Abstract
The influenza virus PB1-F2 protein is a novel protein previously shown to be involved in induction of cell death. Here we characterize the expression and the function of the protein within the context of influenza viral infection in tissue culture and a mouse model. We show that the C-terminal region of the protein can be expressed from a downstream initiation codon and is capable of interaction with the full-length protein. Using this knowledge, we generated influenza viruses knocked out for the expression of PB1-F2 protein and its downstream truncation products. Knocking out the PB1-F2 protein had no effect on viral replication in tissue culture but diminished virus pathogenicity and mortality in mice. The viruses replicated to similar levels in mouse lungs by day 3 postinfection, suggesting that the knockout did not impair viral replication. However, while the PB1-F2 knockout viruses were cleared after day 5, the wild-type viruses were detectable in mouse lungs until day 7, implying that expression of PB1-F2 resulted in delayed clearance of the viruses by the host immune system. Based on our findings and on the fact that the PB1 genomic segment was always newly introduced into some pandemic influenza viruses of the last century, we speculate that the PB1-F2 protein plays an important role in pathogenesis of influenza virus infection and may be an important contributor to pathogenicity of pandemic influenza viruses.
In the course of a systematic search for influenza virus antigenic peptides presented by major histocompatibility complex class I on the surface of infected cells, a cytotoxic T lymphocyte (CTL) peptide that did not correspond to any of the known standard viral open reading frames was identified (4). Further screening of the influenza virus genome revealed that the peptide corresponded to residues 62 to 70 of an 87-amino-acid-long protein encoded by an alternate reading frame within the PB1 gene (4). The translation of the novel protein starts from nucleotide position 120 in the PB1 genomic segment and is believed to be initiated by ribosomal scanning (4, 13). In view of the fact that the protein is expressed from a second open reading frame (+1) of the PB1 gene, it was named PB1-F2 (4).
Further work showed that PB1-F2 is a relatively short-lived protein which is maximally expressed about 5 hours postinfection (4). The protein localizes to both inner and outer mitochondrial membranes, resulting in alteration of mitochondrial morphology, dissipation of mitochondrial membrane potential, and cell death. Knocking out the PB1-F2 open reading frame attenuated the ability of the A/Puerto Rico/8/34 virus to induce apoptosis in immune cells (4). Subsequently, the basic amphipathic helix in the C-terminal region of the PB1-F2 protein was shown to be responsible for its inner mitochondrial membrane targeting (9, 26) and peptides derived from the C-terminal domain were shown to have a cytotoxic effect and to induce formation of nonspecific pores in synthetic bilayer membranes (2).
We further recently showed that PB1-F2 protein interacts with the mitochondrial apoptotic mediators ANT3 and VDAC1 and sensitizes cells to apoptotic stimuli through the mitochondrial pathway (27). Our studies again highlighted the importance of the C-terminal region of the protein, as it was responsible for the interaction with the inner mitochondrial membrane protein ANT3 and induced mitochondrial permeabilization in an ANT3-dependent fashion (27).
Despite the studies outlined above, the precise role of the PB1-F2 protein within the context of viral infection remains unknown. It was shown previously that there was no substantial effect of the PB1-F2 protein on viral gene expression or virus-induced apoptosis in the epithelial cell lines MDCK, MDBK, A549, and HeLa, the first three of which support productive influenza virus infections (4). Furthermore, while the PB1-F2 protein was shown to be more apoptotic in immune cells (4), implying its possible role in modulation of immune response, the significance of this finding was never demonstrated within the context of infection of an animal host.
We decided to further elucidate the role of the PB1-F2 protein in viral infection by determining its contribution to viral pathogenicity in a mouse model. During the course of our studies, we found that the PB1-F2 knockout strategy described previously was not sufficient, as it allowed for expression of the PB1-F2 C-terminal region from a downstream initiation codon. Our new knockout strategy ensured termination of the expression of the downstream truncation product. We show that while the PB1-F2 protein does not enhance viral replication in tissue culture, it contributes to viral pathogenesis in mice by preventing efficient viral clearance. We speculate that the proapoptotic role of the PB1-F2 protein in immune cells may be responsible for the observed effect. This finding suggests a role of the PB1-F2 protein in pathogenicity of pandemic influenza viruses, as novel PB1 segments were introduced into the 1957 and 1968 pandemic viruses as well as into swine viruses circulating since 1998 (12, 19, 28).
MATERIALS AND METHODS
Cell lines, antibodies, and reagents.
293T and MDCK cells were obtained from ATCC and were maintained in Dulbecco's modified Eagle's medium and minimal essential medium, respectively (Gibco), supplemented with 10% fetal calf serum (HyClone) and penicillin-streptomycin (Gibco). Monoclonal anti-PB1-F2 antibody (clone 26D3) has been described previously (27). Monoclonal antibodies to Flag and hemagglutinin (HA) epitopes were obtained from Sigma.
Constructs and cloning.
The pCAGGS vector for the expression of proteins under the control of a chicken β-actin promoter has been described previously (15). An amino-terminal HA or Flag tag was introduced into each construct by PCR with 5′ gene-specific primers possessing the tag sequences. Each tagged gene was introduced into the pCAGGS vector for mammalian expression. HA-tagged constructs expressing either C- or N-terminally truncated PB1-F2 proteins (HA-nF2 and HA-cF2, respectively) were generated by insertion of PCR-amplified C- or N-terminal domains into the pCAGGS vector. The pPolI vectors (PolI is polymerase I) encoding viral genomic RNAs of WSN and PR8 strains have been described previously (8). To generate PB1-F2 knockout mutants, pPolI plasmids encoding the PB1 gene of either PR8 or WSN virus were subjected to three rounds of site-directed mutagenesis (see Results) by use of a Stratagene site-directed mutagenesis kit. To generate a pPolI vector encoding the chimeric WSN-PR8 PB1 segment, a novel asymmetric SapI restriction site was introduced into position 411 of the PR8 PB1 gene (sense strand) and into position 411 of the WSN gene (antisense strand). As SapI cuts downstream of its recognition sequence, this allowed us to subclone the digested pPolI WSN segment of the first 411 nucleotides into the PR8 segment digested with the same enzymes. The PB1 gene of the A/Hong Kong/156/97 (A/HK/156/97) virus (HK PB1) was cloned into pPolI vector by reverse transcriptase PCR from genomic RNA with PB1 gene-specific primers. HK PB1 knocked out for PB1-F2 expression was generated using the strategy described above. Sequences of each generated construct were confirmed by automated sequencing performed at the Mount Sinai sequencing core facility. All primer sequences and vector maps are available upon request.
