Abstract
We have used feline immunodeficiency virus (FIV) protease (PR) as a mutational system to study the molecular basis of substrate-inhibitor specificity for lentivirus PRs, with a focus on human immunodeficiency virus type 1 (HIV-1) PR. Our previous mutagenesis studies demonstrated that discrete substitutions in the active site of FIV PR with structurally equivalent residues of HIV-1 PR dramatically altered the specificity of the mutant PRs in in vitro analyses. Here, we have expanded these studies to analyze the specificity changes in each mutant FIV PR expressed in the context of the natural Gag-Pol polyprotein ex vivo. Expression mutants were prepared in which 4 to 12 HIV-1-equivalent substitutions were made in FIV PR, and cleavage of each Gag-Pol polyprotein was then assessed in pseudovirions from transduced cells. The findings demonstrated that, as with in vitro analyses, inhibitor specificities of the mutants showed increased HIV-1 PR character when analyzed against the natural substrate. In addition, all of the mutant PRs still processed the FIV polyprotein but the apparent order of processing was altered relative to that observed with wild-type FIV PR. Given the importance of the order in which Gag-Pol is processed, these findings likely explain the failure to produce infectious FIVs bearing these mutations.
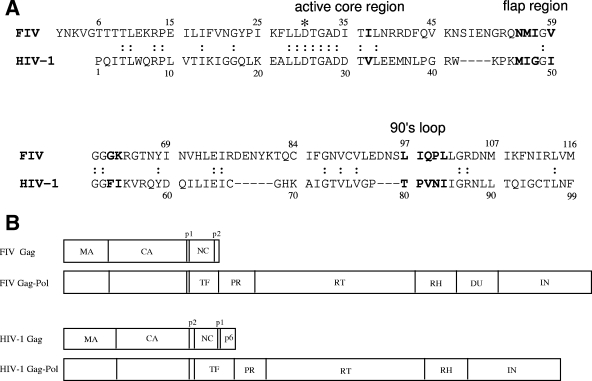
We have used feline immunodeficiency virus (FIV), a member of the lentivirus family, as a small-animal model to develop intervention strategies against lentiviral infection (11, 12, 16). One of our goals is to understand the molecular basis of human immunodeficiency virus type 1 (HIV-1) and FIV protease (PR) substrate and inhibitor specificity in order to develop broad-spectrum PR inhibitors that will inhibit wild-type and drug-resistant PRs. This approach has led to the development of TL-3, an inhibitor that is capable of inhibiting FIV, simian immunodeficiency virus, HIV-1, and several drug-resistant HIV-1 strains ex vivo (5, 25, 26), as well as other potential inhibitors with broad efficacy (24, 33, 34). FIV PR, like HIV-1 PR, is a homodimer, but each monomer is composed of 116 amino acids, as opposed to 99 amino acids for HIV-1 PR (Fig. 1A). The structure of FIV PR has been determined and compared to that of HIV-1 PR (23, 53). FIV PR is very similar to HIV-1 PR, particularly in the active core region, but only shares 27 identical amino acids (23% identical at the amino acid level) and exhibits distinct substrate and inhibitor specificity (1, 30, 31, 44, 53). FIV and HIV-1 PR each prefer their own matrix (MA)-capsid (CA) junction substrate, and FIV PR prefers a longer substrate than HIV-1 PR. Current clinical drugs against HIV-1 PR are poor inhibitors of FIV PR, primarily because of a smaller S3 substrate binding site in FIV PR (25, 26).
FIG. 1.
(A) Amino acid sequence alignment of FIV and HIV-1 PRs. FIV PR monomer is composed of 116 residues, as opposed to 99 residues for HIV-1 PR. There are 27 identical residues between the FIV and HIV-1 PRs. *, catalytic aspartic acid, D25 for FIV PR and D30 for HIV-1 PR. The substitutions made in this study are in boldface. (B) Schematic representation of FIV and HIV-1 Gag and Gag-Pol polyprotein. Cleavage sites and individual mature proteins are shown. HIV-1 has an additional spacer protein, p1, between NC and p6 in the Gag polyprotein, whereas FIV has an additional enzyme, DU, between RH and IN compared to HIV-1. TF, transframe region.
FIV PR is responsible for processing FIV Gag and Gag-Pol polyprotein into 10 individual functional proteins, including MA, CA, p1, nucleocapsid (NC), and p2 from the Gag polyprotein and PR, reverse transcriptase (RT), RNase H (RH), dUTPase (DU), and integrase (IN) from the Pol polyprotein (13) (Fig. 1B). Distinctions relative to HIV-1 Gag-Pol include an additional small spacer protein, p1, between NC and p6 of the HIV-1 Gag polyprotein and the lack of DU in HIV-1. FIV PR, similar to HIV-1 PR, regulates its own activity through autoproteolysis at four cleavage sites in PR (22). The processing sequence of Gag and Gag-Pol precursor proteins, highly regulated by PR, is critical for producing mature viruses for infection and replication (40, 48). Thus, PR is an attractive target for development of antiretroviral drugs and PR inhibitors have drastically slowed the progression of disease and reduced the mortality rate in HIV-1-infected patients (2, 19, 21, 47). However, the high error rate of reverse transcription and high levels of viral replication, combined with a lack of adherence to medication regimens, have led to the development of drug-resistant strains. Additional strategies are therefore needed for drug design to minimize resistance and cross-resistance problems.
We have compared the properties of FIV PR and HIV-1 PR to better understand the molecular basis of retroviral PR substrate and inhibitor specificity. In our previous studies, we replaced up to 24 amino acid residues in and around the active site of FIV PR with positionally equivalent residues of HIV-1 PR and examined the specificity of mutant PRs in vitro (1, 25, 30, 31). Substrate specificity of mutant FIV PRs was analyzed by examining their cleavage efficiency on peptides representing HIV-1 and FIV cleavage sites. The inhibitor specificities of mutant PRs were assessed by measuring the 50% inhibitory concentrations (IC50s) and Ki values of potent HIV-1 PR inhibitors. Two substitutions, I3530D (HIV-1-equivalent numbering is shown as a superscript) of the active core and I5748G of the flap, were intolerant to change in FIV PR. The lost activity of I5748G was rescued by additional secondary substitutions, G6253F and K6354I, at the top of the flap, whereas the recovery of lost activity of the I3530D mutation was evident but limited with additional mutations. In addition, we have learned that some mutants, such as I3732V in the active core, N5546M, M5647I, and V5950I in the flap region, and L9780T, I9881P, Q9982V, P10083N, and L10184I in the “90's loop” region, retained comparable activity against FIV substrates while substantially changing substrate and inhibitor specificity compared with that of HIV-1 PR (30, 31).
Our previous studies of FIV PR specificity have measured in vitro cleavage of fluorogenic substrates by using purified PR, which has been informative but does not address either cleavage or inhibitor specificity of PR in the context of the natural polyprotein substrate. In this study, we employed an ex vivo tissue culture system coupled with expression of wild-type and mutant PRs in the context of the Gag-Pol polyprotein to address several pertinent questions. First of all, we wished to determine the relative substrate specificity of wild-type and mutant FIV PRs in the context of Gag-Pol polyprotein by measuring the processing sequence and pattern of Gag polyprotein cleavage by each PR. The processing sequence and efficiency of HIV-1 Gag polyprotein have been studied in great detail and are critical for viral replication (39, 40). Although the processing sites of the FIV Gag-Pol polyprotein have been identified, little is known about the processing sequence or efficiency of cleavage at each site. Secondly, we wanted to determine how inhibitors affect the processing of the Gag-Pol polyprotein. Finally, we wanted to assess whether such a cell-based system could serve as an accurate screening venue for PR inhibitors. The analysis of the substrate and inhibitor specificity of mutant FIV PR was facilitated by the use of the pCFIVΔorf2Δenv expression plasmid (17) transfected into the 293T human kidney cell line in order to express Gag and Gag-Pol polyprotein and produce viral particles. The processing capability of the mutant PRs was then assessed by Western blot analysis of MA and CA processing, as well as release of RT activity, in the presence and absence of a panel of potent HIV-1 PR inhibitors. The data show that the processing efficiency of the Gag polyprotein and release of RT activity were significantly affected by substitutions of specific HIV-1 amino acid residues in FIV PR. Specifically, the NC-p2 cleavage junction was processed efficiently by the wild type but poorly by the “HIVinized” FIV mutants. In addition, several potent HIV-1 PR inhibitors markedly inhibited the processing of the Gag polyprotein in mutants ex vivo, with specificities similar to that observed against HIV-1 PR and to a greater extent than observed with wild-type FIV PR. This study will further aid the development of the next generation of broad-spectrum PR inhibitors against native and drug-resistant retroviral PRs.
