Abstract
Psittacid herpesvirus 1 (PsHV-1) is the causative agent of Pacheco's disease, an acute, highly contagious, and potentially lethal respiratory herpesvirus infection in psittacine birds, while infectious laryngotracheitis virus (ILTV) is a highly contagious and economically significant avian herpesvirus which is responsible for an acute respiratory disease limited to galliform birds. The complete genome sequence of PsHV-1 has been determined and compared to the ILTV sequence, assembled from published data. The PsHV-1 and ILTV genomes exhibit similar structural characteristics and are 163,025 bp and 148,665 bp in length, respectively. The PsHV-1 genome contains 73 predicted open reading frames (ORFs), while the ILTV genome contains 77 predicted ORFs. Both genomes contain an inversion in the unique long region similar to that observed in pseudorabies virus. PsHV-1 is closely related to ILTV, and it is proposed that it be assigned to the Iltovirus genus. These two avian herpesviruses represent a phylogenetically unique clade of alphaherpesviruses that are distinct from the Marek's disease-like viruses (Mardivirus). The determination of the complete genomic nucleotide sequences of PsHV-1 and ILTV provides a tool for further comparative and functional analysis of this unique class of avian alphaherpesviruses.
Avian herpesviruses comprise a wide range of pathogens and infect a wide variety of hosts. They are also remarkable for their relatively narrow host range. Herpesviruses have been isolated from a large number of avian species. Until now, only infectious laryngotracheitis virus (ILTV) (Iltovirus), the Marek's disease-like viruses (Mardivirus), and Psittacid herpesvirus 1 (unassigned genus) have been classified as avian members of the Alphaherpesvirinae subfamily by the International Committee on Taxonomy of Viruses. Marek's disease virus (MDV) (Gallid herpesvirus 2) and herpesvirus of turkeys (HVT) (Meleagrid herpesvirus 1), although initially classified as gammaherpesviruses based on their lymphotrophic biological properties, were reclassified as alphaherpesviruses based on their genetic structure (9). The genomic sequences of all three serotypes of MDV have been reported (3, 33, 61).
Avian Betaherpesvirinae and Gammaherpesvirinae have not been taxonomically classified to date. However, taxonomically unassigned avian herpesviruses have been identified from a wide variety of avian species, including the bald eagle (Acciptrid herpesvirus 1), duck (Anatid herpesvirus 1), black stork (Ciconiid herpesvirus 1), pigeon (Columbid herpesvirus 1), falcon (Falconid herpesvirus 1), crane (Gruid herpesvirus 1), bobwhite quail (Perdicid herpesvirus 1), cormorant (Phalacrocoracid herpesvirus 1), owl (Strigid herpesvirus 1), penguin (Sphenicid herpesvirus 1), cardinal, finch, canary, vulture, and pheasant (1, 15, 20, 24, 26, 31, 32).
ILTV (Gallid herpesvirus 1) is a highly contagious and economically significant avian herpesvirus (6). Natural infections of ILTV are limited to galliform birds and cause an acute respiratory disease, which can be responsible for significant mortality and loss of productivity in the poultry industry (12, 40). In its epizootic form, the disease caused by ILTV is characterized by signs of respiratory distress accompanied by gasping and expectoration of bloody exudates (27). Infectious laryngotracheitis has been controlled through the use of live-virus vaccines, but vaccination is usually limited to areas where the disease is endemic, due to the relatively high pathogenicity of the vaccines. DNA sequencing efforts in numerous laboratories have generated the complete sequence of the ILTV genome (17, 18, 25, 28, 29, 30, 34, 49, 66, 67, 68), but to date these data have not been assembled and analyzed on a genome-wide scale.
Psittacid herpesvirus 1 (PsHV-1) is the causative agent of Pacheco's disease, an acute, highly contagious, and potentially lethal respiratory herpesvirus infection in psittacine birds (48). Amazon parrots, macaws, and cockatoos are highly susceptible to PsHV-1, which is a disease of great concern to the companion bird markets and exotic bird breeders (11, 22, 51). PsHV-1 targets hepatocytes and lymphocytes and slowly forms syncytial plaques in tissue culture. It has been previously classified both as a betaherpesvirus and as a gammaherpesvirus (31). Although there is limited PsHV-1 sequence available, phylogenetic studies based on the sequences of the PsHV-1 UL16 and UL30 genes have shown that the viruses that cause Pacheco's disease can be discriminated into four major genotypes and that PsHV-1 is most closely related to the alphaherpesvirus ILTV (or gallid herpesvirus 1) (59, 60, 62).
The genetic basis and underlying molecular mechanisms responsible for determining the pathogenicity, virulence, and host range specificity of ILTV and PsHV-1 are poorly understood. The only other avian herpesvirus genomes to be completely sequenced, the three serologically distinct members of the Mardivirus genus (MDV serotype 1 [MDV-1], MDV-2, and HVT), are biologically unique. MDV-1 can induce T-cell lymphomas and may not be representative of other broadly disseminated classes of avian herpesviruses. Comparative genome analysis can offer insight into the biological properties and phylogenetic differences of these viruses. Here we present the first complete genome sequence and analysis of both PsHV-1 and ILTV and demonstrate that they represent a phylogenetically unique class of alphaherpesviruses.
MATERIALS AND METHODS
Virus.
The reference PsHV-1 (isolate 97-0001) was kindly supplied by David Phalen (Texas A&M University). PsHV-1 was isolated from the liver of an Amazon parrot (Amazona oratrix) of unknown age, with hepatic and splenic lesions characteristic of Pacheco's disease.
Virus purification and DNA isolation.
PsHV-1 was propagated on confluent monolayers of primary chicken embryo fibroblast cells (CEF). Fibroblasts were isolated from 11-day-old chicken embryos by the warm trypsinization method and plated in Dulbecco's modified essential medium (DMEM/F-12) (Invitrogen Corp., Gaithersburg, MD) supplemented with penicillin (50 μg/ml), streptomycin (50 μg/ml), and 10% fetal bovine serum. Twenty hours postplating, fibroblast monolayers were infected with a 1:100 dilution of PsHV-1. Six days (144 h) postinfection, CEF monolayers exhibited 90 to 95% cytopathic effect (CPE). PsHV-1 virions were concentrated from the supernatant of infected CEF monolayers by the addition of 7% PEG 8000 and pelleted through a 30% sucrose cushion. Virions were lysed in 10% sodium dodecyl sulfate and deproteinized by incubation at 65°C in the presence of 10 mg/ml proteinase K followed by five extractions with phenol-chloroform-isoamyl alcohol (25:24:1). Virus DNA was precipitated in 100% EtOH for 48 h at −80°C and pelleted by centrifugation at 14,000 rpm for 20 min at 26°C. The virus DNA pellet was resuspended in 500 μl of TE (10 mM Tris HCl-1 mM EDTA, pH 8.0).
