Abstract
The cytoplasmic tail of the influenza A virus M2 protein is highly conserved among influenza A virus isolates. The cytoplasmic tail appears to be dispensable with respect to the ion channel activity associated with the protein but important for virus morphology and the production of infectious virus particles. Using reverse genetics and transcomplementation assays, we demonstrate that the M2 protein cytoplasmic tail is a crucial mediator of infectious virus production. Truncations of the M2 cytoplasmic tail result in a drastic decrease in infectious virus titers, a reduction in the amount of packaged viral RNA, a decrease in budding events, and a reduction in budding efficiency. The M1 protein binds to the M2 cytoplasmic tail, but the M1 binding site is distinct from the sequences that affect infectious virus particle formation. Influenza A virus strains A/Udorn/72 and A/WSN/33 differ in their requirements for M2 cytoplasmic tail sequences, and this requirement maps to the M1 protein. We conclude that the M2 protein is required for the formation of infectious virus particles, implicating the protein as important for influenza A virus assembly in addition to its well-documented role during virus entry and uncoating.
A productive virus infection begins with the release of the viral genome into a permissive host cell. The viral genome can then be replicated, transcribed, and translated. To complete a productive life cycle, a virus must temporarily package its genome into a particle that facilitates the transport and release of the viral genetic material into another permissive cell. For most human viral pathogens, the mechanisms responsible for genome incorporation and virus assembly are yet to be clearly defined.
Budding of enveloped viruses requires the action of a viral protein capable of mediating curvature of the lipid bilayer, followed by fission of the new viral membrane from the cellular membrane (43, 61). The Gag protein of human immunodeficiency virus (12, 19); the matrix proteins of influenza A virus (20, 31), vesicular stomatitis virus (VSV) (33), and simian virus 5 (63); and the VP40 proteins of Ebola (36, 51) and Marburg viruses (71) have been shown to mediate budding of virus-like particles (VLPs) in the absence of other viral proteins.
While matrix proteins alone can drive the process of VLP formation, coexpression of other viral proteins can improve the efficiency of VLP formation. In the case of Ebola and Marburg viruses, coexpression of the viral glycoprotein with VP40 enhances the efficiency of VLP formation over that observed with VP40 alone (3, 35, 71). The extracellular, membrane-proximal region of the VSV G protein has been shown to increase the budding efficiency of VSV (58). The budding of a fully infectious viral particle requires the coordinated action of multiple viral proteins with the lipid bilayer, viral genome, and most likely host proteins. This concerted event increases the efficiency of particle release over that demonstrated in most VLP systems (35, 58).
Influenza A viruses are classified in the Orthomyxoviridae family and have a genome consisting of eight negative-sense, single-stranded RNA segments (30). The viral RNA is packaged into virions as a ribonucleoprotein (RNP) complex consisting of the RNA segment encapsidated by the nucleoprotein (NP) and bound by the polymerase complex proteins PB2, PB1, and PA. Each RNA segment contains specific RNA sequences present in coding and noncoding regions that are important for incorporation into virus particles (17, 18, 34, 80). There appears to be a coordinated packaging of RNPs into virus particles, and the segments may interact to facilitate incorporation of a full complement of genomic RNA segments into an individual virus particle (46, 52).
Assembly of influenza A virus particles is thought to occur through interaction of the viral matrix protein M1 with the viral integral membrane proteins and RNP complexes (62). The M1 protein drives influenza A virus particle budding through late or L domains (26, 27) and affects virion morphology via a number of amino acid residues located throughout the protein (5, 8, 15). The hemagglutinin (HA) and neuraminidase (NA) proteins target to lipid raft microdomains at the apical membrane of polarized epithelial cells where virus budding occurs (2, 60, 72, 83). The cytoplasmic tails of HA and NA are believed to interact with M1 to drive the budding process (1, 10, 16, 29, 83).
The 97 amino acid integral membrane protein M2 is a proton-specific ion channel that is necessary for the efficient release of the viral genome during influenza A virus entry (6, 7, 23, 56, 73, 79). The influx of protons into the virion interior mediated by M2 is believed to disrupt interactions between the viral RNPs (vRNPs), M1 protein, and lipid bilayers, thereby freeing the viral genome from interactions with viral proteins and enabling the viral RNA segments to migrate to the host cell nucleus, where influenza virus RNA replication and transcription occur (38, 77, 84). Genetic evidence suggests an interaction between M2 and M1 (81).
The cytoplasmic tail of M2 is highly conserved among virus strains but is not required for the ion channel activity of the protein (14, 75). Unlike the HA and NA proteins, M2 is not present in lipid rafts, which may account for the low amount of M2 within virions (32, 82, 83). A role for the M2 protein in virus assembly has been postulated, based on the effects of anti-M2 monoclonal antibodies on virus budding (25, 57) and reduced packaging of vRNPs in cells infected with influenza A viruses bearing truncations of the M2 cytoplasmic tail (40). This dual role for M2 is supported by studies on the influenza B virus protein BM2, which also has ion channel activity (45) and is involved in genome packaging (22, 28). In this study, we define specific residues in the M2 cytoplasmic tail necessary for infectious virus production, identify a role for the M2 cytoplasmic tail in virus particle budding, and demonstrate an interaction between the cytoplasmic tail of M2 and M1. Taken together, these results support a new model of influenza virus assembly that places the M2 protein in a central role as a facilitator of infectious particle formation.
MATERIALS AND METHODS
Plasmids.
The plasmid pCAGGS (pC) was used for all mammalian expression studies, and the influenza A/Udorn/72 virus M2 cDNA was used to generate all M2 cDNA constructs. The pC M2, pC M2Stop90, pC M2Stop82, and pC M2Stop70 vectors have been described previously (40). The cDNA encoding M2Stop90 was altered using PCR mutagenesis so the indicated residues would be alanines. The constructs generated were pC M2Stop90ala86-89, pC M2Stop90ala82-85, pC M2Stop90ala82-89, pC M2Stop90ala78-81, pC M2Stop90ala74-77, and pC M2Stop90ala70-73. The M2Stop90 protein supports virus replication in a manner identical to that of full-length M2 (40) and allowed for the differentiation of transcomplementing protein from full-length M2 based on electrophoretic mobility. To construct M2-M1 fusion proteins, the cDNAs encoding M2 and M2Stop70 were altered using PCR mutagenesis to eliminate the M2 stop codon and insert the cDNA encoding either the M1 protein from A/WSN/33 (M1W) or the M1 protein from A/Udorn/72 (M1Ud) generating the plasmids pC M2-M1W, pC M2Stop70-M1W, pC M2-M1Ud, and pC M2Stop70-M1Ud.
The cDNAs encoding the M2 cytoplasmic tail residues 45 to 97, 45 to 89, 45 to 81, 45 to 69, or 70 to 97 were amplified by PCR and cloned into the BamHI and SalI cloning sites in pGEX-4T-1 (Amersham Biosciences), generating the plasmids pGEX-M2Tail, pGEX-M2Tail90, pGEX-M2Tail82, pGEX-M2Tail70, and pGEX-M2Tail70-97, respectively. The cDNA for M2Tail90 was altered using PCR mutagenesis so the indicated residues of the expressed protein would be alanines. The constructs generated were pGEX-M2Tail90ala70-73 and pGEX-M2Tail90ala74-77.
The pBABE plasmid, which expresses puromycin N-acetyltransferase, was used in the generation of stable cell lines as previously described (42).
The plasmid pHH21 M-Udorn encodes the A/Udorn/72 M segment under control of the human RNA polymerase I promoter and murine RNA polymerase I terminator (40, 73). The pHH21 M-Udorn plasmid was altered using PCR mutagenesis to change M2 codons 25 and 26 to stop codons (pHH21 M1Udorn M2Stop). To generate pHH21 M1WSN M2Stop, the DNA between the NheI and StuI restriction enzyme sites of pHH21 M1-Udorn M2Stop was replaced with the corresponding fragment from pHH21 M-WSN. This DNA fragment contains all the M1 amino acid coding changes between the A/WSN/33 and A/Udorn/72 viruses.
