Abstract
Promyelocytic leukemia (PML) nuclear bodies (also known as ND10) are nuclear substructures that contain several proteins, including PML itself, Sp100, and hDaxx. PML has been implicated in many cellular processes, and ND10 are frequently associated with the replicating genomes of DNA viruses. During herpes simplex virus type 1 (HSV-1) infection, the viral regulatory protein ICP0 localizes to ND10 and induces the degradation of PML, thereby disrupting ND10 and dispersing their constituent proteins. ICP0-null mutant viruses are defective in PML degradation and ND10 disruption, and concomitantly they initiate productive infection very inefficiently. Although these data are consistent with a repressive role for PML and/or ND10 during HSV-1 infection, evidence in support of this hypothesis has been inconclusive. By use of short interfering RNA technology, we demonstrate that depletion of PML increases both gene expression and plaque formation by an ICP0-negative HSV-1 mutant, while having no effect on wild-type HSV-1. We conclude that PML contributes to a cellular antiviral repression mechanism that is countered by the activity of ICP0.
Promyelocytic leukemia (PML) nuclear bodies (also known as ND10) are discrete nuclear substructures that are defined by the presence of the promyelocytic leukemia protein, PML. ND10 have been implicated in a great variety of processes, including oncogenesis, apoptosis, viral infection, the stress and interferon responses, DNA repair, the regulation of gene expression, and certain aspects of chromatin structure (for reviews see references 2, 3, 8, 28, and 45). ND10 become intricately associated with the parental genomes of nuclear-replicating DNA viruses, and they are modified by several viral regulatory proteins; structural modification of ND10 induced by these regulatory proteins frequently correlates with the efficiency of viral infection (for reviews, see references 9, 24, and 25). These observations have given rise to the hypothesis that ND10 structures have a repressive effect on viral infection, and viral regulatory proteins that disrupt these structures do so to relieve this repression. However, the evidence in support of this hypothesis is controversial.
PML itself has been implicated in the regulation of infection by a variety of RNA viruses (34), adenoviruses (7, 35), and human cytomegalovirus (HCMV) (1). In the case of herpes simplex virus type 1 (HSV-1), the issues of the roles of PML protein and ND10 are complex. HSV-1 immediate-early regulatory protein ICP0 greatly increases the probability that the virus will enter lytic infection, and this activity correlates very well with ICP0-induced degradation of PML and disruption of ND10 (reviewed in references 10, 11, and 18). These observations have encouraged the hypothesis that PML and/or ND10 have a repressive effect on the development of lytic HSV-1 infection and that through its targeting of these structures ICP0 relieves this repression. Indeed, in the absence of ICP0, HSV-1 genomes have a greatly increased probability of becoming repressed (11, 31, 32, 37). Such repressed genomes are subsequently maintained in a quiescent state that could have similarities to the status of the viral genome during latency. However, high-level expression of PML has no detrimental effect on HSV-1 infection (6, 23), and the replication of neither wild-type (wt) nor ICP0-negative HSV-1 viruses is improved in mouse PML−/− fibroblasts (5). These studies have cast doubt on the functional significance of the association of viral genomes with ND10-like structures and of the degradation of PML and disruption of ND10 that are brought about by ICP0.
The mouse and human PML proteins have significant sequence diversity and therefore may differ in their interactions with other proteins (16). Furthermore, PML is not a single protein but rather a family of related isoforms that arise from extensive alternative splicing and posttranslational modification (21). Differences in the relative abundances of these isoforms between mouse and human cells could also affect overall PML function in a species-specific manner. If the interplay between PML and ICP0 is involved in regulation of the balance between lytic infection and latency, it would be expected that such a critical virus-regulatory mechanism would have evolved in close concert with its natural host. Therefore, despite the negative evidence in mouse PML−/− fibroblasts (5), it was attractive to question whether removal of PML from human cells could affect the efficiency of HSV-1 infection, particularly in the absence of ICP0, and particularly in cell types in which the ICP0-negative defect is most pronounced. We have used retrovirus and lentivirus vectors that express anti-PML short hairpin RNAs (shRNAs) to create human fibroblast (HF) cell lines in which PML expression is reduced to low or negligible levels. We studied the effect of the ablation of PML on the distribution and properties of two other major ND10 proteins, Sp100 and hDaxx, in uninfected cells and the behavior of these proteins in response to HSV-1 infection in both the presence and the absence of ICP0. Although extensive depletion of PML had no effect on the efficiency of wt HSV-1 infection, it increased both the plaque-forming and gene expression efficiencies of an ICP0-negative HSV-1 mutant. We conclude that PML contributes to a repression mechanism that targets HSV-1 genomes and that is countered by the activities of ICP0.
MATERIALS AND METHODS
Viruses and cells.
HSV-1 strain 17+ was the wt strain used, from which was derived the ICP0-null mutant dl1403 (40). All viruses were grown in baby hamster kidney (BHK) cells and titrated in U2OS cells, in which ICP0 is not required for efficient replication of HSV-1 (42). All viruses were used at a multiplicity of infection (MOI) on the basis of their PFU titers in U2OS cells, regardless of the cell type used (11). U2OS cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. BHK cells were grown in Glasgow modified Eagle's medium supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. Human fetal foreskin fibroblast cells (HFFF-2 cells; European Collection of Cell Cultures) and primary human fibroblasts isolated from human foreskin tissue (HF cells) (Department of Urology, University of Erlangen) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. All cell growth media were supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin, except for HF cells, for which 5 μg/ml gentamicin was used. Retrovirus- and lentivirus-transduced cells were maintained in the appropriate medium with puromycin selection (0.5 to 2 μg/ml).
shRNA methods.
