Abstract
The psbB-psbT-psbH-petB-petD operon of higher plant chloroplasts is a heterogeneously composed transcriptional unit that undergoes complex RNA processing events until the mature oligocistronic RNAs are formed. To identify the nucleus-encoded factors required for the processing and expression of psbB-psbT-psbH-petB-petD transcripts, we performed mutational analysis using Arabidopsis. The allelic nuclear mutants hcf152-1 and hcf152-2 were identified that are affected specifically in the accumulation of the plastidial cytochrome b6f complex. In both mutants, reduced amounts of spliced petB RNAs (encoding the cytochrome b6 subunit) were detected, thus explaining the observed protein deficiencies. Additionally, mutant hcf152-1 is affected in the accumulation of transcripts cleaved between the genes psbH and petB. As a result of a close T-DNA insertion, the HCF152 gene was cloned and its identity confirmed by complementation of homozygous mutant plants. HCF152 encodes a pentatricopeptide repeat (PPR) protein with 12 putative PPR motifs that is located inside the chloroplast. The protein shows a significant structural, but not primary, sequence similarity to the maize protein CRP1, which is involved in the processing and translation of the chloroplast petD and petA RNAs. In addition, we found that HCF152 is an RNA binding protein that binds certain areas of the petB transcript. The protein possibly exists in the chloroplast as a homodimer and is not associated with other proteins to form a high molecular mass complex.
INTRODUCTION
Many chloroplast genes are organized in multiple transcription units that are transcribed into polycistronic transcripts. The translatable RNAs of these genes are generated by extensive RNA processing. The functional relevance of these maturation steps and the many auxiliary and regulatory factors that are necessary to catalyze these reactions (Barkan and Goldschmidt-Clermont, 2000) are mostly unknown. Some data indicate that the exonucleolytic and endonucleolytic processing of RNA 5′ ends is important to increase the translational efficiency of chloroplast RNAs (Reinbothe et al., 1993; Barkan et al., 1994; Hirose and Sugiura, 1997; Felder et al., 2001). RNA 3′ end maturation often is coupled with folding the RNA into stem and loop structures that bind specific proteins, thus preventing 3′ exonucleolytic degradation (Monde et al., 2000). In addition, little is known about the functional significance of plastid RNA splicing.
Between 17 and 20 genes (depending on species) of higher plant chloroplasts contain group II intron sequences that must be removed during the processing of the corresponding RNAs. These introns are characterized by a conserved secondary structure that is assumed to enable the intron RNA to fold into a catalytically active form. Group II introns are capable of in vitro autocatalytic splicing under nonphysiological high-salt conditions (Michel and Ferat, 1995). However, genetic evidence indicates that additional factors are required for efficient intron splicing in vivo. Some of them are encoded in an open reading frame within the intron and are designated as maturases. In higher plants and green algae, most of the splicing cofactors are encoded by nuclear genes, several of which have been identified recently. The CRS2 maize protein is involved in the splicing of several chloroplast introns and appears to be a general splicing factor of a subgroup of plastid introns (Jenkins et al., 1997; Vogel et al., 1997; Jenkins and Barkan, 2001). By contrast, CRS1 is necessary solely for the removal of the atpF intron (Jenkins et al., 1997; Till et al., 2001). Mutations in the Chlamydomonas reinhardtii Maa1 and Raa3 nuclear genes result in disturbed trans-splicing of the split intron in the psaA chloroplast gene (Goldschmidt-Clermont et al., 1990; Perron et al., 1999; Rivier et al., 2001). However, once a mature mRNA is produced inside the chloroplast, efficient translation also depends on the turnover rate of this mRNA. Molecular and biochemical analyses have shown that ribonucleoprotein complexes are assembled at the 5′ untranslated regions, and there is evidence that these structures, together with the 3′ stem-and-loop structures, determine the stability of the corresponding mRNA (Salvador et al., 1993; Nickelsen et al., 1994; Hotchkiss and Hollingsworth, 1999; Vaistij et al., 2000; Anthonisen et al., 2001).
We are interested in understanding the functional significance of chloroplast RNA processing for the expression of plastid genes. A genetic screen in Arabidopsis revealed the nuclear gene HCF152, which is involved in the processing and/or stabilization of petB- and psbH-containing transcripts. These genes belong to the psbB-psbT-psbH-petB-petD operon, which is a typical plastidial, polycistronic transcription unit that encodes polypeptides for two different thylakoid membrane complexes (Barkan, 1988; Westhoff and Herrmann, 1988). The psbB, psbT, and psbH genes encode the chlorophyll a apoprotein CP47 and the T and H subunits of photosystem II (PSII). petB and petD encode cytochrome b6 and subunit IV of the cytochrome b6f complex.
The hcf152 mutation is represented by two alleles, one of which is induced by T-DNA insertional mutagenesis (hcf152-1) and the other by ethyl methanesulfonate (EMS) treatment (hcf152-2). The mutants show reductions in the cytochrome b6f complex. At the RNA level, our analysis revealed that the accumulation of spliced petB RNA is impaired in both mutants. Moreover, the T-DNA hcf152-1 mutant also shows a defect in endonucleolytic cleavage between the genes psbH and petB. Molecular cloning of the HCF152 gene revealed that it encodes an 80-kD pentatricopeptide repeat (PPR) protein containing 12 putative PPR motifs. HCF152 is a soluble protein located in the chloroplast without being associated with other proteins to form a high molecular mass complex. We show also that HCF152 is an RNA binding protein that could bind with high affinity to certain sequences of the petB transcript. No significant sequence similarities with other proteins were found. However, HCF152 shows structural similarity to the maize protein CRP1, an accessory factor necessary for processing of the petD transcript and translation of the petA mRNA (Fisk et al., 1999).
RESULTS
Two Allelic, Recessive Nuclear Mutations Lead to a Block in the Photosynthetic Electron Transport Chain
The high-chlorophyll-fluorescence hcf152 mutant (formerly designated CRM3) was selected under standardized screening conditions (Meurer et al., 1996b) in a T-DNA–mutagenized collection of Arabidopsis (Bechtold et al., 1993). Homozygous mutant plants were found to be seedling lethal under autotrophic growth conditions on soil but grew similar to wild-type plants on sucrose-containing medium. An analysis of the ratio of mutant to wild-type seedlings in a segregating F2 generation revealed that hcf152 behaved genetically as a single, recessive mutation. The mutant phenotype and Basta resistance conferred by the T-DNA cosegregated, suggesting that the mutation was induced by the insertion of the T-DNA (data not shown). A second, phenotypically similar mutant (hcf119) was selected from an EMS mutant population (Meurer et al., 1996b). Crosses of heterozygous hcf152 to heterozygous hcf119 mutant plants resulted in a 3:1 segregation ratio of wild-type to mutant phenotypes (data not shown). This finding demonstrated that the two mutations were allelic; consequently, the mutants were named hcf152-1 and hcf152-2.
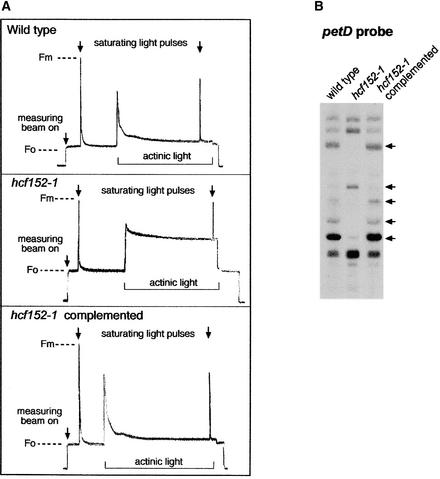
Chlorophyll fluorescence analysis revealed a normal variable-to-maximal fluorescence ratio, indicating that PSII activity was unaffected. However, both mutants exhibited reduced fluorescence quenching under illumination with actinic light (hcf152-1 [see Figure 5] and hcf152-2 [Meurer et al., 1996b]). The P700 redox kinetics showed that the electron flow to photosystem I (PSI) was inhibited but that this photosystem can be oxidized (hcf152-1 [data not shown] and hcf152-2 [Meurer et al., 1996b]). Therefore, both mutants possess normal PSII and PSI activities, but the interconnecting electron flow through the cytochrome b6f complex is disturbed.
Figure 5.
Characterization of Complemented hcf152 Transformants.
(A) Chlorophyll fluorescence induction kinetics of the wild type, hcf152-1 mutants, and hcf152-1 transformants complemented with the open reading frame of the At3g09650 gene. A representative curve for each plant is shown (for details, see Methods). Fm, maximal fluorescence; Fo, initial fluorescence.
(B) Transcript pattern of the psbB-psbT-psbH-petB-petD operon in the T1 generation of complemented hcf152-1. The RNA gel blot was hybridized with the probe against the petD gene. Arrows indicate significant differences between the mutants and the transformants.
Reduced Levels of the Cytochrome b6f Complex Components in hcf152-1 and hcf152-2
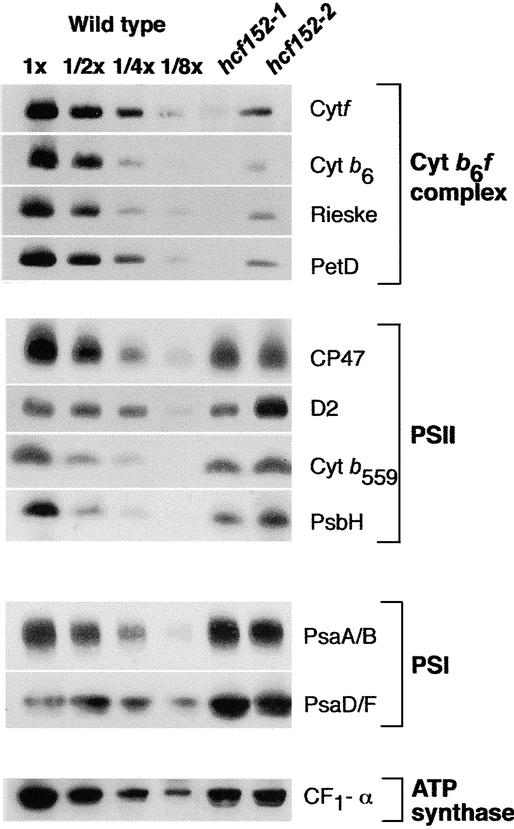
The block in the intersystem electron transport chain found in hcf152-1 and hcf152-2 could be the result of reduced levels of the cytochrome b6f complex. To prove this assumption, immunoblot analyses with antisera against the four major subunits of this photosynthetic complex were performed (Figure 1). It was found that cytochrome f, cytochrome b6, subunit IV of the cytochrome b6f complex (PetD), and the Rieske FeS protein accumulated to <12.5% in hcf152-1. The reduction of cytochrome b6f complex proteins in hcf152-2 was less drastic, and protein amounts reached ∼25% of wild-type levels. Subunits of PSII (CP47, D2, cytochrome b559, and PsbH), PSI (PsaA/B and PsaD/F), and the α-subunit of the ATP synthase accumulated to wild-type levels when the plants were grown under very low light conditions (5 to 10 μmol·m−2·s−1) (Figure 1). Under standard light intensities (20 to 50 μmol·m−2·s−1), decreased amounts of PSII (50%) and PSI (25%) subunits were observed in hcf152-1 but not in hcf152-2.