Virus DNA was purified by fractionation on a 0 to 40% sucrose gradient as previously described (55) and visualized by agarose gel electrophoresis. Fractions containing virus DNA were pooled, and virus DNA was precipitated and resuspended in TE. Purified virus DNA was examined for purity by restriction endonuclease digestion (data not shown) and comparison to previously published results (26).
PsHV-1 library construction, sequencing, and gap closing.
Purified PsHV-1 genomic DNA (10 μg) was sheared by nebulization at 30 lb/in2 for 50 s. Fragment ends were repaired with the End-It DNA end repair kit (Epicenter, Madison, WI) according to the manufacturer's instructions. Agarose gel-purified DNA fragments of 1.5 to 2.5 kb in length were ligated at the EcoRV site of pBluescript SK+ (Stratagene, La Jolla, CA) and transformed into SURE Escherichia coli electrocompetent cells (Stratagene, La Jolla, CA) by electroporation. Glycerol archive stocks of the transformed cells were prepared with a Q-Bot (Genetix Ltd., Dorset, United Kingdom) robot to pick bacterial colonies. Cultures were grown overnight in deep-well plates containing LB plus ampicillin (100 μg/ml). Plasmid DNA was purified by alkaline lysis according to the manufacturer's instructions (Perfectprep-96 Robotic Workstation; Brinkmann-Eppendorf 5 Prime, Boulder, CO). Virus DNA inserts were sequenced from both ends with M13 forward and reverse universal primers, using BigDye dideoxy chain terminator sequencing chemistry, version 3.0 (PE Biosystems, Foster City, CA), and Applied Biosystems PRISM 3700 automated DNA sequencers (PE Biosystems, Foster City, CA).
Gaps in the PsHV-1 sequence were closed by PCR amplification using viral genomic DNA as the template. Each 20-μl reaction mix contained 50 ng of genomic PsHV-1 DNA, 25 pmol of each primer (designed from flanking PsHV-1 sequence), 0.1 mM concentrations of each of the four deoxynucleoside triphosphates, 2.5 mM magnesium chloride, 0.75 U of Taq, and 1× buffer A (Promega, Madison, WI). Amplifications were performed on a PE Biosystems 9700 thermal cycler using an initial denaturation step of 94°C for 5 min, followed by 30 cycles of 94°C for 15 s, 55°C for 30 s, and 60°C for 4 min. PCR products were purified and sequenced as described above.
DNA sequence analysis.
PsHV-1 DNA sequences were assembled with Phrap (16) using Phred quality files and default settings to produce a consensus sequence. Consensus sequences were manually edited within Consed (23). The ILTV DNA sequence was assembled from 14 published ILTV sequences (Table 1). The strategies used to identify PsHV-1 and ILTV genes were based on those used in the sequence analyses of other herpesviruses (3, 7, 33, 36, 52). The primary criteria for identifying a coding sequence was the presence of an open reading frame (ORF) of >100 nt. The identification of ORFs was performed with ORF Finder (21) and Vector NTI (InforMax, Inc.). Searches of predicted PsHV-1 and ILTV proteins for homology to known proteins were performed by applying BLAST (Basic Local Alignment Search Tool) (4) and PSI-BLAST (5).
TABLE 1.
Previously published sequences used for the assembly of the ILTV genome
| GenBank accession no.a | Reference | Size (bp)b |
|---|---|---|
| U80762 | Johnson et al. (29) | 13,700 |
| AJ249803 | Johnson et al. (30) | 4,465 |
| Y14300 | Ziemann et al. (67, 68) | 15,276 |
| D00565* | Griffin and Boursnell (25) | 5,395 |
| X56093* | Poulsen et al. (49) | 3,065 |
| AY033142 | Kehu et al. (unpublished data) | 1,360 |
| AF168792 | Johnson (unpublished data) | 31,332 |
| AY033143 | Kehu et al. (unpublished data) | 378 |
| U06635 | Kingsley and Keeler (35) | 1,807 |
| Y14301 | Ziemann et al. (67, 68) | 1,854 |
| AJ131832 | Fuchs and Mettenleiter (18) | 24,140 |
| X97256 | Fuchs and Mettenleiter (17) | 10,265 |
| L32139* | Johnson et al. (29) | 8,364 |
| U28832 | Wild et al. (66) | 18,900 |
| L32139 | Johnson et al. (29) | 8,364 |
| Total length | 148,665 |
*, reverse complement used in assembly.
Lengths of sequence used in assembly.
Phylogenetic trees were constructed with ClustalW (58) and Treeview (47) within the Vector NTI software suite and calculated with the neighbor-joining algorithm of Saitou and Nei (54). Initially, alignments were processed by the Seqboot program, which generated the requested number (100) of resampled alignments for bootstrapping. The resampled alignments were then processed by the Protdist program, which created distance matrices for each alignment. Each of these matrices was then used by the Neighbor program to create a neighbor-joining tree.
Nucleotide sequence accession numbers.
The complete PsHV-1 and ILTV genome sequences have been deposited in the GenBank database under accession no. AY372243 and NC_006623, respectively.
RESULTS
Genome organization.
