Abstract
Preferential integration into transcriptionally active regions of genomes has been observed for retroviral vectors based on gamma-retroviruses and lentiviruses. However, differences in the integration site preferences were detected, which might be explained by differences in viral components of the preintegration complexes. Viral determinants of integration site preferences have not been defined. Therefore, integration sites of simian immunodeficiency virus (SIV)-based vectors produced in the absence of accessory genes or lacking promoter and enhancer elements were compared. Similar integration patterns for the different SIV vectors indicate that vif, vpr, vpx, nef, env, and promoter or enhancer elements are not required for preferential integration of SIV into transcriptionally active regions of genomes.
Integration of the retroviral genome into the genome of the target cell is an essential step in the retroviral replication cycle. In contrast to what was found for adeno-associated viruses, preferential integration sites could not be detected initially for retroviruses (4). However, since the transcriptional activity of, for example, human immunodeficiency virus (HIV) strongly depends on the integration site (15), selection of appropriate integration sites might favor efficient virus replication. Experimental modulation of the transcriptional activity of a gene, however, revealed reduced frequency of integration of avian leukosis virus into this gene at high transcriptional activity (20, 29). The availability of the human genome sequence allowed Schröder et al. to pursue a different experimental approach (25). They sequenced 436 integration sites of HIV-1, localized these integration sites on the human genome map, and classified the transcriptional activities of the integration sites. This revealed that 69% of the integration sites are in transcriptional units, although they constitute only 33% of the human genome. Frequency of integration into transcriptional units generally correlated with their transcriptional activity in the infected cells. Only genes with the highest expression levels were targeted less frequently. Subsequently, murine leukemia virus (MLV), and to a limited extent avian leukosis virus, was also found to preferentially integrate into transcriptional units (17, 21, 23, 30). MLV preferentially integrated in the vicinities of transcriptional start sites, while HIV integration sites were distributed more evenly through entire transcriptional units. This difference in integration site preference might also have functional consequences regarding the safety of the respective vectors. Using different promoter-trapping vectors based on MLV and HIV-1, expression of the MLV vector driven by cellular promoters of the integration site occurred at higher frequency than for the corresponding HIV-1 vector (7). Recently, simian immunodeficiency virus (SIV) integration sites in a chronically SIV-infected human T-cell line and in peripheral blood mononuclear cells derived from rhesus monkey bone marrow stem cells transduced with an SIV-based vector were also mapped (6, 14). The integration site preferences of SIV mirrored those of HIV-1, indicating a conserved mechanism of target site selection. The interactions mediating the integration site preferences on the molecular level are unknown. Since the integration site preferences differ between members of different genera of retroviruses, differences in the interaction of the preintegration complexes of different retroviruses with cellular components of the nucleus should contribute to integration site preferences. Cellular transcription factors binding to viral promoter and enhancer elements of reverse-transcribed DNA might direct the preintegration complex to actively transcribed regions of the genome. In contrast to gammaretroviruses, lentiviral genomes encode accessory proteins. Since they either are part of the viral particle (Vpr, Vpx) (5, 16, 18, 31) or have an influence on the composition of the viral particle (Vif) (reviewed in reference 13), they could also contribute to target site selection. In a first attempt to define lentiviral determinants of preferential integration into transcriptionally active regions of the genome, we mapped integration sites of SIV-based vectors produced in the absence of accessory genes or lacking promoter and enhancer elements.
MATERIALS AND METHODS
Production of lentiviral vectors.
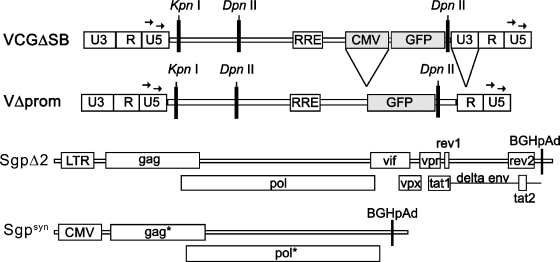
VCGΔSB and the numbering used are based on the SIVmac239 genome (GenBank accession number M33262) with mutations in gag (nucleotide [nt] 1331, T→A; and nt 1334, C→G); deletions in gag-pol, regulatory genes, and env (nt 2031 to 8014); and deletions in nef (nt 9500 to 9687 and nt 9732 to 10056). A cytomegalovirus (CMV)-green fluorescent protein (GFP) expression cassette (1.1-kb BamH1-Xho1 fragment) from HIV-CS-CG (22) was inserted into the upstream nef deletion. VΔprom contains an additional deletion of the entire U3 region of the 3′ long terminal repeat (LTR) (nt 9732 to 10231) and lacks the CMV promoter upstream of the GFP region due to deletion of a SmaI-Eco47III fragment. The mutations in gag and the deletions in nef and the U3 region have been previously characterized (12, 24).