Cells.
Madin-Darby canine kidney (MDCK) cells and human embryonal kidney cells (293T) were cultured in Dulbecco's modified Eagle medium (DMEM; Sigma) containing 10% fetal bovine serum (Atlanta Biologicals, Inc.), 100 U of penicillin/ml (Gibco), 100 μg of streptomycin/ml (Gibco), and 1 mM sodium pyruvate (Mediatech, Inc.). The cells were incubated at 37°C in a humidified 5% CO2 environment.
The procedure for generating stably transfected MDCK cells has been described previously (40). Briefly, MDCK cells were cotransfected with pBABE and the desired M2 expression vector, selected with puromycin (Sigma), and then sorted for M2-expressing cells using flow cytometry. The stable cells were maintained in media containing 7.5 μg/ml puromycin and 5 μM amantadine (Sigma).
Viruses.
The viruses used in this study were rWSN (a recombinant virus derived from A/WSN/33), rWSN M2Stop, rWSN M-Udorn (rWSN with the M segment replaced with that of influenza A/Udorn/72 virus), rWSN M1-Udorn M2Stop, rUdorn (a recombinant virus derived from A/Udorn/72), and rUdorn M2Stop. The M2Stop viruses encode a truncated M2 protein as a result of codons 25 and 26 being altered to stop codons. A similar recombinant virus has been previously described (79).
The parental viruses (rWSN, rWSN M-Ud, and rUd) have been described previously (49, 73). To rescue the M2Stop viruses, the 12-plasmid reverse genetics system and M2-expressing cell line were used as described previously (40).
Low-multiplicity-of-infection (MOI) growth curves were performed by infecting cells at an MOI of 0.01 PFU per cell. The cells were infected for 1 h at room temperature with rocking, washed extensively with phosphate-buffered saline (PBS), and maintained at 37°C in DMEM containing 5 μg/ml acetyl trypsin (Sigma), 0.1% bovine serum albumin (Calbiochem), penicillin, streptomycin, and sodium pyruvate. At the indicated times, an aliquot of medium was removed, stored at −70°C, and replaced with fresh medium.
High-MOI infections were performed by infecting cells at an MOI of 1.0 to 5.0. The cells were infected for 1 h at room temperature with rocking, washed extensively with PBS, and incubated in DMEM containing penicillin, streptomycin, and sodium pyruvate for 12 h. To purify viral particles, the medium was harvested, and the cell debris was removed by low-speed centrifugation. The supernatant was then layered onto a 20% sucrose in PBS solution and centrifuged at 178,000 × g for 1 h in a Beckman ultracentrifuge (model L-80) using an SW41 rotor. The supernatant was removed, and the pellet was suspended in either PBS or 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer.
For metabolic labeling experiments, MDCK cells were infected at a high MOI (1.0 to 5.0) with rWSN or rUdorn for 12 h. The cells were incubated for 30 min in DMEM lacking methionine and cysteine (DMEM Met/Cys−), 30 min in DMEM Met/Cys− supplemented with 50 μCi of [35S]methionine/cysteine (Promix; Amersham Biosciences), and 1 h in chase media (DMEM containing 10% fetal bovine serum, penicillin, streptomycin, and sodium pyruvate). All incubations were performed at 37°C. The cells were lysed in PBS containing 1% NP-40 (Calbiochem), 50 mM iodoacetamide (Sigma), and a protease inhibitor cocktail (54). Cell debris was removed by centrifugation at 16,000 × g for 10 min.
Influenza A virus particles were quantified by hemagglutination of 0.5% chicken red blood cells (54). The infectious viral titer was determined by plaque assay or 50% tissue culture infectious dose (TCID50) assay (40, 54).
Flow cytometry.
MDCK cells were removed from the tissue culture plate by trypsinization. The cells were stained for M2 surface expression using the monoclonal antibody 14c2. The secondary antibodies used were either goat anti-mouse immunoglobulin G conjugated to fluorescein isothiocyanate (Jackson Laboratories) or goat anti-mouse immunoglobulin G (Alexa Fluor 647; Molecular Probes). The specific staining was quantified using a FACSCalibur dual laser flow cytometer (Becton-Dickinson).
Western blotting.
Samples were analyzed by Western blotting as described previously (40). Polyacrylamide gels were either poured immediately prior to use or precast (Ready Gel; Bio-Rad). The polypeptides were separated by SDS-PAGE and transferred onto a polyvinylidene fluoride membrane (Immobilin-FL; Millipore) using a tank transfer system (Mini Trans-Blot; Bio-Rad). The blots were incubated with 14c2 (anti-M2), anti-M1 (HB-64; American Type Culture Center), anti-NP (HB-65; American Type Culture Center), V-314-511-157 (anti-H0 A/PR/8/34; National Institute of Allergy and Infectious Diseases), or anti-A/Udorn/72 virus (goat anti-Udorn structural proteins) antibodies. The primary antibodies were detected using species-specific secondary antibodies coupled to either horseradish peroxidase (Jackson laboratories) or Alexa Fluor 647 (Molecular Probes). Horseradish peroxidase activity was detected using ECL Plus (Amersham) and the chemifluorescent signal was detected using an FLA-5000 (FujiFilm) phosphorimager. The Alexa Fluor 647 signal was also detected on the FLA-5000.
Real-time reverse transcription (RT)-PCR.
The RNA from purified viral particles was extracted using the QIAamp viral RNA minikit (QIAGEN). The amount of M segment was quantified using an ABI 7000 sequence detection system (Applied Biosystems) (40, 78) as described previously, except the annealing temperature was reduced from 60°C to 50°C.
Glutathione S-transferase (GST) pull-down assay.
The pGEX-4T-1 vectors encoding the proteins of interest were transformed into BL21(DE3) pLysS (Stratagene) competent cells for protein expression. An overnight culture, grown in terrific broth at 37°C, was diluted 1:100 into fresh terrific broth and grown at 37°C until the optical density at 600 nm was approximately 0.5. The culture was moved to 30°C, and 0.8 mM isopropyl β-d-thiogalactopyranoside (IPTG; Gene Choice) was added to induce protein expression. The cultures were incubated at 30°C, with shaking, for 3 h, and then the bacteria were harvested by centrifugation. The cells were suspended in PBS and sonicated for 10 s on level 8, 10 s on level 9, and twice for 10 s each on level 10 (550 Sonic Dismembranator; Fisher Scientific). The samples were kept on ice during sonication with a 30-s pause between sonications. After sonication, Triton X-100 (Sigma) was added to a concentration of 1%, and the samples were incubated on ice for 30 min. The cell debris was removed by centrifugation at 12,000 × g. The protein was purified from the supernatant using glutathione Sepharose 4B (Amersham Biosciences). The glutathione Sepharose 4B was equilibrated with 10× the bed volume with PBS two times, the lysate was passed over by gravity flow, and the glutathione Sepharose 4B was washed with 10× the bed volume with PBS two times. The purified protein was eluted and stored in 50 mM Tris, pH 8.0, and 10 mM glutathione (Sigma).
A pull-down assay was performed using a metabolically labeled infected cell lysate. [35S]methionine/cysteine-labeled cell lysates were precleared using glutathione Sepharose 4B bound to GST and rocked at 4°C for 2 h. The supernatant was then incubated with glutathione Sepharose 4B bound to fusion protein for 1 h at 37°C. The beads were washed four times with lysis buffer and one time with PBS and suspended in 2× SDS-PAGE loading buffer. The proteins were resolved by SDS-PAGE and imaged using a phosphorimager.
Immunofluorescence confocal microscopy.