MATERIALS AND METHODS
Construction of pCR2.1-S/N-based plasmids encoding mutant PRs.
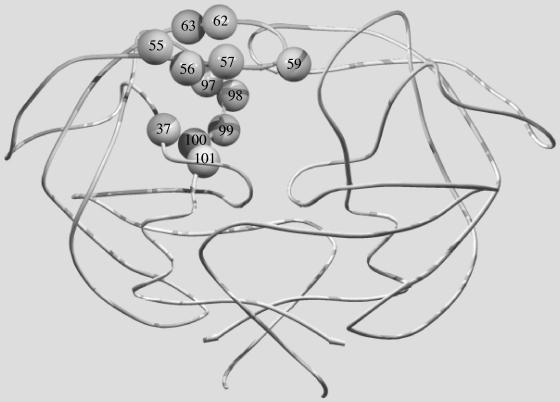
To facilitate the mutagenesis of FIV PR in the Gag-Pol region, the unique SacI- and NsiI-digested fragment encoding wild-type FIV PR of clone 34TF10 (49) was transferred into vector pCR2.1 (Invitrogen Corporation, Carlsbad, CA) to generate the pCR2.1S/N plasmid. PCR-mediated megaprimer mutagenesis (43) of the FIV PR gene was performed on the EcoRI/NsiI fragment of the pCR2.1-S/N plasmid. The sequence of the 5′ primer encoding the EcoRI site was 5′-GGAAATAGAAAGAATTCGGGAAACTGGAAGG-3′. The sequence of the 3′ primer encoding the NsiI site was 5′-CAAGAGGAATGGTGAAATATGCATCCCCTATATC-3′. Sequences of mutagenic primers for FIV PR have been described in detail previously (30, 31). Substitutions for equivalent residues of HIV-1 PR (FIV numbering with HIV-1 numbering in superscript) included I3530D, I5748G, 4s (I3732V/N5546M/M5647I/V5948I), 7s (I3732V/N5546M/M5647I/I5748G/V5950I/G6253F/K6354I), 8s (N5546M/M5647I/V5948I/L9780T/I9881P/Q9982V/P10083N/L10184I), 9s (I3732V/N5546M/M5647I/V5948I/L9780T/I9881P/Q9982V/P10083N/L10184I), and 12s (I3732V/N5546M/M5647I/I5748G/V5950I/G6253F/K6354I/L9780T/I9881P/Q9982V/P10083N/L10184I). Alignment of the amino acid sequences of the FIV and HIV-1 PRs is shown in Fig. 1A. The structural locations of the above substitutions in FIV PR are shown in Fig. 2. Mutation I3530D is located in the active core, and I5748G is in the flap region. I3530D and I5748G were shown to have no detectable activity in a fluorogenic assay in vitro (30, 31). I3732V is in the active core, whereas N5546M, M5647I, V5950I, G6253F, and K6354I are in the flap region. L9780T, I9881P, Q9982V, P10083N, and L10184I are in the 90's loop region. Two single-substitution mutants, D3025N and N5546D, were also constructed. D3025N (equivalent to D25N of HIV-1 PR) is an inactive PR mutant in which the catalytic Asp of the active core is replaced with Asn (23). N5546D is an active flap mutant with biochemical characteristics similar to those of wild-type PR (9). In order to assess the relative substrate specificity between FIV mutant PR and HIV-1 PR against the FIV Gag polyprotein, two chimeric constructs containing FIV Gag/HIV-1 PR were also prepared. The sequences of the 5′ mutagenic primers used to construct FIV-HIV-1 chimeras are as follows: FHIV-5A, 5′-GGAGGAGAAACTATTGGATTTGTAAGCTTTAGCTTCCCTCAGATCACTCTTTGG-3′; FHIV-5B, 5′-ACCTCCAATGGAGGAGAAACTATTGGATTTGTAAATCCTCAGATCACTCTTTGGCAGCGACCC-3′. The sequence of the 3′mutagenic primer is 5′-GGCTGCACTTTAAATTTTTAAATGCATAGAGGG-3′. The primer contains a stop codon at the end of HIV-1 PR; therefore, the rest of pol was not transcribed in both chimeric constructs. All substitutions and mutations were verified by DNA sequencing.
FIG. 2.
Structural locations of substitutions in FIV PR. Substituted residues are shown on one chain of dimeric PR. Substitutions include I3732V in the active core; N5546M, M5647I, I5748G, V5950I, G6253F, and K6354I in the flap; and L9780T, I9881P, Q9982V, P10083N, and L10184I in the 90's loop.
Construction of mutant pCFIV expression plasmids containing mutant PRs.
The strategy for examining the processing efficiency and specificity of FIV Gag or Gag-Pol polyprotein by mutant PRs was to use the pCFIVΔorf2Δenv packaging expression vector (17). The high expression of FIV Gag and Gag-Pol polyprotein is driven by the strong cytomegalovirus promoter. The system is useful for investigating the biological relevance of PR function, since PR is expressed as part of the whole Gag-Pol polyprotein in cell culture. To generate mutant pCFIV clones containing mutant PRs, the wild-type unique PflFI/NsiI restriction fragment was replaced with a fragment from pCR2.1-S/N clones that contained mutant PR as described above. The pCFIV mutants F4s, F7s, F8s, F9s, and F12s, containing 4, 7, 8, 9, and 12 substitutions, respectively, were constructed, and mutations were verified by DNA sequencing. In addition to F4s, F7s, F8s, F9s, and F12s, four single-point mutants, D3025N, I3530D, N5546D, and I5748G (23, 30, 31), were also constructed.
Cell culture and transfection.
Human 293T cells (10), used for transfection, were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1 mM l-glutamine, and 1 mM sodium pyruvate. The pCFIV expression plasmids were transfected into 293T cells for expression of Gag and Gag-Pol polyprotein. Fresh 293T cells were plated at 150,000/35-mm dish or 300,000/60-mm dish overnight. The HIV-1 PR inhibitors saquinavir, ritonavir (RTV), and TL-3 (25, 26) and new compounds (3, 4, 29) APV-1, AB-2, AB-6, and AB-8 were used to study inhibitor specificity by examining the effects of the inhibitors on the processing of Gag polyprotein in the context of expression in 293T cells. Fresh medium with or without inhibitor was exchanged 3 h before transfection 1 day following initial seeding. FuGene6 transfection reagent (9 μl; Roche Diagnostics) and 3 μg of plasmid pCFIV were used for transfection according to the manufacturer's instructions. Fresh PR inhibitor was replenished 24 h after transfection at the same concentration. The expression and processing of Gag polyprotein of cell-associated, viral particle-associated, and RT activities of cell-free supernatant were analyzed 48 h after transfection.
Western blot and RT assays.