293T (8) cells were kept in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml), and 100 U/ml streptomycin (DMEM plus 10% FCS). The cells were transfected in 175-cm2 flasks using the calcium phosphate coprecipitation method essentially as described previously (26). Wild-type SIV vector particles (SIVwt) were prepared by cotransfection of 40 μg of the gag-pol expression plasmid SgpΔ2 (24), 16 μg of the expression plasmid for the G protein of vesicular stomatitis virus (pHIT-G [10]), and 40 μg of the SIV vector VCGΔSB. SIV vector particles in the absence of vif, vpr, vpx, and nef (SIVΔfrxn) were prepared by cotransfection of the codon-optimized SIV gag-pol expression plasmid Sgpsyn (40 μg) (28), pcTat (24 μg) (19), pcRev (24 μg) (19), pHIT-G, and VCGΔSB. Cotransfection of 40 μg VΔprom, 40 μg SgpΔ2, and 16 μg pHIT-G was used to generate SIVΔprom particles. The supernatant of the transfected cells was harvested 48 h and 60 h after transfection and was passed through a 0.45-μm filter. After a centrifugation step (12,000 × g, 4°C, 1 h), the precipitated vector particles were resuspended in DMEM and stored at −80°C.
Transduction of target cells.
On day 1, 293A cells (Quantum Biotechnologies, Montreal, Canada) were plated in 24-well plates at a density of 50,000 cells/well in DMEM plus 10% FCS. One day later, the medium was replaced by 200 μl of concentrated vector supernatants and incubated for 3.5 h prior to the addition of 800 μl medium. One day later, the infection procedure was repeated. On day 4, cells were split into a 25-cm2 flask, and on the next day, cells were transduced with 1 ml of vector supernatant. Cells were infected repeatedly to increase the number of integration events without overloading the cells with a high dose of vector particles. Once the 25-cm2 flasks became confluent, cells were trypsinized and plated into 96-well plates at a density of 5 to 20 cells/well. When the wells became dense, cells were transferred to a 24-well plate. As soon as these were dense, they were lysed in 500 μl buffer K (100 μg/ml protease K, 100 mM Tris, 5 mM EDTA, 0.2% sodium dodecyl sulfate, 200 mM NaCL [pH 8.5]) and incubated overnight at 37°C. An equal volume of isopropanol was added, and precipitated genomic DNA was washed once in 70% ethanol and then transferred to a fresh Eppendorf tube. The DNA was resuspended in 50 μl TE (Tris-EDTA) buffer at 60°C.
LM-PCR.
Genomic DNA of transduced cells was digested with DpnII and subsequently purified and concentrated with Millipore montage PCR centrifugal filter devices. Overnight ligation was performed with 500 ng DNA in 2× ligation buffer (NEB) with preannealed oligonucleotides “Adapter long G” (5′ GTAATACGACTCACTATAGGGCTCCGCTTAAGGGAC 3′) and “Adapter short G” (5′ GATCGTCCCTTAAGCGGAG 3′). KpnI digestion of the ligation reaction was used to avoid amplification of an internal vector fragment by ligation-mediated (LM)-PCR.
A nested PCR was performed with the outer primers “adapter 1 G” (5′ GTAATACGACTCACTATAGGGC 3′) and “U5 SIV LTRs 0” (5′ CTGGTCAACTCGGTACTCAATA 3′) under the following conditions: 300 nM primer, 2 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, and 1 U Platinium Taq DNA polymerase (Invitrogen) in 1× PCR buffer. Cycling conditions were as follows: 95°C for 2 min; 40 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min; and 1 cycle at 72°C for 10 min. For the inner PCR, the primers “adapter 2 G” (5′ AGGGCTCCGCTTAAGGGAC 3′) and “U5 SIV LTRs 1” (5′ TTCTGCTTTGGGAAACCGA 3′) were used under the following conditions: 300 nM primer, 2.5 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, and 1 U Platinium Taq DNA polymerase (Invitrogen) in 1× PCR buffer. Cycling conditions were as follows: 95°C for 2 min; 40 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min; and 1 cycle at 72°C for 10 min. PCR products were separated by agarose gel electrophoresis, and the DNA bands were excised and the DNA purified with a BioGene Geneclean III kit or the freeze-and-squeeze method (27). Purified fragments were sequenced with primer U5 SIV LTRs 1 by Geneart (Regensburg, Germany) using standard procedures.