MDCK cells were plated onto glass coverslips and infected at a high MOI with either rWSN M1Ud M2Stop or rUd M2Stop when 100% confluent. At 12 h postinfection, cells were fixed in PBS containing 1% methanol-free formaldehyde (Polysciences, Inc.). The cells were stained with sera V-314-511-157 (anti-H0 A/PR/8/34; National Institute of Allergy and Infectious Diseases) or goat anti-Udorn, followed by species-specific secondary antibodies coupled to Alexa Fluor 488 (Molecular Probes). The coverslips were mounted in Prolong antifade (Molecular Probes), and the cells were imaged using a Zeiss LSM 510 Meta confocal microscope.
Transmission electron microscopy (TEM).
MDCK cells were plated onto an Aclar disc (Ted Pella Inc., Redding, CA) and infected at 75 to 100% confluence with either rWSN M2Stop or rWSN M1Ud M2Stop at a high MOI. At 12 h postinfection, cells were fixed in 2% paraformaldehyde-2.5% glutaraldehyde (Polysciences Inc., Warrington, PA) in 100 mM phosphate buffer, pH 7.2, for 1 h at room temperature. Following three washes in phosphate buffer, cells were postfixed in 1% osmium tetroxide (Polysciences, Inc.) for 1 h at room temperature. The samples were then rinsed extensively in distilled H2O prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA) for 1 h at room temperature. Following several rinses in distilled H2O, cells were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Sections of 70 to 80 nm were cut, stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA, Inc., Peabody, MA).
RESULTS
M2 expression in stable cell lines and from recombinant viruses.
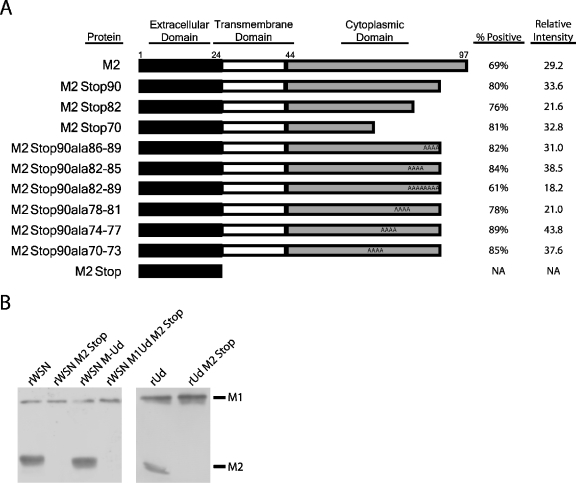
Truncating the M2 cytoplasmic tail by 8 residues does not affect infectious virus particle production, while truncating the cytoplasmic tail by 28 residues decreases infectious virus production 1,000-fold (40, 79). To identify the specific residues in the M2 cytoplasmic tail that are required for infectious virus production, stable MDCK cell lines were generated that expressed M2 proteins (influenza A/Udorn/72 virus strain) with C-terminal truncations and alanine substitutions (Fig. 1A). The percentage of cells expressing the recombinant proteins and the relative expression level were determined by flow cytometry. Only a small amount of M2 is required for infectious virus production (40), and all of the cell lines used exceed that minimum amount. Therefore, any variability in expression between the cell lines should not adversely affect virus assembly. Truncations of the M2 cytoplasmic tail have been demonstrated to have little or no effect on ion channel activity (40, 75).
FIG. 1.
Stable expression of cytoplasmic tail-deleted and mutated forms of the M2 protein in cell lines and recombinant viruses. (A) Schematic depicting the truncated and alanine-substituted M2 proteins. Flow cytometry was used to quantify the percentage of cells expressing the M2 proteins at the cell surface. The relative intensity represents the mean channel fluorescence of cells incubated with an M2-specific monoclonal antibody that recognizes the M2 extracellular domain divided by the mean channel fluorescence of cells incubated with secondary antibody alone. A cell line expressing the M2Stop protein was not generated; the schematic depicts the protein encoded by the M2Stop recombinant viruses. NA, not applicable. (B) MDCK cells were infected at an MOI of 1.0, lysed at 18 h postinfection, and analyzed by Western blotting with monoclonal antibodies against the M1 and M2 proteins.
To determine if there are strain-specific requirements for the M2 cytoplasmic tail, the recombinant strains rWSN (subtype H1N1), rUd (H3N2), and a reassortant strain rWSN M-Ud containing the M segment from rUd and the remaining 7 segments from rWSN were analyzed. The parental viruses and viruses encoding only the M2 ectodomain (referred to as M2Stop) were examined for M2 and M1 expression in infected cell lysates. The M2Stop viruses did not express a detectable amount of M2 protein and, therefore, are effectively M2 knockout viruses (Fig. 1B). M1 expression was not altered between the parental viruses and the M2Stop viruses (Fig. 1B).
Truncating the M2 cytoplasmic tail has general and virus strain-specific effects on influenza A virus replication.
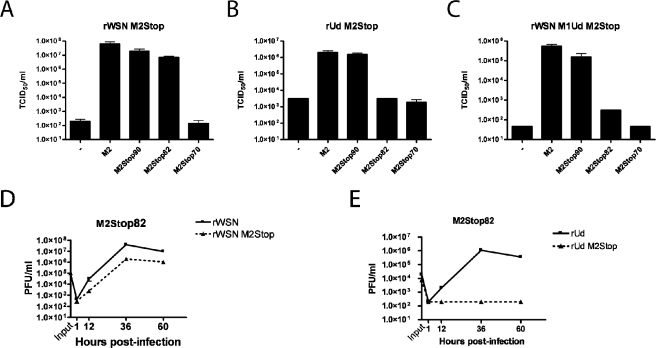
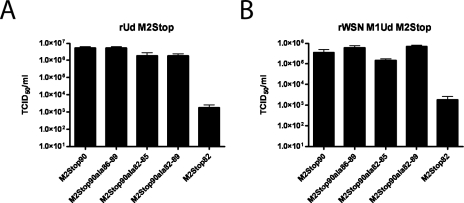
The ability of cells expressing various truncated forms of the M2 protein to support infectious virus production was analyzed with a modified tissue culture infectious dose assay. Influenza A virus strains that do not encode the M2 protein were grown on MDCK cells expressing full-length M2. These transcomplemented viruses were then used to infect MDCK cells expressing various truncated forms of the M2 protein. Cells expressing M2Stop70 or no M2 were unable to support significant amounts of infectious virus production after infection with rWSN M2Stop (Fig. 2A). However, infectious virus production for rWSN M2Stop was approximately 100,000-fold higher in cell lines expressing full-length M2, M2Stop90, or M2Stop82 (Fig. 2A). For rUd M2Stop, infectious virus production was approximately 1,000-fold higher for M2 and M2Stop90 than for cells expressing M2Stop82, M2Stop70, or no M2 protein MDCK cells (Fig. 2B). Since MDCK-M2Stop82 cells supported infectious virus production following infection with rWSN M2Stop but not rUd M2Stop, there appears to be a strain-specific requirement for amino acids 82 to 89 (compare Fig. 2A to 2B). An interaction between the M2 and M1 proteins could be important for virus budding (5, 15, 25, 57, 81). To determine if the rUdorn-specific requirement for amino acids 82 to 89 of the M2 protein was dependent upon the M1 protein, we infected cells with a reassortant virus strain, rWSN M1Ud M2Stop, which is identical to rWSN M2Stop except the M1 protein sequence has been replaced with that of rUd. Infectious virus production for rWSN M1Ud M2Stop was approximately 1,000-fold higher for cells expressing M2 or M2Stop90 cells than cells expressing M2Stop82, M2Stop70, or wild-type MDCK cells (Fig. 2C). The inability of MDCK-M2Stop82 cells to support infectious virus production for rWSN M1Ud M2Stop indicates that sequences in the M1 protein are responsible for the strain-specific requirements for amino acids 82 to 89 of the M2 cytoplasmic tail.
FIG. 2.