Pulse-chase analysis.
MDCK cells were infected with influenza viruses for 12 h, pulse-labeled with [35S]methionine for 30 min, and chased for another 4 h. At each time point, the cells were scraped off the dish and spun down and the cell pellet was flash frozen on liquid nitrogen. Cells were subsequently lysed in coimmunoprecipitation buffer: 0.5% NP-40, 150 mM NaCl, 20 mM HEPES, pH 7.4, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 10% glycerol, and Complete protease inhibitor cocktail and phenylmethylsulfonyl fluoride (Roche). Protein lysates were incubated at 4°C overnight with 3 μg of the 26D3 monoclonal antibody recognizing the N terminus of the PB1-F2 protein. Protein complexes were precipitated with protein G agarose beads (Roche) for 2 h. Beads were washed five times in lysis buffer and resuspended in protein sample buffer. Proteins were subsequently separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by autoradiography.
Western blotting.
Cell extracts were made in the coimmunoprecipitation lysis buffer. Cell lysates were loaded onto a 12% sodium dodecyl sulfate-polyacrylamide gel. Separated proteins were transferred to nitrocellulose membrane and probed with specific antibodies.
Reverse genetics for recombinant viruses.
The reverse genetics for generation of recombinant influenza viruses has been described previously (8). Briefly, for generation of recombinant viruses, 293T cells were transfected with eight pPolI vectors encoding the viral genomic RNA segments and four pCAGGS protein expression vectors encoding the subunits of viral polymerase and the nucleocapsid protein. The transfected 293T cells were cocultured with MDCK cells to amplify the generated viruses. Each of the rescued viruses was further plaque purified and grown in MDCK cells, and titers were determined by utilizing the procedures described previously (22). The mutations were confirmed by sequencing the PB1 genes of the newly generated viruses.
Infections of mice.
Female C57BL/6 mice, 6 to 7 weeks old (Jackson Laboratories), or BALB/c mice (Charles River) were anesthetized with an intraperitoneal injection of 0.1 ml of ketamine-xylazine (0.15 mg ketamine and 0.03 mg xylazine), and 50 μl of infectious virus diluted in phosphate-buffered saline (PBS) was inoculated intranasally. To determine virus pathogenicity, five mice from each group were inoculated with an appropriate dose and were monitored daily for weight loss for 12 days. Mice that lost more than 25% of body weight were euthanized according to the institutional guidelines. To determine viral replication in the lungs, 24 mice were inoculated with each virus and 12 mice were inoculated with PBS. Lungs and sera were collected on days 3, 4, 5, 6, 7, and 9 postinfection from four mice from each virus group and from two mice in the PBS group. The lungs were homogenized and processed for determination of virus titers, which was done by immunofluorescence of infected MDCK cells as described previously (18).
RESULTS
PB1-F2 protein expression in the context of viral infection.
During previous studies, a PR8 influenza virus knocked out for PB1-F2 protein was generated (4). The knockout strategy included modification of the PB1-F2 start codon, without altering the PB1 open reading frame, and introduction of a stop codon after PB1-F2 amino acid 8, which resulted in a Met→Ile substitution in the PB1 at position 40. These mutations resulted in complete abolition of PB1-F2 protein expression when tested in infected cells with an antibody recognizing the N terminus of the protein (4).
Surprisingly, when the PB1-F2 knockout virus was tested in mice, we were not able to show any attenuation of the virus (data not shown). We initially speculated that the knockout strategy described above still allowed for translation of an N-terminally truncated PB1-F2 product(s) from a downstream initiation codon.
Previous studies showed that PB1-F2 protein migrates as more than one band when immunoprecipitated from radiolabeled infected cells (4). This observation was initially attributed to degradation products of the PB1-F2 protein or possible protein posttranslational modifications. Interestingly, only a single protein species was detected when PB1-F2 was immunoblotted and probed with an antibody specific to the N terminus of the protein (4).
