Abstract
The human cytidine deaminases APOBEC3G (hA3G) and APOBEC3F (hA3F) are intracellular antiretroviral factors that can hypermutate nascent reverse transcripts and inhibit the replication of human immunodeficiency virus type 1 (HIV-1). Both enzymes have two cytidine deaminase motifs, although only the C-terminal motif is catalytic. Current models of APOBEC protein function imply editing is the principal mechanism of antiviral activity. In particular, hA3G is a more potent inhibitor of HIV-1 infectivity than hA3F and also induces a greater frequency of mutations in HIV-1 cDNA. We used hA3G/hA3F chimeric proteins to investigate whether cytidine deaminase potential reflects antiviral potency. We show here that the origin of the C-terminal deaminase motif is sufficient to determine the degree of mutation induced in a bacterial assay that measures mutations in chromosomal DNA. In contrast, this was not the case in the context of HIV-1 infection where the N-terminal deaminase motif also modulated the editing capabilities of the chimeras. Surprisingly, although three of the chimeric proteins induced levels of mutation that approximated those of parental hA3F, they displayed lower levels of antiviral activity. Most importantly, real-time PCR experiments revealed that the quantity of reverse transcripts detected in target cells, rather than the mutational burden carried by such DNAs, corresponded closely with viral infectivity. In other words, the antiviral phenotype of APOBEC proteins correlates with their ability to prevent the accumulation of reverse transcripts and not with the induction of hypermutation.
Several members of the apolipoprotein B mRNA-editing enzyme catalytic polypeptide (APOBEC) family of mammalian polynucleotide cytidine deaminases have been shown to have antiretroviral activity (1, 6, 16, 18, 19, 24, 34, 35, 39). These proteins are packaged into retroviral particles where they mediate the deamination of cytidines (C) to uridines (U) in nascent minus-strand viral cDNAs during reverse transcription, resulting in guanosine (G)-to-adenosine (A) hypermutation (or editing) of the viral plus-strand DNA (1, 8, 16, 18, 34, 38, 39). We have also shown that it is possible for at least one of these enzymes to edit human immunodeficiency virus type 1 (HIV-1) RNA, producing C-to-T (thymidine) changes on the positive strand (2). Of the various human enzymes, APOBEC3G (hA3G) and APOBEC3F (hA3F) are both potent inhibitors of HIV-1 infection in the absence of the viral Vif protein and are thought to be responsible for the G-to-A hypermutation of proviruses described in HIV-1-infected individuals (1, 6, 12, 15, 16, 18, 19, 24, 29, 30, 34, 35, 38, 39). Vif prevents the incorporation of hA3G and hA3F into assembling virus particles by recruiting a Cullin5-ElonginB-ElonginC E3 ubiquitin ligase, leading to their polyubiquitination and proteasomal degradation (17, 36).
Initially, C-to-U deamination was thought to be the principal mechanism through which the APOBEC proteins exert their antiviral effects. For instance, we showed that, in general, there was concordance between increasing mutational burden in reverse transcripts and the loss of viral infectivity imparted by different APOBEC family members (1). However, a series of more recent observations have indicated that this may not necessarily be the case and that inhibitory effects can be exerted in the absence of observable editing. First, it has been demonstrated that hA3G and hA3F inhibit hepatitis B virus replication in culture in the absence of discernible editing, seemingly by preventing the association of viral RNA with viral cores (23, 28), although it should be noted that other groups have detected very low levels of viral G-to-A hypermutation (22, 27). Second, APOBEC3A has been reported to inhibit the retrotransposition of intracisternal A-type particles, MusD, and LINE-1 retroelements and also infection by the parvovirus adeno-associated virus by unknown, nonediting, mechanisms (3, 4). Third, it has also been shown that hA3G expressed in unstimulated primary CD4+ T cells prevents infection by incoming HIV-1, without inducing hypermutation in newly synthesized viral cDNAs (5). Fourth, we have reported that engineered, catalytically inactive human APOBEC3G mutant proteins can still inhibit HIV-1 infection, again by an unknown mechanism (21).