Truncation in the M2 protein cytoplasmic tail can reduce infectious virus production. MDCK cells stably expressing the indicated proteins were used to determine the TCID50/ml of rWSN M2Stop (A), rUd M2Stop (B), and rWSN M1Ud M2Stop (C). (D and E) A multistep growth curve was performed on MDCK-M2Stop82 cells with rWSN and rWSN M2Stop (D) or rUd and rUd M2Stop (E). Infectious virus titers were determined on MDCK-M2 cells.
A multistep growth curve was performed to further analyze the replication of rWSN M2Stop and rUd M2Stop on MDCK-M2Stop82 cells. rWSN M2Stop titers were 10- to 20-fold lower than those of rWSN on MDCK-M2Stop82 cells (Fig. 2D), while rUd M2Stop replication was at or below the limit of detection (Fig. 2E), confirming the rUd strain-specific requirement for M2 cytoplasmic tail sequences 82 to 89.
Role for the C-terminal 28 residues of M2 during influenza particle budding.
The small interfering RNA-mediated reduction of M2 protein levels or truncations of the M2 cytoplasmic tail decrease total viral particle production (39, 40, 79). MDCK-M2Stop70 cells released fewer virus particles than MDCK-M2 cells after infection with either rWSN M2Stop, rWSN M1UdM2Stop, or rUd M2Stop (determined by hemagglutination assay; data not shown), supporting the observation that the M2 protein cytoplasmic tail plays a role in virus particle assembly or release.
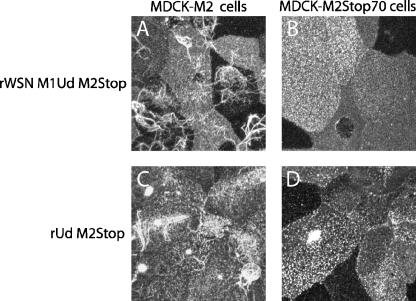
MDCK cells infected with either rWSN M-Ud or rUd produce filamentous particles that can be visualized by confocal microscopy (5, 11, 15, 57, 66). Filamentous particles have a diameter similar to spherical particles but can be up to 10 μm in length (44). A role for the M2 cytoplasmic tail in filamentous particle assembly was examined by infecting MDCK-M2 or MDCK-M2Stop70 cells with either rWSN M1Ud M2Stop or rUd M2Stop and determining the presence of filamentous virus particles by confocal microscopy at 12 h postinfection (Fig. 3). For rWSN M1Ud M2Stop and rUd M2Stop, expression of M2 in trans supported the assembly of filamentous particles (Fig. 3A and 3C). However, infection of MDCK-M2Stop70 cells with either strain resulted in a defect in filamentous particle assembly (Fig. 3B and 3D). These data suggest that the M2 cytoplasmic tail plays a role in the assembly of filamentous influenza A virus particles.
FIG. 3.
Production of filamentous influenza virus particles is dependent on the M2 cytoplasmic tail. MDCK-M2 (A and C) and MDCK-M2Stop70 (B and D) cells were infected at an MOI of 1.0, fixed at 12 h postinfection, and stained with antibodies specific for HA. Cells were infected with rWSN M1Ud M2Stop (A and B) or rUd M2Stop (C and D).
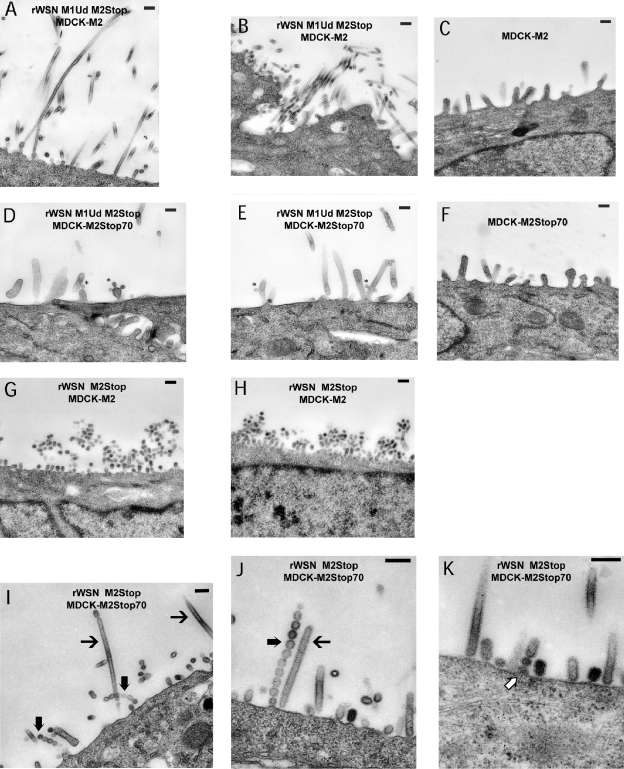
To investigate the effect of the M2 cytoplasmic tail on virus particle budding, we performed thin-section TEM on MDCK cells expressing either M2Stop70 or full-length M2 protein that were infected with either rWSN M2Stop or rWSN M1Ud M2Stop. Filamentous influenza particles were detected in MDCK-M2 cells infected with rWSN M1Ud M2 Stop (Fig. 4A and 4B), while no filaments were detected in mock-infected MDCK-M2 cells (Fig. 4C). In contrast, filamentous particles were absent from MDCK-M2Stop70 cells infected with rWSN M1Ud M2Stop (Fig. 4D and 4E). Infrequently, we were able to detect spherical particles budding from the cell surface (Fig. 4D and 4E), but most of the projections from the infected cell surface were microvilli which have a greater diameter than filamentous particles and were present in mock-infected MDCK-M2Stop70 cells (Fig. 4F). These data are consistent with the data in Fig. 3, and we therefore conclude that the influenza M2 cytoplasmic tail is important for the formation of filamentous influenza A virus particles.
FIG. 4.
Thin-section transmission electron microscopy images of budding influenza particles. MDCK-M2 (A, B, C, G, and H) and MDCK-M2Stop70 (D, E, F, I, J, and K) cells were infected at an MOI of 5.0 with rWSN M2Stop or rWSN M1Ud M2Stop. The cells were infected for 12 h and then processed and imaged by TEM. MDCK-M2 cells were infected with rWSN M1Ud M2Stop (A and B) or rWSN M2Stop (G and H). MDCK-M2Stop70 cells were infected with rWSN M1Ud M2Stop (D and E) or rWSN M2Stop (I to K). →, elongated filamentous particles; ➞, linear spherical particles; ⇒, budding linear spherical particles; *, budding particles. Bars, 200 nm. (C and F) Mock-infected cells. The text in the figures indicates the virus (top line) and cell line (bottom line) used in that panel. Two panels for each condition are shown to demonstrate infected cells from independent experiments and multiple fields.
To investigate the budding of spherical influenza virus particles, MDCK-M2 cells were infected with rWSN M2Stop virus and analyzed by thin-section TEM. Spherical particles were present abundantly at the surface of infected MDCK-M2 cells (Fig. 4G and 4H) compared to mock-infected MDCK-M2 cells (Fig. 4C). In contrast, we were able to detect very little virus particle budding in MDCK-M2Stop70 cells infected with rWSN M2Stop (Fig. 4I-K). The budding events that were detected resembled filamentous influenza virus particles—something not normally observed with viruses encoding the rWSN M1 protein—or linear chains of spherical particles projecting from the plasma membrane in a manner reminiscent of retroviruses bearing mutations in late domains (55, 65, 68). Assembly of the linear chains of spherical particles may occur as a result of virus assembly at discrete sites on the plasma membrane (32) coupled with a failure of the viral buds to be released (Fig. 4J). These results provide evidence that the cytoplasmic tail of M2 participates in virus particle budding.
Requirement for M2 residues 82 to 89 in infectious virus production.