The transfected cells and cell-free supernatants were harvested after 48 h. The cells were lysed in 50 mM Tris-HCl (pH 8.0)-150 mM NaCl-1% NP-40. The viral particles were pelleted from the cell-free supernatant by ultracentrifugation at 50,000 rpm for 30 min in a Beckman TLA-100.3 rotor. The viral particles were lysed in 8 M urea-20 mM Tris-HCl (pH 8.0)-5 mM EDTA. The cleared lysate of cells and viral particles was subjected to 10 to 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and separated protein bands were transferred to nitrocellulose membrane. Rabbit antisera against MA (∼15 kDa), CA (∼24 kDa), and PR (∼13 kDa) were used at a 1:2,000 dilution to examine the processing of Gag and Gag-Pol polyprotein in a Western blot assay. Horseradish peroxidase-conjugated secondary antibodies at a 1:10,000 dilution and SuperSignal West Dura enhanced chemiluminescent substrate (Pierce Biotechnology, Inc.) were used to reveal the protein bands of unprocessed and processed Gag-Pol polyprotein. ImageQuant software (GE Healthcare) was used for quantitation of the relative intensities of protein bands. Viral particle production 48 h after transfection was also monitored by release of RT activity into the culture supernatant as previously described (8). Briefly, cell-free supernatants (50 μl) were mixed with 10 μl of lysis buffer (0.75 M KCl, 20 mM dithiothreitol, 0.5% Triton X-100) and incubated for 10 min at room temperature. A 40-μl volume of a mixture containing 125 mM Tris-HCl (pH 8.1), 12.5 mM MgCl2, 1.25 μg of poly(rA)-poly(dT)12-18 (Amersham Biosciences/GE Healthcare, Piscataway, NJ), and 1.25 μCi of [3H]dTTP (Dupont NEN, Boston, MA) was added to the lysate, and the mixture was incubated for 2 h at 37°C. The mixture was spotted onto DE81 paper and then washed in 0.1 M sodium pyrophosphate, followed by three washes in 0.3 M ammonium formate and one wash in 95% ethanol. The filter papers were then dried, and the radioactivity was determined with a scintillation counter.
PR assay, IC50s, and Ki values.
The activity of FIV PR was assayed in 50 mM sodium citrate-100 mM phosphate buffer (pH 5.25)-200 mM NaCl-1 mM dithiothreitol with the fluorogenic substrate ALT(2-aminobenzoic acid)KVQ/(p-NO2)FVQSKG (14). The data were obtained continuously at an excitation wavelength of 325 nm and an emission wavelength of 410 nm with an F-2000 fluorescence spectrophotometer (Hitachi Inc.). The IC50, the inhibitor concentration that inhibits PR activity by 50%, was determined with the Grafit 4 program (Erithacus Software Ltd.). The Ki (inhibition constant) value was derived from the IC50 by using the following equation for a competitive inhibitor: Ki = IC50/(1 + [S]/Km).
RESULTS
Expression and processing of Gag polyprotein in pCFIV constructs.
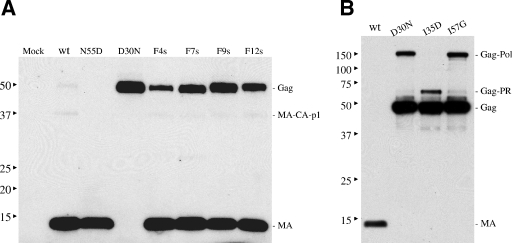
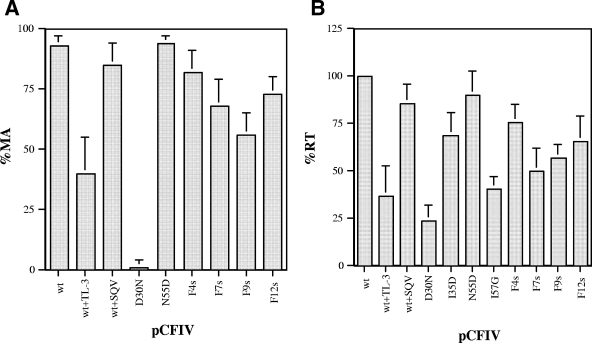
The combined substitutions in the mutant constructs were selected on the basis of our previous studies, wherein all substitutions except I3530D and I5748G generated functionally active PRs in vitro (30, 31). The lost activity of I5748G could be restored successfully when combined with additional G6253F and K6354I substitutions, whereas the lost activity of I3530D could be recovered only slightly with other secondary substitutions (31). F4s, F9s, and F12s were shown to have comparable activities against a fluorogenic substrate in previous studies (31). Expression levels of Gag polyprotein in the cells and in the viral particle lysate from cell-free supernatant (viral particle associated) were examined in a Western blot assay with anti-MA antibodies. The expression levels of Gag polyprotein in the cells were comparable between the wild type and the different mutant pCFIV constructs, and most of the Gag polyprotein remained unprocessed in the cell cytoplasm, as typically observed with cells infected with wild-type FIV (data not shown). Also, the total amounts of Gag polyprotein and processed MA in the viral particles were similar between the wild type and the mutants, indicating that the assembly, budding, and release of Gag-Pol-bearing viral particles were not affected by PR mutations and processing efficiency of mutant PRs (data not shown). However, the processing efficiency of Gag polyprotein by mutant PRs differed from that observed with wild-type FIV PR, as shown by Western blot analysis (Fig. 3A). Processing of Gag polyprotein (∼50 kDa) expressing either wild-type PR or N5546D PR was very efficient, with only residual unprocessed Gag polyprotein remaining in viral particles produced from these two constructs. However, efficiency of cleavage was affected by various other substitutions in PR. Little or no processing of Gag was evident in virus particles expressing mutant D3025N, as expected, since D30 is the catalytic aspartate in FIV PR. Mutants I3530D and I5748G, which have no detectable activity in a PR assay in vitro against a fluorogenic synthetic substrate (30, 31), also failed to efficiently process the Gag polyprotein in this ex vivo assay (Fig. 3B). However, some level of cleavage of Gag-Pol did occur with the I3530D mutant, as the full-length band disappeared and another band of ∼60 kDa appeared. On the basis of the size of the observed product and antibody reactivities, cleavage likely occurred at the junction of PR and RT to produce a Gag-PR polyprotein. MA processing efficiency, quantified by densitometry (Fig. 4A), indicated that mutant F4s had the most efficient processing among the multiple-substitution mutants. However, other multiple-substitution mutants, including F7s, F9s, and F12s, had reasonable processing efficiencies, with 50 to 75% MA processing under the same conditions that yielded nearly 100% cleavage by wild-type PR. Processing of Gag polyprotein by mutant F8s was the least efficient, with less than 50% cleavage under the same conditions (data not shown). This mutant was thus not included in further studies. A faint intermediate band with a molecular mass of >37 kDa was also observed in wild-type PR constructs (Fig. 3A). This band was also detected by anti-CA antibodies, consistent with this species corresponding to an MA-CA-p1 intermediate polyprotein product from cleavage at the p1-NC junction. Similar results for relative processing efficiency were noted when using anti-CA to identify Gag cleavage intermediates (not shown). However, as indicated below, altered processing patterns of Gag polyprotein cleavage were noted with “HIVinized” mutants.
FIG. 3.
(A) Expression and processing of FIV Gag polyprotein in viral particles produced by 293T cells transfected with pCFIV constructs are shown in a Western blot assay probed with anti-MA antibodies. (B) Expression and processing of Gag polyprotein in mutants D3025N, I3530D, and I5748G are shown in a Western blot assay probed with anti-MA antibodies. wt, wild type. The values on the left are molecular sizes in kilodaltons.
FIG. 4.
(A) Processing efficiency of Gag polyprotein in wild-type and mutant pCFIV constructs. Average percent MA (percent processed MA/total unprocessed Gag) was quantified by densitometry. (B) Average percent RT activity (fraction of wild-type activity) of virus-associated supernatant in FIV mutants. wt, wild type. SQV, saquinovir.