Figure 1.
Immunoblot Analysis of Photosynthetic Membrane Proteins from hcf152-1, hcf152-2, and the Wild Type.
Chloroplast proteins (20 μg or the indicated dilution of the wild-type sample) of 3-week-old plants were used for the analysis. Plants were grown under low light (5 to 10 μmol·m−2·s−1) in a growth chamber. Samples were fractionated on SDS-polyacrylamide gels, transferred to nitrocellulose, and immunodetected with the indicated antisera.
Together, the results of the spectroscopic analysis and the immunoblot data indicated that the hcf152-1/2 mutations resulted in a specific reduction of the cytochrome b6f complex. The lesser reduction of the complex in the EMS hcf152-2 mutant correlated with the observation that some of the homozygous hcf152-2 mutants were able to survive and set seeds under autotrophic growth conditions, unlike hcf152-1.
Altered Processing Pattern of psbB-psbT-psbH-petB-petD Transcripts in hcf152-1 and hcf152-2
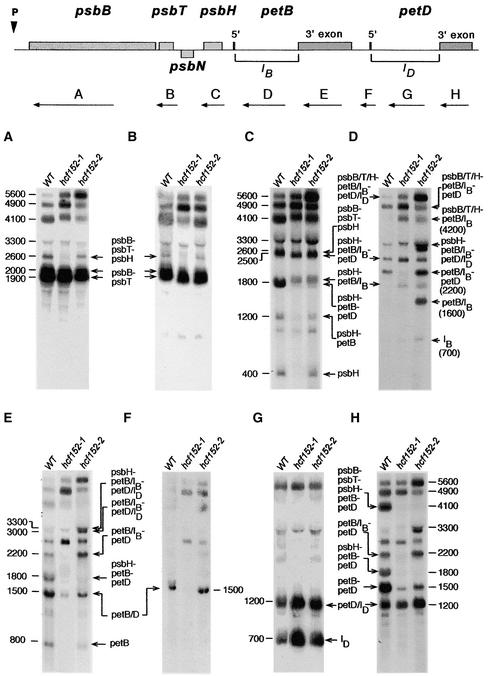
The reduced levels of the cytochrome b6f complex could be attributable to a deficiency in the accumulation of RNAs that encode the subunits of this complex. Therefore, we performed RNA gel blot hybridization experiments with probes for the plastid-encoded petA, petB, petD, and petG genes and the nucleus-encoded petC gene. petA, petG, and petC transcripts accumulated to wild-type levels in both mutants (data not shown). However, qualitative and quantitative differences with respect to the wild type were found for the petB and petD RNAs, which were transcribed as part of the polycistronic transcription unit psbB-psbT-psbH-petB-petD (Barkan, 1988; Westhoff and Herrmann, 1988). This finding suggested that altered RNA processing and/or RNA stability could be responsible for the observed mutant phenotypes. To analyze the processing defects of these RNAs precisely, all genic and extragenic regions of this transcription unit of Arabidopsis were cloned and used as probes in RNA gel blot hybridization experiments (Felder et al., 2001).
Figure 2 shows that the RNA pattern of the psbB/T part of the operon essentially was not affected in either mutant. Specifically, the dicistronic psbB-psbT RNAs of 1900 and 2000 nucleotides accumulated to levels similar to those of the wild type (Figures 2A and 2B). This RNA doublet reflects two different psbB RNA 5′ ends, one arising from transcriptional initiation and the other generated by RNA 5′ processing (Westhoff, 1985; Westhoff and Herrmann, 1988). In contrast to the psbB/T part, the pattern of oligocistronic petB/D RNAs in both mutants differed from that in the wild type. The tricistronic psbH-petB-petD RNA of 1800 nucleotides, the dicistronic petB-petD transcript of 1500 nucleotides, and the monocistronic petB RNA of 800 nucleotides were reduced significantly in hcf152-1 and hcf152-2 (Figures 2E and 2H). These RNAs were fully spliced (i.e., both the petB and petD introns were removed). The amount of RNA reduction differed for the two mutants: whereas the spliced products were reduced drastically in hcf152-1, all spliced transcripts accumulated to detectable amounts in hcf152-2.
Figure 2.
Transcript Pattern of the psbB-psbT-psbH-petB-petD Operon in the Mutants hcf152-1 and hcf152-2.
Eight micrograms of total leaf RNA from 3-week-old mutant and wild-type (WT) seedlings was analyzed by RNA gel blot hybridization. The nylon filters were hybridized with the gene-specific probes indicated with arrows and letters (A to H). Introns of the petB and petD genes are designated IB and ID, respectively. The sizes (in nucleotides) and identities of the transcripts are indicated.
The levels of other petB/D-containing RNAs were increased in hcf152-2 but not in hcf152-1 (Figures 2D and 2E). These were the 3300-nucleotide psbH-petB(IB)-petD(ID) RNA, the 3000-nucleotide petB(IB)-petD(ID) RNA, the 2200-nucleotide petB(IB)-petD RNA, and the 1600-nucleotide monocistronic petB(IB) RNA. Common to all of these RNAs was the continued presence of the petB intron, suggesting that the hcf152-2 mutation affects the splicing of this intron. However, the excised 700-nucleotide petB intron RNA accumulated to nearly similar amounts in the wild type and the mutants (Figure 2D). This finding is in contrast to the supposed splicing defect and indicates that the loss of the spliced petB RNAs also could be the result of a decrease in their stability. The splicing of the petD intron was not impaired, as shown by the accumulation of the 2500-nucleotide psbH-petB(IB)-petD RNA and the 2200-nucleotide petB(IB)-petD transcript in hcf152-2 (Figures 2D and 2H). The spliced monocistronic petD RNA was below the detectable level in both the mutants and the wild type. However, endonucleolytic processing between petB and petD occurred in the wild type and the mutants, because high levels of unspliced monocistronic petD(ID) RNA were detected (Figures 2G and 2H). A lack of monocistronic petD transcripts also was described for spinach chloroplasts (Westhoff and Herrmann, 1988). However, it accumulated in maize to considerable amounts and was clearly detectable (Barkan, 1988).
Impaired accumulation of spliced petB RNAs was not visible immediately in hcf152-1, because this mutant also was disturbed in RNA processing between psbH and petB. RNAs processed between these genes were depleted in hcf152-1. Therefore, unlike the situation in hcf152-2, the 2600-nucleotide psbB-psbT-psbH RNA, the 400-nucleotide monocistronic psbH RNA, and the dicistronic 2200-nucleotide petB(IB)-petD RNA did not accumulate in this mutant (Figures 2A, 2C, and 2E). In addition, the 1600-nucleotide petB(IB) transcript that accumulated in hcf152-2 was not detectable in hcf152-1 (Figure 2D).
The loss of spliced petB transcripts also was visible at the level of the pentacistronic precursor RNAs. Hybridization with the petD 3′ exon probe (Figure 2H) revealed that the fully spliced 4100-nucleotide psbB-psbT-psbH-petB-petD RNA was practically nonexistent in both mutants. A slightly larger transcript of ∼4200 nucleotides was detected in both mutants by all other genic hybridization probes (Figures 2A, 2B, 2C, and 2E). The hybridization behavior of this RNA and its size suggest that it contains psbB, psbT, psbH, and unspliced petB sequences. Accordingly, this RNA could be visualized by hybridization with a petB intron probe in the two mutants but not in the wild type (Figure 2D).
In maize, a mutant has been described that is disturbed in the splicing of several group-II plastid introns (Jenkins et al., 1997). Therefore, the transcript patterns of other group-II intron-containing genes (i.e., atpF and ndhA) were analyzed by RNA gel blot hybridization (data not shown). No significant qualitative and quantitative differences between the two mutants and the wild type were detected, suggesting that the hcf152 mutations specifically affect petB splicing or stabilization of the splicing products and the endonucleolytic cleavage of the psbH-petB segment.
RNase Protection Experiments Confirmed the Processing Defects in hcf152-1 and hcf152-2
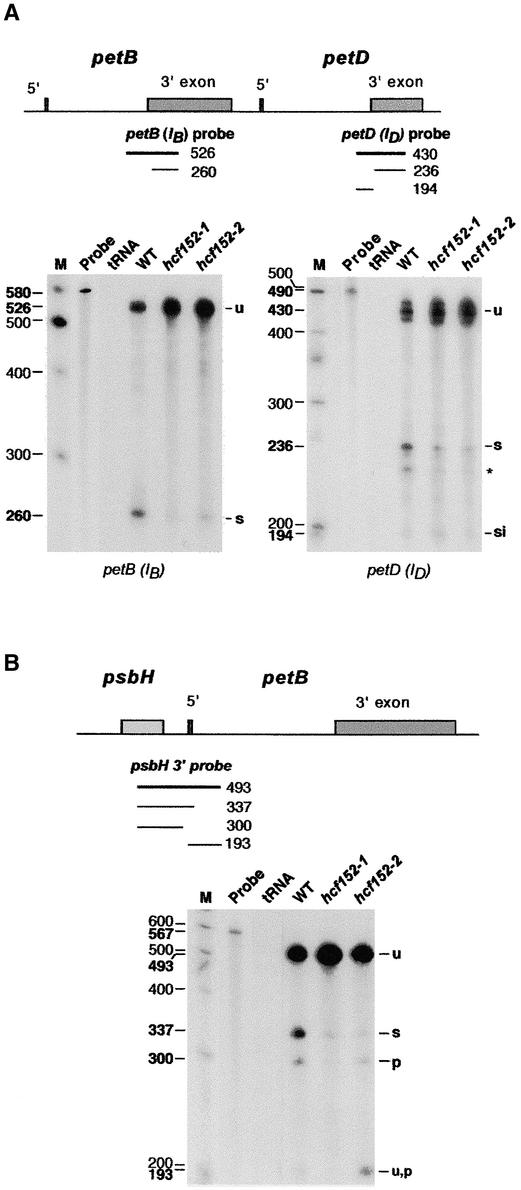
As a result of the complex RNA pattern of the gel blot hybridization experiments, not all of the transcripts were resolved from each other. Therefore, ribonuclease protection experiments were performed with RNA using wild-type and hcf152-1/2 seedlings to confirm the RNA gel blot hybridization results.