MDCK-M2Stop90 cells supported infectious virus production for rUd M2Stop and rWSN M1Ud M2Stop, while the MDCK-M2Stop82 cells did not. To determine which amino acid residues in this region are critical for infectious virus production, mutated M2 proteins containing alanine substitutions (M2Stop90ala82-85, M2Stop90ala86-89, and M2Stop90ala82-89) were generated and stably expressed in MDCK cells (Fig. 1A). For both rUd M2Stop and rWSN M1Ud M2Stop, the alanine substitutions did not affect infectious virus production (Fig. 5A and 5B). This suggests that there may not be a specific sequence requirement in this region of the cytoplasmic tail but perhaps there is a length or structural requirement.
FIG. 5.
Effect of alanine substitution at M2 residues 82 to 89 on infectious virus production. MDCK cells stably expressing the indicated proteins were used to determine the TCID50/ml of rUd M2Stop (A) and rWSN M1Ud M2Stop (B).
Requirement for M2 residues 70 to 81 in infectious virus production.
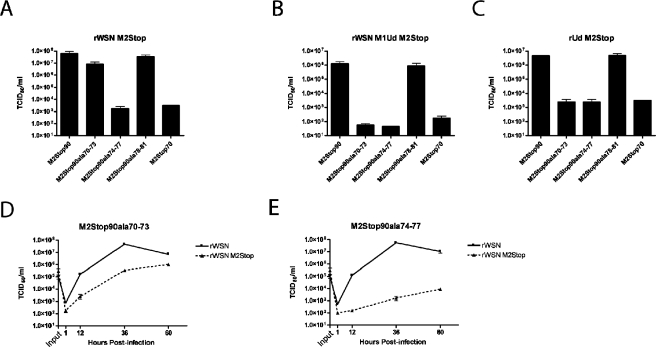
The inability of MDCK-M2Stop70 cells to support infectious virus production for both influenza virus strains rWSN and rUdorn suggested that residues 70 to 81 may be critical for both viruses. Site-directed mutagenesis was used to generate M2 proteins with alanine substitutions (M2Stop90ala70-73, M2Stop90ala74-77, and M2Stop90ala78-81) (Fig. 1A), and MDCK cells stably expressing these proteins were derived.
MDCK cells expressing M2Stop90ala74-77 did not support rWSN M2Stop infectious virus production (virus titers similar to those of MDCK M2Stop70 cells), indicating that residues 74 to 77 are critical for rWSN M2Stop (Fig. 6A). The cell lines expressing M2Stop90, M2Stop90ala70-73, and M2Stop90ala78-81 supported infectious virus production to equivalent amounts for rWSN M2Stop (Fig. 6A). However, some M2Stop90ala70-73 cells were still attached and visible at times when M2Stop90 cells were no longer attached to the tissue culture dish (data not shown), indicating that M2Stop90ala70-73 cells were not capable of supporting infection with rWSN M2Stop as efficiently as M2Stop90 cells. A multistep growth curve was performed with rWSN and rWSN M2Stop on M2Stop90ala70-73- and M2Stop90ala74-77-expressing cells (Fig. 6D and 6E). rWSN M2Stop replicated to 100-fold lower titers at 36 h postinfection than rWSN on MDCK-M2Stop90ala70-73 cells, while on MDCK-M2Stop90ala74-77 cells, virus titers were decreased 10,000-fold. These data indicate that M2 protein residues 74 to 77 are critical for infectious virus production, while residues 70 to 73 are important but not essential.
FIG. 6.
Effect of alanine substitution at M2 residues 70 to 81 on infectious virus production. MDCK cells stably expressing the indicated proteins were used to determine the TCID50/ml of rWSN M2Stop (A), rWSN M1Ud M2Stop (B), and rUd M2Stop (C). A multistep growth curve was performed on MDCK-M2Stop90ala70-73 (D) and MDCK-M2Stop90ala74-77 (E) with rWSN and rWSN M2Stop.
The role of residues 70 to 81 was also analyzed for rWSN M1Ud M2Stop and rUd M2Stop. M2Stop90ala70-73- and M2Stop90ala74-77-expressing cells did not support infectious virus production for either virus (Fig. 6B and 6C), while infection of M2Stop90ala78-81 yielded virus titers similar to that observed after infection of M2Stop90-expressing cells (Fig. 6B and 6C). These results indicate that M2 residues 74 to 77 and 70 to 73 are essential for rUdorn virus infection and that the M1 protein is a key determinant for this requirement.
Requirement for M2 residues 70 to 77 in infectious particle production.
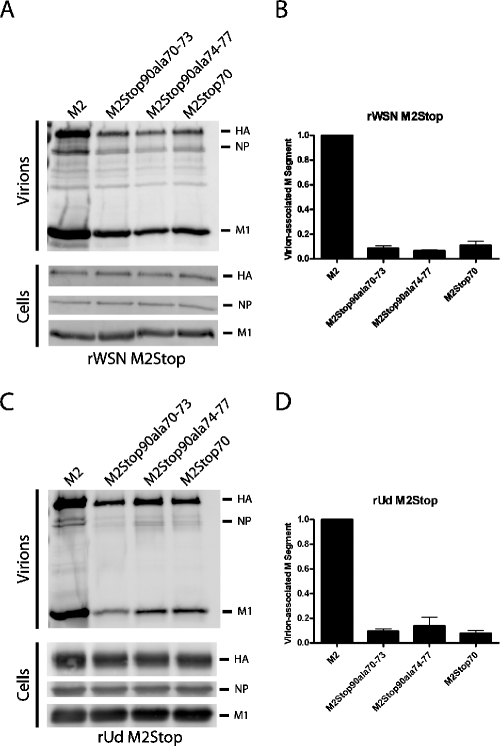
M2 residues 70 to 77 were identified as being important for the production of rWSN M2Stop and rUd M2Stop infectious particles. To determine if residues 70 to 77 are important during virus assembly and/or budding, cell lines were infected with rWSN M2Stop or rUdM2Stop, and the released virions were purified and analyzed by Western blotting (Fig. 7A and 7C). The amount of viral particles released from cell lines expressing the alanine-substituted M2 or M2Stop70 proteins was lower than the amount released from M2-expressing cells, as measured by hemagglutination assay (data now shown). Western blotting for HA, NP, and M1 indicated less viral protein was released from the cell lines expressing either the alanine-substituted M2 or M2Stop70 cell lines than was released from the M2-expressing cells (Fig. 7A and 7C). In three independent experiments, the amount of NP in the supernatant of cells expressing the M2 proteins with altered cytoplasmic tails was reduced by at least 80% compared to cells expressing full-length M2. The cytosolic amounts of NP varied by less than 10% between infected cell lines. M2 virion incorporation was not affected by the alanine substitution or truncation mutations (40; data not shown). The expression of viral proteins was equivalent between virus-infected cells, suggesting that a defect in virus assembly, as opposed to a reduction in viral protein synthesis, was responsible for the observed defect in virus particle secretion (Fig. 7A and 7C). To determine if genome release was also affected by the alanine-substituted M2 proteins, viral RNA was extracted from the purified virions and the M segment was quantified using quantitative RT-PCR. The amount of M segment released from cell lines expressing the alanine-substituted M2 or M2Stop70 proteins was decreased by approximately 10-fold compared to cells expressing the full-length M2 protein (Fig. 7B and 7D). These results suggest that the cytoplasmic tail of M2, specifically residues 70 to 77, is important for the release of infectious influenza virus particles but has a more limited effect on the overall production of virus particles.
FIG. 7.
Expression and incorporation of viral proteins and viral RNA into virions. The indicated cells were infected at an MOI of 5 with rWSN M2Stop (A and B) or an MOI of 1 with rUd M2Stop (C and D). At 12 h postinfection, the medium was collected and viral particles were purified by ultracentrifugation through a 20% sucrose cushion. An infected cell lysate was prepared by adding 1% SDS to the cells. (A and C) The protein composition of the virions and cells was analyzed by Western blotting with antibodies against HA, NP, and M1. (B and D) The viral RNA was extracted from purified particles, and the M segment was quantified by quantitative RT-PCR.