RT activity of pCFIV mutants.
Antibodies to Pol polyprotein, including RT, DU, and IN, were also employed to examine the processing of the Pol region in these experiments. However, the efficiency of detection of the much lower (approximately 20-fold lower) expression of the Pol portion of the Gag-Pol polyprotein was poor by Western assay (data not shown). Thus, in order to assess the processing of the Pol region, we measured the level of RT activity in viral particles released by each construct (Fig. 4B). Different substitution mutants yielded different levels of RT activity, ranging from near wild-type levels with the N5546D single mutant to basal RT values of around 25% to 30% for the inactive D3025N mutant. Interestingly, since there was no significant processing of Gag and Gag-Pol polyprotein in this mutant, the results indicated that unprocessed FIV Pol polyprotein had measurable RT activity, similar to the observations in other retroviral systems (7, 36, 37, 46, 50). Moreover, both the I3530D and I5748G mutants had significant RT activity (∼70% and ∼40% of wild-type levels, respectively), despite having little or no detectable activity in previous PR assays in vitro against synthetic substrates (31). These findings suggest that these two mutants, particularly I3530D, exhibit some level of PR activity in the context of the polyprotein and are able to process Gag-Pol polyprotein above the basal level. There is little full-length Gag-Pol polyprotein (∼170-kDa band) remaining in virus particles expressing the I3530D PR mutant compared to those in D3025N or I5748G (Fig. 3B), consistent with the notion that I3530D has residual activity sufficient to process the Pol portion of the Gag-Pol polyprotein and generate RT activity above baseline levels. Further studies on the processing of the Pol polyprotein are needed to clarify the observation. Of the multiple-substitution mutants, F4s had the highest RT activity, followed by F12s. These findings correlate with the processing efficiency of the Gag polyprotein detailed above.
IC50s and Ki values of HIV-1 PR inhibitors against FIV wild-type and mutant PRs in vitro.
To examine inhibitor specificity for the FIV and HIV mutant PRs, several HIV-1 PR inhibitors, including TL-3 and several new compounds, APV-1, AB-2, AB-5, AB-6, and AB-8, were tested for the ability to inhibit polyprotein processing in virus particles expressing the wild-type, F4s, F9s, and F12s PRs. The new compounds have core structures similar to that of amprenavir and have been shown to be potent inhibitors for HIV-1 PR by in vitro assay against a fluorogenic substrate (3, 4, 29) (Fig. 5). The inhibition potencies (IC50s and Ki values) of these compounds for HIV-1, FIV, and FIV mutant PRs were measured in vitro (Table 1). All inhibitors were highly efficacious against HIV-1 PR, with IC50s below 10 nM. However, most of these inhibitors were not effective against FIV wild-type PR, with the exceptions of TL-3 (IC50 = 90 nM) and compound AB-2 (IC50 = 1,100 nM). Of importance, the efficacy of the other inhibitors was substantially better against multiple-substitution mutants F4s, F9s, and F12s, with markedly improved inhibition constants (Table 1). TL-3 also showed improved inhibition constants against mutants. These results are consistent with the previous findings that these mutant PRs have inhibitor specificities very similar, but not identical, to that of wild-type HIV-1 PR in vitro (31).
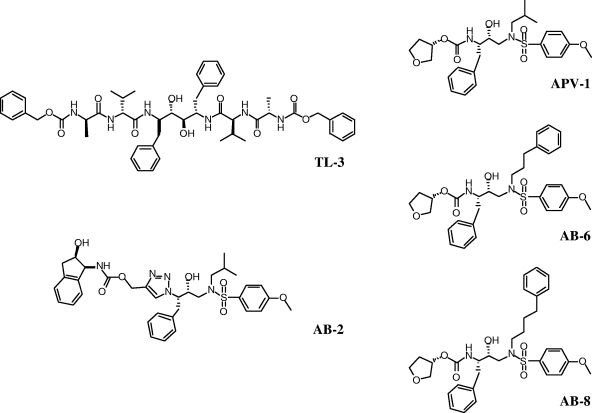
FIG. 5.
Chemical structures of HIV-1 PR inhibitors used to assess the inhibitor specificities of mutants in vitro and ex vivo.
TABLE 1.
IC50s and Ki values of potent HIV-1 PR inhibitors for FIV and HIV-1 PRs
| Inhibitor | IC50 (Ki value)a (nM) for:
|
||||
|---|---|---|---|---|---|
| HIV-1 | FIV | F4sb | F9sc | F12sd | |
| TL-3 | 6 (2) | 90 (41) | 108 (21) | 115 (15) | 71 (10) |
| APV-1 | 8 | >1,000 | 133 (26) | 109 (14) | NAe |
| AB-2 | 5 | 1,100 | 97 (19) | 84 (11) | 111 (16) |
| AB-6 | 6 | >1,000 | 130 (25) | 130 (17) | 81 (11) |
| AB-8 | 6 | >8,000 | 92 (18) | 80 (10) | 93 (13) |
| RTV | 4 | >45,000 | 129 (25) | 231 (30) | 192 (27) |
Ki were derived from IC50s by using the equation Ki = IC50/(1 + [S]/Km).
F4s, I3732V/N5546M/M5647I/V5950I.
F9s, I3732V/N5546M/M5647I/V5950I/L9780T/I9881P/Q9982V/P10083N/L10184I.
F12s, I3732V/N5546M/M5647I/I5748G/V5950I/G6253F/K6354I/L9780T/I9881P/Q9982V/P10083N/L10184I.
NA, not assayed.
Effect of HIV-1 PR inhibitors on the processing of Gag polyprotein in pCFIV mutants.
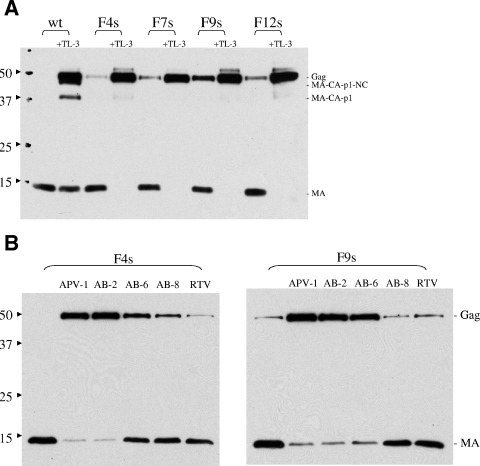
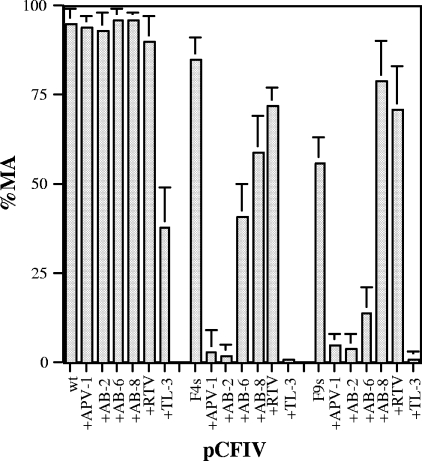
To assess the effect of inhibitors on the Gag polyprotein processing in pCFIV in cell culture, TL-3 and selected compounds mentioned above were tested. Inhibition of processing in F4s, F9s, or F12s constructs by TL-3 at 2.5 μM was much greater than against the wild-type pCFIV construct (Fig. 6A). The results were correlated with the inhibitor potencies (IC50s and Ki values) assayed in vitro (Table 1). TL-3, tested at concentrations ranging from 1 to 10 μM, inhibited polyprotein processing by wild-type PR, but relatively inefficiently, with maximal inhibition of MA processing to approximately 60% at the highest concentrations. However, TL-3 was very potent against F4s, F9s, and F12s, even at lower concentrations, and completely inhibited the processing of the Gag polyprotein in the mutants at 1 μM (data not shown). Inhibition by the FDA-approved HIV-1 PR inhibitors saquinavir and nelfinavir was also evaluated at concentrations ranging from 2.5 μM to 20 μM and showed little inhibition of wild-type FIV or the FIV-HIV mutants, even at the highest concentrations (data not shown), consistent with previous in vitro findings (31). As with the above drugs, the new compounds APV-1, AB-2, AB-6, and AB-8, although exhibiting low nanomolar efficacy against wild-type HIV-1 PR in vitro (3, 4, 29), were poor inhibitors of FIV wild-type PR in vitro (Table 1). However, both APV-1 and AB-2 strongly inhibited ex vivo processing of the Gag polyprotein of the F4s and F9s mutants. AB-6 also strongly inhibited the Gag polyprotein processing of F9s but not that of F4s (Fig. 6B). Under the same conditions, RTV showed poor inhibition of processing. The quantitation data (percent processed MA) for wild-type, F4s, and F9s processing in the presence of inhibitors are summarized in Fig. 7 and correlate well with the low IC50s and Ki values of TL-3, APV-1, and AB-2 against the respective PRs in vitro (Table 1). However, AB-8, although exhibiting a similar low inhibition constant against these mutants, was not a good inhibitor ex vivo, consistent with presumed lower bioavailability in cell culture.