An RNase protection assay with an RNA probe spanning the petB 3′ splice junction revealed a larger fragment representing unspliced RNA (526 nucleotides) and a smaller one representing spliced RNA (260 nucleotides) (Figure 3A). The larger fragment was protected by wild-type and hcf152-1/2 RNA. In accordance with the RNA gel blot data, an increased amount of this fragment, protected by unspliced RNA, was visible in both mutants. By contrast, the signal of the partially protected fragment of 260 nucleotides was reduced drastically in hcf152-1 and reduced moderately in hcf152-2. This result confirmed that decreased amounts of spliced petB transcripts accumulated in both mutants, as deduced from the RNA gel blot analysis.
Figure 3.
RNase Protection Assay for Processed and Spliced RNAs of the psbB-psbT-psbH-petB-petD Operon in hcf152-1, hcf152-2, and the Wild Type.
RNase protection assay for spliced and processed petB, petD (A), and psbH (B) RNAs. Total leaf RNA isolated from the wild type (WT) or the mutants (hcf152-1 and hcf152-2) was annealed with uniformly labeled probes spanning the petB, petD, and psbH splice junction and processing sites. The transcribed probes included a small amount of vector sequence on both sides to be separated from the protected fragment of the unspliced RNA. A parallel control reaction contained no leaf RNA but an equivalent amount of yeast tRNA (tRNA). The RNA was digested with RNase T1, and the protected fragments were analyzed on sequencing gels and by autoradiography. The locations of the probes are indicated in the schemes (thick lines). The sizes of the protected fragments (thin lines and numbers in boldface to the left of the lanes) were determined by coelectrophoresis with a DNA sequence ladder. For subsequent identification of protected bands, a 100-bp DNA sequence ladder was used that was radioactively labeled (M) (sizes of the marker fragments are indicated with lightface numbers). Protected probe fragments correspond to unspliced (u), spliced (s), spliced intron (si), unspliced processed (u,p), or processed (p) RNA and are indicated at right. A protected band of the petD probe of ∼220 nucleotides of unknown origin is indicated with an asterisk. S1 protection experiments revealed a single protected fragment of 430 nucleotides for the unspliced petD RNAs, indicating that the triplet detected here reflects heterogeneous cutting by RNase T1.
To analyze the splicing of the petD intron, we performed RNase protection experiments using a probe spanning the 3′ splice junction of this intron (Figure 3A). Four probe fragments were found to be protected by the wild-type and mutant RNAs: the largest 430-nucleotide fragment (which appeared in this assay as three bands) corresponding to the unspliced transcript; the 236-nucleotide fragment representing the spliced petD RNA; a 220-nucleotide fragment whose nature is unknown; and the 194-nucleotide fragment corresponding to the petD intron. The probe fragment corresponding to the unspliced transcripts accumulated above the wild-type level in both mutants. The detected amount of the probe fragment protected by spliced petD RNAs was reduced in hcf152-1 and hcf152-2. This result revealed that although correct petD intron splicing occurred, the amount of spliced petD RNA was reduced in both mutants, in agreement with the RNA gel blot hybridization data.
To analyze RNA processing between the psbH and petB genes, an RNase protection assay with a probe spanning the psbH-petB intergenic region was performed (Figure 3B). The detected fragments correspond to the fully unprocessed RNA (493 nucleotides), the transcripts with the spliced petB intron (337 nucleotides), the transcripts with the free psbH 3′ end (300 nucleotides), and the processed but unspliced petB 5′ end (193 nucleotides). Protection of the 337-nucleotide fragment was reduced significantly in both mutants, again confirming the impaired accumulation of spliced petB transcripts. Only traces of the 300-nucleotide probe fragment protected by the free psbH 3′ ends were detectable in hcf152-1. This result shows that the processing between the psbH and petB genes was disturbed drastically in this mutant. The 300-nucleotide signal in hcf152-2 was reduced only slightly. Thus, RNAs with a free psbH 3′ end accumulated in hcf152-2 to a considerable level. This result reflects the RNA gel blot data from hcf152-2, in which psbH transcripts with a free 3′ end were found in close to wild-type amounts (Figures 2B and 2C, psbB-psbT-psbH and psbH RNAs). The protected fragment corresponding to the processed but unspliced petB 5′ end (193 nucleotides) was not detectable in hcf152-1 but accumulated above the wild-type level in hcf152-2. This finding also reflects the RNA gel blot data (Figure 2D, the 1600-nucleotide band petB/IB) and confirms that processing between psbH and petB was disturbed in hcf152-1 but not in hcf152-2.
Together, the results of the RNA gel blot analysis and the RNase protection experiments indicate that the hcf152 mutations cause pleiotropic effects in the expression of petB-containing RNAs. The T-DNA allele HCF152-1 impaired both the endonucleolytic cleavage between psbH and petB and petB intron splicing or stabilization of the splicing products. The EMS mutation resulted in the reduced accumulation of spliced petB transcripts but did not affect processing between psbH and petB. These phenotypes could have resulted directly from the lack or change of the protein encoded by the HCF152 gene. Alternatively, the HCF152 protein could be involved in another related function (e.g., in the translation of these genes) that when impaired results in defective RNA processing.
Identification of the HCF152 Gene
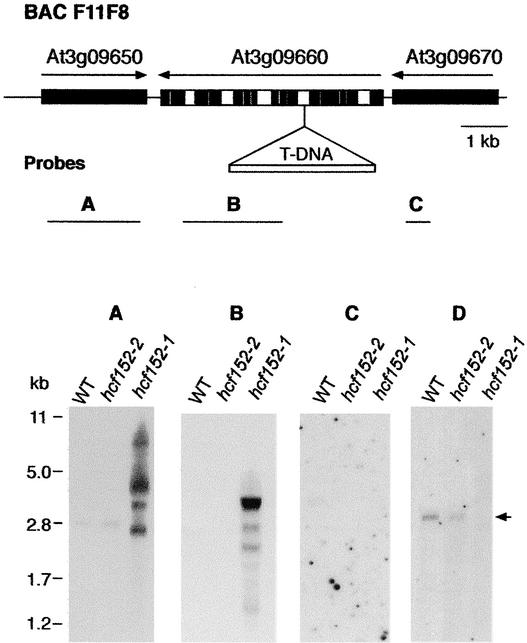
Segregation analysis of the mutant phenotype and the phosphinotricin resistance showed that the mutated gene in hcf152-1 was tagged by the T-DNA. DNA gel blot analysis revealed a single T-DNA insertion with a complex structure of tandem and inverted repeats (data not shown). The genomic region flanking the left border of the T-DNA was isolated by inverse PCR (Ochman et al., 1993) and then sequenced. Database searches revealed that the DNA sequence obtained belonged to one intron of the putative At3g09660 gene (Figure 4). This gene was assigned to encode a putative minichromosome maintenance factor (MIPS Arabidopsis database). The suggested nature of this gene had no obvious relevance to the observed mutant phenotype. Furthermore, the predicted At3g09660 protein did not show the signature of a protein that is transported into plastids. Because it is known that T-DNA insertions may affect neighboring genes (Tax and Vernon, 2001), the two flanking genes, At3g09650 and At3g09670, were investigated for possible involvement.
Figure 4.
Map of the Genomic Region Around the T-DNA Insertion in hcf152-1 and Analysis of Transcript Levels of the Corresponding Genomic Region.
In the scheme at top, the T-DNA insertion is located in a genomic segment of BAC F11F8. Genes adjacent to the insertion point are shown and designated according to the MIPS database code. Black bars represent exon sequences, and white bars represent intron sequences. Arrows indicate gene orientation. The probes used in RNA gel blot hybridization experiments are depicted below the scheme.
(A) to (C) Transcript levels of the At3g09650 (A), At3g09660 (B), and At3g09670 (C) genes in wild-type (WT) and hcf152-1/2 detected with double-stranded probes. Five micrograms of poly(A) RNA was loaded in each lane.
(D) Wild-type and mutant RNA hybridized with an antisense probe of the At3g09650 gene. The transcript of HCF152 is indicated by the arrow. Two micrograms of poly(A) RNA was loaded in each lane.
To determine which of the three genes are expressed in the photosynthetic tissue of wild-type plants and to compare the expression pattern with that of the mutant plants, poly(A) RNA was isolated from leaves and examined by RNA gel blot analysis (Figure 4). Hybridization with double-stranded probes for each of the three genes revealed that only At3g09650 was expressed in wild-type leaves, resulting in a transcript of 2800 nucleotides (Figure 4A), whereas RNAs from At3g09660 (Figure 4B) and At3g09670 (Figure 4C) were not detected. The RNA of At3g09650 also was found in the EMS-induced hcf152-2 mutant but was absent in the T-DNA insertion hcf152-1 mutant (Figures 4A and 4D). Instead, several different transcripts ranging from 7000 to 1000 nucleotides were detectable in the T-DNA mutant using a double-stranded probe (Figure 4A). An antisense probe for the At3g09650 gene detected none of these transcripts (Figure 4D). This finding indicates that these artificial RNAs are antisense transcripts of the At3g09650 gene that might be caused by the read-through transcription from the 35S promoter of the Basta pGKB5 gene (Bouchez et al., 1993) into the adjacent genomic DNA. Consequently, the At3g09660 probe also detected a multiple RNA binding pattern ranging from 4000 to 1000 nucleotides in the T-DNA mutant (Figure 4B). This finding indicates that the T-DNA insertion into At3g09660 has an inhibitory effect on At3g09650 expression, which moreover suggests that the drastic reduction in the gene expression level could be responsible for the mutant phenotype of hcf152-1.