Mapping of integration sites.
Sequences were first viewed using Chromas 2.23 software (Technelysium Pty Ltd., Tewantin, Australia) to detect the 5′ LTR sequence and the adapter sequence. Sequences were also checked for preservation of the terminal CA nucleotides of the LTR.
For fast and standardized analysis of sequences obtained from the LM-PCRs, we used the novel IntegrationMap task (F. A. Giordano, A. Hotz-Wagenblatt, D. Lauterborn, J.-U. Appelt, K. Fellenberg, K. Z. Nagy, W. J. Zeller, S. Suhai, S. Fruehauf, and S. Laufs, submitted). This task (available online at http://genius.embnet.dkfz-heidelberg.de/menu/biounit/open-husar/ or, for all services after registration for an account, at http://genome.dkfz-heidelberg.de) retrieves detailed information about whether integrations are located in or close to a gene, the name of the gene, the exact localization in the transcriptional units, and further parameters, such as distance from transcription start site to integration. Since only those genes annotated in the Ensembl (version 30.35c, 22 March 2005 freeze; http://www.ensembl.org/index.html) or RefSeq (refseq_RNA9.0) database are found by IntegrationMap, the data describe existing and validated genes.
Hits outside Ensembl genes were examined with NCBI BLAST (http://www.ncbi.nlm.nih.gov/blast/), yielding pictures of chromosomal localization. Gene density at 0.5 Mbp in each direction from the integration site was determined using the NCBI map viewer (http://www.ncbi.nlm.nih.gov/mapview/). Targeted RefSeq gene numbers were used to retrieve frequencies of sequential analysis of gene expression (SAGE) tags from the 293 SAGE library (SAGE_Kidney_embryonic_CL_293-control at http://cgap.nci.nih.gov/SAGE/AnatomicViewer). Frequencies of expressed tags per 200,000 identified tags were taken as gene expression levels.
Random insertion site generation.
For comparison of data in terms of preferential integration patterns, we created a random sequence set consisting of 2,000 sequences (F. A. Giordano, B. Fehse, A. Hotz-Wagenblatt, S. D. V. C. Jonnakuty, J.-U. Appelt, Z. Nagy, K. Kuehlcke, S. Naundorf, A. R. Zander, W. J. Zeller, S. Fruehauf, and S. Laufs, submitted). Random numerical values following a uniform distribution were generated such that the values are evenly spread over a given interval. The first interval (1 to 3,121) was then divided into 24 parts, corresponding to the chromosomes with partition sizes according to the sizes of the individual chromosomes. The generated values then give the chromosome numbers with respect to their sizes. Another random number (interval, 100 to 200) was generated to determine the length of the random sequence and a third one (interval, first base of chromosome to last base of chromosome) for the start position within the chromosome. With the resulting values for chromosome number, start position, and length, the sequences were fetched from the Ensembl human genome database (http://www.ensembl.org/index.html) using the same Ensembl draft that IntegrationMap uses.
In addition, 200 RefSeq genes were randomly selected from the 24,772 genes of the database (database hg17, May 2004 freeze; http://genome.ucsc.edu), and the transcriptional activities of the 200 genes were estimated using the SAGE approach described above. The product of the percentage of randomly selected RefSeq genes that are transcriptionally active (32.5%) and the percentage of hit RefSeq genes in the whole-genome random sequence set (28.5%) was taken as the frequency of random integration into transcriptionally active genes.
Statistical analyses.
GraphPad Prism (Prism 4 for Windows; GraphPad Software, Inc., San Diego, CA) and statistics calculator (Statpac, Inc., Bloomington, MN) software were used for statistical analysis. The χ2 test was performed with the statistics calculator to compare the frequencies of integration of different SIV vectors within the RefSeq genes. Furthermore, the nonparametric Kruskal-Wallis test was performed with GraphPad Prism to compare the transcriptional activities of the genes targeted by the different SIV vectors used.