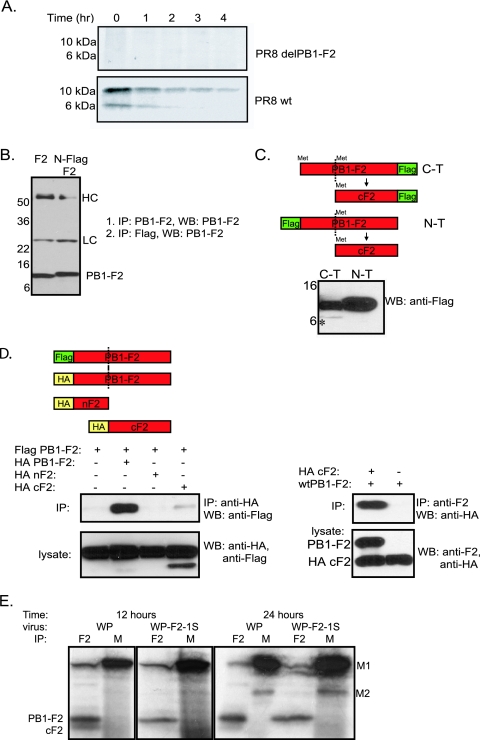
To better understand the mechanism of PB1-F2 protein expression within the cell, we initially set out to elucidate the causes of the observed differences in numbers of protein species between the radiolabeled and immunoblotted lysates. To confirm the previous observations, we infected cells with PR8 virus and, 12 h later, labeled the cells with [35S]methionine for 30 min and then chased the cells for another 4 h (Fig. 1A). The PR8 virus, knocked out for PB1-F2 protein expression as described above, was used as a control (4). At 1-h time intervals, the cells were collected and lysed, and immunoprecipitations were performed with anti-PB1-F2 monoclonal antibody 26D3 recognizing the N terminus of the protein. Pulse-chase experiments revealed that the PB1-F2 protein possessed a half-life of about 1 h. Interestingly, two major species of the protein were detected in the lysates (Fig. 1A). Since the antibody used for immunoprecipitations recognizes the N terminus of the PB1-F2 protein, we initially speculated that the protein was an N-terminal degradation product. However, similarly to previous observations, the band did not appear when nonradiolabeled immunoprecipitates of PB1-F2-infected cells were immunoblotted with the same antibody (Fig. 1B). We concluded that the observed lower band could not be the N-terminal portion of the protein. Moreover, the lower band was present at time zero after the pulse, suggesting that it was most likely not a degradation intermediate of the full-length protein but rather another protein which coimmunoprecipitated with PB1-F2 (Fig. 1A). Sequence analysis of the PB1-F2 protein further revealed that the majority of methionines (responsible for the radiolabeled products) are located in the C terminus of the protein (PB1-F2 positions 39, 46, and 51), which led us to hypothesize that the observed lower band is the C terminus of the PB1-F2 which coimmunoprecipitated with the full-length protein.
FIG. 1.
The C-terminal region of PB1-F2 interacts with the full-length protein and can be expressed from a downstream initiation site. (A) Pulse-chase for PB1-F2 in wt PR8 virus- and PR8delF2 virus-infected MDCK cells. MDCK cells were infected with the corresponding virus at an MOI of 1 for 12 h, pulse-labeled for 10 min, and chased for another 4 h. (B) Immunoprecipitation (IP) and Western blotting (WB) with the 26D3 PB1-F2 antibody recognizing the N terminus of the protein do not reveal additional bands of PB1-F2. HC, immunoglobulin heavy chain; LC, immunoglobulin light chain. (C) The PB1-F2 C terminus (C-T) can be expressed separately from a plasmid from a downstream initiation site. Plasmids encoding either N- or C-terminally Flag-tagged PB1-F2 were transfected into 293T cells for 24 h, and lysates were probed by Western blotting with monoclonal anti-Flag antibody. The diagram depicts the expected proteins and their downstream truncation products. N-T, N terminus; *, C-terminal domain of the protein. In panels B and C, numbers to the left of blots are molecular size markers in kilodaltons. (D) Coimmunoprecipitation of PB1-F2 N and C termini with full-length PB1-F2 protein. (Left) IP against HA-tagged N and C termini and WB against Flag-tagged full-length PB1-F2. (Right) IP against PB1-F2 with anti-F2 antibody and WB against HA-tagged C terminus. (E) Expression of PB1-F2 from the WP virus and the corresponding virus knocked out for downstream PB1-F2 initiation codons (WP-F2-1S). Cell lysates from virus-infected cells were immunoprecipitated with either anti-PB1-F2 polyclonal antiserum (F2) or anti-M1 monoclonal antibody (M), which recognizes M1 and M2 proteins.
To formally demonstrate that a downstream initiation codon accounts for the appearance of the lower band, the downstream methionines at PB1-F2 positions 39, 46, and 51 were mutated to threonines, and infectious WSN viruses, possessing either wild-type (wt) PR8 PB1 (WP) or PR8 PB1 with the mutated downstream PB1-F2 initiation codons, were generated. 293T cells were infected with each of the viruses at a multiplicity of infection (MOI) of 2 for 12 or 24 h and radiolabeled for 30 min with [35S]methionine. After radiolabeling, the cells were lysed and immunoprecipitations were performed with anti-PB1-F2 polyclonal serum. As can be seen in Fig. 1E, elimination of the downstream initiation codons resulted in disappearance of the lower band (and a decreased signal of the mutated full-length protein). This is compatible with the hypothesis that the C terminus of the PB1-F2 protein can be expressed independently.
To confirm that the C terminus of the PB1-F2 protein interacts with the full-length protein, we performed a series of coimmunoprecipitation experiments utilizing PB1-F2 proteins tagged with Flag epitope on either the N or the C termini (Fig. 1C and D). Western blotting for lysates expressing either one of the constructs revealed two protein species in the cells expressing the C-terminally tagged PB1-F2 but not in the cells expressing the N-terminally tagged protein (Fig. 1C). Furthermore, when expressed separately, the C-terminal region of PB1-F2 interacted with the full-length protein in immunoprecipitation experiments (Fig. 1D).
This finding is supported by the recent nuclear magnetic resonance studies showing that the C-terminal region of the PB1-F2 protein is capable of interacting, resulting in oligomers (11). Furthermore, other studies showed that the C-terminal region of the protein possesses an amphipathic helix which by itself is capable of inducing permeabilization of the inner mitochondrial membrane (2, 9) and interaction with the inner mitochondrial membrane ANT3 protein (2, 9, 27).
These findings had important implications for our further work since they suggested that the C terminus of the protein can be expressed separately from a downstream initiation codon. As the C-terminal region has been shown to be responsible for targeting the protein to mitochondria and the induction of apoptosis (9, 26, 27), this finding was important in our further designs of strategies of knocking out the PB1-F2 protein and its downstream initiation product.