The following short interfering RNA sequences were incorporated into retrovirus vector pSIREN-RetroQ (BD Biosciences): siC, GTGCGTTGCTAGTACCAAC; siLuci, GTGCGTTGCTAGTACCAAC; and siPML2, AGATGCAGCTGTATCCAAG. The sequences were built into duplex oligonucleotides using the design tool on the BD Biosciences website. These contained the following elements: a BamHI cloning site at the 5′ end, the coding strand sequence of the hairpin (as above), a loop region, the complementary noncoding strand sequence of the hairpin, an RNA polymerase III termination sequence, a marker NheI restriction site, and a 3′ EcoRI cloning site. In the siC clone, the complementary strand was omitted. Plasmid clones were identified by the presence of the NheI restriction site and then confirmed by DNA sequence analysis. The names of the inserted sequences are used to identify the derived cell lines as follows: siC expresses a single-stranded luciferase coding region RNA; siLuci expresses an antiluciferase shRNA; siPML2 expresses an anti-PML shRNA directed against codons 394 to 400 in PML exon 4 which is conserved in all major PML isoforms (21); siV cells are transduced with the empty vector. Retrovirus stocks were prepared either by cotransfecting a pSIREN-RetroQ plasmid with pVSV-G (BD Biosciences) into GP2-293 cells or by cotransfecting 293FT cells (BD Biosciences) with plasmid pHIT60 (coding for murine leukemia virus gag/pol) together with pVSV-G and a pSIREN-RetroQ plasmid. Both procedures produced replication-deficient retroviruses, pseudotyped with the vesicular stomatitis virus coat protein and including the shRNA transcription unit and a puromycin resistance gene. Cell supernatants were harvested 72 h after transfection, clarified, filtered, and stored as recommended by the supplier.
Anti-PML lentivirus plasmids, based on pLKO.1puro and part of the MISSION shRNA lentivirus vector collection (26), were purchased from Sigma-Aldrich. The shRNAs included the indicated coding strand sequences: 3865, CAATACAACGACAGCCCAGAA (starting at PML codon 469); 3869, GTGTACGCCTTCTCCATCAAA (starting at PML codon 454); and green fluorescent protein (GFP), CAACAAGATGAAGAGCACCAA.
Further details of these clones are available from the Sigma-Aldrich website. Plasmids were cotransfected into 293T cells with pVSV-G and helper plasmid pCMVDR8.91 (kindly supplied by Didier Trono; http://tronolab.com/index.php). Virus supernatants were harvested 48 h after transfection and then filtered before use. HF cells derived from these lentiviruses are named HF-3865, HF-3869, and HF-GFP.
Target cells were infected with retrovirus or lentivirus supernatants, and then cells with integrated viral sequences were selected using puromycin at 2 μg/ml and then passaged with continuous puromycin selection. The degree of PML silencing was regularly monitored by both fluorescence and Western blotting. Control cell cultures were generated with the control retroviruses and lentiviruses in parallel. All experiments were repeated a number of times on several different independent batches of cell isolates.
Virus plaque assays in transduced cells.
Cells were seeded into 24-well dishes at 1 × 105 cells per well and then infected the following day with appropriate sequential threefold dilutions of wt HSV-1 or mutant dl1403. After virus adsorption, the cells were overlaid with medium containing 1% human serum, and then the cells were stained for plaque counts 2 days later. The number of plaques on PML-depleted cells was calculated in comparison to that on control cells at the equivalent dilution.
Infections and Western blot analysis.
Cells were seeded into 24-well dishes at 1 × 105 cells per well and then infected the following day with wt HSV-1 or mutant dl1403 as stated in the relevant figure legend. Cell monolayers were washed twice with 1 ml of phosphate-buffered saline before being harvested in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Proteins were resolved on 7.5% sodium dodecyl sulfate gels and then transferred to nitrocellulose membranes by Western blotting. ICP0, ICP4, and UL42 were detected using anti-ICP0 monoclonal antibody (MAb) 11060, anti-ICP4 MAb 10176, and anti-UL42 MAb Z1F11 as previously described (4). PML was detected with monoclonal antibody 5E10 (41), and Sp100 and hDaxx with rabbit sera SpGH (38) and r1866 (30), respectively.
Immunofluorescence.
Cells on 13-mm glass coverslips were infected with either wt or ICP0-null mutant HSV-1 at the chosen multiplicity and harvested at times as detailed in the figure legends. The cells were fixed and prepared for immunofluorescence. ICP4 was detected with monoclonal antibody 58S, PML with rabbit serum r8, Sp100 with rabbit serum SpGH or rat serum r26, and hDaxx with rabbit serum r1866, as previously described (14). In some instances PML was detected using monoclonal antibody 5E10 (41). The secondary antibodies used were fluorescein isothiocyanate-conjugated sheep anti-mouse immunoglobulin G (Sigma) or Cy3-conjugated goat anti-rabbit immunoglobulin G (Amersham). The samples were examined using a Zeiss LSM 510 confocal microscope, with 488-nm and 543-nm laser lines, scanning each channel separately under image capture conditions that eliminated channel overlap. The images were exported as TIF files and then processed using Photoshop.
RESULTS
Generation of human fibroblast cells with severely reduced PML expression.
Primary HF cells were transduced with a retrovirus vector expressing shPML2 to produce cell populations in which PML protein levels were highly suppressed. Control cells containing integrated copies of either the vector without an shRNA insert (siV cells) or the vector with control sequences (siC and siLuci cells) were produced in parallel. PML levels in siPML2 cells were undetectable by Western blot analysis using monoclonal antibody 5E10 (Fig. 1), an antibody that recognizes an epitope in exon 5 of PML (22) and hence recognizes all major alternatively spliced PML isoforms (21). Analysis by immunofluorescence demonstrated that 1 to 2% of the cells in the siPML2 population contained apparently normal ND10 structures, while in a further 5 to 8% there was evidence of one or a few punctate structures giving a signal that was far weaker than that from ND10 in control cells. The great majority of siPML2 cells gave a PML signal that was not detectable by the image capture conditions used for images of control siC cells (Fig. 2, compare rows 1 and 2). We conclude that the retrovirus expressing shPML2 provides an effective means of massively reducing PML expression levels in human fibroblasts. The siC and siPML2 cells could be passaged for several generations without any obvious differences in their growth rates and propensity to undergo apoptosis under normal cell culture conditions.
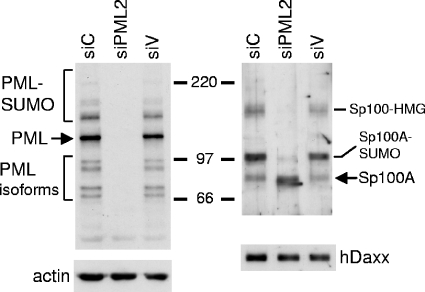
FIG. 1.