To confirm this conclusion, two experimental lines were pursued. First, complementation of the mutant defect in hcf152-1 was attempted by transformation with a wild-type At3g09650 gene. Second, the entire At3g09650 gene was sequenced in hcf152-1 and hcf152-2. Because the T-DNA mutation was induced in the Wassilewskija ecotype, the gene from the wild-type plants of this ecotype also was sequenced.
The At3g09650 sequence in the Arabidopsis database revealed no intron sequences for this gene. Therefore, to restore the mutant phenotype of hcf152-1 mutants, we constructed a clone of At3g09650 containing the complete open reading frame using PCR (see Methods). The sequence in the database revealed that the assigned putative ATG start codon is preceded by a second ATG in the same reading frame, 41 amino acids in front of the first one. Because we did not know which of these two start codons is recognized by the ribosomes, the transformation construct included the sequence with the upstream start codon to ensure that the complete coding sequence of At3g09650 was used to complement the mutant. The open reading frame of At3g09650 was fused to the 35S promoter of Cauliflower mosaic virus (see Methods), and the chimeric gene was introduced into homozygous hcf152-1 mutant plants via Agrobacterium tumefaciens using a root transformation protocol (Valvekens et al., 1988). Twelve independent transformants were regenerated, and the chlorophyll fluorescence kinetics of these plants were measured (Figure 5A). Eleven of the transformants showed normal wild-type fluorescence behavior; only in one regenerated plant was the mutant phenotype still visible (data not shown). Four independent transformants were cultivated in soil and set seeds. The obtained T1 plants segregated, as expected, into wild-type and mutant phenotypes. T1 seedlings with the wild-type phenotype were selected and investigated for the pattern of petB-petD–containing transcripts by means of RNA gel blot hybridization experiments (Figure 5B). In all four lines investigated, a wild-type petB-petD RNA pattern was observed. The complementation test thus demonstrated that the RNA-processing defect of hcf152-1 was restored by introducing the At3g09650 gene into the genome of this mutant.
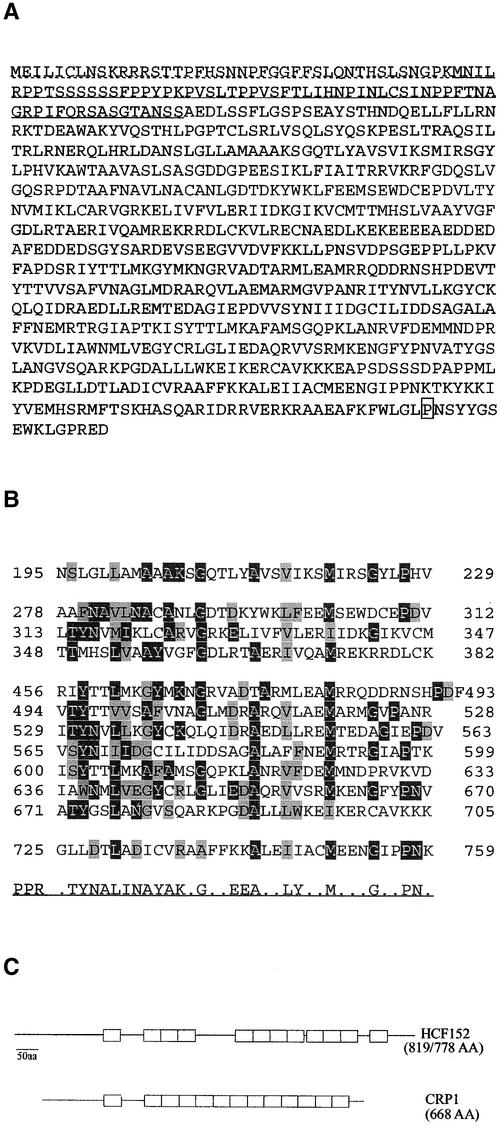
DNA sequence analysis of the entire open reading frame of At3g09650, of 1600 bp of the 5′ untranslated region, and of 600 bp downstream of the 3′ end of the T-DNA mutant allele revealed no differences compared with the sequence of the Wassilewskija wild type. We also sequenced the At3g09650 gene of the allelic EMS hcf152-2 mutant. The open reading frame of HCF152-2 possesses a point mutation at the 3′ end that is a C-to-T exchange at nucleotide 2288. This mutation changes a Pro codon in the distal C terminus to a Leu codon (Figure 6A). Both the existence of this point mutation and the complementation analysis demonstrated that the observed mutant phenotype of hcf152-1 and hcf152-2 is caused by the inactivation/mutation of the At3g09650 gene. Therefore, we have designated this gene HCF152.
Figure 6.
Predicted Amino Acid Sequence of HCF152 and Alignments of Its PPR Motifs.
(A) Protein sequence of HCF152 with the putative plastid transit sequence underlined. The dotted line indicates the sequence translated from the upstream ATG start codon. The amino acid that is altered in the EMS allele HCF152-2 (Pro → Leu) is boxed.
(B) Alignment of the 12 putative PPR motifs. Residues identical to the consensus motif (shown at bottom) are shaded in black, and similar residues are shaded in gray.
(C) Comparison of the PPR motif arrangement in HCF152 with that of CRP1 of maize. The PPR motifs are indicated as open boxes. aa, amino acids.
HCF152 Is a PPR Protein Imported into the Chloroplast
The open reading frame of HCF152 encodes a polypeptide of 778 amino acids (or, including the upstream ATG, of 819 amino acids) with a calculated molecular mass of 87 (or 92) kD. Database searches revealed that HCF152 is a PPR protein. PPR motifs consist of a degenerate 35–amino acid consensus sequence with a characteristic distribution of hydrophobic, hydrophilic, and small side-chain amino acids ending in a Pro-induced turn (Figure 6B) (Small and Peeters, 2000). Twelve PPR motifs with varying degrees of conservation were identified in HCF152 (Figure 6B), but only the 2 to 4 and 5 to 11 repeats were contiguous. It is possible that a 13th PPR domain exists in HCF152 between motifs 4 and 5 (amino acids 364 to 401), but the sequence is not well conserved, so we omitted this PPR domain in Figures 6B and 6C. Database searches with the deduced HCF152 sequence showed weak similarities (24% identity) to an Arabidopsis hypothetical protein (At1g03100) containing the so-called clathrin sevenfold repeat (Smith and Pearse, 1999) and to a large group of proteins designated salt-inducible proteins. All of these proteins contain several PPR domains, but none was characterized. Interestingly, HCF152 was found to be 22% identical to the CRP1 protein of maize. CRP1 also is a PPR protein and is necessary for processing of the monocistronic petD mRNA from a polycistronic precursor and translating the chloroplast petA transcripts (encoding cytochrome f) (Barkan et al., 1994; Fisk et al., 1999). Despite the low sequence identity, the arrangement of the PPR repeats is comparable in HCF152 and CRP1 (Figure 6C). Besides the PPR domains, no other known protein motifs were found in HCF152.
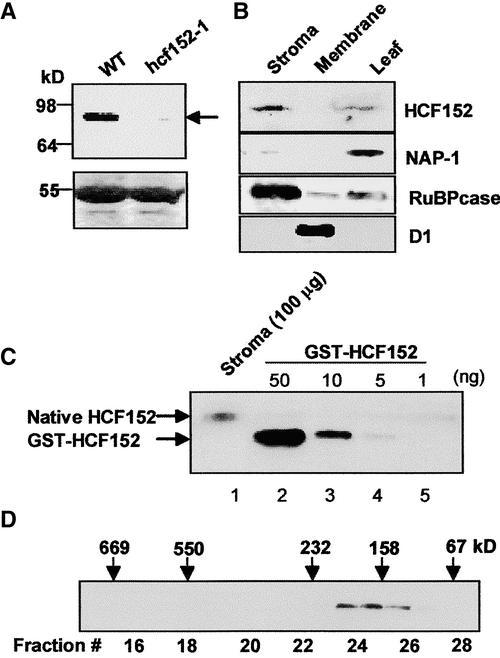
The N-terminal sequence of HCF152, starting with the first identified Met, contains many charged and hydroxylated amino acids, which is characteristic of chloroplast transit sequences (Cline and Henry, 1996), suggesting that the protein is located in this organelle. To test this assumption and to detect the mature HCF152 protein in Arabidopsis, specific antibodies to the recombinant protein were generated and used to decorate an immunoblot of total leaf proteins extracted from wild-type and hcf152-1 seedlings. The antibodies revealed a band at ∼85 kD that was not present in the mutant plants, indicating that this protein is either absent or is produced at a very low level in the mutant (Figure 7A). The HCF152 protein contains 778 (819) amino acids, 64 (105) of which are predicted to function as the chloroplast transit peptide that is cleaved after translocation into the chloroplast, resulting in a molecular mass of 80 kD. Therefore, the detected protein migrates slightly higher than its molecular mass predicts.
Figure 7.
Detection of HCF152 in Arabidopsis Chloroplasts.
(A) Detection of HCF152 in the wild type (WT) and hcf152-1 mutants. The protein was detected by immunoblot analysis using a specific HCF152 antibody (top gel). Total leaf proteins (50 μg) were analyzed. Pounceau S staining of the large subunit of ribulose bisphosphate carboxylase at 55 kD is presented in the bottom gel.
(B) Immunolocalization of HCF152. Chloroplasts were purified from Arabidopsis leaves and were fractionated further into the stroma and membrane fractions. Proteins (100 μg) of these fractions and total soluble leaf proteins were examined by immunoblot analysis using the specific antibodies to HCF152, the cytoplasm- and nucleus-located NAP-1, the stroma-located large subunit of ribulose bisphosphate carboxylase (RuBPcase), and the thylakoid protein D1.