Interaction between the cytoplasmic tail of M2 and M1.
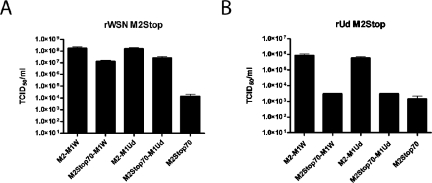
Previous data have implicated an interaction between the M2 cytoplasmic tail and M1 during virus assembly (5, 15, 25, 57, 81). To determine if an interaction between the M2 cytoplasmic tail and the M1 protein was critical for infectious virus production, expression vectors containing genetic fusions between M2 and M1 as well as between M2Stop70 and M1 were constructed (Table 1). Both the WSN M1 protein and the Udorn M1 protein were used in these constructs. Stably transfected MDCK cells expressing these constructs were generated, and the expression of the fusion proteins at the plasma membrane was confirmed by flow cytometry with a monoclonal antibody, 14c2, that recognizes the extracellular domain of the M2 protein (Table 1). Fusing the M1 open reading frame to M2Stop70 restored the replication of rWSN M2Stop compared to cells expressing M2Stop70 (Fig. 8A). The M1 proteins from rUdorn and rWSN were both able to support virus infection to comparable levels. Fusing M1 to the full-length M2 protein led to a slight increase in virus production compared to cell lines expressing M2Stop70-M1, but the virus titers were comparable to cell lines expressing full-length M2 alone (Fig. 2A), indicating that fusion of full-length M1 to the full-length M2 protein did not adversely affect the ability of M2 to function in this transcomplementation assay. However, the M2Stop70-M1 cell lines were unable to support infectious virus production after infection with rUd M2Stop (Fig. 8B). Again, fusion of full-length M1 to the full-length M2 protein led to virus titers comparable to those of cell lines expressing full-length M2 alone (Fig. 2B). The ability of M2Stop70-M1 fusions to support infectious virus production for rWSN M2Stop supports a model that requires an interaction between the M2 and M1 proteins; however, this interaction may be necessary, but not sufficient, to support rUd M2Stop replication, as this virus requires additional regions of the M2 cytoplasmic tail for virus infectivity (Fig. 2). Clearly, these results reaffirm the virus strain-specific requirements for M2 cytoplasmic tail sequences.
TABLE 1.
Stable expression of M2-M1 fusions in MDCK cells
| Protein | % Positivea | Relative intensityb |
|---|---|---|
| M2-M1W | 31 | 41.8 |
| M2Stop70-M1W | 37 | 25.2 |
| M2-M1Ud | 76 | 25.2 |
| M2Stop70-M1Ud | 47 | 25.4 |
The percent positive was determined by staining unpermeabilized cells with 14C2 followed by Alexa Fluor 647 goat anti-mouse IgG and analysis by flow cytometry.
The relative intensity represents the mean channel fluorescence for the M2-M1-expressing cells divided by the mean channel fluorescence of the cells incubated with secondary antibody alone.
FIG. 8.
Ability of an M2-M1 fusion protein to support infectious virus production. MDCK cells stably expressing the indicated proteins were used to determine TCID50/ml of rWSN M2Stop (A) and rUd M2Stop (B).
Direct physical interaction between the M2 cytoplasmic tail and the M1 protein.
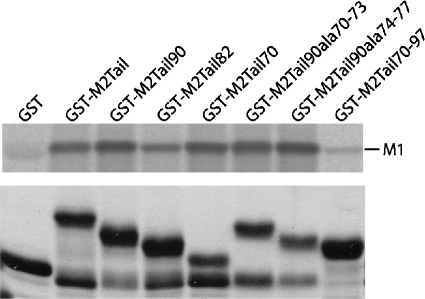
The ability of M2Stop70-M1 fusion proteins to support rWSN M2Stop replication provides further support for the hypothesis that the M2 and M1 proteins interact during virus assembly. To determine if there is a physical interaction between the proteins, the full-length M2 cytoplasmic tail was fused to GST and used in a pull-down experiment. A metabolically labeled rWSN-infected cell lysate was generated and incubated with each of the GST proteins. The M1 protein was pulled down by the GST-M2Tail protein but not by GST alone, providing evidence for a physical interaction between the proteins (Fig. 9). GST fusion proteins containing the M2Tail70, M2Tail90Ala70-73, or M2Tail90Ala74-77 sequences were able to interact with M1 (Fig. 9). Since rWSN M2Stop and rUdorn M2Stop viruses were not efficiently complemented by M2 proteins bearing these sequences (Fig. 6), we conclude that interactions between the M2 cytoplasmic tail and the M1 protein—at least as judged by this assay—are not responsible for the this phenotype. To confirm that M2 residues 70 to 97 were not involved in M1 binding, the GST-M2Tail70-97 protein was expressed and purified. GST-M2Tail70-97 did not pull down a substantial amount of M1, suggesting that this region of the cytoplasmic tail may not be directly involved in M1 binding. Similar results were obtained using an rUdorn-infected cell lysate (data not shown). These data provide evidence that the M2 cytoplasmic tail does interact with the M1 protein and that the binding site is between residues 45 and 69.
FIG. 9.
Interaction of M1 with the cytoplasmic tail of M2. A cell lysate from rWSN-infected MDCK cells metabolically labeled with [35S]methionine and cysteine was incubated with the indicated GST fusion proteins. The bound M1 protein was resolved by SDS-PAGE and imaged using a phosphorimager. A Coomassie-stained gel (bottom panel) shows the amount of purified GST fusion protein used for each pull-down assay.
DISCUSSION
The ion channel activity of the M2 protein (56) plays a key role during the uncoating process by disrupting interactions between the vRNPs, the M1 protein, and lipid bilayers, allowing the vRNPs to leave the site of virus-cell membrane fusion and move into the nucleus, where RNA transcription and replication occur (23, 37, 73). The ion channel activity is also responsible for altering the pH of the Golgi apparatus and preventing premature, low pH-induced conformational changes in HA proteins that are cleaved in the trans-Golgi by furin-like proteases (23, 59, 67, 69). We expand our knowledge about the role of the M2 protein in the influenza virus life cycle by presenting data that implicate M2 in the production of infectious virus particles by enhancing the efficiency of infectious virus production.
M2 has been shown to be dispensable for the formation of influenza A virus VLPs (20, 31), and our data are consistent with these findings. While infectious virus titers were reduced drastically in the absence of the M2 cytoplasmic tail, viral protein secretion was still detected. The M2 protein is necessary to promote the efficient transfer of a reporter vRNP in a VLP assay (21, 41, 50), but this was believed to be because uncoating required the M2 protein ion channel activity, not because M2 was important in packaging RNPs. The RNP reporter gene constructs used in these experiments also lacked RNA packaging signals (17, 18, 34, 80) and were assessed as single vRNPs, not with the additional 7 vRNPs that together comprise the influenza A virus genome. Our data imply that the packaging of a full complement of RNPs requires the M2 cytoplasmic tail, and in this context, the M2 protein enhances viral particle budding as well as infectious virus particle production. In the absence of the M2 cytoplasmic tail, virus particle budding occurs, but the particles are not infectious due to a lack of or reduced genomic RNA packaging. In the presence of the M2 cytoplasmic tail, the budding particles now contain a full complement of genomic RNA segments, as judged by increased infectivity and increased NS segment incorporation. Our data (Fig. 7) (40) suggest that at least 2 genomic RNA segments, NS and M, are not packaged efficiently in the absence of the M2 cytoplasmic tail. The use of techniques that can assess the packaging of all eight viral genomic RNA segments will provide further insight into the amount of genomic RNA segments packaged into influenza A virus particles bearing altered M2 cytoplasmic tails (52).