Effect of shPML2 expression on PML protein levels and SUMO modification of Sp100 in retrovirally transduced HF cells. Whole-cell extracts from 5 × 104 untreated cells were analyzed by Western blotting for PML, Sp100, hDaxx, and actin. The left panels show PML protein levels in siC, siPML2, and siV cells compared to actin. The major PML isoform is shown by an arrow, with the presumed SUMO-modified species migrating more slowly. The faster-migrating bands detected by 5E10 are definitively identified as lower-molecular-weight PML isoforms by this approach. The lower right panel shows that the levels of hDaxx are unaffected by ablation of PML. The upper right panel illustrates that there are several differences in the pattern of Sp100 isoforms in PML-depleted cells, as discussed in detail in the text. Numbers between the panels show molecular masses in kilodaltons.
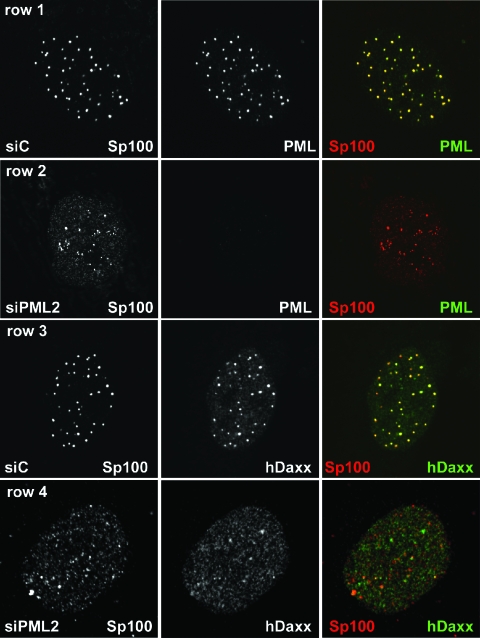
FIG. 2.
Distributions of PML, Sp100, and hDaxx in siPML2 cells and control fibroblasts. Rows 1 and 2: siC (row 1) and siPML2 (row 2) cells were costained for PML (5E10; green) and Sp100 (SpGH; red). The images were captured using identical microscope settings. Rows 3 and 4: siC (row 3) and siPML2 (row 4) cells were costained for hDaxx (r1866; green) and Sp100 (rat26; red).
Loss of PML alters the localization of hDaxx and Sp100 and the pattern of Sp100 isoform expression.
To determine the effect of ablation of PML on the level of expression and intracellular distribution of selected other PML nuclear body proteins in human fibroblasts, whole-cell extracts were analyzed by Western blotting for hDaxx and Sp100. In contrast to the absence of an effect on hDaxx, depletion of PML caused substantial changes to the expression of the various Sp100 isoforms (Fig. 1). These bands were confirmed as being isoforms of Sp100 by the use of an alternative antibody (supplied by Chemicon) and by short interfering RNA-mediated depletion (data not shown). Sp100 exhibits several isoforms as a result of alternative splicing and posttranslational modification, namely, Sp100-A, Sp100-B (or Sp100-Alt212), Sp100-C, and Sp100-HMG (17, 39). By comparison with the published literature (17, 39), the lowermost band, of approximately 100 kDa, is Sp100-A (which has a mobility very similar to that of Sp100-C), while the major band above is SUMO-modified Sp100-A. These identifications were supported by the results of a transfection experiment using an Sp100-A expression plasmid (data not shown). Sp100-B (apparent molecular mass, 145 kDa) migrates more slowly than SUMO-modified Sp100-A and is difficult to detect because it is expressed in minor amounts (17). The uppermost major Sp100 band in Fig. 1 has a mobility expected of isoform Sp100-HMG (apparent molecular mass, 175 kDa). Therefore, it appears that depletion of PML causes substantial changes to the relative expression and/or stability of different isoforms of Sp100, in particular an apparent reduction in Sp100-HMG and a reduction in the SUMO-modified form of Sp100-A, with a concomitant increase in unmodified Sp100-A. These observations demonstrate that the expression and SUMO modification status of Sp100 are to some extent regulated, either directly or indirectly, by PML.
Whereas both hDaxx and Sp100 are tightly associated with PML in nuclear bodies in siC cells (Fig. 2, rows 1 and 3), loss of PML led to changes in the distribution of both of the other proteins. Although both still formed discrete foci in the absence of PML, in the case of hDaxx these were fewer in number and there was an increase in the intensity of the nuclear-diffuse hDaxx signal (Fig. 2, row 4). The Sp100 foci were not generally reduced in number in siPML2 cells, but they were smaller, less intense, and less evenly distributed than in siC cells (Fig. 2, rows 2 and 4). Costaining for Sp100 and hDaxx demonstrated that the great majority of the Sp100 foci did not contain distinct accumulations of hDaxx, and indeed, in contrast to their extensive colocalization in PML-positive cells, in siPML2 cells the two proteins showed little association (Fig. 2, row 4).
These results are consistent with previous observations using PML-null mouse cells (20, 43, 44), but the availability of anti-Sp100 reagents for use in Western blot analysis of human proteins has revealed that lack of PML results in substantial changes to the pattern of expression of the various Sp100 isoforms. In addition, depletion of PML causes the intracellular localizations of hDaxx and Sp100 to become largely unlinked.
Changes in the expression profile of Sp100 during HSV-1 infection may be an indirect effect caused by degradation of PML.
The changes in the expression profile of Sp100 in siPML2 cells (Fig. 1) are reminiscent of those that occur during wt HSV-1 infection (29). Therefore, it is possible that the ICP0-dependent loss of the various lower-mobility Sp100 isoforms during HSV-1 infection is not due to a direct effect of ICP0 on Sp100 itself but rather is an indirect effect of the actions of ICP0 that lead to degradation of PML and disruption of ND10 (12). To test this hypothesis, siC and siPML2 cells were infected with wt HSV-1 and cell extracts were analyzed for Sp100, actin, and the viral proteins ICP0 and UL42. Viral gene expression by wt HSV-1 was comparable in the two cell lines (see also Fig. 7).