(C) Quantification of HCF152 in the stroma fraction. A total of 100 μg of chloroplast stromal proteins as well as the indicated amounts of glutathione S-transferase fused to the N terminus of the HCF152 protein (GST-HCF152) was examined by immunoblot analysis. The native HCF152 and the GST-fused HCF152 are indicated by arrows. The HCF152 antibodies do not cross-react with the GST part of the fusion protein used for quantification.
(D) Fractionation of HCF152 by size-exclusion chromatography. Chloroplast-soluble proteins were fractionated on a Superdex 200 size-exclusion column. The proteins of each fraction were examined for the presence of HCF152 by immunoblot analysis with specific antibodies. The elution profiles of several molecular mass markers are indicated at top. Extensive treatment with ribonucleases did not change the elution profile.
To locate the HCF152 protein in the plant cell, chloroplasts were purified and fractionated further into stroma and membrane fractions. The HCF152 protein was located exclusively in the stroma fraction of the chloroplast and was enriched compared with the total soluble leaf protein fraction (Figure 7B). To quantify the amount of HCF152 in the stromal proteins of the chloroplast, the signal obtained with 100 μg of stromal proteins was compared with the signal of known amounts of the purified recombinant protein. The amount of HCF152 was estimated to be ∼5 ng in 100 μg of stromal protein (0.005%) (Figure 7C). In comparison, the amounts of two other proteins from spinach chloroplasts involved in RNA metabolism, the exoribonuclease polynucleotide phosphorylase and the RNA binding protein 28 RNP, were found to be present in higher amounts of 0.10 and 0.35%, respectively (Lisitsky et al., 1995; Nakamura et al., 2001; Yehudai-Resheff et al., 2001). This calculation provides an estimated 1000 to 1500 molecules of HCF152 in each chloroplast, which is approximately the number of petB transcripts found in a chloroplast of dark-grown barley (Rapp et al., 1992).
HCF152 Is Eluted at 180 kD When Fractionated on a Size-Exclusion Column
Several proteins described previously as involved in chloroplast gene expression were found to be associated in high molecular mass complexes of ∼300 to 1700 kD (Fisk et al., 1999; Boudreau et al., 2000; Vaistij et al., 2000; Jenkins and Barkan, 2001; Rivier et al., 2001; Till et al., 2001; Auchincloss et al., 2002). To determine whether HCF152 also is associated in such a complex, chloroplast-soluble proteins were fractionated through a size-exclusion column, and the presence of HCF152 was detected with specific antibodies. HCF152 was eluted in one peak at ∼180 kD (Figure 7D), and, unlike CRP1 of maize, was not associated in a high molecular mass complex of several hundred kilodaltons. Treatment of the chloroplast proteins with ribonuclease before fractionation to remove any RNA molecules that may bind and modify the elution profile did not change the migration pattern (data not shown), indicating that the HCF152 complex did not contain RNA components. Because the molecular mass of a mature HCF152 is 85 kD, the result of the size fractionation suggested that either the protein was not associated in a complex and was fractionated at this aberrant molecular mass or that HCF152 was associated with other chloroplast proteins in a complex or, alternatively, formed a homodimer.
Together, these results indicated that HCF152 is a relatively low-abundance soluble protein located in the chloroplast that is not associated in a high molecular mass complex.
RNA Binding Characteristics of HCF152
hcf152-1 was isolated as a nonphotosynthetic high-chlorophyll-fluorescence mutant. Examination of the pattern of chloroplast transcripts by RNA gel blot analysis revealed differences in the psbB-psbT-psbH-petB-petD polycistronic transcriptional unit and, more precisely, in the vicinity of petB. This observation led to the hypothesis that the HCF152 gene product is required, either directly or indirectly, for the correct endonucleolytic processing between psbH and petB and for splicing of the petB intron or stabilization of the splicing products. The involvement of HCF152 in these processes by enzymatic activity such as degradation or cleavage of the RNA seems unlikely, because no such activity was detected when the recombinant, bacteria-produced protein was incubated with different kinds of RNA molecules (data not shown). Therefore, we next decided to analyze whether the HCF152 harboring the 12 PPR motifs is an RNA binding protein.
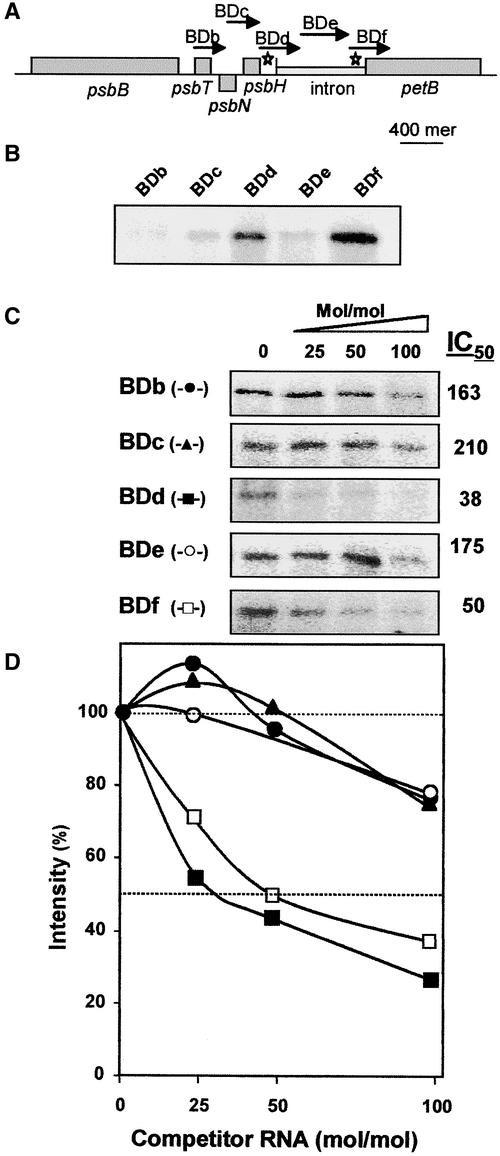
First, an RNA binding, UV cross-linking experiment was performed to analyze several RNA molecules of ∼400 nucleotides spanning the psbB multicistronic transcript (Figures 8A and 8B). To prevent nonspecific binding of HCF152 to RNA, an extra amount of ∼330-fold yeast tRNA was included in the reaction mixture. Two RNA molecules corresponding to the 5′ and 3′ borders of the petB intron, and to the corresponding parts of the related exons, were found to bind the recombinant HCF152 (BDd and BDf in Figure 8B). The UV cross-linking assay gave very low signals, with RNAs corresponding to the sequences of psbT, psbN, and the middle of the petB intron (BDb, BDc, and BDe). Therefore, HCF152, a PPR family protein, is an RNA binding protein that may bind in vitro certain RNA sequences with higher affinity than others. Moreover, the two RNAs that gave the high UV cross-linking signal represent the intron-exon junctions of the petB transcript, whose splicing was possibly impaired, and the psbH-petB intergenic region. However, because the sequence of nucleotides differed within the RNA molecules involved, the lack of a UV cross-linking signal does not necessarily indicate that there is no binding. To verify the binding properties of HCF152, we analyzed the binding of these RNAs using the UV cross-linking competition method. In this method, a single RNA is radioactively labeled to provide the UV cross-linking signal when binding the protein, whereas extra amounts of the tested RNA molecule are added and compete for the binding of the radioactive RNA. An RNA that efficiently competes for the binding binds HCF152 with high affinity. The parameter IC50 was defined as the concentration of the competitor RNA that resulted in a reduction of 50% of the radioactive UV cross-linking signal (Lisitsky et al., 1995). The lowest IC50 was for certain RNA; the highest was for the affinity of this RNA to the protein.
Figure 8.
RNA Binding Characteristics of HCF152.
(A) Scheme of the Arabidopsis psbB-petB region. The RNA probes for the binding assays are indicated by arrows and letters (BDb to BDf). The length of each arrow indicates the length of the probe, and a scale bar for 400 nucleotides is indicated. Stars indicate the high-affinity binding sites for HCF152.
(B) RNA binding of HCF152-F to several RNAs derived from the psbB-petD operon was analyzed with the UV cross-linking assay.
(C) Competition UV cross-linking assay. The indicated RNAs competed with the BDd RNA binding to the recombinant HCF152-F. The experiments were performed with radiolabeled BDd RNA and 25-, 50-, or 100-fold molar excess of the nonradioactive RNAs BDb to BDf. The IC50 values calculated from the graphs shown in (D) of at least three independent experiments are shown.
(D) Intensities of the bands in (C) were quantified and plotted. The intensity without a competitor RNA was defined as 100%. Closed circle, BDb; closed triangle, BDc; closed square, BDd; open circle, BDe; open square, BDf.
Taking this finding into account, competitive UV cross-linking assays were performed. The UV cross-linking assay was repeated using 32P-BDd RNA, the protein, the ∼330-fold yeast tRNA, and the corresponding nonradioactive RNAs in molar excess as indicated in Figure 8. As shown in Figures 8C and 8D, the results of this assay confirmed that the two RNAs, BDd and BDf, bound HCF152 with relatively high affinity, whereas the other RNAs derived from the psbB multicistronic transcript displayed very low affinities.
Together, the experiments presented here identified HCF152 as a nucleus-encoded protein containing 12 PPR domains that has low abundance, is located in the stroma fraction of the chloroplast, and is capable of binding the petB pre-RNA in vitro.
DISCUSSION
All available evidence suggests that most primary transcripts of plastid-encoded genes have to be processed to function efficiently as templates for translation. Processing includes endonucleolytic cleavage in the intercistronic regions, exonucleolytic trimming of RNA 5′ and 3′ ends, and splicing of intron sequences. If mature transcripts are produced, specific stabilization factors often are required to ensure the efficient translation of the RNA. Only limited data are available at present regarding the proteins involved in RNA maturation and stabilization inside the chloroplast. Nucleus-encoded factors play an important role during these processes; therefore, the isolation of nuclear mutants with defects in the metabolism of chloroplast RNAs provides a suitable means to identify these factors. Here, we describe the two allelic high-chlorophyll-fluorescence mutants of Arabidopsis, hcf152-1 and hcf152-2, which are defective in distinct steps of the expression of the polycistronic psbB-psbT-psbH-petB-petD transcription unit. We further report the identification of the nuclear gene that is responsible for the observed phenotypes of the mutants.