Knocking out the PB1-F2 protein from the highly virulent mouse-adapted WSN virus.
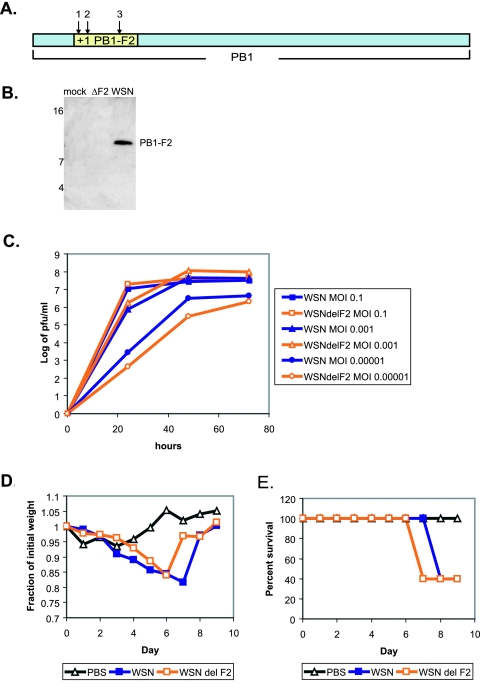
In order to evaluate the contribution of the PB1-F2 protein to viral pathogenicity, we initially generated a recombinant influenza virus of the highly pathogenic mouse-adapted WSN strain knocked out for the PB1-F2 protein (Fig. 2A). To ensure complete abolition of PB1-F2 protein expression, the start codon of the gene was mutated from ATG to ACG (T120C) and two stop codons were introduced, one at position 12 (C153G) and one at position 58 (G291A) of the protein. The latter stop codon was generated to prevent synthesis of a truncated protein from a downstream position. The mutations described above were silent in the open reading frame of the PB1 gene. The virus possessing the mutated PB1 segment (WSNdelF2) and the corresponding wt control virus were generated utilizing a 12-plasmid reverse genetics system as described previously (8). The resultant viruses were initially tested for the expression of PB1-F2 protein in the infected MDCK cells. As can be seen from Fig. 2B, cells infected with WSNdelF2 virus were negative for the expression of PB1-F2 protein compared to the cells infected with the wt control.
FIG. 2.
Knocking out PB1-F2 from a highly virulent mouse-adapted WSN virus. (A) PB1-F2 knockout strategy. Arrows indicate the positions of mutations: 1, T120C; 2, C153G; 3, G291A. (B) Expression of PB1-F2 by virus. MDCK cells were infected at an MOI of 5 for 12 h, and lysates were probed by Western blotting with 26D3 antibody. Numbers to the left of blots are molecular size markers in kilodaltons. (C) Growth of WSN and WSNdelF2 viruses in MDCK tissue culture. Cells were inoculated at indicated MOIs, and infection supernatants were collected at 24, 48, and 72 h. (D) Pathogenesis of WSN and WSNdelF2 viruses in mice. Five mice per virus group were inoculated with 5 × 102 PFU of the indicated viruses, and mouse weights were assessed every day after infection. (E) Survival of mice infected with WSN and WSNdelF2 viruses as described for panel D.
The recombinant viruses were then tested for the ability to replicate in the MDCK cell culture. As can be seen from Fig. 2C, the WSNdelF2 virus replicated equally well or better than its wt WSN counterpart (at 72 h, P was >0.3 at an MOI of 0.1, P was <0.005 at an MOI of 0.001, and P was >0.06 at an MOI of 0.00001). Moreover, when intranasally inoculated into 6-week-old C57BL/6 mice, the viruses were equally pathogenic, as exemplified by mouse weight loss and percent survival upon infection with a low dose (5 × 102 PFU) of each virus (Fig. 2D and E).
Based on these results, we hypothesized that within a highly pathogenic mouse-adapted influenza viral strain such as WSN, the contribution of the PB1-F2 protein might not be significant. We decided to turn to a virus model which is attenuated in mice to see whether viral pathogenicity could be influenced by the expression of PB1-F2 protein. Our previous work revealed that substitution of the WSN PB1 gene with the PB1 gene of PR8 virus attenuates the virus in the mouse model (data not shown).
Generation of a model influenza virus to study contribution of the PB1-F2 protein.
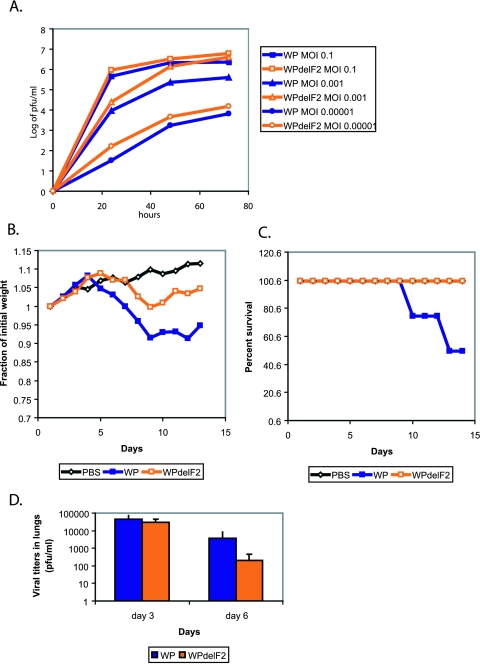
Utilizing the mutagenesis strategy described above, we generated a plasmid encoding the PR8 PB1 segment knocked out for PB1-F2 expression and rescued WSN viruses possessing either a wt or a mutant PR8 PB1 segment (WP and WPdelF2 viruses, respectively). Similarly to the results with the wt WSN virus, the WPdelF2 virus replicated equally well or better than its WP counterpart (Fig. 3A) (at 72 h, P was >0.1 at an MOI of 0.1, P was <0.05 at an MOI of 0.001, and P was <0.04 at an MOI of 0.00001). Both viruses were nonpathogenic in mice when used at doses up to 1 × 106 PFU. We speculate that poor compatibility between the PB1 protein of PR8 virus and the PB2 and PA proteins of WSN virus is responsible for the observed attenuation.