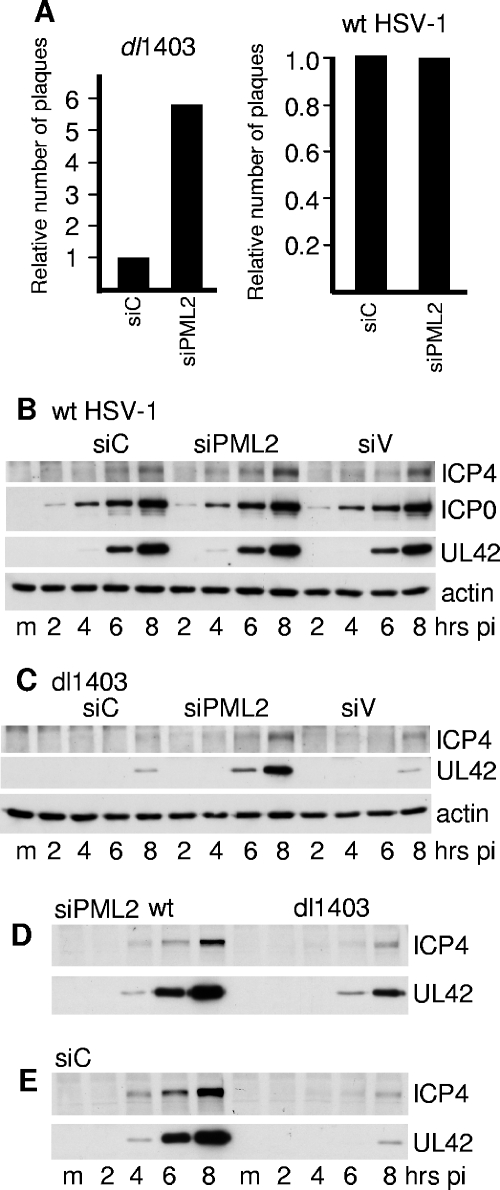
FIG. 7.
(A) Relative efficiencies of wt and ICP0-null mutant HSV-1 dl1403 plaque formation in cells transduced with retroviruses expressing anti-PML or control shRNAs. The increase of dl1403 titers in siPML2 compared to siC cells varied from 2.4- to 10.0-fold, the average of eight experiments being 5.8-fold. In a typical example of an experiment, the titers of a stock of dl1403 in siC, siPML2, and U2OS cells were 9.2 × 104, 5.2 × 105, and 4.8 × 107, respectively. The titers were determined by a series of threefold dilutions from an appropriate starting point. For wt HSV-1, the average relative titers on siPML2 cells compared to siC cells over three experiments were 0.82, 1.2, and 0.92 (average, 0.98). In a typical example of an experiment, the titers of a stock of wt HSV-1 in siC, siPML2, and U2OS cells were 6.7 × 108, 4.9 × 108, and 5.8 × 108, respectively. (B to E). Increase in gene expression of ICP0-null but not wt HSV-1 in cells depleted for PML. siC, siPML2, and siV cells were seeded into 24-well dishes. The following day, the cells in one well of each cell type were resuspended in trypsin and counted. Cells were then infected at an MOI of 2 with either wt HSV-1 or dl1403, and then samples were harvested at 2, 4, 6, and 8 h after infection using volumes of gel loading buffer to give equal total cell protein concentrations. Actin levels and expression of ICP4 and UL42 (and ICP0 in the wt HSV-1 samples) were analyzed by Western blotting. (B) wt HSV-1 infection of siC, siPML2, and siV cells. (C) dl1403 infection of siC, siPML2, and siV cells. (D) The wt HSV-1 and dl1403 samples from the siPML2 infection analyzed on the same gel. (E) The wt HSV-1 and dl1403 samples from the siC infection analyzed on the same gel. pi, postinfection. m, mock infected.
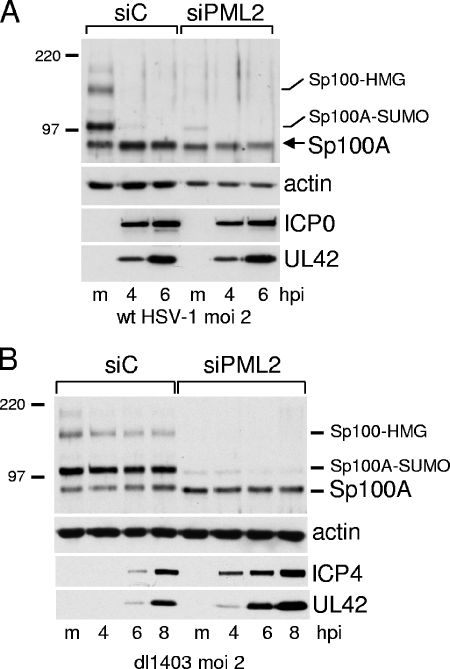
Infection of siC cells with wt HSV-1 caused the loss of both SUMO-modified and unmodified forms of PML (data not shown) in the same way as previously established in other human fibroblast cells (29). However, the wt virus had no effect on the unmodified form of Sp100A in siPML2 cells (Fig. 3A), and dl1403 infection had little effect on any of the Sp100 isoforms in both siC and siPML2 cells (Fig. 3B). The greater efficiency of dl1403 gene expression in siPML2 than in siC cells (Fig. 3B) is investigated in more detail later in this paper (see Fig. 7 and 8). Therefore, while it has been established previously that ICP0 causes the loss of the SUMO-modified Sp100A in transfected and infected cells (27, 29), it is now apparent that ICP0 has no effect on unmodified Sp100A, and the ICP0-induced loss of SUMO-modified Sp100A and the higher-molecular-weight isoforms of Sp100 can be replicated simply by shRNA-mediated depletion of PML. This result illustrates the principle that through its induction of PML degradation, ICP0 could have effects on the diverse cellular proteins and pathways that can be regulated by PML. These observations do not eliminate the possibility that ICP0 has a direct effect on the SUMO-modified and other higher-molecular-weight species of Sp100, but an indirect effect caused by inducing the degradation of PML is the simpler hypothesis.
FIG. 3.
(A) ICP0 has no effect on unmodified Sp100 during wt HSV-1 infection of cells lacking PML. siC and siPML2 cells were infected with wt HSV-1 (MOI of 2), and then samples were harvested at 4 and 6 h after infection for analysis by Western blotting for Sp100, actin, ICP0, and UL42 as indicated. (B) The loss of the higher-molecular-weight isoforms of Sp100 during wt HSV-1 infection does not occur in ICP0-null mutant dl1403-infected cells. siC and siPML2 cells were infected with dl1403 at an MOI of 2, and samples were taken at the indicated times after infection for analysis by Western blotting for Sp100, actin, ICP4, and UL42. hpi, hours postinfection. Numbers at left are molecular masses in kilodaltons. m, mock infected.
FIG. 8.