HCF152 Is Involved Directly or Indirectly in the Accumulation of petB Splicing Products and in Endonucleolytic Cleavage between psbH and petB
The homozygous hcf152-1/2 mutants showed a significant deficiency of cytochrome b6f complex subunits, whereas components of PSII, PSI, and the ATP synthase accumulated to nearly wild-type levels in both mutants. However, in the EMS hcf152-2 mutant, all cytochrome b6f complex proteins clearly were detectable compared with those in hcf152-1. Therefore, the phenotype of the EMS mutant can be characterized as the weaker of the two. The accumulation behavior of photosynthetic proteins was reflected by chlorophyll fluorescence measurements, which showed drastically reduced intersystem electron transport but normal PSII and PSI activity. Together, these results showed that the nuclear mutations in hcf152-1/2 affect primarily the accumulation of the cytochrome b6f complex.
At the RNA level, the reduced amounts of the cytochrome b6f complex were correlated with defects in the processing or stabilization of petB-containing transcripts of the psbB-psbT-psbH-petB-petD transcription unit. In both mutants, decreased amounts of spliced petB transcripts were detected, indicating that the HCF152 protein could be essential for the removal of this intron from the precursor RNA. Accordingly, a significant overaccumulation of unspliced petB-containing transcripts was observed in the EMS hcf152-2 mutant. However, a splicing defect also should result in reduced levels of free intron RNA, but similar amounts of the petB intron were detected in the wild type and the mutants. Therefore, it is possible that HCF152 is not necessary for the removal of the petB intron but may function as a specific stabilizing factor that binds and stabilizes only the processed and spliced petB transcripts. No defects in the splicing of the atpF and ndhA introns were detected. Furthermore, the mutant phenotype was restricted to the cytochrome b6f complex. Therefore, one possibility is that HCF152 encodes a nuclear factor that is involved directly in the splicing or stabilization of petB-containing RNAs without being involved in the splicing of other plastidial group-II introns. Alternatively, HCF152 also could function indirectly as a specific translation factor of petB transcripts that, when absent, result in the rapid degradation of spliced petB RNAs.
The T-DNA insertion hcf152-1 mutant shows an additional processing defect besides the lesion described above. RNA gel blot analysis and nuclease protection experiments revealed that intercistronic cleavage between psbH and petB was disturbed in hcf152-1 (Figure 3B) (Felder et al., 2001). Because a significant overaccumulation of unprocessed RNA was not observed in the mutant, we assume that the cleaved transcripts were degraded. The RNA accumulation behavior of the EMS mutant hcf152-2 indicates normal processing between psbH and petB. These differences between the mutants suggest that the endonucleolytic cleavage between psbH and petB is impaired in hcf152-1, in which the protein is absent, but is not disturbed in hcf152-2, in which the protein is produced with one amino acid change. We conclude from these data that HCF152 plays a dual role: it could be involved specifically in intercistronic processing between psbH and petB and in splicing of the petB intron. Alternatively, it also could be required to stabilize the spliced, processed transcripts, either directly or indirectly.
HCF152 Encodes a PPR Protein Targeted to the Chloroplast
The N-terminal sequence of HCF152 possessed the attributes of a chloroplast transit peptide, indicating that the protein is located in this organelle. This inference was confirmed by import studies with spinach chloroplasts (data not shown). The experiments revealed that the 95-kD HCF152 preprotein was imported into spinach chloroplasts with concomitant cleavage of the transit sequence, leading to a mature protein with a relative molecular mass of ∼85 kD. Chloroplast fractionation and immunoblot analysis showed that HCF152 was localized within the chloroplast stromal compartment.
HCF152 contains no strong similarities to proteins with known functions. A protein motif search with the Pfam database (Sonnhammer et al., 1998; Bateman et al., 1999) identified 12 PPRs. PPR proteins constitute a large family of plant proteins having >200 genes in the complete Arabidopsis genome (Small and Peeters, 2000). Two-thirds of these proteins are predicted to be targeted to chloroplasts or mitochondria. PPR protein genes are restricted largely to the plant kingdom, with only a few genes found in human, yeast, and Neurospora (Small and Peeters, 2000). The PPR motif is predicted to resemble the structure of the tetratricopeptide repeat (TPR) motif that consists of a protein sequence constituting two antiparallel α-helices. A tandem array of TPR motifs is able to form a superhelix with a central groove, which very likely is involved in protein–protein interaction (Das et al., 1998).
Concerning the number and distribution of PPR motifs, HCF152 showed high similarities to the RNA-processing factor CRP1 of maize. CRP1 possesses 13 PPR domains: we identified 12 domains in HCF152 with significant similarities to the PPR consensus sequence and a possible 13th motif with a low degree of conservation. Despite the lack of significant sequence similarities, HCF152 and CRP1 show this structural correspondence, suggesting that both proteins might be involved in functionally comparable steps during RNA processing. CRP1, which is required for the processing of the petD RNA and the translation of petA transcripts, is, like HCF152, a soluble component of the chloroplast stroma and part of a multisubunit protein complex of ∼350 kD (Fisk et al., 1999). HCF152 was fractionated on a size-exclusion column at 180 kD, suggesting that two monomers are associated to form a homodimer complex. Therefore, unlike CRP1, HCF152 is not associated in a high molecular mass complex. In addition, we found HCF152 to be an RNA binding protein that binds to the petB intron-exon junctions and the region between psbH and petB. In the hcf152-1 mutant, this protein was not produced, resulting in no processing between psbH and petB and drastically impaired petB intron splicing or stabilization of splicing products (Figures 2 and 3).
However, in hcf152-2, the protein was produced with one amino acid change located near the C terminus but not in a PPR domain (Figure 6). This point mutation in the produced protein leads to reduced accumulation of spliced petB RNAs in the mutant but does not influence the processing between psbH and petB. The significance of this amino acid change is unclear, because database searches revealed no HCF152 orthologs in other species. Unfortunately, all attempts to produce HCF152-2 as a soluble protein failed, and we were not able to analyze RNA binding properties. However, preliminary results suggest that HCF152 forms a homodimer and that the formation of the homodimer is impaired in HCF152-2 (data not shown). Therefore, it is possible that the modified protein produced in the hcf152-2 mutant is not assembled correctly into the homodimer form and that in the absence of homodimer formation, the HCF152 function is impaired and unspliced RNAs accumulate. Together, these results favor the model that HCF152 is a processing factor that binds as a homodimer to RNA sequences in the petB intron vicinity, resulting in the accumulation of the spliced, processed petB transcripts either directly or indirectly.
Besides CRP1 and HCF152, several other PPR proteins have been characterized in detail: PET309 of Saccharomyces cerevisiae and CYA-5 of Neurospora crassa are required for the production or stabilization of intron-containing coxI mRNA and the translation of the coxI mRNA (Coffin et al., 1997; Manthey et al., 1998). In petunia plants, a PPR protein required to restore the fertility of cytoplasmic male-sterile plants was identified recently (Bentolila et al., 2002). A striking similarity of CRP1, PET309, and CYA-5 is their involvement in the organellar RNA metabolism. This observation led to the assumption that PPR domains are putative RNA binding motifs. The predicted three-dimensional structure of the PPR proteins indicates that an interaction with a single RNA strand in the central groove might be possible (Small and Peeters, 2000). RNA binding studies with the recently identified PPR protein P67 from radish support this assumption and show the binding of P67 to a fragment of the radish nuclear pre-rRNA (Lahmy et al., 2000). However the function of this protein, which is located inside the chloroplast, is not known. As described above, the HCF152 protein was found to be a specific RNA binding protein. Additional biochemical analysis will reveal whether the PPR domains are responsible for the RNA binding specificity and affinity.
HCF152 Is a Chloroplast RNA Processing and Splicing Factor
The precise function of HCF152 remains unclear. This is true as well for the other plastidial RNA-processing factors that were identified in Chlamydomonas, maize, and Arabidopsis by genetic means. These proteins may be divided into four classes. HCF152 and its distant relative CRP1 of maize are members of the PPR protein family and are involved in RNA splicing or stabilization, endonucleolytic RNA processing, and presumably RNA translation. HCF107 and its evolutionary ortholog Mbb1 belong to the TPR proteins that are required for endonucleolytic processing and/or the stability of RNAs and, in addition, also appear to be involved in the translation of their respective RNAs (Vaistij et al., 2000; Felder et al., 2001). The third group of proteins is more heterogeneous with respect to its primary structure. This group encompasses Maa2 (Perron et al., 1999) and presumably Raa3 (Rivier et al., 2001) of Chlamydomonas, both of which are involved in trans-splicing of the psaA introns. Maa2 shows similarities to pseudouridine synthases, and Raa3 possesses a small region that has similarities with pyridoxamine 5′-phosphate oxidases. CRS2 (Jenkins and Barkan, 2001), a general group-II splicing factor of maize that is homologous with peptidyl-tRNA hydrolases, also belongs to this group of proteins that appear to be derived from enzymes of the general RNA metabolism. CRS1 of maize (Till et al., 2001), which is involved in atpF intron splicing, constitutes the founding member of a novel class of RNA splicing factors dating back to eubacterial and archaeal bacterial homologs.
Together with HCF107/Mbb1 (Felder et al., 2001), CRP1, and CRS2 of maize (Jenkins et al., 1997), HCF152 is the fourth nuclear factor that has been identified to be involved in the correct processing of psbB-psbT-psbH-petB-petD transcripts. These data indicate that a complex enzymatic machinery is required to process the psbB-psbT-psbH-petB-petD primary transcript into mature RNAs. Biochemical studies will provide greater insight into the mechanism by which HCF152 is required for the accumulation of spliced petB-containing RNAs and, concomitantly, the processing between psbH and petB.