An interaction between the M1 and M2 proteins was implicated to play a role in virus budding and virion morphology based on the effect of M2 monoclonal antibodies on influenza A virus budding (25, 57, 81). Amino acid changes in the M1 (positions 31 and 41) and M2 (positions 71 and 78) proteins were identified in viruses that were resistant to the effects of M2 monoclonal antibodies (81). Our data also suggest that this region of the M2 cytoplasmic tail is important in virus assembly in conjunction with the matrix protein. The M2 cytoplasmic tail can affect the morphology of virus particles budding from infected cells (Fig. 3 and 4). Curiously, when we isolate virus particles from rWSN M-Ud-infected cells, we find very few filamentous particles (40), despite the presence of long filaments on the surfaces of cells expressing the same M segment proteins (Fig. 3). This may be due to reduced release of filamentous virus particles from these cells, but we cannot rule out technical issues pertaining to the isolation of filamentous virus particles.
The M1 protein has been shown to be important in virus budding (4, 10, 20, 31) as well as in the formation of filamentous and spherical virus particles (5, 15). We have separated the M2 cytoplasmic tail domains required for M1 binding (amino acids 45 to 69) from domains that affect the efficiency of infectious virus production (amino acids 70 to 77), implying that binding of M1 alone may not be enough to stimulate infectious virus production. However, based on the GST pull-down data alone, we cannot rule out that amino acids 70 to 77 enhance M1 binding to the M2 cytoplasmic tail. The region of the M2 cytoplasmic tail that binds M1 overlaps with a domain predicted to be involved in cholesterol binding (64); however, the significance of cholesterol binding by the M2 protein during influenza replication has not be elucidated, and for rUdorn, the virus strain used in all complementation assays in this study, the cholesterol binding motif is absent (64). We suggest that the M2 cytoplasmic tail has an M1 binding domain (amino acids 45 to 69) as well as an effector domain (amino acids 70 to 77) that mediates efficient infectious virus formation and virus morphology. The amino acids in the effector domain play essential as well as virus strain-specific roles in infectious virus particle assembly and morphology. In addition to residues 70 to 77, we also demonstrated an rUdorn strain-specific requirement for residues 82 to 89 for infection virus production.
The M2 cytoplasmic tail is modified by phosphorylation primarily at residue 64 (24) and palmitoylation at position 50 (70, 76), but both of these modifications have been shown to be dispensable for virus replication (9, 74). In light of our data on strain-specific roles for the M2 cytoplasmic tail, it will be important to reproduce these studies in additional influenza A virus strains.
Influenza A virus assembly occurs at discreet sites of the plasma membrane that are thought to be coalesced glycolipid rafts (32, 60, 72). Unlike the HA and NA proteins, the M2 protein does not localize to glycolipid rafts when expressed in virus-infected or cDNA-transfected cells (32, 83), and only a small amount of M2 expression is needed to restore replication to an M2-truncated virus (40). A precise mechanism for M2 incorporation into virus particles has not been demonstrated to date (53, 64); however, the M2 cytoplasmic tail amino acids 70 to 97 are not essential for virion incorporation of M2 (40). Therefore, we believe virion incorporation signals—if they exist—reside somewhere in the first 70 amino acids of the protein. It is interesting to speculate that the interaction between amino acids 45 to 69 of the M2 cytoplasmic tail and the M1 protein may be responsible for incorporation of M2 into virus particles.
We speculate that the influenza virus M2 protein mediates infectious virus production in a two-step process. First, the membrane-proximal region of the M2 cytoplasmic tail binds to the M1 protein. The M1 protein, specifically M1 that is in the vRNP complex, then interacts with additional M2 cytoplasmic tail sequences to mediate infectious virus budding. The requirement for the downstream sequences of the M2 cytoplasmic tail is dependent on the virus strain or, more precisely, the nature of the M1 protein encoded by the virus. For some virus strains (rWSN), there is only a core sequence requirement, but for other viruses (rUdorn), multiple amino acids are required for infectious particle formation, as shown by the M2-M1 fusion protein data (Fig. 8). The requirement for various regions of the M2 cytoplasmic tail may reflect the ability of the M1 protein to mediate viral budding or be dependent on the morphology of the particles. The incorporation of the viral genome, perhaps through interactions with M2, may be necessary to promote the interactions required for particle assembly. A similar assembly mechanism has been identified for the retrovirus murine leukemia virus (47, 48). The assembly of retroviruses is promoted by contact between the nucleocapsid domain of Gag and the viral genome, which promotes the Gag-Gag interactions necessary for particle formation (13). Influenza A virus may utilize the viral genome during assembly to facilitate interactions necessary to drive particle assembly. While speculative, this model is eminently testable using a combination of structural, biochemical, and reverse genetics approaches.
Acknowledgments
We thank the Molecular Microbiology Imaging Facility and the members of the Pekosz laboratory for stimulating discussions and critical evaluations.
M.F.M. was supported by grant T32 AI053629 and the Kauffman Fellowship Program in Life Sciences Entrepreneurship. This work was supported by a Young Investigator Award from the Whitaker Foundation (A.P.) and Public Health Services grant R01 AI053629 (A.P.).
REFERENCES
- 1.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barman, S., A. Ali, E. K. Hui, L. Adhikary, and D. P. Nayak. 2001. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 77:61-69. [DOI] [PubMed] [Google Scholar]
- 3.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourmakina, S. V., and A. Garcia-Sastre. 2005. The morphology and composition of influenza A virus particles are not affected by low levels of M1 and M2 proteins in infected cells. J. Virol. 79:7926-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinskaya, A. G., N. K. Vorkunova, G. V. Kornilayeva, R. A. Narmanbetova, and G. K. Vorkunova. 1982. Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol. 60:49-59. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinskaya, A. G., N. K. Vorkunova, and N. L. Pushkarskaya. 1982. Uncoating of a rimantadine-resistant variant of influenza virus in the presence of rimantadine. J. Gen. Virol. 60:61-66. [DOI] [PubMed] [Google Scholar]
- 8.Burleigh, L. M., L. J. Calder, J. J. Skehel, and D. A. Steinhauer. 2005. Influenza A viruses with mutations in the M1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 79:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castrucci, M. R., M. Hughes, L. Calzoletti, I. Donatelli, K. Wells, A. Takada, and Y. Kawaoka. 1997. The cysteine residues of the M2 protein are not required for influenza A virus replication. Virology 238:128-134. [DOI] [PubMed] [Google Scholar]
- 10.Chen, B. J., M. Takeda, and R. A. Lamb. 2005. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 79:13673-13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, J. C., A. W. Hampson, and R. C. Hamilton. 1980. An immunofluorescence study of influenza virus filament formation. Arch. Virol. 63:275-284. [DOI] [PubMed] [Google Scholar]
- 12.Delchambre, M., D. Gheysen, D. Thines, C. Thiriart, E. Jacobs, E. Verdin, M. Horth, A. Burny, and F. Bex. 1989. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 8:2653-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643-655. [DOI] [PubMed] [Google Scholar]
- 14.Duff, K. C., and R. H. Ashley. 1992. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology 190:485-489. [DOI] [PubMed] [Google Scholar]
- 15.Elleman, C. J., and W. S. Barclay. 2004. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 321:144-153. [DOI] [PubMed] [Google Scholar]
- 16.Enami, M., and K. Enami. 1996. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J. Virol. 70:6653-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii, K., Y. Fujii, T. Noda, Y. Muramoto, T. Watanabe, A. Takada, H. Goto, T. Horimoto, and Y. Kawaoka. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 79:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii, Y., H. Goto, T. Watanabe, T. Yoshida, and Y. Kawaoka. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. USA 100:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 20.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Puertas, P., I. Mena, M. Castillo, A. Vivo, E. Perez-Pastrana, and A. Portela. 1999. Efficient formation of influenza virus-like particles: dependence on the expression levels of viral proteins. J. Gen. Virol. 80(Pt 7):1635-1645. [DOI] [PubMed] [Google Scholar]
- 22.Hatta, M., H. Goto, and Y. Kawaoka. 2004. Influenza B virus requires BM2 protein for replication. J. Virol. 78:5576-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holsinger, L. J., M. A. Shaughnessy, A. Micko, L. H. Pinto, and R. A. Lamb. 1995. Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 69:1219-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughey, P. G., P. C. Roberts, L. J. Holsinger, S. L. Zebedee, R. A. Lamb, and R. W. Compans. 1995. Effects of antibody to the influenza A virus M2 protein on M2 surface expression and virus assembly. Virology 212:411-421. [DOI] [PubMed] [Google Scholar]
- 26.Hui, E. K.-W., S. Barman, D. H.-P. Tang, B. France, and D. P. Nayak. 2006. YRKL sequence of influenza virus M1 functions as the L domain motif and interacts with VPS28 and Cdc42. J. Virol. 80:2291-2308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Hui, E. K.-W., S. Barman, T. Y. Yang, and D. P. Nayak. 2003. Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motifs. J. Virol. 77:7078-7092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Imai, M., S. Watanabe, A. Ninomiya, M. Obuchi, and T. Odagiri. 2004. Influenza B virus BM2 protein is a crucial component for incorporation of viral ribonucleoprotein complex into virions during virus assembly. J. Virol. 78:11007-11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1532. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams, & Wilkins, Philadelphia, Pa.
