Our data also indicate the requirement for a functional SAND domain in the localization of Sp100 isoforms outside of ND10 and for the repression of the ICP0 promoter. SAND domains of AIRE, NUDR, DEAF-1, and GMEB-1 proteins, which are all transcriptional regulators, bind DNA. DNA binding properties have not been demonstrated for the Sp100 SAND domain, but the structure of the SAND domain was recently determined and allows speculation about such properties (5). A highly conserved tryptophan occurs at the DNA binding interface of the SAND domain of each of these proteins. Unlike wild-type Sp100B, the Sp100B variant, constructed with a mutation of this tryptophan (W655Q), lost its ability to repress expression from transfected DNA or infected virus. Further studies are needed to define the mechanisms of viral repression by the SAND domains in the individual Sp100 isoforms.
Initial transcription and replication of DNA viruses, such as HSV-1, human cytomegalovirus, adenovirus 5, and simian virus 40, is closely associated with ND10 or ND10-associated protein aggregates (13, 23, 35), which contain IFN-upregulated proteins such as PML and Sp100. Paradoxically, most of these viruses have developed gene products that either disperse ND10 or lead to the hydrolysis of ND10-associated proteins. In the case of HSV-1, the IE protein ICP0 initially accumulates in ND10 and induces the dispersal of ND10 and its associated proteins (12, 25, 34) and the degradation of PML and Sp100 proteins at 2 to 4 h postinfection (7, 18, 40) at a time when IE transcripts can be detected at sites adjacent to ND10 or ND10-associated protein aggregates (35). ICP0 is necessary to prevent IFN-β from silencing HSV-1 IE gene expression (20, 38), in part by counteracting an IFN-induced block to virus transcription (38). These observations raise the question of why these viruses start transcription and replication at a site with an abundance of proteins that have IFN-upregulated antiviral properties. This paradox can be resolved if one assumes that nuclear domains or the proteins migrating to viral genomes containing the nonrepressive Sp100A isoform are or generate environments that do not interfere with transcriptional activation of HSV-1 IE genes, whereas other dispersed Sp100 isoforms suppress viral genomes in other parts of the nuclear space. The greater likelihood that HSV-1 starts transcription in association with ND10 may therefore be a consequence of a less repressive environment at such sites or of the newly formed HSV genome-based sites that attract these proteins (13, 14). At later times of infection, the virus accumulates sufficient amounts of IE proteins and switches to the next stage of the viral transcription cascade. The accumulated ICP0 destroys PML and all Sp100 isoforms. IFN helps to suppress these steps by increasing the levels of Sp100 isoforms with SAND domains throughout the nucleoplasm. This IFN-dependent increase of repressive Sp100 isoforms should take place preemptively in cells adjacent to the infected ones, raising the bar to successful infection. Sp100B might lead to repression of IE genes through recognition of viral DNA. The other isoforms may recruit different corepressors, such as histone deacetylases and/or HMTases, dependent on their respective domains, including the plant homebox domain and Bromo domains in Sp100C or the HMG domain in the Sp100-HMG isoform, a context in which HP1 binding becomes relevant. The differential splicing after IFN induction in the respective cell types may suggest that different cellular environments will produce different levels of viral suppression or permissiveness.
Acknowledgments
We thank W. Yen for help with cell cultures; Ingrid Schwienhorst for original observations with 293-S cells; P. Hemmerich, J. L. Taylor, and A. Dejean for supplying plasmids; and members of G.G.M.'s laboratory for many helpful discussions.
This study was supported by funds from NIH AI 41136, NIH GM 57599, NIH HD 34612, and the Robert Leet and Clara Guthrie Patterson Trust. Support for the microscopy and sequencing facility came from NIH Core grant CA-10815. This project is funded in part by a grant from the Pennsylvania Department of Health.