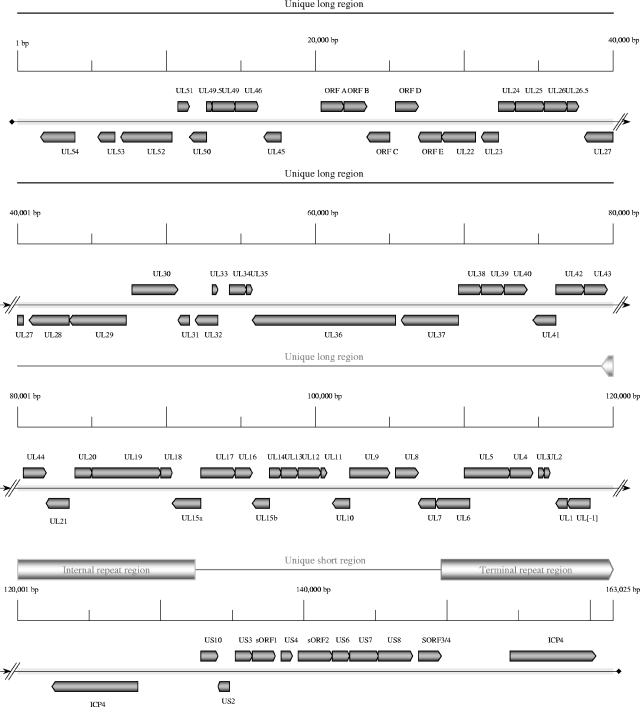
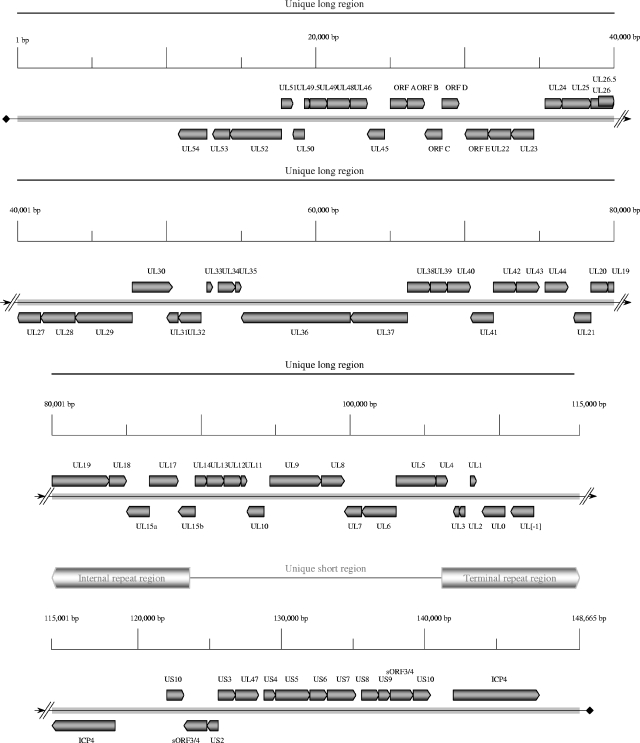
Both PsHV-1 and ILTV exhibit the structural characteristics of class D herpesvirus genomes, such as pseudorabies virus (PRV) and varicella zoster virus (VZV), which contain two domains of unique sequences (53). In this class of herpesvirus genomes, only the shorter unique sequence (US) is flanked by inverted repeats (IR and TR). The US region can then invert relative to the larger unique domain (UL), and the genome can exist in two equimolar isomeric forms. The PsHV-1 genome is 163,025 bp in length, with a base composition of 60.95% G+C. The UL sequence is 119,146 bp in length, and the US sequence is 16,405 bp (Table 2). The inverted repeat elements are each 13,737 bp in length. The ILTV genome, as assembled from 14 different published accessions (Table 1), is 148,665 bp in length, with a G+C content of 48.16%. Two sequences used in the assembly (accession no. L32139 and U28832) do not overlap. PCR amplification of ILTV DNA across the junction of these sequences indicated that there was no additional ILTV sequence present. The ILTV genome is also organized into UL (113,039 bp) and US (13,232 bp) regions, with IR and TR repeats (11,202 bp) flanking the US region (Table 2). Unlike the MDV family of avian alphaherpesviruses, which exhibit genome structures similar to those of herpes simplex virus type 1 (HSV-1) and HSV-2, the UL regions of the ILTV and PsHV-1 genomes are not bracketed by inverted repeats and the US regions are considerably larger (Table 2).
TABLE 2.
Comparison of avian alphaherpesvirus genome organizations
| Virus | Length (bp)a
|
Accession no. | ||||||
|---|---|---|---|---|---|---|---|---|
| TRL | UL | IRL | IRS | US | TRS | Total | ||
| PsHV-1 | 119,146 | 13,737 | 16,405 | 13,737 | 163,025 | AY372243 | ||
| ILTV | 113,039 | 11,202 | 13,232 | 11,202 | 148,665 | NC_006623 | ||
| MDV-1 | 12,584 | 113,563 | 12,584 | 12,120 | 10,847 | 12,120 | 173,818 | AF147806 |
| MDV-2 | 11,951 | 109,932 | 11,951 | 9,164 | 12,109 | 9,164 | 164,271 | AB049735 |
| HVT | 7,072 | 110,694 | 7,072 | 13,610 | 8,615 | 13,610 | 160,673 | AF282130 |
TRL, terminal repeat, long region; IRL, internal repeat, long region; IRS, internal repeat, short region; TRS, terminal repeat, short region.
Gene identification.
The PsHV-1 genome contains 73 predicted ORFs (Table 3; Fig. 1), while the ILTV genome contains 77 predicted ORFs (Fig. 2 and Table 3). Sixty-one PsHV-1 genes are located within the UL region, 10 genes are within the US region, and a single copy of 1 gene (ICP4) is located within each of the repeats. The UL region of ILTV contains 62 genes, and the US region contains 9 genes. One major structural difference between the PsHV-1 and ILTV genomes occurs in the size and genetic makeup of the inverted repeat regions. The ILTV inverted repeat regions are 2,535 bp (18.5%) shorter than the PsHV-1 inverted repeats and contain two copies of three genes: ICP4, US10, and a homolog of the MDV sORF4/3 gene. Like that of HVT, neither the ILTV nor the PsHV-1 genome contains a region comparable to that in MDV-1 that has been implicated in oncogenesis.
TABLE 3.
ORFs identified within the PsHV-1 and ILTV genomes
| PsHV1
|
ILTV
|
Length (aa)a
|
% Identity (to ILTV) | Putative functionb | |||
|---|---|---|---|---|---|---|---|
| ORF | Nucleotide position | ORF | Nucleotide position | PsHV1 | ILTV | ||
| UL54 | 4295-2376 | UL54 | 12082-10787 | 639 | 420 | 38 | Posttranslational regulator of gene expression |
| UL53 | 6461-5385 | UL53 | 13916-12855 | 358 | 337 | 31 | Glycoprotein K; exocytosis |
| UL52 | 10335-6454 | UL52 | 17176-13847 | 1,293 | 1,110 | 42 | DNA helicase-primase |
| UL51 | 10319-11101 | UL51 | 17189-17875 | 260 | 229 | 55 | Unknown |
| UL50 | 12476-11232 | UL50 | 19182-17935 | 414 | 416 | 46 | Deoxyuridine triphosphatase |
| UL49.5 | 12667-13014 | UL49.5 | 19336-19686 | 140 | 266 | 50 | Putative viral membrane protein |
| UL49 | 13184-14038 | UL49 | 19749-20546 | 283 | 117 | 50 | Viral tegument protein |
| Not present | UL48 | 20695-21882 | 396 | Viral tegument protein (α-TIF) | |||
| UL46 | 14500-16323 | UL46 | 21888-23558 | 606 | 539 | 44 | Tegument phosphoprotein; α-TIF modulation |
| UL45 | 17426-16521 | UL45 | 24559-23663 | 301 | 281 | 26 | Tegument/envelope protein |
| ORF A | 21070-22260 | ORF A | 25275-26405 | 396 | 376 | 27 | Hypothetical protein |
| ORF B | 22726-23829 | ORF B | 26448-27467 | 367 | 340 | 30 | Hypothetical protein |
| ORF C | 25174-24056 | ORF C | 28529-27528 | 372 | 334 | 33 | Hypothetical protein |
| ORF D | 25554-26654 | ORF D | 28639-29760 | 366 | 374 | 38 | Hypothetical protein |
| ORF E | 28424-27054 | ORF E | 31067-29838 | 456 | 410 | 28 | Hypothetical protein |
| UL22 | 31254-28834 | UL22 | 33539-31128 | 828 | 779 | 34 | Glycoprotein H; fusion complexes with gL |
| UL23 | 32569-31550 | UL23 | 34667-33576 | 339 | 363 | 40 | Thymidine kinase |
| UL24 | 32533-33489 | UL24 | 34556-35416 | 318 | 287 | 41 | Unknown |