- 31.Latham, T., and J. M. Galarza. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leser, G. P., and R. A. Lamb. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215-227. [DOI] [PubMed] [Google Scholar]
- 33.Li, Y., L. Luo, M. Schubert, R. R. Wagner, and C. Y. Kang. 1993. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J. Virol. 67:4415-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, Y., Y. Hong, and T. G. Parslow. 2005. cis-acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 79:10348-10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Licata, J. M., R. F. Johnson, Z. Han, and R. N. Harty. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 78:7344-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117-130. [DOI] [PubMed] [Google Scholar]
- 38.Martin, K., and A. Helenius. 1991. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 65:232-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCown, M., M. S. Diamond, and A. Pekosz. 2003. The utility of siRNA transcripts produced by RNA polymerase I in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology 313:514-524. [DOI] [PubMed] [Google Scholar]
- 40.McCown, M. F., and A. Pekosz. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79:3595-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mena, I., A. Vivo, E. Perez, and A. Portela. 1996. Rescue of a synthetic chloramphenicol acetyltransferase RNA into influenza virus-like particles obtained from recombinant plasmids. J. Virol. 70:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 44.Mosley, V. M., and R. W. G. Wyckoff. 1946. Electron micrography of the virus of influenza. Nature 157:263. [DOI] [PubMed] [Google Scholar]
- 45.Mould, J. A., R. G. Paterson, M. Takeda, Y. Ohigashi, P. Venkataraman, R. A. Lamb, and L. H. Pinto. 2003. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell 5:175-184. [DOI] [PubMed] [Google Scholar]
- 46.Muramoto, Y., A. Takada, K. Fujii, T. Noda, K. Iwatsuki-Horimoto, S. Watanabe, T. Horimoto, H. Kida, and Y. Kawaoka. 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 80:2318-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muriaux, D., S. Costes, K. Nagashima, J. Mirro, E. Cho, S. Lockett, and A. Rein. 2004. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 78:12378-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neumann, G., T. Watanabe, and Y. Kawaoka. 2000. Plasmid-driven formation of influenza virus-like particles. J. Virol. 74:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noda, T., H. Sagara, A. Yen, A. Takada, H. Kida, R. H. Cheng, and Y. Kawaoka. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439:490-492. [DOI] [PubMed] [Google Scholar]
- 53.Park, E. K., M. R. Castrucci, A. Portner, and Y. Kawaoka. 1998. The M2 ectodomain is important for its incorporation into influenza A virions. J. Virol. 72:2449-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. J. Davison and R. M. Elliott (ed.), Molecular virology: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 55.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinto, L. H., L. J. Holsinger, and R. A. Lamb. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517-528. [DOI] [PubMed] [Google Scholar]
- 57.Roberts, P. C., R. A. Lamb, and R. W. Compans. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127-137. [DOI] [PubMed] [Google Scholar]
- 58.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakaguchi, T., G. P. Leser, and R. A. Lamb. 1996. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J. Cell Biol. 133:733-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 61.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt, A. P., and R. A. Lamb. 2005. Influenza virus assembly and budding at the viral budozone. Adv. Virus Res. 64:383-416. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt, A. P., G. P. Leser, E. Morita, W. I. Sundquist, and R. A. Lamb. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schroeder, C., H. Heider, E. Moncke-Buchner, and T. I. Lin. 2005. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur. Biophys. J. 34:52-66. [DOI] [PubMed] [Google Scholar]
- 65.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simpson-Holley, M., D. Ellis, D. Fisher, D. Elton, J. McCauley, and P. Digard. 2002. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 301:212-225. [DOI] [PubMed] [Google Scholar]
- 67.Steinhauer, D. A., S. A. Wharton, J. J. Skehel, D. C. Wiley, and A. J. Hay. 1991. Amantadine selection of a mutant influenza virus containing an acid-stable hemagglutinin glycoprotein: evidence for virus-specific regulation of the pH of glycoprotein transport vesicles. Proc. Natl. Acad. Sci. USA 88:11525-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sugrue, R. J., G. Bahadur, M. C. Zambon, M. Hall-Smith, A. R. Douglas, and A. J. Hay. 1990. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 9:3469-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugrue, R. J., R. B. Belshe, and A. J. Hay. 1990. Palmitoylation of the influenza A virus M2 protein. Virology 179:51-56. [DOI] [PubMed] [Google Scholar]
- 71.Swenson, D. L., K. L. Warfield, K. Kuehl, T. Larsen, M. C. Hevey, A. Schmaljohn, S. Bavari, and M. J. Aman. 2004. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immunol. Med. Microbiol. 40:27-31. [DOI] [PubMed] [Google Scholar]
- 72.Takeda, M., G. P. Leser, C. J. Russell, and R. A. Lamb. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 100:14610-14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takeda, M., A. Pekosz, K. Shuck, L. H. Pinto, and R. A. Lamb. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 76:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas, J. M., M. P. Stevens, N. Percy, and W. S. Barclay. 1998. Phosphorylation of the M2 protein of influenza A virus is not essential for virus viability. Virology 252:54-64. [DOI] [PubMed] [Google Scholar]
- 75.Tobler, K., M. L. Kelly, L. H. Pinto, and R. A. Lamb. 1999. Effect of cytoplasmic tail truncations on the activity of the M2 ion channel of influenza A virus. J. Virol. 73:9695-9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veit, M., H. D. Klenk, A. Kendal, and R. Rott. 1991. The M2 protein of influenza A virus is acylated. J. Gen. Virol. 72(Pt 6):1461-1465. [DOI] [PubMed] [Google Scholar]
- 77.Wang, C., K. Takeuchi, L. H. Pinto, and R. A. Lamb. 1993. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 67:5585-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward, C. L., M. H. Dempsey, C. J. Ring, R. E. Kempson, L. Zhang, D. Gor, B. W. Snowden, and M. Tisdale. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 29:179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe, T., S. Watanabe, H. Ito, H. Kida, and Y. Kawaoka. 2001. Influenza A virus can undergo multiple cycles of replication without M2 ion channel activity. J. Virol. 75:5656-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe, T., S. Watanabe, T. Noda, Y. Fujii, and Y. Kawaoka. 2003. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 77:10575-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zebedee, S. L., and R. A. Lamb. 1989. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc. Natl. Acad. Sci. USA 86:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zebedee, S. L., and R. A. Lamb. 1988. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 62:2762-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhirnov, O. P. 1992. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology 186:324-330. [DOI] [PubMed] [Google Scholar]