METHODS
Growth Conditions and Mutant Selection
The hcf152-1 mutant (originally designated CRM3) of Arabidopsis thaliana was identified in a collection of T-DNA insertion lines (Bechtold et al., 1993; Bouchez et al., 1993), whereas hcf152-2 (formerly designated hcf119) was selected from an ethyl methanesulfonate–induced mutant collection (Meurer et al., 1996b). Mutant plants exhibiting a high-chlorophyll-fluorescence phenotype were selected in the dark under UV light (Meurer et al., 1996b). The seedlings were grown on sucrose-supplemented Gelrite medium, as described previously (Meurer et al., 1998). For the selection of T-DNA–induced Basta resistance, phosphinotricin was added to the medium to a final concentration of 10 mg/L.
Isolation of Chloroplast Membrane Proteins and Immunoblot Analysis
For the detection of photosynthetic membrane proteins, the isolation of chloroplast proteins was performed as described by Lennartz et al. (2001). SDS-PAGE (Schägger and von Jagow, 1987) and immunodecoration of electroblotted proteins followed the methods described by Meurer et al. (1996b)(1998).
RNA Gel Blot Hybridization Analysis
RNA isolation and RNA gel blot analysis of total leaf RNA was performed as described previously (Westhoff et al., 1991; Meurer et al., 1996a). Hybridization probes were made by cloning either PCR fragments or restriction fragments, as described by Felder et al. (2001). The probes were labeled by random priming. For the isolation of poly(A) RNA, total RNA was treated with Oligotex (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the protocol supplied by the manufacturer. DNA templates of each of the three genes located around the T-DNA insertion site were prepared for the synthesis of double- and single-stranded DNA probes for RNA gel blot hybridization. The following primers were used to amplify the sequences: Atg309650, 5′-TGTAAATTCGGAAAACGAATG-3′ and 5′-GTTGTTCACTCACACCTACTG-3′; Atg309660, 5′-CGGAATCTGTAAATGGAGAGTTAGA-3′ and 5′-TTCTGAGTCTTGATCTTCTGATAG-3′; Atg309670, 5′-CGGAATCTGTAAATGGAGAGTTAG-3′ and 5′-TTCTGAGTCTTGATCTTCTGATAG-3′. Radioactively labeled single-stranded probes against the gene transcripts were generated according to Felder et al. (2001). After PCR, the labeled probe was used directly for RNA gel blot hybridization.
RNase Protection Experiments
RNase protection experiments were performed as described by Rott et al. (1999). The antisense probes for analyzing the psbH 3′ end and the RNAs spanning the petB and petD intron-exon splice junctions were derived from plasmids (pBluescript II KS+; Stratagene) containing the corresponding fragments of the psbB transcription unit. To clone the fragments, they were amplified by conventional PCR (Dieffenbach and Dveksler, 1995) using the following pairs of primers: psbH, 5′-GGAAGGCCTGTTCTAGATCTGGTC-3′ and 5′-GGCGGATCCTAAGTATGAATCATAA-3′; petB, 5′-CGGGATCCAAGAGGCCTGTAACGA-3′ and 5′-GCTCTAGACCATCGATGAACTGA-3′; petD, 5′-CCATCGATACCTGTAAGAATGGA-3′ and 5′-GCTCTAGAAATACCATTCAGGTA-3′. Restriction enzyme cleavage sites, created at the 5′ end of each primer, were used for the insertion of these fragments into the vector. Uniformly labeled antisense RNAs were generated by T7 RNA polymerase transcription of the linearized plasmids. All probes are diagrammed in Figure 3.
Inverse PCR, Cloning, and Sequence Analysis of the HCF152 Open Reading Frame
Genomic DNA was isolated from 3-week-old mutant plants according to Dellaporta et al. (1983), restricted with PstI, and self-ligated. Standard PCR with the primers LB-TAG5 (5′-CTACAAATTGCCTTTTCTTATCGA-3′) and LB-TAG11 (5′-CACAACAGACAATCGGCTGCTCTG-3′) resulted in a 660-bp product. This product was reamplified with the nested primers LB-TAG14 (5′-GGTAATAGGACACTGGGATTCGTC-3′) and LB-TAG13 (5′-TTGTCAAGACCGACCTGTCCGGTG-3′), yielding a 450-bp DNA fragment. The PCR product was cloned into the EcoRV site of pBluescript II KS+, resulting in clone pnAt-HCF152, which was sequenced. The complete open reading frame for the Atg309650 gene was constructed using the primers 152M (5′-TGTAAATTCGGAAAACGAATG-3′) and 152F (5′-GTTGTTCACTCACACCTACTG-3′) and cloned into pBluescript II KS+.
Complementation of the hcf152 Mutant Phenotype
The generated open reading frame of HCF152 was used for the complementation of homozygous hcf152-1 plants by Agrobacterium tumefaciens–mediated root transformation essentially as described by Meurer et al. (1998). Independent transgenic plants were recovered and analyzed by chlorophyll fluorescence induction to measure photosynthetic efficiency in the plant leaves (Meurer et al., 1996b). The transformed plants were selfed to produce the T1 generation of the complemented hcf152-1 mutants. In addition, the genotype of the regenerated plants and the presence of the transgene were verified using PCR.
Immunolocalization Studies of HCF152 with Protein Gel Blots
Chloroplasts were isolated as described previously (Yehudai-Resheff et al., 2001) with minor modifications. Arabidopsis leaves (10 to 50 g) were homogenized with a juice mixer in buffer GR (0.33 M sorbitol, 50 mM Hepes-NaOH, pH 8.0, 5 mM MgCl2, 2.5 mM EDTA, 0.1% [w/v] BSA, and 3 mM DTT) and filtered through several layers of gauze. The filtrate was centrifuged at 6000g for 1 min, and the pellet, consisting of a chloroplast-enriched fraction, was washed with buffer GR. Chloroplasts were suspended in buffer E (20 mM Hepes-NaOH, pH 7.9, 12.5 mM MgCl2, 60 mM KCl, 0.1 mM EDTA, 2 mM DTT, and 17% glycerol) and were disrupted by freezing, thawing, and mild sonication, followed by separation of the soluble proteins (stroma) from the membrane fraction by centrifugation. The total soluble leaf protein fraction was prepared from homogenized Arabidopsis leaves in buffer E after centrifugation to pellet the membrane fraction. For subcellular localization of HCF152, 100 μg of stroma, membrane proteins, and total soluble leaf proteins was separated by SDS-PAGE using a 10% polyacrylamide gel and blotted to a nitrocellulose membrane that was decorated with specific antibodies. Antibodies to NAP-1 (Yoon et al., 1995), D1 (Schuster et al., 1988), and ribulose bisphosphate carboxylase were used to identify nuclear/cytoplasm-, thylakoid-, and stroma-specific proteins.
Production of Recombinant HCF152
To produce the mature protein without the chloroplast transit peptide, the ChloroP program (Emanuelsson et al., 1999) was used to define the transit peptide and the cleavage site. The corresponding DNA sequence of the mature protein was amplified by PCR using primers 5′-CCGAATTCCGCCGAAGACCTCTCGTCT and 3′-CGCGTCGACTTCTCTTGGACCTAACTTCC (underlined nucleotides are restriction sites used for cloning) and the plasmid pcAT152, which contains the complete open reading frame of HCF152. For expression in Escherichia coli, the PCR product was inserted in frame into the pBAD/Thio-TOPO vector (Invitrogen, Carlsbad, CA). Thus, the mature protein was fused with thioredoxin (18 kD) and six His residues at the N and C termini, respectively. Expression and purification of the bacterially produced protein were performed according to the manufacturer's protocol. Briefly, the LMG194 strain of E. coli containing the vector described above was grown in RM medium (40 mM Na2HPO4, 20 mM KH2PO4, 10 mM NaCl, 20 mM NH4Cl, 2% NZ-Amine A (Difco, Detroit, MI), 0.2% glucose, and 10 mM MgCl2) at 37°C to an OD600 of 0.5. Expression was induced by 0.2% arabinose, and the incubation was continued for 12 h at 16°C. Cells were harvested by centrifugation and lysed in 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM imidazole, 0.5% Triton X-100, 10 mM MgCl2, 10% glycerol, and 1 mg/mL lysozyme. After incubation for 30 min at 4°C, sonication, and freezing, the soluble proteins were collected by centrifugation at 15,000g for 15 min. HCF152-F was purified further by binding to nickel–nitrilotriacetic acid agarose resin (Qiagen, Valencia, CA), eluted with imidazole (Fluka, Milwaukee, WI), and dialyzed against buffer E.
Size-Exclusion Chromatography
Size-exclusion chromatography was performed by applying chloroplast-soluble proteins to a Superdex 200 column in buffer E at a flow rate of 0.5 mL/min. Proteins were precipitated by cold acetone and analyzed by SDS-PAGE and immunoblotting using specific antibodies. For digestion of the RNA, the extract was incubated with RNase A (1 mg/mL) and 3 units/μL RNase T1 at 37°C for 1 h. The Superdex 200 column was calibrated with the following protein standards: thyroglobulin (669 kD), catalase (232 kD), aldolase (158 kD), BSA (67 kD), and casein (30 kD).
Production of Antibodies
For the production of antibodies, the HCF152 open reading frame was fused to glutathione S-transferase using the expression vector pGEX4T3 (Amersham Bioscience). The bacterially expressed protein was purified by affinity chromatography on the column and injected into a rabbit (Lisitsky et al., 1997). The antibodies were checked against an unrelated glutathione S-transferase fusion protein and gave no signal in immunoblot analysis.
Preparation of RNA Probes
The following fragments of Arabidopsis chloroplast DNA were amplified by PCR using the appropriate primers and used as templates for the transcription of the corresponding RNA by the T7 RNA polymerase primed with the T7 promoter sequence attached to the 5′ end of the forward primer. For BDb RNA (nucleotides 74106 to 74373 [Sato et al., 1999]), the primers 5′-AATACGACTCACTATAGCTCTTAGTATCCACTTTAGGG-3′ and 5′-CAGCAACCCTAGTCGCC-3′ were used (underlined letters show the promoter sequences for the T7 RNA polymerase). For BDc RNA (nucleotides 74357 to 74700), the primers were 5′-AATACGACTCACTATAGGCGACTAGGGTTGCTG-3′ and 5′-CACTGAAATTCCATCCAG-3′. For BDd RNA (nucleotides 74683 to 75110), the primers were 5′-AATACGACTCACTATAGCTGGATGGAATTTCAGTG-3′ and 5′-GCGTTTGTCCTTTGATCC-3′. For BDe RNA (nucleotides 75093 to 75478), the primers were 5′-AATACGACTCACTATAGGATCAAAGGACAAACGC-3′ and 5′-GAGTCAAAATGCGAAAGC-3′. For BDf RNA (nucleotides 75461 to 75808), the primers were 5′-AATACGACTCACTATAGCTTTCGCATTTTGACTC-3′ and 5′-GTCATAGCAAATCCCGTAGC-3′. The PCR product was purified from gels using the QIAquick gel extraction kit (Qiagen), and the radiolabeled RNA probe was transcribed as described previously (Lisitsky et al., 1996). For the production of nonradioactive RNA, the transcription reaction mixture included 5 mM of each nucleotide (Lisitsky et al., 1995).