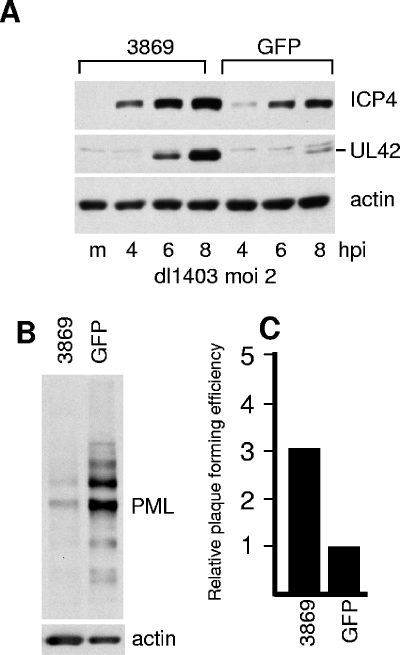
Enhancement of dl1403 gene expression and plaque formation in cells expressing a different anti-PML shRNA. HF cells were transduced with lentiviruses expressing control GFP or anti-PML shRNA 3869. m, mock infected. (A) Cells were infected with dl1403 at an MOI of 2, and samples were harvested at the indicated time points. Expression of ICP4, UL42, and actin was detected by Western blotting. (B) PML expression in HF-3869 and HF-GFP cells compared to actin. (C) Relative plaque-forming efficiency of dl1403 in HF-3869 cells, relative to that in HF-GFP cells. The increase in plaque formation in HF-3869 cells averaged 3.0-fold from four replicate experiments, with a range of 2.0 to 3.6.
Loss of PML does not abrogate recruitment of Sp100 and hDaxx to HSV-1 genome-associated foci in the absence of ICP0.
At the early stages of infection by HSV-1 and other DNA viruses, the parental viral genomes and their ensuing viral replication compartments become associated with ND10 proteins and structures (9, 24, 25). In the case of HSV-1, it has been demonstrated that a major factor in this process is the recruitment of PML, Sp100, and hDaxx into structures that resemble ND10 and that become associated with or juxtaposed to viral nucleoprotein complexes (13, 14). In wt HSV-1 infections, this process is very difficult to visualize because of the rapid degradation of PML that is induced by ICP0, but in ICP0-null mutant HSV-1 infections the phenomenon becomes obvious, especially in human fibroblasts in the early stages of infection. The recruitment of cellular proteins to viral nucleoprotein complexes is most easily detected in cells at the edges of developing viral plaques because in this case the viral genomes are frequently localized in a highly asymmetric pattern (13, 14). Since PML is required for the assembly of ND10 (20, 44) and Sp100 and hDaxx do not colocalize in siPML2 cells (Fig. 2), we questioned whether PML was required for the recruitment of Sp100 and hDaxx into foci associated with HSV-1 nucleoprotein complexes.
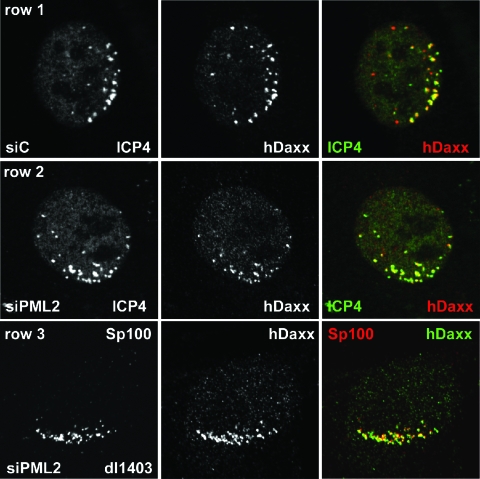
siC and siPML2 cells were infected at a low multiplicity with HSV-1 ICP0-null mutant virus dl1403 and then stained the following day for either PML, Sp100, or hDaxx and for ICP4 to detect viral nucleoprotein complexes. The controls for the distributions of these proteins in uninfected siC and siPML2 cells are shown in Fig. 2. The recruitment of PML (Fig. 4, row 1), Sp100 (Fig. 4, row 3), and hDaxx (Fig. 5, row 1) into foci that were associated but not precisely colocalized with accumulations of ICP4 was prominent in dl1403-infected siC cells. Surprisingly, recruitment of Sp100 (Fig. 4, row 4) and hDaxx (Fig. 5, row 2) also occurred in dl1403-infected siPML2 cells. This result was strikingly illustrated by costaining cells at the edges of dl1403 plaques for Sp100 and Daxx (Fig. 5, row 3). Although these two proteins are largely separately distributed in siPML2 cells (Fig. 2, row 4, and data not shown), in such cells in the early stages of infection by dl1403 they become markedly associated (Fig. 5, row 3).
FIG. 4.
Redistribution of ND10 proteins in PML-depleted and control cells at the edges of plaques of ICP0-null mutant virus dl1403. siC cells (rows 1 and 3) and siPML2 cells (rows 2 and 4) were infected with dl1403 at an MOI of 0.1 and then stained the following day for ICP4 (58S; green) and PML (r8; red) (rows 1 and 2) or ICP4 (58S; green) and Sp100 (SpGH; red). Cells in the early stages of infection at the edges of plaques displaying the characteristic asymmetric distributions of early ICP4 foci were selected for imaging without prior visual inspection of the PML or Sp100 signals. The image capture settings for the control cells (rows 1 and 3) were reused without adjustment for the siPML2 cells (rows 2 and 4).
FIG. 5.
Redistribution of ND10 proteins in PML-depleted and control cells at the edges of plaques of ICP0-null mutant virus dl1403. siC (row 1) and siPML2 (row 2) cells were infected with dl1403 at an MOI of 0.1 and then stained the following day for ICP4 (58S; green) and hDaxx (r1866; red). Cells in the early stages of infection at the edges of plaques displaying the characteristic asymmetric distributions of early ICP4 foci were selected for imaging without prior visual inspection of the hDaxx signal. The image capture settings for the control cells were reused without adjustment for the siPML2 cells. Row 3 shows a similarly infected siPML2 cell costained for Sp100 (rat26; red) and hDaxx (r1866; green). Although not stained for a viral antigen, viral plaques in this sample could be readily identified by the characteristic changes that occur in the distributions of Sp100 and hDaxx compared to neighboring uninfected cells (compare with Fig. 2, row 4).