FIG. 6.
(A) Processing of Gag polyprotein by wild-type and mutant PRs in the presence and absence of 2.5 μM TL-3. Processed bands are shown by a Western blot assay probed with anti-MA antibodies. (B) Processing of Gag polyprotein by F4s and F9s PRs in the presence of additional inhibitors including APV-1, AB-2, AB-6, AB-8, and RTV at 2.5 μM. Western blot assays were probed with anti-MA antibodies. wt, wild type. The values on the left are molecular sizes in kilodaltons.
FIG. 7.
Inhibition of Gag polyprotein processing in the presence and absence of different inhibitors (2.5 μM). Inhibition (% MA) of processing in the wild type (wt), F4s, and F9s was quantified by densitometry.
Altered processing pattern of FIV Gag polyprotein in mutants.
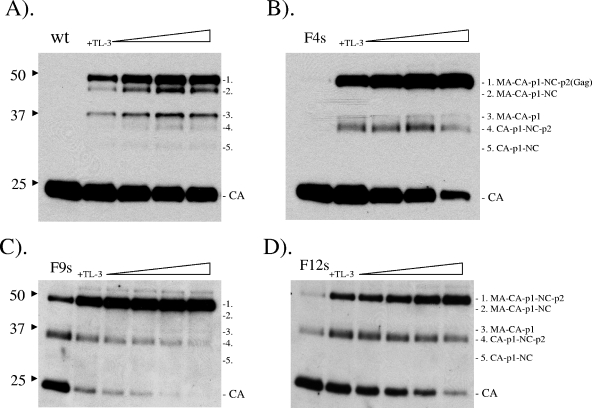
The cleavage sites and mature products of FIV Gag and Gag-Pol polyprotein have been mapped and identified in previous studies (13, 44). However, there has been no study of the processing sequence or pattern of FIV Gag or Gag-Pol polyprotein. To examine the processing pattern of the FIV Gag polyprotein by the wild type and the F4s, F9s, and F12s mutants in the pCFIV system, various concentrations of TL-3 were used to modulate the activity of PRs expressed in the context of the Gag-Pol polyprotein in order to reveal the intermediate cleavage products generated during Gag processing (Fig. 8). Although TL-3 is the best-known inhibitor for FIV PR (Ki = 41 nM), it was not potent enough to completely inhibit the processing of the wild-type FIV Gag polyprotein in cell culture, even at the solubility limits of the compound in tissue culture medium (approximately 10 to 15 μM). Under these partial-cleavage conditions, intermediate cleavage products can be detected by anti-CA antibodies in particles expressing wild-type FIV PR, including an approximately 48-kDa band (Fig. 8A, second band from top) corresponding to MA-CA-p1-NC and an approximately 39-kDa band (Fig. 8A, third band from the top) corresponding to MA-CA-p1. Of particular interest, the 48-kDa band (band 2) was missing in all of the mutants (Fig. 8B, C, and D), indicating that there is little or no cleavage at the NC-p2 junction in the FIV-HIV mutants under conditions in which wild-type FIV PR elicits that cleavage. In addition to these two bands, there were two additional bands detected by anti-CA antibodies in the construct expressing wild-type PR. One is an approximately 35-kDa species (Fig. 8A, fourth band from top) which represents CA-p1-NC-p2 (not detected by anti-MA antibodies), and the other is a lower band of approximately 32 kDa corresponding to CA-p1-NC. Consistent with the above observations, the latter band was not detected in the mutants, since cleavage did not occur at the NC-p2 junction in the “HIVinized” mutants. The equivalent HIV-1 site (p2-NC site) is the most rapidly cleaved site among all of cleavage sites of the HIV-1 Gag polyprotein (39, 40). Likewise in this study, p1-NC cleavage occurred efficiently with wild-type FIV PR. In contrast, the data indicate that cleavage at the MA-CA site occurs more efficiently in all of the mutants, with band 4 more intense than band 3 (Fig. 8B, C, and D). Given that the order of cleavage is critical for generation of infectious particles, this may explain our failure to generate infectious viruses encoding the F4s, F9s, and F12s FIV-HIV mutant PRs (data not shown).
FIG. 8.
(A) Pattern of FIV Gag polyprotein processing by wild-type FIV PR in pCFIV constructs, shown by Western blot assays probed with anti-CA antibodies. Various concentrations of TL-3 (1 to 5 μM) were used (note the increase in inhibitor sensitivity) to temper processing and reveal Gag polyprotein processing intermediates. Altered patterns of Gag polyprotein processing in F4s (B), F9s (C), and F12s (D) are shown in Western blot assays probed with anti-CA antibodies. Various concentrations of TL-3 (25 to 100 nM) were used (note the increase in inhibitor sensitivity). Four Gag polyprotein processing intermediates are indicated. The values on the left are molecular sizes in kilodaltons.
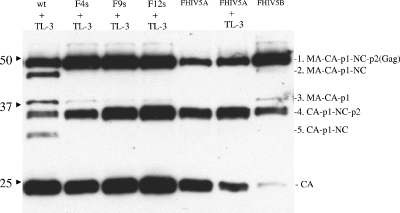
Processing pattern of FIV Gag polyprotein by HIV-1 PR in pCFIV chimeric constructs.
To determine if wild-type HIV-1 PR could process the FIV Gag polyprotein in a manner similar to that noted with the mutants, two chimeric pCFIV constructs that contained HIV-1 PR in place of FIV PR were generated. One construct (FHIV-5B) contained only HIV-1 PR substituted for FIV PR, whereas the other (FHIV-5A) also contained four amino acids (P4 to P1 [SFSF]) within the HIV-1 TF-PR cleavage junction (Fig. 1B) in order to facilitate the efficient release of HIV-1 PR. Both constructs contained a stop codon at the end of HIV-1 PR so that the rest of the Pol polyprotein was not expressed. The processing efficiency of FIV Gag polyprotein by FIV and HIV-1 PRs was examined in a Western blot assay probed with anti-CA antibodies (Fig. 9). TL-3 at various concentrations was used to reveal the processing intermediates in wild-type and mutant FIVs. The findings showed that the FIV Gag polyprotein was processed, but only by the FHIV-5A construct encoding the entire TF-PR cleavage junction of HIV-1, consistent with inefficient cleavage of the chimeric junction in FHIV-5B. These data were consistent with the interpretation that blocking of the release of HIV-1 PR decreased the processing efficiency of the Gag polyprotein (54). These results also showed that the pattern of processing was altered in a manner similar to that observed with the FIV-HIV F4s, F9s, and F12s mutant constructs; i.e., there was little or no processing at the FIV NC-p2 site, since band 2 (MA-CA-p1-NC) was missing. In addition, the results indicated that there was better processing at the MA-CA site by HIV-1 PR, as the fourth band (CA-p1-NC-p2) is more intense than the third band. Thus, the overall findings indicate that the discrete mutations in FIV PR to introduce HIV-1 character yielded both cleavage phenotype and inhibitor specificities similar to those of HIV-1 PR.