RESULTS
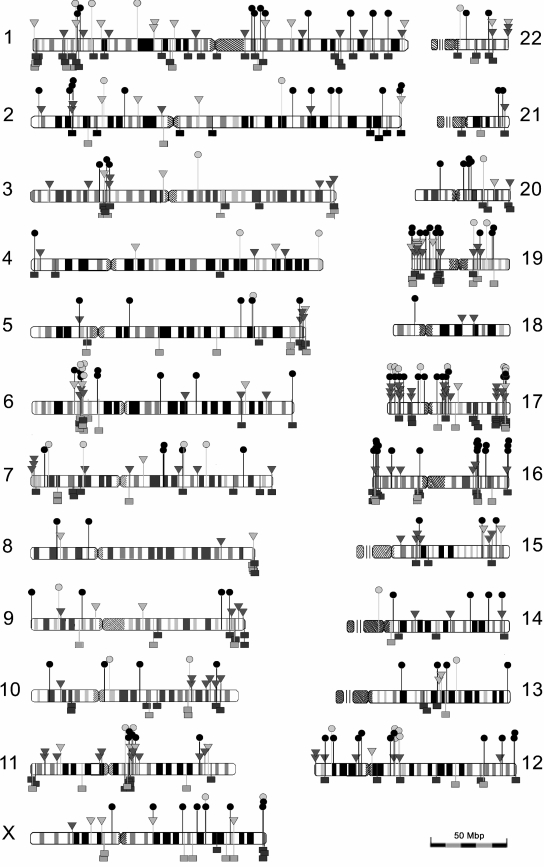
To study the effects of accessory genes of SIV and the role of promoter and enhancer elements on integration site preferences, three different SIV-based vector particles were produced. SIVwt contained a prototypic SIV vector construct (VCGΔSB), in which large regions of the SIV genome are deleted (Fig. 1). The GFP reporter gene is driven by the immediate-early promoter of human CMV. This vector was packaged by cotransfection with SgpΔ2 (24), an expression plasmid carrying SIVmac239 gag-pol, tat, rev, vif, vpr, and vpx. To produce SIVΔfrxn, VCGΔSB was cotransfected with a codon-optimized expression plasmid for SIVmac239 gag-pol (Sgpsyn in reference 28) and expression plasmids for tat and rev. A self-inactivating mutant of VCGΔSB also lacking the CMV promoter of the GFP expression cassette (VΔprom) was packaged with SgpΔ2, resulting in SIVΔprom particles. All three vector particles were pseudotyped with the G protein of vesicular stomatitis virus. The 293A target cells were transduced with the different vector particles. Five to 20 cells were plated in 96-well plates and subsequently expanded into 24-well plates. Genomic DNA was extracted and analyzed for integration sites by ligation-mediated PCR. Totals of 221, 191, and 174 unique integration sites could be mapped for SIVwt, SIVΔprom, and SIVΔfrxn, respectively (Fig. 2). Integration sites were found on all chromosomes but seemed to cluster on some chromosomes and particular loci. Six loci spanning approximately 53.5 Mbp (1.8% of the haploid genome) harbored 17.4% of all integration events (Table 1). No cluster formation on chromosomal level was detected for a data set representing random integration events. The gene densities at the clustered integration sites of SIV (34 to 46 genes/Mbp [Table 1]) were high compared to that for the same chromosome (data not shown) or the whole genome (9.5 genes/Mbp).
FIG. 1.
Map of SIV vector constructs and gag-pol expression plasmids. Viral open reading frames and intact regulatory elements are marked by white boxes. GFP, coding region for the green fluorescent protein; CMV, human cytomegalovirus immediate-early promoter. The arrows mark the primer binding sites of U5 SIV LTRs 0 and U5 SIV LTRs 1; KpnI and DpnII restriction sites relevant for the LM-PCR are also shown (not all DpnII sites are shown).
FIG. 2.
Integration sites of the different vectors. Circle, SIVΔprom; triangle, SIVΔfrxn; square, SIVwt; light gray symbols, integration outside RefSeq genes; black and dark gray symbols, integration within RefSeq genes.
TABLE 1.