Although we cannot exclude the possibility that trace amounts of PML remain in the illustrated siPML2 cells, we note that the cells were selected for imaging on the basis of ICP4 distribution alone and that all cells with ICP4 foci at the edge of the nucleus exhibited the characteristic asymmetric distribution of Sp100 and hDaxx. At the very least, the amounts of Sp100 and hDaxx that are recruited to sites juxtaposed to HSV-1 genomes are grossly disproportionate to the amount of PML in the cell. It appears either that PML, Sp100, and hDaxx individually contain determinants that enable their relocation to sites associated with viral nucleoprotein complexes or that there is some other factor to which all three proteins respond. In the case of Sp100, and taken with the results of Fig. 3, the recruitment must also be independent of the SUMO modification status of the protein.
Redistribution of Sp100 and hDaxx in PML-depleted cells infected with wt HSV-1.
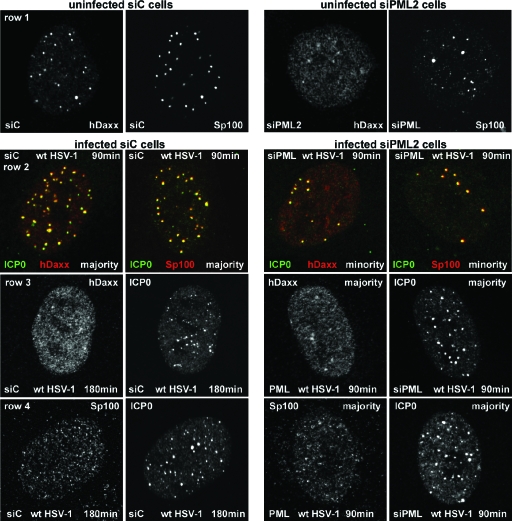
At the early stages of wt HSV-1 infection ICP0 accumulates at ND10 structures and then brings about their disruption. To investigate where ICP0 is localized with respect to Sp100 and hDaxx in the apparent absence of PML, siPML2 and siC control cells were infected with wt HSV-1 and samples were harvested at 90 and 180 min after infection for immunofluorescence analysis. In siC cells the results were as expected from many previous studies: ICP0 initially forms distinct foci that colocalize with Sp100 and hDaxx (Fig. 6, row 2, two left panels). Once ICP0 levels had accumulated sufficiently, both Sp100 and hDaxx exhibited a mainly diffuse staining pattern (Fig. 6, two left panels of rows 3 and 4). Since PML is required to nucleate ND10 (20, 44) and since Sp100 and hDaxx do not colocalize in distinct foci in cells depleted for PML (Fig. 2), it was possible that ICP0, Sp100, and hDaxx would be independently located in siPML2 cells. However, in siPML2 cells expressing very low levels of ICP0, it was clear that both Sp100 and hDaxx became prominently recruited into foci that also contain ICP0 (Fig. 6, row 2, two right panels; compare with the uninfected siPML2 cells in Fig. 2, row 4, and Fig. 6, row 1, right panels). Therefore, ICP0, Sp100, and hDaxx respond to a stimulus in the early stages of HSV-1 infection that leads to the assembly of complexes containing all three proteins, even in the absence of detectable levels of PML. The distinct differences between the distributions of Sp100 and hDaxx in uninfected siPML2 cells and those in the early stages of infection can be interpreted only in terms of substantial, active recruitment of the two cellular proteins into novel, ICP0-containing foci.
FIG. 6.
ICP0 initially colocalizes with and then disperses Sp100 and hDaxx in both siC and siPML2 cells. siC and siPML2 cells were infected with wt HSV-1 (MOI of 2), and samples were analyzed by immunofluorescence after 90 and 180 minutes of infection. The panels are appropriately labeled to indicate the cell type, the time point, and the staining regimen used. Row 2 shows individual cells dual labeled for the indicated proteins. Rows 1, 3, and 4 show the separated red and green channels for either single siC cells (left of panels) or siPML2 cells (right of panels). Row 1 shows control uninfected siC and siPML2 cells, and rows 2 to 4 show infected siC and siPML2 cells as indicated.
This association between ICP0, Sp100, and hDaxx in siPML2 cells was only transitory. In the 90-minute time point sample of this experiment, there were a minority of siPML2 cells (on the order of 10%) expressing very low levels of ICP0 (as illustrated in Fig. 6, row 2), but in cells with greater amounts of ICP0, Sp100 and hDaxx were mainly diffusely distributed and no longer colocalized with ICP0 (Fig. 6, rows 3 and 4, right panels). Although only about 10% of the infected siPML2 cell population contained foci with colocalizing ICP0, Sp100, and hDaxx, all the cells in which ICP0 expression was very low exhibited this phenotype. Therefore, the ICP0-Sp100-hDaxx association was short-lived in siPML2 cells. In contrast, in the presence of PML in siC cells, the majority (on the order of 90%) of cells at the 90-minute time point expressing relatively high levels of the viral protein contained punctate ICP0 foci that included both Sp100 and hDaxx (Fig. 6, row 1, left panels). Therefore, the presence of PML reduces the rate at which ICP0 disrupts these virally induced ND10-like structures. By 180 min, the great majority of infected siC cells displayed mainly diffuse Sp100 and hDaxx staining (Fig. 6, rows 3 and 4, left panels) and in this regard were very similar to siPML2 cells at the 90-minute time point.
Taken with other recently published observations (13), these observations imply that (i) ICP0, Sp100, hDaxx, and PML colocalize in both preexisting ND10 and novel ND10-like structures at the early stages of infection; (ii) ICP0 brings about the destruction of these foci; (iii) endogenous PML reduces the rate at which this disruption occurs; and (iv) any distinct foci that Sp100 and hDaxx form in the apparent absence of PML are also susceptible to disruption by ICP0, even in the absence of any apparent biochemical effects of ICP0 on these proteins. All these observations strongly suggest that, while PML is a major orchestrator of ND10 structures in uninfected cells, either undetectable levels of PML can have the same effect as the normal amount of the protein or there are other factors that can allow the assembly of complexes containing ICP0, Sp100, and hDaxx in the early stages of HSV-1 infection. These factors are sensitive to the effects of ICP0. This conclusion parallels that from the observation of efficient recruitment of Sp100 and hDaxx into complexes associated with HSV-1 nucleoprotein complexes in cells at the edges of developing plaques of an ICP0-null mutant virus (Fig. 4 and 5).
An ICP0-negative HSV-1 mutant is partially complemented in PML-negative cells.