A number of studies have reported that hA3G is a more potent inhibitor of HIV-1 than hA3F (1, 16, 34, 37), and we have previously noted that hA3G induces ∼10-fold more mutations in HIV-1 reverse transcripts than does hA3F (1). We therefore wanted to investigate whether these two manifestations of APOBEC function are inextricably linked. In order to study this, we created chimeric proteins between hA3G and hA3F and then characterized them in a number of different ways. Of note, these two enzymes share a domain organization in which there are two predicted cytidine deaminase core domains, but only the carboxy-terminal domain mediates hypermutation (7, 21). As expected, we found that the dinucleotide substrate preferences for each chimera matched that for the APOBEC protein from which the C-terminal domain had been derived: CpC for hA3G, and TpC for hA3F (the mutated residues are underlined). Several of the chimeras were able to edit DNA, both in the context of a bacterial assay and as viral reverse transcripts but, surprisingly, none were as potent as the parental hA3G and hA3F proteins at inhibiting HIV-1 infection. Indeed, the extent to which each chimera edited viral cDNA did not correlate with the extent of suppression of HIV-1 infection, nor, in fact, with the level of mutation detected in bacteria. Most notably, however, viruses produced in the presence of the chimeric proteins accumulated levels of reverse transcripts in target cells that correlated well with their relative infectivities. Thus, we suggest that the ability to prevent viral DNA accumulation represents an important aspect of APOBEC antiviral function and that this does not correlate with the magnitude of cytidine deaminase activity.
MATERIALS AND METHODS
Plasmid constructs.
Hemagglutinin (HA)-tagged hA3G and hA3F constructs have been described before (1, 24). Our hA3F cDNA differs from database entry AAH38808 at two residues: our clone expresses an alanine instead of threonine at residue 272 and a glycine instead of glutamic acid at position 370. Both changes have been recorded as natural polymorphisms. HA-tagged hA3G/hA3F chimeric cDNAs were generated by overlapping extension PCR, using hA3G and hA3F expression plasmids as templates. 5′ and 3′ cDNA fragments were amplified separately, by using a primer specific for the overlap region and, for 5′ fragments, a primer specific for the 5′ of hA3G/hA3F, KB 22 (5′-CGCGAAGCTTATGAAGCCTCACTTCAGAAACACA), or, for 3′ fragments, a primer specific for the 3′ end of hA3G, KB 52 (5′- CGCTCTAGAGTTTTCCTGATTCTGGAGAAT), or hA3F, KB 24 (5′- CGCTCTAGACTCGAGAATCTCCTGCAGCTT). The matching 5′ and 3′ fragments were then mixed, and full-length genes were amplified by using the two terminal primers. The resulting products were then cloned into pCMV4-HA, a vector encoding an HA tag as a 3′ fusion to the inserted cDNA (24). All constructs were verified by DNA sequencing. Each chimera was also subcloned from pCMV4-HA into the Escherichia coli expression plasmid pTrc99A after digestion with the restriction enzymes Asp718 and SmaI.
Expression vectors for wild-type and Δvif HIV-1 (pIIIB and pIIIB/Δvif, respectively) were modified by site-directed mutagenesis (Stratagene) at nucleotide 567 of the provirus to create a G-to-A substitution in the U5 region of the 5′-long terminal repeat (LTR) that would copy to the 3′-LTR during reverse transcription (1). This modification was introduced to ensure that all DNA sequencing data reflected the products of reverse transcription and not DNA from contaminating transfection cocktails.
Viral infections and cells.