Clustered integrations
| Chromosomal locus or loci | Size of clustera (Mbp) | No. of integration events | Integration density (events/Mbp) | Gene density of cluster (genes/Mbp) |
|---|---|---|---|---|
| 6p21.33-6p21.31 | 6 | 17 | 2.8 | 44 |
| 11q13.1-11q13.4 | 8 | 15 | 1.9 | 39 |
| 16p13.3 | 5 | 15 | 3 | 46 |
| 17p13.3-17p13.1 | 7 | 17 | 2.4 | 37 |
| 17q25.1-17q25.3 | 7 | 23 | 3.3 | 34 |
| 19p13.3 | 6.5 | 15 | 2.3 | 36 |
| Haploid genome | 3,011 | 586 | 0.2 | 9.5 |
Largest distance between two integration events within the chromosomal loci.
Preferential integration into gene-rich regions of the genome was also evident by analysis of integration into RefSeq genes (Table 2). Of all 586 SIV integration sites, 370 (64%) mapped to RefSeq genes, for which mRNA genes have been identified. This RefSeq gene insertion frequency was more than twofold higher than that for a randomly generated data set (P < 0.001), where we obtained a frequency of hit RefSeq genes of 28.5%. Percentage of integration into RefSeq genes did not differ significantly between SIVwt, SIVΔfrxn, and SIVΔprom (Table 2). The preference of SIV vectors for integration into RefSeq genes is comparable to that of HIV-1 but differs from that of MLV (Table 2) (6, 14), considering that the data for HIV-1 and MLV given in Table 2 were obtained with a 2002 database freeze containing 18,214 annotated RefSeq genes instead of 24,194 RefSeq genes as in the current database.
TABLE 2.
Characterization of SIV integration sites within RefSeq genesa
| Vector particle | % of integration events within:
|
||
|---|---|---|---|
| RefSeq genes | ±5 kb of transcription start site | ±5 kb of CpG islands | |
| SIVwt | 59.3 | 17.6 | 1.8 |
| SIVΔprom | 67.0c | 12.6c | 2.1c |
| SIVΔfrxn | 66.7c | 13.2c | 0c |
| SIV total | 64.0 | 14.7 | 1.4 |
| Random | 28.5d | 11.2e | 4.0d |
| HIVb | 57.8 | 10.8 | 2.1 |
| MLVb | 34.2 | 20.2 | 16.8 |
| Randomb | 22.4 | 4.3 | 2.1 |
The total number of integration sites for SIVwt, SIVΔprom and SIVΔfrxn are 221, 191 and 174, respectively.
Data were obtained from reference 30, which used a 2002 freeze of the database with only 18,214 RefSeq genes, compared to 24,194 RefSeg genes in the present database.
No significant difference from SIVwt value.
Highly significant difference from SIV total (P = 0.002).
Significant difference from SIV total (P = 0.022). A χ2 test was used for all statistical analyses.
Given the differences in integration frequencies in the vicinities of transcriptional start sites and CpG islands between lentiviruses and MLV, we also determined these parameters for the different SIV vectors (Table 2). Integration within ±5 kb of transcriptional start sites was observed with frequencies similar to those for the random data set, while integration of the SIV vectors within the vicinities of CpG islands was underrepresented compared to that in the random data set (Table 2). No significant differences were detected between the different SIV vectors.
Integration into a RefSeq gene does not imply that this gene is actually transcribed in the vector-transduced cells. To determine whether the SIV vectors preferentially integrate into transcriptionally active genes, we assigned each targeted gene a transcriptional activity based on expression profiles determined for the target cell line by SAGE. Approximately 28.8% of the integrations were into RefSeq genes, for which expression is predicted by SAGE (Table 3). For random integration, the frequency of hitting transcriptionally active RefSeq genes is estimated to be 9%, indicating preferential integration of the SIV vectors into transcriptionally active RefSeq genes. Neither deletion of the promoter nor omission of the accessory genes reduced the frequency of integration of the SIV vector into transcriptionally active RefSeq genes significantly. Median expression levels for the targeted RefSeq genes did not differ significantly between the SIV vectors and the randomly picked RefSeq genes (Table 3).
TABLE 3.
Transcriptional activities of targeted genesa
| Vector | % Integration within active RefSeq genes | Median frequency of tags of hit RefSeq genesc | 25th percentile | 75th percentile |
|---|---|---|---|---|
| SIVwt | 25.3 | 6.5 | 4 | 14 |
| SIVΔprom | 28.8 | 9 | 4 | 23 |
| SIVΔfrxn | 33.3 | 9 | 4 | 23 |
| SIV total | 28.8 | 9 | 4 | 19 |
| Random | 9.0b | 9 | 4 | 23 |
As an estimate for the transcriptional activity of RefSeq genes in 293 target cells, the representation of the RefSeq genes in a SAGE library was retrieved from the SAGE Genie website (http://cgap.nci.nih.gov/SAGE, library SAGE_Kidney_embryonic_CL_293-control). RefSeq genes represented in this library at least once were considered transcriptionally active. The median frequency of the tags of transcriptionally active hit genes per 200,000 tags in this SAGE library was compared to those of 200 randomly picked RefSeq genes. There is no significant difference between the different SIV vector particles.