The role of PML protein and PML nuclear bodies in HSV-1 infection has been a long-standing question. The availability of untransformed human fibroblast cells that express severely reduced amounts of PML allowed this question to be revisited in cells in which the requirement for ICP0 for efficient HSV-1 replication is most pronounced. We found that the plaque-forming efficiency of dl1403 was greater in siPML2 than in siC cells, whereas wt HSV-1 exhibited similar plaque numbers in both cell types (Fig. 7A). The plaque-forming efficiency of dl1403 in siC cells was similar to that in several control cell lines (including si-Luci, siV, and the parental human fibroblast cells) and on the order of 500-fold lower than that in U2OS cells (data not shown), in which ICP0 is not required for efficient HSV-1 infection (11, 42). In addition, another human fibroblast cell line (HFFF-2) transduced with siPML2 gave similar increases in dl1403 plaque formation (data not shown). Therefore, the lack of PML enhances the plaque-forming ability of ICP0-null mutant HSV-1 while having no effect on the wt virus. Note that the complementation of dl1403 in terms of plaque formation in siPML2 cells was limited, as a further substantial increase in plaque formation would be expected if complementation had been complete.
Western blot analysis indicated that the expression of ICP0, ICP4, and UL42 by wt HSV-1 was equally efficient in siC, siPML2, and siV cells (Fig. 7B). UL42 is a typical early viral gene product indicative of progression into the early phase of HSV-1 infection, and its expression is representative of other viral early proteins. In contrast to the result in wt HSV-1 infection, expression of UL42 by dl1403 was significantly enhanced in siPML2 cells (by 17-fold at the 8-h time point in this example) compared to the controls (Fig. 7C; also Fig. 3B). However, viral gene expression by wt HSV-1 and that by dl1403 were not equally efficient in siPML2 cells (Fig. 7D), although the differential between the two viruses was greatly decreased in siPML2 compared to siC cells (compare Fig. 7D and 7E). These results are consistent with the plaque assay data, allowing the conclusion that depletion of PML increases the efficiency of viral gene expression by an ICP0-null mutant HSV-1 but that the ICP0-negative phenotype is not completely overcome in this situation. Similar dl1403 gene expression results were obtained using HFFF-2 human fibroblasts and HeLa cells transduced with the retrovirus expressing siPML2, while dl1403 gene expression was never enhanced in control cells transduced with siC, si-Luci, or siV vectors (data not shown).
To guard against off-target effects of siPML2, the above results were confirmed using a different shRNA and a different vector system. HF cells were transduced with lentivirus vectors expressing anti-PML (shRNA 3869) or control anti-GFP shRNA sequences. The resultant HF-3869 cell line exhibited decreased levels of PML compared to the HF-GFP control cells, although in this case the depletion was not as efficient as that in siPML2 cells. Both plaque formation and particularly viral protein expression by dl1403 were more efficient in HF-3869 cells than in the control cells (Fig. 8). A third anti-PML shRNA sequence, 3865, also resulted in depletion of PML and an increase in dl1403 gene expression and plaque formation in transduced cell lines, although all three of these effects were to lesser extents than those in HF-3869 cells (data not shown). These data confirm the conclusion that PML contributes to an antiviral repression mechanism that is counteracted by ICP0.
DISCUSSION
We have demonstrated that depletion of PML from human fibroblast cells leads to increased gene expression and plaque-forming efficiency by ICP0-null mutant but not wt HSV-1. These results provide direct evidence that PML contributes to a cellular repression mechanism that targets HSV-1 and that is inactivated by ICP0. Therefore, the degradation of PML that is induced through the RING finger ubiquitin E3 ligase activity of ICP0 can be concluded to be involved directly in the regulation of HSV-1 gene expression. While previous studies have shown that ICP0 requires the ubiquitin E3 ligase activity of its RING finger to enhance viral gene expression and induce reactivation of quiescent viral genomes in cultured cells (10, 11, 31), the observation that this activity targets not only PML but also several other cellular proteins has until now precluded the conclusion that the degradation of any one of the known target proteins is relevant for viral infection. In other words, one or more of the documented targets of ICP0 could be “innocent bystanders” that are degraded because they share some property with the biologically relevant target(s). The data described here demonstrate that PML is a biologically relevant target of ICP0. Furthermore, it is likely that the apparent effects of ICP0 on the various isoforms of Sp100 during wt HSV-1 infection are a consequence of ICP0-induced degradation of PML, rather than being a direct effect of ICP0 on Sp100 itself. This latter observation illustrates the principle that through inducing the degradation of PML, ICP0 could affect all those pathways in which PML has been shown to have a role.
Previous work on the role of PML in HSV-1 infection has concentrated on the approaches of high-level exogenous expression of PML and the use of PML-null mice and fibroblast cells derived from such mice (5, 6, 23). These studies have found no evidence that high-level expression of PML reduces either wt or ICP0-null mutant HSV-1 infection and that ICP0-null mutant viruses replicate no better in mouse PML−/− fibroblasts than in control cells. Our own unpublished data are in agreement with these conclusions. How can the differences between these previous studies and the work presented here be reconciled? We note that high-level expression of PML is unable to neutralize the effect of ICP0 on the disruption of ND10 structures during HSV-1 infection (15). This observation provides a plausible explanation for the lack of repression of wt HSV-1 by overexpressed PML. In the case of the absence of increased replication of ICP0-null mutant HSV-1 in mouse PML-null fibroblasts (5), there may be species-specific differences resulting from the marked sequence divergence between mouse and human PML proteins (16), and from potential differences in the relative abundances and expression of the various PML isoforms. It is also possible that the mouse cells, being derived from viable animals, have undergone adaptive changes that compensate for the loss of PML in terms of repression of “foreign” DNA sequences.
It is important to emphasize that extensive reductions in PML protein levels do not completely overcome the plaque-forming and gene expression defects of dl1403. Indeed, a considerable proportion of the repression of HSV-1 replication that occurs in the absence of ICP0 remains active in PML-depleted cells. There are several factors that could contribute to this situation: (i) despite being reduced to levels undetectable by Western blotting, sufficient PML may remain in many cells to cause a degree of repression; (ii) PML contributes to the repression by interacting with or orchestrating other proteins that are the actual repressors—in the absence of PML, these other proteins will still be present and may still be able to function, albeit less efficiently; (iii) the other ND10 proteins (and almost certainly other cellular proteins not yet analyzed) that are recruited to dl1403 nucleoprotein complexes in the apparent absence of PML may also be involved in repression of viral gene expression in the absence of ICP0; (iv) degradation of PML is likely to be only one aspect of the important functions of ICP0—these other functions remain defective in dl1403-infected PML-depleted cells. Whatever the origin of the remaining defect of ICP0-null mutant HSV-1 in PML-depleted human fibroblasts, the results reported here clearly establish that PML has an antiviral role during HSV-1 infection.