All cell lines were maintained under standard conditions. Stocks of wild type or Δvif HIV-1 were prepared in the presence of individual APOBEC proteins by cotransfection of 1:1 ratios of APOBEC cDNA to HIV plasmid DNA into 293T cells, quantitated by enzyme-linked immunosorbent assay (ELISA) for p24CA content, and stored at −80°C. For quantitative PCR, virus-containing supernatants were treated with 20 μl of RQ1-DNase (Promega)/ml before quantitation. Single-cycle infectivities were determined by challenging the TZM-β-gal indicator cell line (33) (105 cells) with viruses corresponding to 3 to 5 ng of p24CA and measuring the induced expression of β-galactosidase activity in cell lysates after ∼28 h, using the Galacto-Star system from Applied Biosystems.
E. coli mutation assays.
The KL16 strain of E. coli was transformed with IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible APOBEC expression vectors or with pTrc99A empty vector as a control. Individual colonies were picked and grown to saturation in LB medium containing 100 μg of ampicillin/ml and 1 mM IPTG. Appropriate dilutions were plated onto agar plates containing either 100 μg of ampicillin/ml to measure cell viability or 100 μg of rifampin/ml to select for rifampin resistant colonies and then incubated at 37°C overnight. Mutation frequencies were reported as the number of rifampin-resistant colonies per 109 viable cells.
Immunoblot analysis.
Virions were concentrated by centrifugation through a 20% (wt/vol) sucrose cushion before immunoblot analysis. HA epitope-tagged APOBEC proteins, heat shock protein 90 (HSP 90), 14-3-3γ, or viral p24CA proteins were detected in whole-cell lysates of transfected 293T cells or in virion lysates by using the mono-HA.11 (Covance), HSP 90α/β (H-114; Santa Cruz Biotechnology), anti-14-3-3γ (C-16; Santa Cruz Biotechnology) or 24-2 monoclonal anti-CA antibodies, respectively; horseradish peroxidase-conjugated secondary antibodies; and enhanced chemiluminescence.
Retroviral sequence analysis.
Total DNA was purified from vif-deficient HIV-1-infected cells by using the DNeasy kit from QIAGEN. After treatment with DpnI for 2 h at 37°C, viral DNA was subjected to high-fidelity PCR (Advantage-HF 2 Polymerase; BD Biosciences) using HIV-1-specific oligonucleotides: EcoR1.nef.s (5′-CCGAATTCAGGCAGCTGTAGATCTTAGCCACTT) and BamH1.U5.a (5′-CAGGATCCGGTCTGAGGGATCTCTAGTTAC), gel purified, and cloned into pBluescript using EcoRI and BamHI restriction sites. The resulting fragments encoded a 650-bp nef-U5 region. All pBluescript clones were sequenced on an ABI 3730 Sequencer and analyzed.
Quantitative PCR.
DNase-treated viral preparations containing 30 ng of p24CA were added to 106 SupT1 cells rotating at 4°C for 2 h. Cells were then washed, resuspended in fresh RPMI medium, and incubated at 37°C for 0 to 48 h. Total DNA was purified by using the DNeasy kit from QIAGEN and eluted in a total volume of 200 μl. After treatment with DpnI for 2 h at 37°C, 2 μl of DNA was analyzed by real-time quantitative PCR. Early reverse transcription products were detected by using primers that amplify the region between nucleotides 500 and 635 of the provirus: oHC64 (5′-TAACTAGGGAACCCACTGC) and oHC65 (5′-GCTAGAGATTTTCCACACTG) and probe oHC66 (5′-FAM-ACACAACAGACGGGCACACACTA-TAMRA); later products of reverse transcription were detected with primers that amplify the region between nucleotides 500 and 695 of the provirus: oHC64 and gagM661as (5′-CTGCGTCGAGAGAGCTCCTCTGGTT) and the oHC66 probe. Reactions were performed in triplicate, in TaqMan Universal PCR master mix (UNG-less), using 0.9 pmol of each primer/μl and 0.25 pmol of probe/μl. After 10 min at 95°C, reactions were cycled through 15 s at 95°C, followed by 1 min at 60°C for 40 repeats, carried out on an ABI Prism model 7900HT (Applied Biosystems). The Δvif-HIV expression vector, pIIIB/Δvif, was diluted into purified SupT1 cellular DNA to create a series of control samples that were used to confirm the linearity of the assay.