P was <0.001 compared to SIV total (χ2 test).
There is no significant difference between the three constructs and the random data (Kruskal-Wallis test).
DISCUSSION
Mapping of 586 integration sites of SIV-based vectors extends previous reports on SIV integration site preferences. Hematti et al. analyzed integration sites in rhesus monkey peripheral blood mononuclear cells derived from stem cells transduced with an SIV-based vector (14), while Crise et al. determined 148 integration sites in a chronically SIV-infected human T-cell line (6). Consistent with their results, we observed preferential integration of our SIV-based vectors into RefSeq genes. As previously observed for HIV-1 and SIV (6, 21, 30), there was a slight preference for integration into the proximities of transcriptional start sites, while CpG islands seem to be disfavored. Since MLV integrates less frequently into RefSeq genes and more frequently within the vicinities of transcriptional start sites, the integration site preferences of lentiviruses and lentiviral vectors clearly differ from those of MLV. These differences indicate that accessibility of genes in the target cells is not the only determinant of integration site preference.
A striking clustering of integration events was observed with 17.4% of all integrations into six loci spanning only 1.8% of the human genome. All of these clusters had exceptionally high densities of genes and colocalized with regions of increased gene expression as defined previously (1). However, frequency of integration into other regions of increased gene expression was rather variable. In addition, the loci identified in this study differ from clustered integration sites found in other studies (14, 17, 25). Thus, the transcriptional activities at the loci in the particular target cells might well influence integration frequency.
Transcription factors binding to promoter and enhancer elements of the reverse-transcribed viral DNA might capture the viral DNA and redirect its transport to the sites of transcription. To test this hypothesis, all known promoter and enhancer elements were deleted from the vector construct. However, the deletion of these elements did not change integration site preferences. Thus, promoter and enhancer elements do not seem to modulate integration site preferences of SIV-based vectors. If there is a cis-acting DNA element determining integration site preferences of SIV, it should be located either from position 775 to 2030, from position 8015 to 9499, or from position 9689 to 9731 of the SIVmac239 proviral DNA (GenBank entry number M33262), since they are the only SIV sequences retained in the vector construct. Whether promoter and enhancer elements of MLV, which preferentially integrates into the proximities of transcriptional start sites, contribute to integration site preferences of MLV remains to be determined.
Other potential determinants of integration site preferences could be components of the vector particle or the preintegration complex. Ciuffi et al. recently reported that depletion of the cellular LEDGF/p75 protein reduced the frequency of integration of HIV-1 into transcription units (3). Since LEDGF/p75 interacts with HIV-1 integrase (2), this also suggests that the HIV-1 integrase tethers the preintegration complex to LEDGF/p75 and thereby affects integration site selection (3). However, even in LEDGF/p75-depleted cells, HIV-1 integrated more frequently into transcriptional units than would be expected from random integration (3). In a first attempt, to narrow the range of potential viral determinants, we compared integration site preferences of wild-type vector particles with those of vector particles produced in the absence of accessory genes. Although neither Vif, Vpr, nor Vpx is known to be part of the preintegration complex, all three influence the composition of the virus particles. While Vif inhibits incorporation of Apobec (reviewed in reference 13), Vpr and Vpx are incorporated into the particles. Therefore, all three might affect early events after cell entry, affecting integration site selection. In particular, Vpr has been reported to facilitate infection of nondividing cells by coupling the preintegration complex to a nuclear import pathway (9, 11). Comparing integration site preferences of vectors prepared in the absence of vif, vpr, and vpx with those of wild-type vectors revealed no evidence for a change in integration site preference. Thus, the viral determinants of integration site selection must be retained in the gag-pol, tat, or rev genes or in cis-acting DNA elements of the integrating vector construct.
Acknowledgments
We thank Ralf Wagner and Geneart (Regensburg) for providing Sgpsyn. The technical assistance of Hans Jürgen Engel is gratefully acknowledged.