| UL25 | 33628-35511 | UL25 | 35392-37107 | 627 | 572 | 47 | DNA packaging protein |
| UL26 | 35709-37382 | UL26 | 37288-39045 | 557 | 586 | 33 | Capsid protein p40 |
| UL26.5 | 37199-37891 | UL26.5 | 38428-39045 | 230 | 206 | 13 | Virion scaffold protein |
| UL27 | 40995-38260 | UL27 | 41747-39099 | 911 | 873 | 59 | Glycoprotein B |
| UL28 | 43743-41164 | UL28 | 44013-41722 | 859 | 537 | 47 | ICP18.5; cleavage/packaging |
| UL29 | 47426-43860 | UL29 | 47094-44098 | 1,188 | 999 | 59 | Major single-strand DNA binding protein |
| UL30 | 47858-51103 | UL30 | 47271-50291 | 1,081 | 1,007 | 54 | DNA polymerase |
| UL31 | 52171-51134 | UL31 | 51483-50467 | 345 | 339 | 68 | Nuclear phosphoprotein |
| UL32 | 54020-52164 | UL32 | 53233-51479 | 618 | 582 | 48 | Envelope glycoprotein |
| UL33 | 54019-54393 | UL33 | 53190-53579 | 138 | 119 | 58 | DNA packaging |
| UL34 | 54616-55440 | UL34 | 53611-54486 | 300 | 290 | 62 | Membrane-associated phosphoprotein |
| UL35 | 55567-55992 | UL35 | 54515-54886 | 141 | 124 | 50 | Capsid protein |
| UL36 | 65672-56085 | UL36 | 62584-54917 | 3,209 | 2,556 | 36 | Major tegument protein |
| UL37 | 69349-66437 | UL37 | 65881-63212 | 970 | 890 | 34 | Tegument protein |
| UL38 | 69618-71078 | UL38 | 65973-67280 | 486 | 412 | 41 | DNA binding; capsid protein |
| UL39 | 71339-73774 | UL39 | 67618-69972 | 818 | 785 | 50 | Large-subunit ribonucleotide reductase |
| UL40 | 73851-74792 | UL40 | 69827-70915 | 313 | 310 | 69 | Small-subunit ribonucleotide reductase |
| UL41 | 76206-74884 | UL41 | 72176-70983 | 440 | 398 | 73 | Virion host shutoff |
| UL42 | 76680-78182 | UL42 | 72398-73693 | 519 | 432 | 34 | Processivity factor for DNA polymerase |
| UL43 | 78100-79626 | UL43 | 73756-74970 | 508 | 300 | 11 | Unknown |
| UL44 | 80418-81806 | UL44 | 75683-76924 | 462 | 414 | 29 | Glycoprotein C |
| UL21 | 83761-82052 | UL21 | 78611-77016 | 569 | 532 | 28 | Nucleocapsid protein |
| UL20 | 84089-84835 | UL20 | 78782-79477 | 248 | 232 | 30 | Membrane protein |
| UL19 | 85082-89323 | UL19 | 79664-83872 | 1,413 | 1,403 | 65 | Major capsid protein |
| UL18 | 89684-90649 | UL18 | 84059-85015 | 321 | 319 | 64 | Capsid protein |
| UL15a | 92293-90773 | UL15a | 86212-85103 | 513 | 764 | 66 | Terminase; DNA packaging |
| UL17 | 92187-94634 | UL17 | 86355-88505 | 815 | 341 | 38 | Tegument protein |
| UL16 | 94538-95605 | Not present | 355 | Capsid assembly | |||
| UL15b | 96940-95588 | UL15b | 89761-88598 | 450 | 764 | 43 | Terminase; DNA packaging |
| UL14 | 96939-97529 | UL14 | 89595-90353 | 196 | 196 | 45 | Unknown |
| UL13 | 97403-98785 | UL13 | 90212-91606 | 486 | 465 | 45 | Serine/threonine protein kinase |
| UL12 | 99023-100585 | UL12 | 91750-93366 | 566 | 526 | 58 | Alkaline deoxynuclease |
| UL11 | 100585-100728 | UL11 | 93259-93502 | 48 | 80 | 42 | Myristoylated tegument protein |
| UL10 | 102288-101047 | UL10 | 94758-93580 | 413 | 393 | 46 | Glycoprotein M |
| UL9 | 102386-105028 | UL9 | 94653-97382 | 880 | 892 | 53 | Ori binding protein |
| UL8 | 105636-107582 | UL8 | 97378-99762 | 648 | 795 | 34 | Helicase-primase component |
| UL7 | 108856-107690 | UL7 | 100889-99816 | 388 | 358 | 41 | Unknown |
| UL6 | 110909-108561 | UL6 | 102807-100669 | 782 | 713 | 47 | Minor capsid protein |
| UL5 | 110993-113560 | UL5 | 102795-105314 | 855 | 840 | 58 | Helicase-primase component |
| UL4 | 113781-114539 | UL4 | 105403-105936 | 252 | 178 | 81 | Unknown |
| UL3 | 114921-115523 | UL3 | 106948-106349 | 200 | 196 | 66 | Unknown |
| UL2 | 115671-116759 | UL2 | 107950-107060 | 265 | 297 | 50 | Uracil DNA glycosylase |
| UL1 | 117253-116705 | UL1 | 107920-108279 | 182 | 131 | 32 | Glycoprotein L |
| Not present | UL0 | 111514-110171 | 447 | Unknown | |||
| UL[−1] | 118632-117331 | UL[−1] | 111670-112026 | 463 | 501 | 23 | Unknown |
| ICP4a | 127595-121494 | ICP4 | 118888-114500 | 2,033 | 1,463 | 35 | Gene regulation |
| US10 | 133103-133948 | US10 | 122103-122936 | 281 | 278 | 32 | Unknown |
| Not present | sORF4/3 | 124190-123309 | 293 | Unknown | |||
| US2 | 134374-134634 | US2 | 125011-124325 | 85 | 118 | 34 | Unknown |
| US3 | 136263-134785 | US3 | 125100-126527 | 498 | 471 | 48 | Protein kinase |
| sORF1 | 136535-138352 | UL47 | 126616-128484 | 605 | 623 | 45 | UL47 |
| US4 | 138546-139388 | US4 | 128651-129526 | 280 | 292 | 24 | Glycoprotein G |
| sORF2 | 139667-142642 | US5 | 129739-132693 | 991 | 985 | 18 | Glycoprotein J |
| US6 | 142740-143891 | US6 | 132441-133805 | 383 | 434 | 28 | Glycoprotein D |
| US7 | 144089-145315 | US7 | 133916-135001 | 492 | 362 | 34 | Glycoprotein I |
| US8 | 145663-147369 | US8 | 135198-136694 | 568 | 499 | 27 | Glycoprotein E |
| Not present | US9 | 136704-137483 | 259 | Unknown | |||
| sORF4/3 | 148377-149249 | sORF4/3 | 137535-138416 | 290 | 322 | 34 | Unknown |
| Not present | US10 | 138704-139402 | 232 | Unknown | |||
| ICP4b | 154577-160678 | ICP4 | 142837-147225 | 2,033 | 1,463 | 35 | Gene regulation |
aa, amino acids.