FIG. 3.
Knocking out PB1-F2 from WSN virus possessing the PR8 PB1 gene (WP). (A) Growth of WP and WPdelF2 viruses in MDCK tissue culture. Cells were inoculated at the indicated MOIs, and infection supernatants were collected at 24, 48, and 72 h. (B) Pathogenesis of viruses in mice. Five mice per virus group were inoculated with 1 × 106 PFU of the indicated viruses, and mouse weights were assessed every day after infection. (C) Survival of mice infected with WP and WPdelF2 viruses as described for panel B. (D) Lung virus titers from mice infected with WP and WPdelF2 viruses. Ten mice per virus group were infected with 1 × 106 PFU of each virus. Five mice from each group were sacrificed on days 3 and 6, and lung virus titers were assessed by infection of MDCK cell monolayers. Error bars represent standard errors of the means.
When intranasally inoculated into C57BL/6 mice at 1 × 106 PFU, the viruses knocked out for PB1-F2 expression (WPdelF2) failed to induce significant weight loss or lethality in the infected animals compared to the PB1-F2-expressing control (WP) (Fig. 3B and C). Interestingly, while both viruses initially replicated in mouse lungs to similar titers (day 3), WPdelF2 viruses were cleared from the lungs more efficiently by day 6 (Fig. 3D); however, the difference in lung virus titers was not statistically significant at this time point (P > 0.1). Nevertheless, these results suggested that PB1-F2 protein contributes to viral pathogenicity, possibly by delaying clearance of the virus from the lungs.
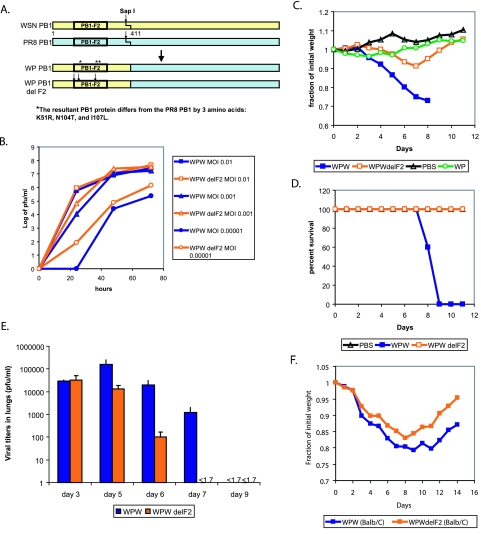
To further evaluate the mechanism of PB1-F2-induced enhancement of pathogenesis, we sought to generate a virus model that was not as severely attenuated as the WP strain. For these purposes, we generated a WSN virus possessing a chimeric PB1 segment, with nucleotides 1 to 411 from the WSN strain and nucleotides 412 to 2341 from the PR8 strain. The resultant PB1 segment encodes a wt WSN PB1-F2 protein and a chimeric WSN-PR8 PB1 protein which differs from the wt PR8 PB1 by three amino acids (K51R, N104T, and I107L) (Fig. 4A). The resultant virus (termed WPW) was intranasally inoculated into C57BL/6 mice and was determined to have a 50% lethal dose (LD50) of 2 × 104 (data not shown), which lies between the LD50s of the highly virulent and the nonpathogenic models. Utilizing the strategy described above, we furthermore generated the PB1-F2 knockout counterpart virus (termed WP-WdelF2) and compared both viruses for pathogenicity in MDCK cell culture and mice. Similarly to the results with WSN and WP viruses, the WP-WdelF2 virus was comparable in growth to the WPW virus at high MOIs and grew better than the WPW virus at low MOIs (Fig. 4B) (at 72 h, P was >0.2 at an MOI of 0.1, P was >0.08 at an MOI of 0.001, and P was <0.003 at an MOI of 0.00001).
FIG. 4.
Contribution of PB1-F2 to pathogenicity of a virus with an intermediate virulent phenotype (WPW). (A) Construction of a chimeric WSN-PR8 PB1 gene (WPW) and a WSN-PR8 PB1 gene knocked out for PB1-F2 (WP-WdelF2). The PB1-F2 protein was knocked out in a manner analogous to that described in the legend for Fig. 2. (B) Replication of WPW and WP-WdelF2 viruses in tissue culture. Cells were inoculated at the indicated MOIs, and infection supernatants were collected at 24, 48, and 72 h. (C) Pathogenesis of viruses in mice. Five mice per virus group were inoculated with 4 × 105 PFU of the indicated viruses, and mouse weights were assessed every day after infection. (D) Survival of mice infected with WPW and WP-WdelF2 viruses as described for panel C. (E) Mouse lung virus titers. Twenty-five mice per virus group were infected with 4 × 105 PFU of each virus. Five mice from each group were sacrificed on days 3, 5, 6, 7, and 9, and lung virus titers were assessed by infection of MDCK cell monolayers. Error bars represent standard errors of the means. (F) Viral pathogenesis in BALB/c mice. Five mice per group were inoculated with 2 × 105 PFU of the indicated viruses, and mouse weights were assessed every day after infection.