REFERENCES
- 1.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloch, D. B., S. M. de la Monte, P. Guigaouri, A. Filippov, and K. D. Bloch. 1996. Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem. 271:29198-29204. [DOI] [PubMed] [Google Scholar]
- 3.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 4.Boisvert, F. M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottomley, M. J., M. W. Collard, J. I. Huggenvik, Z. Liu, T. J. Gibson, and M. Sattler. 2001. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8:626-633. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 7.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 8.Chelbi-Alix, M. K., L. Pelicano, F. Quignon, M. H. Koken, L. Venturini, M. Stadler, J. Pavlovic, L. Degos, and H. de The. 1995. Induction of the PML protein by interferons in normal and APL cells. Leukemia 9:2027-2033. [PubMed] [Google Scholar]
- 9.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, R. D., A. Cross, and A. Orr. 1993. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology 197:751-756. [DOI] [PubMed] [Google Scholar]
- 11.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gongora, C., G. David, L. Pintard, C. Tissot, T. D. Hua, A. Dejean, and N. Mechti. 1997. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem. 272:19457-19463. [DOI] [PubMed] [Google Scholar]
- 16.Gongora, R., R. P. Stephan, Z. Zhang, and M. D. Cooper. 2001. An essential role for daxx in the inhibition of b lymphopoiesis by type i interferons. Immunity 14:727-737. [DOI] [PubMed] [Google Scholar]
- 17.Grotzinger, T., T. Sternsdorf, K. Jensen, and H. Will. 1996. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML). Eur. J. Biochem. 238:554-560. [DOI] [PubMed] [Google Scholar]
- 18.Gu, H., and B. Roizman. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. USA 100:8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guldner, H. H., C. Szostecki, T. Grotzinger, and H. Will. 1992. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol. 149:4067-4073. [PubMed] [Google Scholar]
- 20.Harle, P., B. Sainz, Jr., D. J. Carr, and W. P. Halford. 2002. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293:295-304. [DOI] [PubMed] [Google Scholar]
- 21.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamitani, T., K. Kito, H. P. Nguyen, H. Wada, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273:26675-26682. [DOI] [PubMed] [Google Scholar]
- 27.Kamitani, T., H. P. Nguyen, K. Kito, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 273:3117-3120. [DOI] [PubMed] [Google Scholar]
- 28.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 29.Klotzbucher, A., S. Mittnacht, H. Kirchner, and H. Jacobsen. 1990. Different effects of IFN gamma and IFN alpha/beta on “immediate early” gene expression of HSV-1. Virology 179:487-491. [DOI] [PubMed] [Google Scholar]
- 30.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871-876. [PubMed] [Google Scholar]
- 31.Lengyel, P. 1993. Tumor-suppressor genes: news about the interferon connection. Proc. Natl. Acad. Sci. USA 90:5893-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard, G. T., and G. C. Sen. 1996. Effects of adenovirus E1A protein on interferon-signaling. Virology 224:25-33. [DOI] [PubMed] [Google Scholar]
- 33.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 34.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 35.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 36.Melroe, G. T., N. A. DeLuca, and D. M. Knipe. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittnacht, S., P. Straub, H. Kirchner, and H. Jacobsen. 1988. Interferon treatment inhibits onset of herpes simplex virus immediate-early transcription. Virology 164:201-210. [DOI] [PubMed] [Google Scholar]
- 38.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Negorev, D., A. M. Ishov, and G. G. Maul. 2001. Evidence for separate ND10-binding and homo-oligomerization domains of Sp100. J. Cell Sci. 114:59-68. [DOI] [PubMed] [Google Scholar]
- 43.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 44.Oberman, F., and A. Panet. 1989. Characterization of the early steps of herpes simplex virus replication in interferon-treated human cells. J. Interferon Res. 9:563-571. [DOI] [PubMed] [Google Scholar]
- 45.Oberman, F., and A. Panet. 1988. Inhibition of transcription of herpes simplex virus immediate early genes in interferon-treated human cells. J. Gen. Virol. 69:1167-1177. [DOI] [PubMed] [Google Scholar]
- 46.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeler, J. S., and A. Dejean. 2001. SUMO: of branched proteins and nuclear bodies. Oncogene 20:7243-7249. [DOI] [PubMed] [Google Scholar]
- 48.Seeler, J. S., A. Marchio, R. Losson, J. M. Desterro, R. T. Hay, P. Chambon, and A. Dejean. 2001. Common properties of nuclear body protein sp100 and tif1alpha chromatin factor: role of sumo modification. Mol. Cell. Biol. 21:3314-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seeler, J. S., A. Marchio, D. Sitterlin, C. Transy, and A. Dejean. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA 95:7316-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 51.Sourvinos, G., and R. D. Everett. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 21:4989-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stadler, M., M. K. Chelbi-Alix, M. H. Koken, L. Venturini, C. Lee, A. Saib, F. Quignon, L. Pelicano, M. C. Guillemin, C. Schindler, et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565-2573. [PubMed] [Google Scholar]
- 53.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 54.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, Q., L. Li, A. M. Ishov, V. Revol, A. L. Epstein, and G. G. Maul. 2003. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J. Virol. 77:5821-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, J. L., S. D. Little, and W. J. O'Brien. 1998. The comparative anti-herpes simplex virus effects of human interferons. J. Interferon Cytokine Res. 18:159-165. [DOI] [PubMed] [Google Scholar]
- 57.Wilcox, K. W., S. Sheriff, A. Isaac, and J. L. Taylor. 2005. SP100B is a repressor of gene expression. J. Cell. Biochem. 95:352-365. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]