This work was supported by grants from the German Research Foundation (Ue45-5/1 and FR-1732-3/1).
REFERENCES
- 1.Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M. C. Hermus, R. van Asperen, K. Boon, P. A. Voute, S. Heisterkamp, A. van Kampen, and R. Versteeg. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291:1289-1292. [DOI] [PubMed] [Google Scholar]
- 2.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 3.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 4.Coffin, J. 1996. Retroviridae: the viruses and their replication, p. 1767-1848. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 5.Cohen, E. A., G. Dehni, J. G. Sodroski, and W. A. Haseltine. 1990. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J. Virol. 64:3097-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crise, B., Y. Li, C. Yuan, D. R. Morcock, D. Whitby, D. J. Munroe, L. O. Arthur, and X. Wu. 2005. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 79:12199-12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Palma, M., E. Montini, F. R. de Sio, F. Benedicenti, A. Gentile, E. Medico, and L. Naldini. 2005. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood 105:2307-2315. [DOI] [PubMed] [Google Scholar]
- 8.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher, T. M. I., B. Brichacek, N. Sharova, M. A. Newman, G. Stivahtis, P. M. Sharp, M. Emerman, B. H. Hahn, and M. Stevenson. 1997. Nuclear import and cell cycle arrest function of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIVsm. EMBO J. 15:6155-6165. [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier, R. A. M., B. E. Meyer, J. H. M. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallay, P., V. Stitt, C. Mundy, M. Oettinger, and D. Trono. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunwald, T., F. S. Pedersen, R. Wagner, and K. Überla. 2004. Reducing mobilization of simian immunodeficiency virus based vectors by primer complementation. J. Gene Med. 6:147-154. [DOI] [PubMed] [Google Scholar]
- 13.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 14.Hematti, P., B. K. Hong, C. Ferguson, R. Adler, H. Hanawa, S. Sellers, I. E. Holt, C. E. Eckfeldt, Y. Sharma, M. Schmidt, C. von Kalle, D. A. Persons, E. M. Billings, C. M. Verfaillie, A. W. Nienhuis, T. G. Wolfsberg, C. E. Dunbar, and B. Calmels. 2004. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLOS Biol. 2:e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kappes, J. C., J. S. Parkin, J. A. Conway, J. Kim, C. G. Brouillette, G. M. Shaw, and B. H. Hahn. 1993. Intracellular transport and virion incorporation of vpx requires interaction with other virus type-specific components. Virology 193:222-233. [DOI] [PubMed] [Google Scholar]
- 17.Laufs, S., K. Z. Nagy, F. A. Giordano, A. Hotz-Wagenblatt, W. J. Zeller, and S. Fruehauf. 2004. Insertion of retroviral vectors in NOD/SCID repopulating human peripheral blood progenitor cells occurs preferentially in the vicinity of transcription start regions and in introns. Mol. Ther. 10:874-881. [DOI] [PubMed] [Google Scholar]
- 18.Lu, Y. L., P. Spearman, and L. Ratner. 1993. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 20.Maxfield, L. F., C. D. Fraize, and J. M. Coffin. 2005. Relationship between retroviral DNA-integration-site selection and host cell transcription. Proc. Natl. Acad. Sci. USA 102:1436-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLOS Biol. 2:e234. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi, H., U. Blömer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 78:11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnell, T., P. Foley, M. Wirth, J. Münch, and K. Überla. 2000. Development of a self-inactivating, minimal lentivirus vector based on simian immunodeficiency virus. Hum. Gene Ther. 11:439-447. [DOI] [PubMed] [Google Scholar]
- 25.Schröder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 26.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thuring, R. W., J. P. Sanders, and P. Borst. 1975. A freeze-squeeze method for recovering long DNA from agarose gels. Anal. Biochem. 66:213-220. [DOI] [PubMed] [Google Scholar]
- 28.Wagner, R., M. Graf, K. Bieler, H. Wolf, T. Grunwald, P. Foley, and K. Überla. 2000. Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus type 1 and simian immunodeficiency virus: implications for the safety of lentiviral vectors. Hum. Gene Ther. 11:2403-2413. [DOI] [PubMed] [Google Scholar]
- 29.Weidhaas, J. B., E. L. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 31.Yu, X. F., M. Matsuda, M. Essex, and T. H. Lee. 1990. Open reading frame vpr of simian immunodeficiency virus encodes a virion-associated protein. J. Virol. 64:5688-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]