FIG. 9.
Comparison of processing of FIV Gag polyprotein generated by wild-type (wt) FIV PR, “HIVinized” FIV mutants, and HIV-1 PR expressed in the context of FIV Gag-Pol. Processed bands are revealed by Western blot assays with anti-CA antibodies. F4s, F9s, and F12s represent FIV mutants containing 4, 9, and 12 HIV-equivalent substitutions, respectively. FHIV-5A represents the FIV Gag polyprotein expressing wild-type HIV-1 PR, with the entire N-terminal PR cleavage junction composed of HIV residues; FHIV-5B also expresses wild-type HIV-1 PR, but the N-terminal half (P4 to P1) of the N-terminal PR cleavage junction is FIV derived. The efficiency of cleavage of FHIV-5A is greater than of FHIV-5B, consistent with more-efficient release of PR activity from the former construct. The values on the left are molecular sizes in kilodaltons.
DISCUSSION
We have previously identified residues in the active site of FIV PR that dictate substrate and inhibitor specificities relative to HIV-1 PR by using extensive mutational analysis, enzyme assays, and crystallographic approaches (23, 25, 26, 28, 30, 31). These in vitro findings confirmed and extended studies using similar approaches to demonstrate substrate and inhibitor specificity of Rous sarcoma virus PR substituted with HIV-1 PR-specific residues (6, 15, 27, 42). By inserting certain of the HIV-specific residues into FIV PR, we were able to partially impart HIV cleavage and inhibitor specificities to FIV PR.
In the present study, we extended these experiments to assess the ability of “HIVinized” FIV PRs to facilitate cleavage in the context of the natural Gag-Pol polyprotein substrate by measuring the degree and specificity of polyprotein processing in the context of pseudovirions encoding Gag-Pol. Further, we demonstrated the utility of this ex vivo culture system as a tool for determining the relative efficacy of specific compounds to inhibit processing by the wild-type, chimeric, and drug-resistant PRs. Such analyses complement and extend the in vitro measurements of the cleavage and inhibition properties of purified PRs done with synthetic peptides and fluorogenic substrates. The cell culture system not only allows direct assessments of PR function in the context of the natural substrate but also yields a measurement of the ability of PR inhibitors to enter living cells, as well as an indication of the relative toxicity of such compounds. The efficiency of expression of the Gag polyprotein in this packaging system is comparable between the wild type and the mutants, and the assembly and release of viral particles are not affected by PR mutations, since the proper cleavage and maturation of Gag and Gag-Pol polyprotein are not required for virus production and particle release. Thus, analysis of the Gag-Pol polyprotein in released virions as a function of PR structure and inhibitor treatments can be carried out to assess processing efficiency and ordering of cleavage events. The substrate specificity can be examined by analyzing the processing of Gag and Gag-Pol polyprotein. The inhibitor specificity can be examined by analyzing the temporal degree of processing in the presence or absence of any inhibitor. Of importance, these expression constructs provide a useful system to analyze the substrate and inhibitor specificity of wild-type and mutant FIV or HIV PRs ex vivo without the biohazards associated with the use of infectious virus.
These findings show that FIV PRs into which specific HIV-1 substitutions have been introduced take on cleavage and inhibitor characteristics with increased similarity to those of HIV-1 PR. Further, these studies show that although polyprotein cleavage is facilitated by these mutant PRs, the order of site cleavage is not the same as for wild-type FIV PR. Given the importance of the ordering of site cleavage for generation of infectious, competent virus (18, 20, 39, 52), these findings likely explain the noninfectious nature of chimeric FIVs encoding either wild-type HIV-1 PR or the specific FIV-HIV mutant PRs.
Interestingly, the RT activity of the active-site mutant D30N (D25N in HIV-1), as well as two other inactive point mutants, I3530D and I5748G, remained around 70% to 40% of that of the wild type (Fig. 4B), with little or no processing of Gag polyprotein (no detectable MA or CA in the Western blot assay in Fig. 3B). The latter findings agree with our previous findings that these PRs are active-site mutants and were shown to have no detectable activity in an in vitro PR assay (23, 30) but further indicate that nonprocessed FIV RT in the Gag-Pol polyprotein retains measurable activity, similar to reported findings obtained with HIV-1 RT in the context of the polyprotein (36, 37). Temporal processing of wild-type and mutant FIV Gag polyproteins was controlled by addition of the broad-based PR inhibitor TL-3, which allowed detection of the intermediate cleavage products as a function of PR structure. All of the “HIVinized” mutants tested showed marked changes in substrate specificity against the Gag polyprotein. Processing occurred in each mutant, but the processing pattern and order of Gag polyprotein cleavage were altered relative to those of that generated by wild-type FIV PR. Specifically, there was little or no cleavage at the FIV NC-p2 junction (VNQM-QQAV) with any of the F4s, F9s, and F12s mutants, since the 48-kDa band corresponding to the intermediate cleavage product MA-CA-p1-NC was observed when cleavage was facilitated by wild-type PR but not in the blot patterns generated by any of the mutants (Fig. 8 and 9). The observation of a change in specificity at this cleavage site but not other sites was somewhat surprising, since the mutant PRs, particularly F4s and to a lesser degree F9s and F12s, were shown to have activity comparable to that of wild-type PR against a fluorogenic substrate representing the p1-NC junction in vitro (31). Consistent with the above results, an ∼32-kDa intermediate (band 5) corresponding to CA-p1-NC was also missing from the analysis of the cleavage patterns of the three mutants (Fig. 9). These results indicate that either one or some combination of the four mutations I3732V, N5546M, M5647I, and V5950I might be responsible for the change in specificity at this processing junction. Experiments are under way to construct single-substitution pCFIV mutants and examine the processing patterns of Gag polyprotein by these mutants in order to determine the residue(s) responsible for dictating the observed changes in processing specificity.
The small C-terminal peptide p2 of the FIV Gag polyprotein represents the functionally equivalent p6 domain of the HIV-1 Gag polyprotein and contains conserved PSAP and LLDL motifs that are critical for particle formation (35). The assembly and production of viral particles in all of the FIV mutant constructs were not affected by PR mutations in spite of the resulting poor processing efficiency at the NC-p2 cleavage site, which indicates that release of p2 from the polyprotein is not a prerequisite for its function in particle release. However, the proper ordered and efficient processing of the NC-p2 cleavage junction to generate mature NC and p2 may be crucial for the infectivity of FIV. The amino acid sequences from P4 to P4′of the equivalent cleavage junctions, the NC-p1 junction (RQAN-FLGK) and the p1-p6 junction (PGNF-LQSR), of HIV-1 are very different from that of the NC-p2 junction (VNQM-QQAV) of FIV. Important to the present observations, the P2 side chain of the FIV NC-p2 junction is Gln, which is not found in any HIV-1 cleavage junction or any peptide cleaved by HIV-1 PR (1, 2) and is thus consistent with the failure of the “HIVinized” PRs to cleave at this site. The processing efficiency of equivalent cleavage sites between NC and p6 of the HIV-1 Gag polyprotein has been shown to be important to the fitness of HIV-1 and poor or delayed processing of the NC-p1 or p1-p6 junction was associated with reduced viral infection and replication capacity (32, 41, 45, 51). Overall, these findings suggest that cleavage junctions NC-p1-p6 in HIV-1 and NC-p2 in FIV may offer a valuable structural framework for inhibitor design.