RESULTS
To begin to investigate which regions of the APOBEC proteins are required for antiviral function and whether the same site(s) are responsible for editing, such that the two activities are inseparable, we took two family members with similar properties but different magnitudes of antiviral activity and editing potency, hA3G and hA3F, and constructed chimeric proteins from them.
Generation of hA3G/hA3F chimeric proteins.
The amino acid sequences of hA3G and hA3F are ca. 50% identical. In fact, the first 60 residues of the two enzymes are exactly the same, with the exception of residue 48, which is proline in hA3G and arginine in hA3F. There are several other stretches of identical residues between the two proteins, and we used PCR to synthesize fragments of DNA that coded for the amino terminus of the protein to one of these regions or from the same region to the carboxy terminus of the protein, for both hA3G and hA3F. An hA3G fragment was then combined with the corresponding hA3F fragment with overlapping identical ends and used as the template to PCR amplify a full-length chimeric APOBEC cDNA that was finally cloned into a vector expressing a C-terminal HA epitope tag. Five chimeras were constructed as shown in Fig. 1; chimera A, with the fusion junction between hA3G sequence and hA3F sequence at amino acids 114VTLTI (numbers refer to the position in the hA3G sequence); chimera B, with the junction at 121ARLYY; chimera D, with the junction at 283TSWSPC; and chimeras E and F, both with the protein fusion junction at 191EILR. As mentioned earlier, hA3G and hA3F have two cytidine deaminase domains that appear to have resulted from a duplication of a single domain (13). Based on sequence alignments and a predicted structure of APOBEC3 proteins (11), the fusion junction between chimeras E and F lies exactly at the beginning of the proposed duplication, in the region joining the two repeats. A sixth construct, chimera C, was designed but never synthesized.
FIG. 1.
Illustration of chimeric hA3G and hA3F proteins. Chimeric hA3G and hA3F cDNAs were constructed by using PCR, producing a panel of chimeric open reading frames. Dark gray shading indicates hA3G sequence, and light gray shading indicates hA3F sequence. The black boxes show the positions of the cytidine deaminase motifs.
Effect of hA3G/hA3F chimeric proteins on HIV-1.
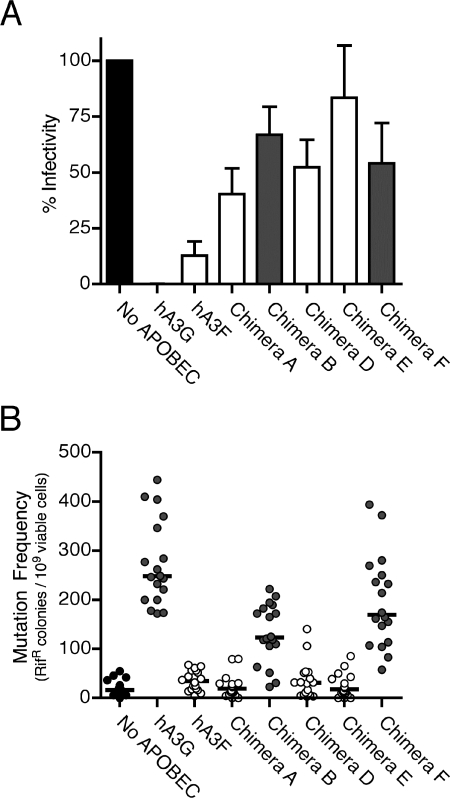
We first examined the effects of the hA3G/hA3F chimeric proteins on HIV-1 infectivity. Vectors expressing each chimera, HA-tagged hA3G, HA-tagged hA3F, or an empty vector were cotransfected into 293T cells, along with a proviral construct coding for either wild-type or vif-deficient (Δvif) HIV-1. The resulting viral stocks were collected, normalized for antigen content, and used to infect a reporter cell line. As expected, like hA3G and hA3F, none of the chimeras affected wild-type HIV-1 infectivity (data not shown). However, whereas both parental proteins markedly reduced the infectivity of Δvif HIV-1 (approximately 10-fold for hA3F and >100-fold for hA3G), the effects of the hA3G/hA3F chimeras were rather more modest (Fig. 2A). The greatest effect was consistently seen with chimera A, which contains the N-terminal deaminase motif