When WPW virus was intranasally inoculated into C57BL/6 mice at 5 × 104 PFU, there was a strong reduction in mouse weight and all animals eventually succumbed to infection by day 10 (Fig. 4C and D). In contrast, only a mild reduction in weight was observed for the mouse group inoculated with WP-WdelF2 virus. All of these animals survived the infection. Lungs of the infected mice were collected on days 3, 4, 5, 6, 7, and 9 postinfection and assessed for total virus titers. Similarly to the results observed for the WP viruses, both WPW and WP-WdelF2 viruses initially replicated to similar titers, but the clearance of the WPW virus from the lungs was delayed by 2 days (P was <0.03 at day 5, P was <0.02 at day 6, and P was <0.03 at day 7) (Fig. 4E). These results led us to speculate that while replication of the PB1-F2 knockout viruses was not impaired, the immune responses responsible for clearance of the viruses were different between the two groups.
Recent studies showed that some influenza viral epitopes can delay viral clearance (7). To confirm that the observed decrease in pathogenicity was due to the loss of PB1-F2 protein function rather than the loss of a CTL epitope, we performed similar experiments with BALB/c mice, since the CD8 cells from BALB/c mice fail to recognize the PB1-F2 epitope (3). WPW and WP-WdelF2 viruses were intranasally inoculated into BALB/c mice at 2 × 105 PFU. While this dose was not sufficient to kill the animals, the mice inoculated with the WPW virus showed a greater degree of weight loss and took longer to recover from the infection than the mice infected with the corresponding virus knocked out for PB1-F2 (Fig. 4F). This suggested that the observed decrease in pathogenicity in PB1-F2 knockout viruses was independent of loss of the CTL epitope.
The PB1-F2 protein from the A/HK/156/97 virus contributes to viral virulence.
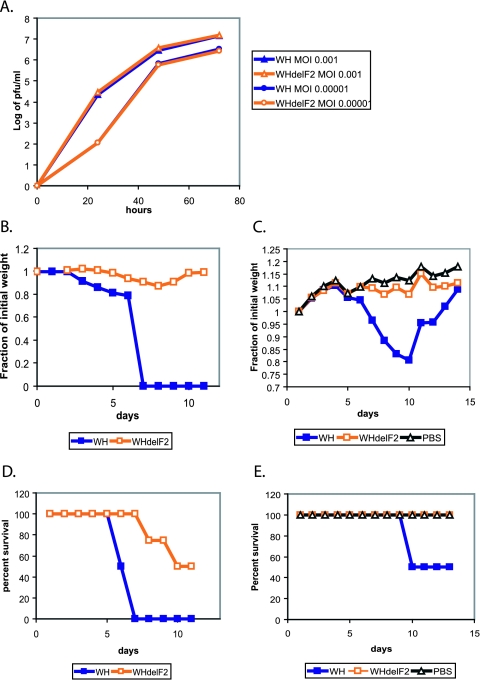
Based on our findings, we conclude that the PB1-F2 protein plays an important role in viral pathogenesis in an animal host and may reverse an attenuating effect of a heterologous PB1 gene. The reassortment of the PB1 genomic segment is not unprecedented and is of particular interest, since, in addition to the glycoprotein genes, PB1 was the only other gene segment that was often exchanged in the reassortant (pandemic) viruses in the past century (12, 28). To test whether PB1-F2 protein contributes to virulence of highly pathogenic influenza viral strains, we cloned the PB1 gene from the A/HK/156/97 virus, a pathogenic H5N1 virus which caused a lethal infection in a 3-year-old boy in Hong Kong in 1997 (5). Interestingly, the PB1 gene of this virus came from a distinct avian source (10, 14). Using the strategy outlined above, we knocked out the PB1-F2 protein open reading frame and generated two viruses: WSN with HK PB1 (WH) and WSN with HK PB1 knocked out for PB1-F2 (WHdelF2). Similarly to the above-described results, knocking out PB1-F2 had no effect on viral replication in MDCK cell culture (at 72 h, P was >0.6 at an MOI of 0.001 and P was >0.3 at an MOI of 0.00001) (Fig. 5A). Upon inoculation of mice, however, the WH virus proved to be more pathogenic and induced more significant weight loss and mortality than its PB1-F2 knockout counterpart (Fig. 5B). The LD50 for the WH virus was calculated to be around 2 × 103 PFU, while the WHdelF2 virus had an LD50 of 4 × 105 PFU. Based on these studies, we conclude that the PB1-F2 protein plays a role in viral virulence and may contribute to pathogenicity of highly virulent influenza viral strains.
FIG. 5.
Contribution of A/HK/156/97 PB1-F2 to viral virulence. (A) Growth of the WH and WHdelF2 viruses in tissue culture. Cells were inoculated at the indicated MOIs, and infection supernatants were collected at 24, 48, and 72 h. (B) Pathogenesis of the WH and the WHdelF2 viruses in mice at 4 × 105 PFU. Eight mice per virus group were inoculated with 4 × 105 PFU of the indicated viruses, and mouse weights were monitored over the next 12 days. (C) Pathogenesis of the WH and the WHdelF2 viruses in mice at 2 × 103 PFU. The experiment was performed as described for panel B, with eight mice per virus group. (D and E) Survival of mice infected with WH and WHdelF2 viruses, as described for panels B and C.
DISCUSSION
Previous studies proposed that within an infected host the PB1-F2 protein may play a role in elimination of immune cells and in suppression of antiviral immune signaling by eliminating the infected cells more effectively (4, 27).
To confirm the role of the protein in pathogenesis of influenza viral infection, we initially sought to generate influenza viral mutants knocked out for PB1-F2 protein expression. In the original studies of the PB1-F2 protein, PB1-F2 knockout mutants were created by elimination of a start codon and introduction of a stop codon several amino acids downstream (4). Our current findings indicate that this virus would still be capable of expressing the C-terminal domain from a downstream initiation codon. Since similar expression patterns of two products were noted when the protein was expressed from a plasmid, we conclude that the downstream initiation codon must be able to translate the C-terminal region of the PB1-F2 protein. A possible initiation site is the second ATG at position 233 of the PB1 gene segment. To further elucidate the role of the protein within the context of viral infection, we generated influenza virus mutants knocked out for the expression of PB1-F2 protein and its downstream products.