In addition to the poor processing at the FIV NC-p2 site by FIV mutants, processing of the MA-CA cleavage site in the Gag polyprotein of the mutants appeared to be enhanced relative to cleavage at the two junctions between CA and NC (CA-p1 and p1-NC). This is demonstrated by an increase in the intensity of the 35-kDa band (band 4) representing CA-p1-NC-p2 (only detected by anti-CA antibodies and not by anti-MA antibodies) in the mutant constructs compared to that generated by wild-type PR (Fig. 9). FIV wild-type PR processed the FIV p1-NC site efficiently, similar to the reported efficient cleavage of the equivalent HIV-1 p2-NC site by HIV-1 PR (38-40). This observation indicated that the efficiency and sequence of processing at the p1-NC cleavage site were also altered by one or some combination of the mutations in FIV PR. The results indicated that the F4s, F9s, and F12s PRs cleaved the FIV MA-CA site (PQAY-PIQT) in the context of the polyprotein with greater efficiency than the p1-NC site (TKVQ-VVQS). These results were surprising, since F4s and the other mutants processed FIV p1-NC efficiently when it was presented as a fluorogenic substrate in vitro (31). However, the efficient cleavage of FIV MA-CA (PQAY-PIQT) by the mutants is consistent with an increased HIV-1 character, since this cleavage site is strikingly similar to the HIV-1 MA-CA (SQNY-PIVQ) site.
TL-3, a broad-based inhibitor, is potent against wild-type FIV and all of the mutants tested in this study by both in vitro and cell-based ex vivo assays. In fact, TL-3 was more efficient in blocking the activity of “HIVinized” PRs, consistent with a 10-fold better activity of the inhibitor against HIV-1 PR in vitro (25, 26). Our results showed that TL-3 was also a more efficient inhibitor ex vivo than the other compounds tested (Fig. 7), consistent with findings in vitro against wild-type FIV (Table 1). However, on the basis of the in vitro findings, we expected better performance against some of the mutants, particularly with AB-2 and APV-1, both of which would have been predicted to perform as well as TL-3 against some mutants. No overt cell toxicity was noted with any of the compounds. These results may indicate either poorer uptake by the target cells of the latter inhibitors or an increased rate of breakdown ex vivo.
Acknowledgments
We thank Sohela de Rozieres and Magnus Sundstrom for review and discussions of the data and Jackie Wold for administrative assistance. We also thank Joel Gottesfeld for aid in densitometric quantitation of polyprotein cleavage products, Philip Dawson and Michael Churchill for synthesis of FIV fluorogenic substrate, and F. S. Liang for synthesis of the APV-1 inhibitor.
This research was supported by grant GM048870 from the National Institute of General Medical Sciences and grant R01 AI040882 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
REFERENCES
- 1.Beck, Z. Q., Y. C. Lin, and J. H. Elder. 2001. Molecular basis for the relative substrate specificity of human immunodeficiency virus type 1 and feline immunodeficiency virus proteases. J. Virol. 75:9458-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, Z. Q., G. M. Morris, and J. H. Elder. 2002. Defining HIV-1 protease substrate selectivity. Curr. Drug Targets Infect. Disord. 2:37-50. [DOI] [PubMed] [Google Scholar]
- 3.Brik, A., J. Alexandratos, Y. C. Lin, J. H. Elder, A. J. Olson, A. Wlodawer, D. S. Goodsell, and C. H. Wong. 2005. 1,2,3-Triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors. ChemBioChem 6:1167-1169. [DOI] [PubMed] [Google Scholar]
- 4.Brik, A., J. Muldoon, Y. C. Lin, J. H. Elder, D. S. Goodsell, A. J. Olson, V. V. Fokin, K. B. Sharpless, and C. H. Wong. 2003. Rapid diversity-oriented synthesis in microtiter plates for in situ screening of HIV protease inhibitors. ChemBioChem 4:1246-1248. [DOI] [PubMed] [Google Scholar]
- 5.Buhler, B., Y. C. Lin, G. Morris, A. J. Olson, C. H. Wong, D. D. Richman, J. H. Elder, and B. E. Torbett. 2001. Viral evolution in response to the broad-based retroviral protease inhibitor TL-3. J. Virol. 75:9502-9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron, C. E., T. W. Ridky, S. Shulenin, J. Leis, I. T. Weber, T. Copeland, A. Wlodawer, H. Burstein, D. Bizub-Bender, and A. M. Skalka. 1994. Mutational analysis of the substrate binding pockets of the Rous sarcoma virus and human immunodeficiency virus-1 proteases. J. Biol. Chem. 269:11170-11177. [PubMed] [Google Scholar]
- 7.Crawford, S., and S. P. Goff. 1985. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J. Virol. 53:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Parseval, A., and J. H. Elder. 2001. Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell-surface heparans, and an unidentified non-CXCR4 receptor. J. Virol. 75:4528-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Rozieres, S., C. H. Swan, D. A. Sheeter, K. J. Clingerman, Y. C. Lin, S. Huitron-Resendiz, S. Henriksen, B. E. Torbett, and J. H. Elder. 2004. Assessment of FIV-C infection of cats as a function of treatment with the protease inhibitor, TL-3. Retrovirology 1:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elder, J. H., G. A. Dean, E. A. Hoover, J. A. Hoxie, M. H. Malim, L. Mathes, J. C. Neil, T. W. North, E. Sparger, M. B. Tompkins, W. A. Tompkins, J. Yamamoto, N. Yuhki, N. C. Pedersen, and R. H. Miller. 1998. Lessons from the cat: feline immunodeficiency virus as a tool to develop intervention strategies against human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 14:797-801. [DOI] [PubMed] [Google Scholar]
- 12.Elder, J. H., and T. R. Phillips. 1995. Feline immunodeficiency virus as a model for development of molecular approaches to intervention strategies against lentivirus infections. Adv. Virus Res. 45:225-247. [DOI] [PubMed] [Google Scholar]
- 13.Elder, J. H., M. Schnolzer, C. S. Hasselkus-Light, M. Henson, D. A. Lerner, T. R. Phillips, P. C. Wagaman, and S. B. Kent. 1993. Identification of proteolytic processing sites within the Gag and Pol polyproteins of feline immunodeficiency virus. J. Virol. 67:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald, M. C., G. S. Laco, J. H. Elder, and S. B. Kent. 1997. A continuous fluorometric assay for the feline immunodeficiency virus protease. Anal. Biochem. 254:226-230. [DOI] [PubMed] [Google Scholar]
- 15.Grinde, B., C. E. Cameron, J. Leis, I. T. Weber, A. Wlodawer, H. Burstein, D. Bizub, and A. M. Skalka. 1992. Mutations that alter the activity of the Rous sarcoma virus protease. J. Biol. Chem. 267:9481-9490. [PubMed] [Google Scholar]
- 16.Henriksen, S. J., O. Prospero-Garcia, T. R. Phillips, H. S. Fox, F. E. Bloom, and J. H. Elder. 1995. Feline immunodeficiency virus as a model for study of lentivirus infection of the central nervous system. Curr. Top. Microbiol. Immunol. 202:167-186. [DOI] [PubMed] [Google Scholar]
- 17.Johnston, J. C., M. Gasmi, L. E. Lim, J. H. Elder, J. K. Yee, D. J. Jolly, K. P. Campbell, B. L. Davidson, and S. L. Sauter. 1999. Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors. J. Virol. 73:4991-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan, A. H., J. A. Zack, M. Knigge, D. A. Paul, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 67:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozal, M. 2004. Cross-resistance patterns among HIV protease inhibitors. AIDS Patient Care STDs 18:199-208. [DOI] [PubMed] [Google Scholar]