Function or property as demonstrated for the ILTV and/or HSV-1 homolog.
FIG. 1.
Organization of the PsHV-1 genome. This map of the PsHV-1 genome shows the locations and sizes of predicted ORFs. Predicted PsHV-1 genes are labeled according to homology with characterized HSV-1 genes. Thick gray bars flanking the unique short region indicate the internal and terminal repeat regions. A “ruler” designates distances in 5,000-bp increments. Arrows on the ORF boxes indicate orientations.
FIG. 2.
Organization of the ILTV genome. This map of the ILTV genome shows the locations and sizes of predicted ORFs. Predicted ILTV genes are labeled according to homology with characterized HSV-1 genes. Thick gray bars flanking the unique short region indicate the internal and terminal repeat regions. A “ruler” designates distances in 5,000-bp increments. Arrows on the ORF boxes indicate orientations.
Gene order is generally conserved within the UL and US regions of these two viral genomes, and they are colinear relative to other alphaherpesviruses. The UL regions of ILTV and PsHV-1 share two striking features. First, both genomes contain a unique block of five ORFs (designated A to E) previously found only in ILTV (64). Secondly, both ILTV and PsHV-1 contain an inversion of the genome from UL22 to UL44, which is similar to an inversion found between UL27 and UL44 in the PRV genome (36). The predicted PsHV-1 genes are 13% to 73% identical at the amino acid level to the corresponding predicted ILTV genes and have 99 to 100% identity to PsHV-1 sequences previously submitted to GenBank (59, 62).
The ILTV and PsHV-1 UL regions also exhibit differences in gene organization. PsHV-1 is predicted to contain a homolog of the UL16 gene, which is absent from the ILTV genome. Conversely, ILTV is predicted to contain the UL48 and UL0 ORFs, neither of which are found in the PsHV-1 genome.
US region.
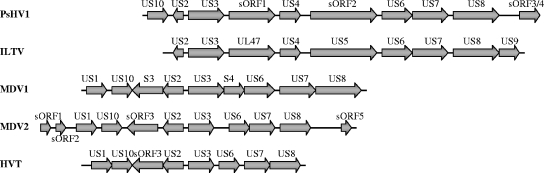
There are ten predicted ORFs within the US region of PsHV-1: US10, US2, US3, sORF1, US4, sORF2, US6, US7, US8, and sORF3/4. Gene arrangement within the PsHV-1 US region is quite similar to the corresponding region of ILTV; however, a homolog of ILTV US9 is not present in PsHV-1, nor does PsHV-1 contain the duplication of the US10 and sORF3/4 genes within the inverted repeat region, as observed in ILTV. Although the organizations of the ILTV and PsHV-1 US regions are very similar, they differ greatly from the corresponding region of the three serotypes of MDV (Fig. 3). Interestingly, neither avian herpesvirus UL region contains a homolog of UL47, a tegument phosphoprotein. However both ILTV (UL47) and PsHV-1 (sORF1) encode an ORF in the US region with weak (18%) identity to the HSV UL47 gene (38, 66).
FIG. 3.
Comparison of the unique short regions of five avian alphaherpesviruses. ORFs identified within the unique short regions of five avian alphaherpesviruses are indicated. Regions are aligned with respect to the conserved US2 and US3 genes. ORFs are labeled according to homology with characterized HSV-1 genes. Arrows on the ORF boxes indicate the direction of transcription.
The US regions of both ILTV and PsHV-1 are predicted to encode five structural glycoproteins. One of these genes was initially designated gp60 (37). Recent studies have adopted the HSV-1 designation of gJ (US5) for this ORF (19). The PsHV-1 US region contains a colinear ORF. The predicted translation product of this gene is a glycoprotein that has only 18% amino acid identity to the gJ glycoprotein of ILTV, although 9 of the 10 cysteine residues are conserved (Fig. 4). The PsHV-1 ORF has been designated sORF2.
FIG. 4.
Amino acid alignment of the predicted PsHV-1 sORF2 and ILTV gJ proteins. Amino acid sequences were aligned with the AlignX program (Vector NTI). Gaps in the alignments are represented as dashes. The consensus sequence is shown below the alignment. Potential glycosylation sites are indicated by shaded boxes, with asparagines (N) indicated in boldface type.
Phylogenetic analysis of PsHV-1 and ILTV.
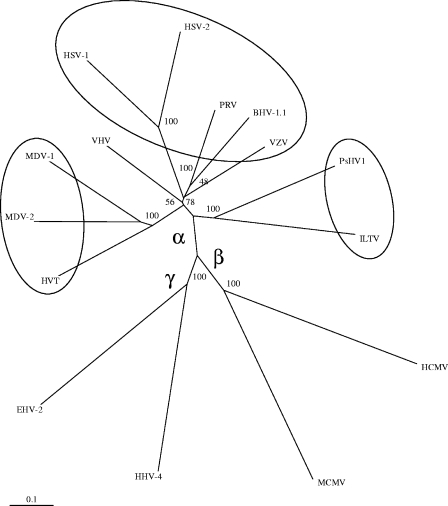
ILTV and PsHV-1 share 70 conserved genes, and BLAST results indicate that PsHV-1 proteins have the strongest similarity to homologs from ILTV (average, 42.5%) compared to other herpesviruses. A phylogenetic analysis of 15 herpesvirus DNA polymerase (UL30) amino acid sequences is depicted in Fig. 5. The herpesvirus DNA polymerase exhibits the highest levels of conservation between Herpesviridae family members and demonstrates a distinct division of the subfamilies Alpha-, Beta-, and Gammaherpesvirinae. In this bootstrapped analysis, the PsHV-1 and ILTV DNA polymerase genes align into the subfamily Alphaherpesvirinae and are distinctly separated from the Marek's disease family of herpesviruses (Mardivirus) and from vulture herpesvirus (VHV). A similar analysis of five additional genes (ICP4, UL27 [gB], UL44 [gC], US8 [gE], and UL23 [thymidine kinase]) reveals the same phylogenetic pattern (data not shown). In all cases the PsHV-1 genomic sequence is most closely related to ILTV, suggesting that ILTV and PsHV-1 form a distinct clade within the Alphaherpesvirinae.