Surprisingly, knocking out PB1-F2 from wt WSN virus had no effect in mice. This is not unprecedented, since in the context of a very pathogenic mouse-adapted virus, an accessory factor like PB1-F2 may play only a small role.
These findings led us to speculate that the protein may be more important within the context of a virus with a reduced replication potential. Within a natural setting, this may be important when influenza viruses cross species barriers or when new viral strains are generated by reassortment. In both cases, viruses may be reduced in their ability to replicate in the new host and may require some degree of immunosuppression in order to allow for several rounds of replication and for adaptive mutations to take place.
To explore this model, we decided to focus on the PB1 gene. We hypothesized that a reassortant PB1 gene might not create a perfect fit with other components of the viral polymerase and within an infected host may require PB1-F2 protein for its accessory, possibly immunosuppressive, function. To test this hypothesis, we initially generated a WSN influenza virus model attenuated by introduction of a heterologous PB1 polymerase protein (from PR8 virus) (LD50 of 1 × 106 PFU). The observed attenuation was most likely due to incompatibility of the PR8 PB1 protein with other components of viral polymerase (PB2 and PA).
When tested in mice, this virus was more pathogenic than its PB1-F2 knockout counterpart (Fig. 3). Interestingly, this virus was not attenuated in tissue culture or in its initial replication in mouse lungs. This suggested that deletion of PB1-F2 had no inherent negative effect on the ability of the influenza virus to replicate. Rather, its delayed clearance from the lungs suggested that the PB1-F2 protein attenuated an immune response to the infection.
As this virus was too attenuated, studies of pathogenesis proved to be difficult. In order to overcome this problem, we created a WSN virus with an intermediate pathogenic phenotype (LD50 of 2 × 104 PFU). This virus possesses a chimeric WSN-PR8 PB1 protein, which may allow it to better interact with the other components of WSN polymerase and thus to promote more efficient viral replication. When tested in mice, this virus induced significantly higher weight loss and mortality than its PB1-F2 knockout counterpart (Fig. 4). Furthermore, as observed for the virus possessing the wt PR8 PB1 gene, the initial replication rates between the two viruses were similar. However, the kinetics of viral clearance were different. While the mutant virus could no longer be detected in mouse lungs after day 5, the wt virus was still present in the lungs on day 7. This suggests that the immune response to the wt virus was suppressed compared to the response to the mutant.
Both of the viruses eventually got cleared from the mouse lungs. However, mice inoculated with the mutant virus recovered, while mice in the wt group succumbed to the infection. This is not surprising, since influenza virus infection is associated with development of immunopathology, which can become irreversible depending on the levels of viral replication in the lung (25). It is likely that the enhancement of viral pathogenicity is due to apoptosis of the infected immune cells, which delays the acquired immune response to the infection. We speculate that, during the initial stages of viral replication, antigen presentation by professional antigen-presenting cells such as macrophages and dendritic cells may be impaired due to a greater degree of apoptosis in those cells (4). Similar findings were observed previously for the highly pathogenic form of lymphocytic choriomeningitis virus (1). The high degree of immunosuppression and lymphopenia induced by the highly pathogenic H5N1 viruses from the 1997 outbreak in Hong Kong could be another example of influenza virus preventing efficient maturation of an immune response through its interference with antigen-presenting cells (24). This could be due to the strong suppression of dendritic cell maturation by the highly effective NS1 protein (20) or perhaps killing of the antigen-presenting cells. Further experiments assessing the immune responses to the wt and PB1-F2 knockout viruses will be needed in order to confirm this hypothesis.
Role of the PB1-F2 protein in pathogenesis of human influenza viral infections: implications for pandemics.
Large-scale sequence analysis of the avian influenza virus isolates recently revealed that, out of all influenza viral genes, the transcript encoding the PB1-F2 gene is under the highest positive-selection pressure for nonsynonymous substitutions (16). In addition, the expression of the protein by the majority of the avian influenza strains implies that the protein plays an important role in the viral life cycle (16).
Sequence analysis of the human influenza viral PB1 genes available in the database revealed that the PB1-F2 protein is encoded by all of the human H3N2 viruses. We find that since the reemergence of the H1N1 virus in 1977, the human and pig H1N1 viruses encode only a C-terminally truncated PB1-F2 protein, lacking the region responsible for mitochondrial targeting and apoptosis. The two human post-1977 H1N1 virus isolates encoding a full-length PB1-F2 protein, A/Kiev/59/79 (17) and A/Wisconsin/10/98 (GenBank accession number AI342823) appear to encode a PB1 gene from an H3N2 virus. In fact, both viruses turned out to be natural reassortants between H3N2 and H1N1 strains. Interestingly, H1N1 viruses are also known to cause disease less frequently than the H3N2 viruses. By the same argument, we speculate that expression of the PB1-F2 protein by the H3N2 viruses contributes to disease severity in humans, though further experimentation with animal models will be necessary.
In our virus model, the highly mouse-adapted WSN virus strain was attenuated by introduction of a heterologous PB1 segment, mimicking gene reassortment in pandemic viruses, since the PB1 segment was introduced from an avian source into the 1957 and 1968 pandemic viruses (12, 28). While changes in the surface glycoproteins allow the reassortant pandemic viruses to overcome a preexisting humoral immune response, they cannot be solely responsible for the high virulence of the pandemic influenza viruses. This indicates that certain highly pathogenic viruses may possess characteristic mutation signatures in genes other than the surface antigens, which may either augment viral replication or modulate the host innate immune responses, thereby enhancing pathogenesis of the disease progression. Indeed, recent reconstruction of the 1918 virus has confirmed that polymerase genes contribute to the high pathogenicity of the recombinant 1918 virus in mice (23). In fact, substitution of the viral polymerase genomic segments with those of a modern H1N1 strain severely attenuated the virus in mice (23). It would be interesting to determine whether the PB1-F2 protein from the 1918 virus contributes to the highly pathogenic phenotype of this virus.