UV Cross-Linking
For UV cross-linking of the HCF152 protein to radiolabeled RNA, the protein was incubated with 32P-RNA in buffer containing 10 mM Hepes-NaOH, pH 7.9, 30 mM KCl, 6 mM MgCl2, 0.05 mM EDTA, 2 mM DTT, 8% glycerol, 0.0067% Triton X-100, and 67 μg/mL yeast tRNA (Sigma) for 15 min. The protein and RNA were cross-linked with 1.8 J of UV irradiation in a UV cross-linker (Hoefer, San Francisco, CA) after digestion of the RNA with 10 μg of RNase A and 30 units of RNase T1 at 37°C for 1 h, fractionation by SDS-PAGE, and analysis by autoradiography. For the competition assay, the protein was mixed with nonradioactive RNA for 5 min and the radiolabeled RNA was added.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The accession number of the CRP1 protein is AAC25599.
Acknowledgments
The authors are grateful to P. Westhoff (Heinrich-Heine-Universität, Düsseldorf, Germany) for helpful advice and discussions. We thank Maria Koczor and Susanne Paradies for their skilled technical assistance and an anonymous reviewer for the protocol for the ribonuclease protection experiment. We thank Shaul Yalovsky and Shimon Gepstein for the Nap-1 and ribulose bisphosphate carboxylase antibodies. This research was supported by grants from the Deutsche Forschungsgemeinschaft to K.M. through SFB 189 at the University of Düsseldorf and by a grant from the German–Israeli Foundation for Scientific Research and Development to P. Westhoff, K.M., and G.S.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010397.
References
- Anthonisen, I.L., Salvador, M.L., and Klein, U. (2001). Sequence specific elements in the 5′ untranslated regions of rbcL and atpB gene mRNAs stabilize transcripts in the chloroplast of Chlamydomonas reinhardtii. RNA 7, 1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchincloss, A.H., Zerges, W., Perron, K., Girard-Bascou, J., and Rochaix, J.D. (2002). Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 157, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7, 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Barkan, A., Walker, M., Nolasco, M., and Johnson, D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13, 3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Finn, R.D., and Sonnhammer, E.L. (1999). Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bentolila, S., Alfonso, A.A., and Hanson, M.R. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris 316, 1188–1193. [Google Scholar]
- Boudreau, E., Nickelsen, J., Lemaire, S.D., Ossenbuhl, F., and Rochaix, J.D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19, 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., and Henry, R. (1996). Import and routing of nucleus-encoded chloroplast proteins. Annu. Rev. Cell Dev. Biol. 12, 1–26. [DOI] [PubMed] [Google Scholar]
- Coffin, J.W., Dhillon, R., Ritzel, R.G., and Nargang, F.E. (1997). The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 32, 273–280. [DOI] [PubMed] [Google Scholar]
- Das, A.K., Cohen, P.W., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Dieffenbach, C.W., and Dveksler, G.S. (1995). PCR Primer: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder, S., Meierhoff, K., Sane, A.P., Meurer, J., Driemel, C., Plucken, H., Klaff, P., Stein, B., Bechtold, N., and Westhoff, P. (2001). The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13, 2127–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene CRP1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M., Girard-Bascou, J., Choquet, Y., and Rochaix, J.D. (1990). Trans-splicing mutants of Chlamydomonas reinhardtii. Mol. Gen. Genet. 223, 417–425. [DOI] [PubMed] [Google Scholar]
- Hirose, T., and Sugiura, M. (1997). Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: A possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 16, 6804–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss, T.L., and Hollingsworth, M.J. (1999). ATP synthase 5′ untranslated regions are specifically bound by chloroplast polypeptides. Curr. Genet. 35, 512–520. [DOI] [PubMed] [Google Scholar]
- Jenkins, B.D., and Barkan, A. (2001). Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, B.D., Kulhanek, D.J., and Barkan, A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy, S., Barneche, F., Derancourt, J., Filipowicz, W., Delseny, M., and Echeverria, M. (2000). A chloroplastic RNA-binding protein is a new member of the PPR family. FEBS Lett. 480, 255–260. [DOI] [PubMed] [Google Scholar]
- Lennartz, K., Plucken, H., Seidler, A., Westhoff, P., Bechtold, N., and Meierhoff, K. (2001). HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13, 2539–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky, I., Klaff, P., and Schuster, G. (1996). Addition of poly(A)-rich sequences to endonucleolytic cleavage sites in the degradation of spinach chloroplast mRNA. Proc. Natl. Acad. Sci. USA 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky, I., Kotler, A., and Schuster, G. (1997). The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: The exoribonuclease 100RNP/PNPase displays high binding affinity for poly(A) sequences. J. Biol. Chem. 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- Lisitsky, I., Liveanu, V., and Schuster, G. (1995). RNA-binding characteristics of a ribonucleoprotein from spinach chloroplast. Plant Physiol. 107, 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey, G.M., Przybyla-Zawislak, B.D., and McEwen, J.E. (1998). The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondria inner membrane. Eur. J. Biochem. 255, 156–161. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Berger, A., and Westhoff, P. (1996. a). A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell 8, 1193–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer, J., Meierhoff, K., and Westhoff, P. (1996. b). Isolation of high chlorophyll fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and Northern hybridisation. Planta 198, 385–396. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Plücken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F., and Ferat, J.L. (1995). Structure and activities of group II introns. Annu. Rev. Biochem. 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Monde, R.A., Schuster, G., and Stern, D.B. (2000). Processing and degradation of chloroplast mRNA. Biochimie 82, 573–582. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Ohta, M., Sugiura, M., and Sugita, M. (2001). Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem. 276, 147–152. [DOI] [PubMed] [Google Scholar]
- Nickelsen, J., van Dillewijn, J., Rahire, M., and Rochaix, J.-D. (1994). Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 13, 3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., Ayaly, F.H., and Hartl, D.L. (1993). Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol. 218, 309–321. [DOI] [PubMed] [Google Scholar]
- Perron, K., Goldschmidt-Clermont, M., and Rochaix, J.D. (1999). A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18, 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp, J.C., Baumgartner, B.J., and Mullet, J. (1992). Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes: Transcription rates and mRNA levels vary over 300-fold; predicted mRNA stabilities vary 30-fold. J. Biol. Chem. 267, 21404–21411. [PubMed] [Google Scholar]
- Reinbothe, S., Reinbothe, C., Heintzen, C., Seidenbecher, C., and Parthier, B. (1993). A methyl jasmonate-induced shift in the length of the 5′ untranslated region impairs translation of the plastid rbcL transcript in barley. EMBO J. 12, 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier, C., Goldschmidt-Clermont, M., and Rochaix, J.D. (2001). Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 20, 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott, R., Livean, V., Drager, R.G., Higgs, D., Stern, D.B., and Schuster, G. (1999). Altering the 3′ UTR endonucleolytic cleavage site of a Chlamydomonas chloroplast mRNA affects 3′-end maturation in vitro but not in vivo. Plant Mol. Biol. 40, 679–686. [DOI] [PubMed] [Google Scholar]
- Salvador, L.M., Klein, U., and Bogorad, L. (1993). 5′ sequences are important positive and negative determinants of the longevity of Chlamydomonas chloroplast gene transcripts. Proc. Natl. Acad. Sci. USA 90, 1556–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., and Tabata, S. (1999). Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 6, 283–290. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1–100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schuster, G., Timberg, R., and Ohad, I. (1988). Turnover of photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur. J. Biochem. 177, 403–410. [DOI] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif: A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47. [DOI] [PubMed] [Google Scholar]
- Smith, C.J., and Pearse, B.M. (1999). Clathrin: Anatomy of a coat protein. Trends Cell Biol. 9, 335–338. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L., Eddy, S.R., Birney, E., Bateman, A., and Durbin, R. (1998). Pfam: Multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26, 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax, F.E., and Vernon, D.M. (2001). T-DNA-associated duplication/translocations in Arabidopsis: Implications for mutant analysis and functional genomics. Plant Physiol. 126, 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B., Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2001). CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Boudreau, E., Lemaire, S.D., Goldschmidt-Clermont, M., and Rochaix, J.D. (2000). Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., van Montagu, M., and van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Hess, W.R., and Borner, T. (1997). Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 25, 2030–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff, P. (1985). Transcription of the gene encoding the 51 kd chlorophyll α-apoprotein of the photosystem II reaction centre from spinach. Mol. Gen. Genet. 201, 115–123. [Google Scholar]
- Westhoff, P., and Herrmann, R.G. (1988). Complex RNA maturation in chloroplasts: The psbB operon from spinach. Eur. J. Biochem. 171, 551–564. [DOI] [PubMed] [Google Scholar]
- Westhoff, P., Offermann-Steinhard, K., Höfer, M., Eskins, K., Oswald, A., and Streubel, M. (1991). Differential accumulation of plastid transcripts encoding photosystem II components in the mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants. Planta 184, 377–388. [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Hirsh, M., and Schuster, G. (2001). Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, H.W., Kim, M.C., Lee, S.Y., Hwang, I., Bahk, J.D., Hong, J.C., Ishimi, Y., and Cho, M.J. (1995). Molecular cloning and functional characterization of a cDNA encoding nucleosome assembly protein 1 (NAP-1) from soybean. Mol. Gen. Genet. 249, 465–473. [DOI] [PubMed] [Google Scholar]