FIG. 5.
Phylogenetic analysis of herpesvirus DNA polymerase (UL30) proteins. A phylogenetic tree of UL30 (DNA polymerase homolog) for representative alpha-, gamma-, and betaherpesviruses is shown. The tree was constructed and bootstrap analysis was performed as described in Materials and Methods. The measure of divergence is presented as a scale at the lower left. Accession numbers of the protein sequences used in tree assembly are as follows: AY372243 (PsHV-1), NC006623 (ILTV), AB024414.1 (MDV-2, strain HPRS24), AF282130.1 (HVT), P04293 (HSV-1), P07918 (HSV-2), P09252 (VZV), NC006151 (PRV), AJ004801 (bovine herpesvirus type 1.1), P08546 (human cytomegalovirus), P27172 (murine cytomegalovirus), P03198 (human herpesvirus 4; Epstein-Barr virus), P52367 (equine herpesvirus type 2); AAT79466 (VHV), AAF66765 (MDV-1, strain GA).
DISCUSSION
Herpesviruses of mammals and birds clearly descend from a common ancestor, but their genomes exhibit significant variation with respect to nucleotide sequence, gene content, and genomic organization (2, 10, 13, 14, 41, 42, 43, 65). Among avian hosts, the herpesviruses also tend to be species specific with respect to susceptibility, pathogenicity, and virulence. Although they differ significantly in G+C content—61% for PsHV-1 and 48% for ILTV—phylogenetically, PsHV-1 and ILTV are closely related. While their genomic structure, content, and organization are similar to other alphaherpesviruses, they represent a unique class of avian alphaherpesviruses. Similarities between PsHV-1 and ILTV can be seen in the UL inversion, the absence of repeats flanking the UL region, the conserved structure of the US region, and a conserved cluster of five unique ORFs in the UL region. The similarity of the PsHV-1 and ILTV genomes suggests that PsHV-1 (psittacid) and ILTV (gallid) may represent a class of avian alphaherpesviruses that diverged early from a common ancestor and are distinct from the Marek's disease family of alphaherpesviruses, as previously suggested by other researchers (33). Based on this evidence, we propose that PsHV-1 be formally assigned to the Iltovirus genus of the Alphaherpesvirinae.
To date, avian herpesviruses have been isolated from the owl (strigid); chicken and pheasant (gallid); turkey (meleagrid); pigeon (columbid); falcon (falconid); vulture and crane (gruiformid); duck (anatid); quail (perdicid); cardinal, finch, and canary (passeriform), and parrot (psittacid) (1, 26). With the advent of rapid and cost-effective sequencing strategies, it is now feasible to sequence representative herpesvirus isolates from different avian genera in order to determine whether the ILTV/PsHV-1 genus of the alphaherpesviruses is broadly represented. As evidenced by the VHV DNA polymerase analysis (Fig. 5), as more avian herpesvirus sequence becomes available additional avian herpesvirus genera may be identified.
Herpesviruses have several common properties, including a large number of conserved enzymes involved in nucleic acid metabolism, DNA synthesis, and protein processing. As expected, PsHV-1 and ILTV resemble other alphaherpesviruses in genome organization and gene content. The ILTV and PsHV-1 genomes also share several unique characteristics with respect to genome arrangement and content, which may provide clues to differences and similarities in their tissue tropism, pathogenicity, and host range. One characteristic that sets PsHV-1 and ILTV apart from the other alphaherpesviruses is the presence of five unique, conserved ORFs in the UL region (ORF A to ORF E), which Veits et al. (64) have found to be dispensable for ILTV replication in tissue culture. They have suggested that these genes may play a role in immune evasion or species specificity.
Herpesvirus-encoded structural glycoproteins play major roles in host range and pathogenicity. Both ILTV and PsHV-1 potentially encode 10 structural glycoproteins. ILTV and PsHV-1 contain structurally similar glycoprotein C (gC) proteins, which appear to lack consensus alphaherpesvirus glycoprotein-encoded heparan binding domains (34). Earlier experiments have shown that the initial attachment step of ILTV to susceptible cells does not involve interactions with heparan- or chondroitin sulfate-containing proteoglycans (35). This observation has also been confirmed for PsHV-1 (data not shown).
The gJ homolog in ILTV encodes an N- and O-linked modified glycoprotein with significant homology to equine herpesvirus type 1 gp2 (19). Veits et al. (63) have indicated that gJ (US5) may be a dominant antigen for the humoral immune response against ILTV in chickens. More-recent studies conducted with gJ-negative ILTV mutants have demonstrated that the gJ gene plays a role in determining the severity of infection and suggest that gJ may be of interest as a target for live-virus vaccine development in ILTV (19). These two gene products (gC and gJ) may determine host and/or tissue preferences for these viruses. However, the potential PsHV-1 homolog of US5 (sORF2) appears to be mainly conserved only by position, exhibiting only 18% amino acid identity to ILTV gJ. A comparison of mutant viruses containing deletions of the gC or gJ genes may help determine their roles in virus growth and replication in vitro and in vivo.
ILTV and PsHV-1 also differ with respect to specific genes known to be involved in virus growth and pathogenicity. ILTV lacks a homolog to UL16, a tegument protein that interacts with UL11 and is involved in nucleocapsid processing (39). Conversely, a homolog to α-TIF (UL48) is not found in the PsHV-1 genome but is present in ILTV. UL48 is best known for its role in enabling virus-specific immediate early (IE) gene transcription (8). In certain isolates of HSV-1 lacking viral α-TIF, amounts of IE proteins sufficient to promote virus growth and replication are made, although CPE in tissue culture progresses more slowly (50). While ILTV infection of primary cell cultures (chicken hepatocyte or chicken kidney) results in >90% CPE after 16 h, PsHV-1 infections to >90% CPE in CEF are considerably slower (∼144 h). It is possible that the lack of an α-TIF gene product in PsHV-1 may result in lower replication efficiency and a subsequently reduced rate of CPE. This slow growth in tissue culture might also account for some early confusion regarding the classification of PsHV-1. In addition, the UL48 protein appears to be involved in the final maturation process of virion formation in the cytoplasm (45, 46). This activity may be modulated by the association of UL48 with UL47, another tegument protein (44). In both PsHV-1 and ILTV, an ORF exhibiting limited homology to UL47 has been identified in the US region (sORF1, UL47).