This conclusion is not restricted to HSV-1 infection. Parallel work provides strong evidence that depletion of PML increases HCMV gene expression and plaque-forming efficiency (41a). In this study, gene expression and plaque formation by both wt and IE72-deficient HCMV were enhanced in PML-depleted cells. There are several similarities between the characteristics of HCMV and HSV-1 infections in PML-depleted cells. For example, although Sp100 and hDaxx are not colocalized in uninfected siPML2 cells, they form colocalizing foci in the virus-infected cells. These foci are associated with accumulations of the major viral immediate-early transactivators (HSV-1 ICP4 and HCMV IE2), and in wt virus infections they are transient and later disperse through the activities of HSV-1 ICP0 and HCMV IE1, the viral regulatory proteins that bring about the disruption of ND10 in normal cells. We did note some differences between the HSV-1 and HCMV studies: firstly, wt HSV-1 infection was not stimulated in PML-depleted cells; secondly, whereas reintroduction of PML isoform VI reversed the effect of PML depletion during HCMV infection (41a) it did not do so in the case of dl1403 (data not shown). Given that adenovirus E4orf3 protein exerts its effect through PML isoform II rather than PML isoform VI (19), it appears that viruses can target different specific isoforms of PML. The role of each of the six major isoforms of PML during HSV-1 infection of PML-depleted cells is currently under study.
Taken together, the observations in these and other recent studies (33, 36) are consistent with the more general hypothesis that ND10 may contribute to an innate defense mechanism that responds to the entry of viral genomes into the nucleus. Especially in the case of the herpesviruses, the viral genome comprises a substantial unit of DNA, which, because it is not initially in a conventional chromatin structure, is quite unlike any resident cellular chromatin. The redistribution of ND10 proteins in response to viral genome entry (13, 14) indicates that the cell responds in a highly dynamic manner to the entry of this foreign material. Recent evidence that HCMV tegument protein pp71 interacts with and induces the degradation of the ND10 component protein hDaxx in order to relieve repression of HCMV gene expression (33, 36) provides strong support for the hypothesis that one function of ND10, and perhaps several components thereof, is in defense from infection by large DNA viruses.
Acknowledgments
This work was supported by the U.K. Medical Research Council, the DFG (SFB473), the IZKF Erlangen, and the BioMedTec International Graduate School BIGSS.
The skilled technical assistance of Regina Muller is gratefully acknowledged. We thank Didier Trono for lentivirus helper plasmid pCMVDR8.91, Roel van Driel for anti-PML antibody 5E10, Thomas Sternsdorf and Hans Will for antibody SpGH, and Carlos Parada and other members of the Glasgow laboratory for helpful discussions.
REFERENCES
- 1.Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi, R., and P. P. Pandolfi. 2003. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 22:9048-9057. [DOI] [PubMed] [Google Scholar]
- 3.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canning, M., C. Boutell, J. Parkinson, and R. D. Everett. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 279:38160-38168. [DOI] [PubMed] [Google Scholar]
- 5.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 7.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 8.Eskiw, C. H., and D. P. Bazett-Jones. 2002. The promyelocytic leukemia nuclear body: sites of activity? Biochem. Cell Biol. 80:301-310. [DOI] [PubMed] [Google Scholar]
- 9.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 10.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 11.Everett, R. D., C. Boutell, and A. Orr. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., and A. Zafiropoulos. 2004. Visualization by live-cell microscopy of disruption of ND10 during herpes simplex virus type 1 infection. J. Virol. 78:11411-11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goddard, A. D., J. Q. Yuan, L. Fairbairn, M. Dexter, J. Borrow, C. Kozak, and E. Solomon. 1995. Cloning of the murine homolog of the leukemia-associated PML gene. Mamm. Genome 6:732-737. [DOI] [PubMed] [Google Scholar]
- 17.Guldner, H. H., C. Szostecki, P. Schroder, U. Matschl, K. Jensen, C. Luders, H. Will, and T. Sternsdorf. 1999. Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif. J. Cell Sci. 112:733-747. [DOI] [PubMed] [Google Scholar]
- 18.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoppe, A., S. J. Beech, J. Dimmock, and K. N. Leppard. 2006. Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J. Virol. 80:3042-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 22.Koken, M. H., F. Puvion-Dutilleul, M. C. Guillemin, A. Viron, G. Linares-Cruz, N. Stuurman, L. de Jong, C. Szostecki, F. Calvo, C. Chomienne, et al. 1994. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 13:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 25.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75.8599237 [Google Scholar]
- 26.Moffat, J., D. A. Grueneberg, X. Yang, S. Y. Kim, A. M. Kloepfer, G. Hinkle, B. Piqani, T. M. Eisenhaure, B. Luo, J. K. Grenier, A. E. Carpenter, S. Y. Foo, S. A. Stewart, B. R. Stockwell, N. Hacohen, W. C. Hahn, E. S. Lander, D. M. Sabatini, and D. E. Root. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283-1298. [DOI] [PubMed] [Google Scholar]
- 27.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pluta, A. F., W. C. Earnshaw, and I. G. Goldberg. 1998. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J. Cell Sci. 111:2029-2041. [DOI] [PubMed] [Google Scholar]
- 31.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 32.Preston, C. M., and M. J. Nicholl. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 71:7807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preston, C. M., and M. J. Nicholl. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87:1113-1121. [DOI] [PubMed] [Google Scholar]
- 34.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 35.Rosa-Calatrava, M., F. Puvion-Dutilleul, P. Lutz, D. Dreyer, H. de The, B. Chatton, and C. Kedinger. 2003. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 4:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternsdorf, T., H. H. Guldner, C. Szostecki, T. Grotzinger, and H. Will. 1995. Two nuclear dot-associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand. J. Immunol. 42:257-268. [DOI] [PubMed] [Google Scholar]
- 39.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 41.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 41a.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong, S., P. Hu, T. Z. Ye, R. Stan, N. A. Ellis, and P. P. Pandolfi. 1999. A role for PML and the nuclear body in genomic stability. Oncogene 18:7941-7947. [DOI] [PubMed] [Google Scholar]
- 44.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]
- 45.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]