of hA3G and the C-terminal deaminase motif of hA3F. Viruses produced in the presence of chimera A were ∼2.5-fold less infectious than those produced in the presence of a control vector (Fig. 2A). It is interesting that its counterpart, chimera B, which contains the N-terminal deaminase motif of hA3F and the C-terminal deaminase motif of hA3G, was less antiviral than chimera A. This pattern was reversed in the chimera E-F pair, where chimera F, with the C-terminal deaminase motif of hA3G was slightly more antiviral than chimera E, containing the C-terminal deaminase motif of hA3F. Thus, there appears to be no correlation between a chimeric APOBEC protein's ability to inhibit HIV-1 infection and the origin of the catalytically active C-terminal deaminase domain (Fig. 2, compare gray bars with white bars). Another interesting observation is that chimera B only differs from hA3G by 37 amino acids toward the N terminus, yet it has severely diminished anti-HIV capabilities. Mutating the arginine at position 48 in chimera B to proline, the residue found in hA3G at that position, had no effect on the antiviral activity of chimera B (data not shown).
FIG. 2.
Activity of hA3G/hA3F chimeras. (A) Effects of hA3G/hA3F chimeras on HIV-1 infectivity. vif-deficient HIV-1 was produced in the presence of either hA3G, hA3F, chimeric proteins, or a control vector. Normalized viral aliquots were used to challenge TZM-β-gal indicator cells, and productive infection was measured as the induction of β-galactosidase activity. Values are presented as percent infectivity relative to virus produced in the absence of any APOBEC protein (black bar). The gray and white bars indicate proteins expressing the C-terminal deaminase motif of hA3G and hA3F, respectively. The data are the means of five independent experiments, and error bars represent the standard deviation. (B) Effects of hA3G/hA3F chimeras on rifampin resistance (Rifr) in E. coli. Mutation frequencies are shown for 18 independent E. coli cultures expressing hA3G, hA3F, chimeric proteins, or a control vector. Each datum point corresponds to the number of Rif-resistant colonies in a single culture. The median mutation rate is shown for each condition (black line). Gray and white circles indicate proteins expressing the C-terminal deaminase motif of hA3G and hA3F, respectively.
Editing of bacterial DNA by hA3G/hA3F chimeric proteins.
Since our chimeric proteins had less antiviral activity than the parental hA3G and hA3F proteins, we next wanted to confirm that this was not simply due to a generalized loss of deaminase function. Accordingly, we tested the intrinsic DNA editing activity of the chimeras by using a previously described E. coli-based assay (9, 21). Bacterial colonies expressing chimeric or wild-type proteins were used to inoculate liquid cultures, which were then divided onto ampicillin- and rifampin (Rif)-containing agar plates. In the presence of a deaminase, the bacterial rpoB gene can be mutated, producing Rif-resistant colonies. The number of colonies on the rifampin-containing plate, divided by the viable cell count calculated from the ampicillin-containing agar plate, gives a measure of the frequency of mutation induced by that particular enzyme.