The PsHV-1 and ILTV genome sequences provide tools for subsequent studies designed to examine the expression and analyze the functions of these virus genomes. In addition, these sequences will aid in the study of these two herpesviruses. Based on a phylogenetic analysis, Styles et al. (57) have identified four “genotypes” of PsHV. Similarly, Sellers et al. (56) have identified the presence of novel ILTV subgroups that may play a role in the propagation and subsequent outbreaks of ILTV. Comparative sequence analysis will aid in understanding the apparent diversity within these related groups of avian alphaherpesviruses. The complete nucleotide sequences of the psittacid herpesvirus 1 (PsHV-1) and gallid herpesvirus 1 (ILTV) genomes offer researchers the opportunity to begin a comparative analysis of a unique class of avian herpesviruses that are biologically and phylogenetically distinct from the Mardivirus genus.
Acknowledgments
This work was supported by Morris Animal Foundation grant 98ZO-26.
We thank the members of the Structural Genomics Group of DuPont Crop Genetics for technical assistance. We also thank Leslie Harvell, Lisa Schwartz, Travis Bliss, Carl Schmidt, and the reviewers of the manuscript for valued contributions and suggestions.
REFERENCES
- 1.Aini, L., L. M. Shih, A. E. Castro, and Y. X. Zee. 1993. Comparison of herpesvirus isolates from falcons, pigeons, and psittacines by restriction endonuclease analysis. J. Wildlife Dis. 29:196-202. [DOI] [PubMed] [Google Scholar]
- 2.Alba, M. M., R. Das, C. A. Orengo, and P. Kellam. 2001. Genomewide function conservation and phylogeny in the Herpesviridae. Genome Res. 11:43-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfonso, L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagust T. J., R. C. Jones, and J. S. Guy. 2000. Avian infectious laryngotracheitis. Rev. Sci. Technol. 19:483-492. [DOI] [PubMed] [Google Scholar]
- 7.Bahr, U., and G. Darai. 2001. Analysis and characterization of the complete genome of tupaia (tree shrew) herpesvirus. J. Virol. 75:4854-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of a gene. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckmaster, A. E., S. D. Scott, M. J. Sanderson, M. G. Boursnell, N. L. J. Ross, and M. M. Binns. 1988. Gene sequence and mapping data from Marek's disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J. Gen. Virol. 69:2033-2042. [DOI] [PubMed] [Google Scholar]
- 10.Cardoso, M., A. Hyatt, P. Selleck, S. Lowther, V. Prakash, D. Pain, A. A. Cunningham, and D. Boyle. 2005. Phylogenetic analysis of the DNA polymerase gene of a novel alphaherpesvirus isolated from an Indian (Gyps) vulture. Virus Genes 30:371-381. [DOI] [PubMed] [Google Scholar]
- 11.Cho, B. R., and T. L. McDonald. 1980. Isolation and characterization of a herpesvirus of Pacheco's parrot disease. Avian Dis. 24:268-277. [Google Scholar]
- 12.Cover, M. S., and W. J. Benton. 1958. The biological variation of the infectious laryngotracheitis virus. Avian Dis. 2:375-383. [Google Scholar]
- 13.Davison, A. 2002. Comments on the phylogenetics and evolution of herpesviruses and other large DNA viruses. Virus Res. 82:127-132. [DOI] [PubMed] [Google Scholar]
- 14.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 15.Ehlers, B., K. Borchers, C. Grund, K. Frolich, H. Ludwig, and H. J. Buhk. 1999. Detection of new DNA polymerase genes of known and potentially novel herpesviruses by PCR with degenerate and deoxyinosine-substituted primers. Virus Genes 18:211-220. [DOI] [PubMed] [Google Scholar]
- 16.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, W., and T. C. Mettenleiter. 1996. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J. Gen. Virol. 77:2221-2229. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, W., and T. C. Mettenleiter. 1999. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL10 gene product as a nonglycosylated and nonessential virion protein. J. Gen. Virol. 80:2173-2182. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, W., D. Wiesner, J. Veits, J. P. Teifke, and T. C. Mettenleiter. 2005. In vitro and in vivo relevance of infectious laryngotracheitis virus gJ proteins that are expressed from spliced and nonspliced mRNAs. J. Virol. 79:705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner, R., J. Wilkerson, and J. C. Johnson. 1993. Molecular characterization of the DNA of anatid herpesvirus 1. Intervirology 36:99-112. [DOI] [PubMed] [Google Scholar]
- 21.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 22.Gómez-Villamandos, J. C., E. Mozos, M. A. Sherra, A. Fernàndez, and F. Diaz. 1991. Mortality in psittacine birds resembling Pacheco's disease in Spain. Avian Pathol. 20:541-547. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:192-202. [DOI] [PubMed] [Google Scholar]
- 24.Gough, R. E., S. E. Drury, R. J. Higgins, and N. H. Harcourt-Brown. 1995. Isolation of a herpesvirus from a snowy owl (Nyctea scandiaca). Vet. Rec. 136:541-542. [DOI] [PubMed] [Google Scholar]
- 25.Griffin, A. M., and M. E. Boursnell. 1990. Analysis of the nucleotide sequence of DNA from the region of the thymidine kinase gene of infectious laryngotracheitis virus: potential evolutionary relationships between the herpesvirus subfamilies. J. Gen. Virol. 71:841-850. [DOI] [PubMed] [Google Scholar]
- 26.Günther, B. M. F., B. G. Klupp, M. Gravendyck, J. E. Lohr, T. C. Mettenleiter, and E. F. Kaleta. 1997. Comparison of the genomes of 15 avian herpesvirus isolates by restriction endonuclease analysis. Avian Pathol. 26:305-316. [DOI] [PubMed] [Google Scholar]
- 27.Hanson, L. E., and T. J. Bagust. 1991. Laryngotracheitis, p. 485-495. In B. W. Calnek, H. J. Barnes, C. W. Beard, W. M. Reid, and H. W. Yoder, Jr. (ed.), Diseases of poultry, 9th ed. Iowa State University Press, Ames, Iowa.