The effect of the “new” introduction of the PB1 gene in pandemic viruses is unknown. We speculate that the newly introduced PB1 gene may play an active role in pathogenesis of a reassortant virus within an animal host, with the PB1-F2 protein being an important contributor. Introduction of a novel PB1 into the 1998 swine reassortant virus further implicates the role of this gene in pathogenesis of animal influenza (19, 28). Indeed, PB1 segments from influenza viruses infecting different hosts have been shown to play a role in determining host range specificity (21). In support of our hypothesis, we find that the PB1-F2 protein of the virulent 1997 H5N1 strain contributes to viral pathogenesis in mice (Fig. 5). We suspect that the delay in viral clearance due to expression of PB1-F2 protein may allow for prolonged viral replication and development of irreversible pulmonary immunopathology, the findings observed with highly pathogenic influenza strains.
Acknowledgments
We thank Adolfo García-Sastre for helpful discussions.
This work was partially supported by NIH grants RO1 AI-18898-25, U54 AI-057158, PO1 AI058113, U19 AI062623, and PO1 AI052106 (P.P.) and NIH training grant AI007647 (D.Z.). Peter Palese is an Ellison Medical Foundation Scholar in Global Infectious Diseases.
REFERENCES
- 1.Borrow, P., C. F. Evans, and M. B. Oldstone. 1995. Virus-induced immunosuppression: immune system-mediated destruction of virus-infected dendritic cells results in generalized immune suppression. J. Virol. 69:1059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanturiya, A. N., G. Basanez, U. Schubert, P. Henklein, J. W. Yewdell, and J. Zimmerberg. 2004. PB1-F2, an influenza A virus-encoded proapoptotic mitochondrial protein, creates variably sized pores in planar lipid membranes. J. Virol. 78:6304-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, W., J. R. Bennink, and J. W. Yewdell. 2003. Systematic search fails to detect immunogenic MHC class-I-restricted determinants encoded by influenza A virus noncoding sequences. Virology 305:50-54. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 5.Claas, E. C., J. C. de Jong, R. van Beek, G. F. Rimmelzwaan, and A. D. Osterhaus. 1998. Human influenza virus A/HongKong/156/97 (H5N1) infection. Vaccine 16:977-978. [DOI] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.Crowe, S. R., S. C. Miller, and D. L. Woodland. 2006. Identification of protective and non-protective T cell epitopes in influenza. Vaccine 24:452-456. [DOI] [PubMed] [Google Scholar]
- 8.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbs, J. S., D. Malide, F. Hornung, J. R. Bennink, and J. W. Yewdell. 2003. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J. Virol. 77:7214-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 96:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henklein, P., K. Bruns, M. Nimtz, V. Wray, U. Tessmer, and U. Schubert. 2005. Influenza A virus protein PB1-F2: synthesis and characterization of the biologically active full length protein and related peptides. J. Pept. Sci. 11:481-490. [DOI] [PubMed] [Google Scholar]
- 12.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb, R. A., and M. Takeda. 2001. Death by influenza virus protein. Nat. Med. 7:1286-1288. [DOI] [PubMed] [Google Scholar]
- 14.Liu, J., K. Okazaki, H. Ozaki, Y. Sakoda, Q. Wu, F. Chen, and H. Kida. 2003. H9N2 influenza viruses prevalent in poultry in China are phylogenetically distinct from A/quail/Hong Kong/G1/97 presumed to be the donor of the internal protein genes of the H5N1 Hong Kong/97 virus. Avian Pathol. 32:551-560. [DOI] [PubMed] [Google Scholar]
- 15.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 16.Obenauer, J. C., J. Denson, P. K. Mehta, X. Su, S. Mukatira, D. B. Finkelstein, X. Xu, J. Wang, J. Ma, Y. Fan, K. M. Rakestraw, R. G. Webster, E. Hoffmann, S. Krauss, J. Zheng, Z. Zhang, and C. W. Naeve. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1562-1563. [DOI] [PubMed] [Google Scholar]
- 17.Petrov, N. A., S. Golovin, L. V. Mamaev, S. V. Netesov, and S. K. Vasilenko. 1987. Primary structure of the full-size DNA copy of the influenza virus A/Kiev/59/79 (H1N1) PB1 gene protein. Bioorg. Khim. 13:1170-1175. (In Russian.) [PubMed] [Google Scholar]
- 18.Quinlivan, M., D. Zamarin, A. Garcia-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richt, J. A., K. M. Lager, B. H. Janke, R. D. Woods, R. G. Webster, and R. J. Webby. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 41:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 21.Snyder, M. H., A. J. Buckler-White, W. T. London, E. L. Tierney, and B. R. Murphy. 1987. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J. Virol. 61:2857-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 24.Tumpey, T. M., X. Lu, T. Morken, S. R. Zaki, and J. M. Katz. 2000. Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J. Virol. 74:6105-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells, M. A., P. Albrecht, and F. A. Ennis. 1981. Recovery from a viral respiratory infection. I. Influenza pneumonia in normal and T-deficient mice. J. Immunol. 126:1036-1041. [PubMed] [Google Scholar]
- 26.Yamada, H., R. Chounan, Y. Higashi, N. Kurihara, and H. Kido. 2004. Mitochondrial targeting sequence of the influenza A virus PB1-F2 protein and its function in mitochondria. FEBS Lett. 578:331-336. [DOI] [PubMed] [Google Scholar]
- 27.Zamarin, D., A. Garcia-Sastre, X. Xiao, R. Wang, and P. Palese. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathogens 1:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K. Yoon, S. Krauss, and R. G. Webster. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]