- 20.Kräusslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutilek, V. D., D. A. Sheeter, J. H. Elder, and B. E. Torbett. 2003. Is resistance futile? Curr. Drug Targets Infect Disord. 3:295-309. [DOI] [PubMed] [Google Scholar]
- 22.Laco, G. S., M. C. Fitzgerald, G. M. Morris, A. J. Olson, S. B. Kent, and J. H. Elder. 1997. Molecular analysis of the feline immunodeficiency virus protease: generation of a novel form of the protease by autoproteolysis and construction of cleavage-resistant proteases. J. Virol. 71:5505-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laco, G. S., C. Schalk-Hihi, J. Lubkowski, G. Morris, A. Zdanov, A. Olson, J. H. Elder, A. Wlodawer, and A. Gustchina. 1997. Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor. Biochemistry 36:10696-10708. [DOI] [PubMed] [Google Scholar]
- 24.Le, V. D., C. C. Mak, Y. C. Lin, J. H. Elder, and C. H. Wong. 2001. Structure-activity studies of FIV and HIV protease inhibitors containing allophenylnorstatine. Bioorg. Med. Chem. 9:1185-1195. [DOI] [PubMed] [Google Scholar]
- 25.Lee, T., G. S. Laco, B. E. Torbett, H. S. Fox, D. L. Lerner, J. H. Elder, and C. H. Wong. 1998. Analysis of the S3 and S3′ subsite specificities of feline immunodeficiency virus (FIV) protease: development of a broad-based protease inhibitor efficacious against FIV, SIV, and HIV in vitro and ex vivo. Proc. Natl. Acad. Sci. USA 95:939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, T., V. D. Le, D. Lim, Y. C. Lin, G. Morris, A. L. Wong, A. J. Olson, J. H. Elder, and C. H. Wong. 1999. Development of a new type of protease inhibitors, efficacious against FIV and HIV variants. J. Am. Chem. Soc. 121:1145-1155. [Google Scholar]
- 27.Leis, J. P., and C. E. Cameron. 1994. Engineering proteases with altered specificity. Curr. Opin. Biotechnol. 5:403-408. [DOI] [PubMed] [Google Scholar]
- 28.Li, M., G. M. Morris, T. Lee, G. S. Laco, C. H. Wong, A. J. Olson, J. H. Elder, A. Wlodawer, and A. Gustchina. 2000. Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease. Proteins 38:29-40. [DOI] [PubMed] [Google Scholar]
- 29.Liang, F. S., A. Brik, Y. C. Lin, J. H. Elder, and C. H. Wong. 2005. Epoxide opening in water and screening in situ for rapid discovery of enzyme inhibitors in microtiter plates. Bioorg. Med. Chem. 14:1058-1062. [DOI] [PubMed] [Google Scholar]
- 30.Lin, Y. C., Z. Beck, T. Lee, V. D. Le, G. M. Morris, A. J. Olson, C. H. Wong, and J. H. Elder. 2000. Alteration of substrate and inhibitor specificity of feline immunodeficiency virus protease. J. Virol. 74:4710-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, Y. C., Z. Beck, G. M. Morris, A. J. Olson, and J. H. Elder. 2003. Structural basis for distinctions between substrate and inhibitor specificities for feline immunodeficiency virus and human immunodeficiency virus proteases. J. Virol. 77:6589-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maguire, M. F., R. Guinea, P. Griffin, S. Macmanus, R. C. Elston, J. Wolfram, N. Richards, M. H. Hanlon, D. J. Porter, T. Wrin, N. Parkin, M. Tisdale, E. Furfine, C. Petropoulos, B. W. Snowden, and J. P. Kleim. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 76:7398-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mak, C. C., A. Brik, D. L. Lerner, J. H. Elder, G. M. Morris, A. J. Olson, and C. H. Wong. 2003. Design and synthesis of broad-based mono- and bi-cyclic inhibitors of FIV and HIV proteases. Bioorg. Med. Chem. 11:2025-2040. [DOI] [PubMed] [Google Scholar]
- 34.Mak, C. C., V. D. Le, Y. C. Lin, J. H. Elder, and C. H. Wong. 2001. Design, synthesis, and biological evaluation of HIV/FIV protease inhibitors incorporating a conformationally constrained macrocycle with a small P3′ residue. Bioorg. Med. Chem. Lett. 11:219-222. [DOI] [PubMed] [Google Scholar]
- 35.Manrique, M. L., M. L. Rauddi, S. A. Gonzalez, and J. L. Affranchino. 2004. Functional domains in the feline immunodeficiency virus nucleocapsid protein. Virology 327:83-92. [DOI] [PubMed] [Google Scholar]
- 36.Peng, C., N. T. Chang, and T. W. Chang. 1991. Identification and characterization of human immunodeficiency virus type 1 gag-pol fusion protein in transfected mammalian cells. J. Virol. 65:2751-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, C., B. K. Ho, T. W. Chang, and N. T. Chang. 1989. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J. Virol. 63:2550-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettit, S. C., J. C. Clemente, J. A. Jeung, B. M. Dunn, and A. H. Kaplan. 2005. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J. Virol. 79:10601-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettit, S. C., N. Sheng, R. Tritch, S. Erickson-Viitanen, and R. Swanstrom. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 436:15-25. [DOI] [PubMed] [Google Scholar]
- 41.Resch, W., N. Parkin, T. Watkins, J. Harris, and R. Swanstrom. 2005. Evolution of human immunodeficiency virus type 1 protease genotypes and phenotypes in vivo under selective pressure of the protease inhibitor ritonavir. J. Virol. 79:10638-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ridky, T. W., D. Bizub-Bender, C. E. Cameron, I. T. Weber, A. Wlodawer, T. Copeland, A. M. Skalka, and J. Leis. 1996. Programming the Rous sarcoma virus protease to cleave new substrate sequences. J. Biol. Chem. 271:10538-10544. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 44.Schnolzer, M., H. R. Rackwitz, A. Gustchina, G. S. Laco, A. Wlodawer, J. H. Elder, and S. B. Kent. 1996. Comparative properties of feline immunodeficiency virus (FIV) and human immunodeficiency virus type 1 (HIV-1) proteinases prepared by total chemical synthesis. Virology 224:268-275. [DOI] [PubMed] [Google Scholar]
- 45.Sheng, N., S. C. Pettit, R. J. Tritch, D. H. Ozturk, M. M. Rayner, R. Swanstrom, and S. Erickson-Viitanen. 1997. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 71:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart, L., and V. M. Vogt. 1991. trans-acting viral protease is necessary and sufficient for activation of avian leukosis virus reverse transcriptase. J. Virol. 65:6218-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanstrom, R., and J. Erona. 2000. Human immunodeficiency virus type-1 protease inhibitors: therapeutic successes and failures, suppression and resistance. Pharmacol. Ther. 86:145-170. [DOI] [PubMed] [Google Scholar]
- 48.Swanstrom, R., and J. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin and S. Hughes (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 49.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanese, N., M. Roth, H. Epstein, and S. P. Goff. 1989. An insertion mutation in the pol gene of Moloney murine leukemia virus results in temperature-sensitive pol maturation and viral replication. Virology 170:378-384. [DOI] [PubMed] [Google Scholar]
- 51.Watkins, T., W. Resch, D. Irlbeck, and R. Swanstrom. 2003. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 47:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Kräusslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wlodawer, A., A. Gustchina, L. Reshetnikova, J. Lubkowski, A. Zdanov, K. Y. Hui, E. L. Angleton, W. G. Farmerie, M. M. Goodenow, D. Bhatt, et al. 1995. Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus. Nat. Struct. Biol. 2:480-488. [DOI] [PubMed] [Google Scholar]
- 54.Zybarth, G., H. G. Kräusslich, K. Partin, and C. Carter. 1994. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. J. Virol. 68:240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]