- 28.Johnson, M. A., C. T. Prideaux, K. Kongsuwan, S. G. Tyack, and M. Sheppard. 1995. ICP27 immediate early gene, glycoprotein K (gK) and DNA helicase homologues of infectious laryngotracheitis virus (gallid herpesvirus 1) SA-2 strain. Arch. Virol. 140:623-634. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, M. A., S. G. Tyack, C. Prideaux, K. Kongsuwan, and M. Sheppard. 1995. Nucleotide sequence of infectious laryngotracheitis virus (gallid herpesvirus 1) ICP4 gene. Virus Res. 35:193-204. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, M. A., S. G. Tyack, C. T. Prideaux, K. Kongsuwan, and M. Sheppard. 1997. Nucleotide sequence of the left-terminus of infectious laryngotracheitis virus (Gallid herpesvirus 1) SA-2 strain. Arch. Virol. 142:1903-1910. [DOI] [PubMed] [Google Scholar]
- 31.Kaleta, E. F. 1990. Herpesviruses of birds: a review. Avian Pathol. 19:193-211. [DOI] [PubMed] [Google Scholar]
- 32.Kaleta, E. F., H. J. Marschall, G. Glunder, and B. Stiburek. 1980. Isolation and serological differentiation of a herpesvirus from bobwhite quail (Colinus virginianus, L. 1758). Arch. Virol. 66:359-364. [DOI] [PubMed] [Google Scholar]
- 33.Kingham, B. F., V. Zelnik, J. Kopacek, V. Majerciak, E. Ney, E., and C. J. Schmidt. 2001. The genome of herpesvirus of turkeys: comparative analysis with Marek's disease viruses. J. Gen. Virol. 82:1123-1135. [DOI] [PubMed] [Google Scholar]
- 34.Kingsley, D. H., J. W. Hazel, and C. L. Keeler, Jr. 1994. Identification and characterization of the infectious laryngotracheitis virus glycoprotein C gene. Virology 203:336-343. [DOI] [PubMed] [Google Scholar]
- 35.Kingsley, D. H., and C. L. Keeler, Jr. 1999. Infectious laryngotracheitis virus, an alpha herpesvirus that does not interact with cell surface heparan sulfate. Virology 10:213-219. [DOI] [PubMed] [Google Scholar]
- 36.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kongsuwan, K., M. A. Johnson, C. T. Prideaux, and M. Sheppard. 1993. Use of lambda gt11 and monoclonal antibodies to map the gene for the 60,000 dalton glycoprotein of infectious laryngotracheitis virus. Virus Genes 7:297-303. [DOI] [PubMed] [Google Scholar]
- 38.Kongsuwan, K., C. T. Prideaux, M. A. Johnson, M. Sheppard, and S. Rhodes. 1995. Nucleotide sequence analysis of an infectious laryngotracheitis virus gene corresponding to the US3 of HSV-1 and a unique gene encoding a 67 kDa protein. Arch. Virol. 140:27-39. [DOI] [PubMed] [Google Scholar]
- 39.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simples virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May, H. G., and R. P. Tittsler. 1925. Tracheo-laryngitis in poultry. J. Am. Vet. Med. Assoc. 67:229-231. [Google Scholar]
- 41.McGeoch, D. J., and A. J. Davison. 1999. The molecular evolutionary history of the herpesviruses, p. 441-446. In E. Domingo, R. Webster, and J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, England.
- 42.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGeoch, D. J., D. Gatherer, and A. Dolan. 2005. On phylogenetic relationships among major lineages of the Gammaherpesvirinae. J. Gen. Virol. 86:307-316. [DOI] [PubMed] [Google Scholar]
- 44.Meredith, D. M., J. A. Lindsay, I. W. Halliburton, and G. R. Whittaker. 1991. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J. Gen. Virol. 72:2771-2775. [DOI] [PubMed] [Google Scholar]
- 45.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mossman, K., R. Sherburne, C. Lavery, J. Duncan, and J. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 48.Panigrahy, B., and L. C. Grumbles. 1984. Pacheco's disease in psittacine birds. Avian Dis. 28:808-812. [PubMed] [Google Scholar]
- 49.Poulsen, D. J., C. R. Burton, J. J. O'Brian, S. J. Rabin, and C. L. Keeler, Jr. 1991. Identification of the infectious laryngotracheitis virus glycoprotein gB gene by the polymerase chain reaction. Virus Genes 5:335-347. [DOI] [PubMed] [Google Scholar]
- 50.Rajcani, J., and V. Durmanova. 2000. Early expression of herpes simplex virus (HSV) proteins and reactivation of latent infection. Folia Microbiol. (Praha) 45:7-28. [DOI] [PubMed] [Google Scholar]
- 51.Randall, D. J., M. D. Dagless, H. G. R. Jones, and J. W. MacDonald. 1979. Herpesvirus infection resembling Pacheco's disease in Amazon parrots. Avian Pathol. 8:229-238. [DOI] [PubMed] [Google Scholar]
- 52.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roizman, B. 1993. The family Herpesviridae, p. 1-9. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), The human herpesviruses. Raven Press, New York, N.Y.
- 54.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Sellers, H. S., M. Garcia, J. R. Glisson, T. P. Brown, J. S. Dander, and J. S. Guy. 2004. Mild infectious laryngotracheitis in broilers in the southeast. Avian Dis. 48:430-436. [DOI] [PubMed] [Google Scholar]
- 57.Styles, D. K., E. K. Tomaszewski, and D. N. Phalen. 2005. A novel psittacid herpesvirus found in African grey parrots (Psittacus erithacus erithacus). Avian Pathol. 34:150-154. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomaszewski, E., V. G. Wilson, W. L. Wigle, and D. N. Phalen. 2001. Detection and heterogeneity of herpesviruses causing Pacheco's disease in parrots. J. Clin. Microbiol. 39:533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomaszewski, E. K., E. F. Kaleta, and D. N. Phalen. 2003. Molecular phylogeny of the psittacid herpesvirus causing Pacheco's disease: correlation of genotype with phenotypic expression. J. Virol. 77:11260-11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tulman, E. R., L. Alfonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.VanDevanter, D. R., P. Warrener, L. Bennett, E. R. Schultz, L. Coulter, R. L. Garber, and T. M. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veits, J., B. Kollner, J. P. Teifke, H. Granzow, T. C. Mettenleiter, and W. Fuchs. 2003. Isolation and characterization of monoclonal antibodies against structural proteins of infectious laryngotracheitis virus. Avian Dis. 47:330-342. [DOI] [PubMed] [Google Scholar]
- 64.Veits, J., T. C. Mettenleiter, and W. Fuchs. 2003. Five unique open reading frames of infectious laryngotracheitis virus are expressed during infection but are dispensable for virus replication in cell culture. J. Gen. Virol. 84:1415-1425. [DOI] [PubMed] [Google Scholar]
- 65.Weir, J. P. 1998. Genome organization and evolution of the human herpesviruses. Virus Genes 16:85-93. [DOI] [PubMed] [Google Scholar]
- 66.Wild, M. A., S. Cook, and M. Cochran. 1996. A genomic map of infectious laryngotracheitis virus and the sequence and organization of genes present in the unique short and flanking regions. Virus Genes 12:107-116. [DOI] [PubMed] [Google Scholar]
- 67.Ziemann, K., T. C. Mettenleiter, and W. Fuchs. 1998. Gene arrangement within the unique long genome region of infectious laryngotracheitis virus is distinct from that of other alphaherpesviruses. J. Virol. 72:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziemann, K., T. C. Mettenleiter, and W. Fuchs. 1998. Infectious laryngotracheitis herpesvirus expresses a related pair of unique nuclear proteins which are encoded by split genes located at the right end of the UL genome region. J. Virol. 72:6867-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]