In this experiment, hA3G induced a large increase in median mutation frequency (15-fold above background), while the expression of hA3F triggered only a modest twofold increase above spontaneous background levels (Fig. 2B). This corresponds with previous reports that hA3F generates lower levels of mutation in viral cDNA than hA3G (1). Provocatively, bacteria expressing chimera B or chimera F, the two chimeras containing the C-terminal deaminase motif of hA3G, displayed elevated mutation frequencies of 7.5- and 10.3-fold, respectively, nearly as high as those of bacteria expressing wild-type hA3G (Fig. 2B, gray circles). The remaining three chimeras, all containing the hA3F C-terminal deaminase motif, induced much lower levels of mutation that resembled those obtained with hA3F (Fig. 2B, white circles). While it is difficult to interpret the editing abilities of chimeras A, D, and E, since they were all relatively inefficient at mutating the rpoB gene, both chimera B and chimera F clearly retained substantial levels of cytidine deaminase activity. It was also apparent, from all of the chimeras, that the extent of mutation induced depended on which deaminase motif was present at the C terminus and that deaminase potential in this assay did not reflect antiviral activity (compare Fig. 2A and B).
Expression and viral packaging of hA3G/hA3F chimeric proteins.
Since chimeras B and F edited bacterial DNA nearly as frequently as hA3G, and markedly more efficiently than hA3F, a lack of intrinsic deaminase function could not account for the reduced antiviral effects of the chimeras compared to the wild-type proteins. One possible explanation for the modest levels of antiviral activity was that the chimeric proteins were expressed at lower levels than hA3G and hA3F in eukaryotic cells or were less stable. However, immunoblot analysis of whole-cell lysates of the 293T cells used to produce virus stocks demonstrated that all of the chimeras were expressed at levels similar to those of the parental proteins in these transient transfections (Fig. 3, upper panels), with the presumed breakdown products corresponding most closely to those seen for the parental APOBEC protein with the same C-terminal deaminase motif. Protein expression was also confirmed in 293T cells by indirect immunofluorescence with an anti-HA tag antibody. All of the chimeric proteins appeared to have the same patterns of cytoplasmic localization as hA3G and hA3F (data not shown).
FIG. 3.
Expression and viral incorporation of hA3G/hA3F chimeras. Immunoblot analysis of HA-tagged APOBEC proteins in 293T cells transfected with APOBEC expression plasmids and concentrated virus produced from these cells was carried out. HSP 90 and HIV-1 CA proteins were also detected to confirm equal cell and virion loading, respectively.
Another possible reason for lower antiviral activity could be that the chimeric proteins are not efficiently packaged into viral particles and thus cannot exert their antiviral potential. Viruses synthesized in the presence of each chimera, wild-type hA3G or hA3F, or no APOBEC were harvested from 293T culture supernatants, purified, and analyzed by immunoblotting. As shown in Fig. 3 (lower panels), all of the chimeras were packaged into HIV-1 virions under these conditions. An abundant cellular protein that has previously been shown to be absent from viral particles (32), called 14-3-3γ, was not detected in any of the virion preparations, although it was readily detectable in the producer cells (data not shown), implying that the observed packaging of the chimeric proteins, as well as hA3G and hA3F, was specific.
Editing of viral DNA by hA3G/hA3F chimeric proteins.
Since at least two of the chimeras (B and F) were able to induce high levels of mutation in bacterial DNA and all of the chimeras were packaged into virions, we assessed the editing of viral reverse transcripts. We recovered viral cDNAs from cells infected with Δvif viruses produced in the presence of wild-type or chimeric APOBEC proteins and cloned and sequenced a 650-bp nef/3′-LTR fragment (the results are summarized in Table 1). Consistent with earlier reports, hA3G induced a high level of G-to-A mutation in HIV-1 reverse transcripts. Specifically, all examined reverse transcripts contained mutations, with an average of approximately 12 changes per 650-bp sequence. As in previous experiments, reverse transcripts from virions produced in the presence of hA3F also contained mutations, but at a much lower rate than for hA3G. As before, these mutations consisted mainly of G-to-A changes, but with some C-to-T changes also detectable. However, only 36% of the analyzed cDNAs harbored mutations, bringing the average number of mutations per 100 bp to 0.11. Surprisingly, we also found mutations in reverse transcripts synthesized by virions produced in the presence of the chimeric proteins. Chimeras A, B, and F all induced mutations, with an average number of mutations per 100 bp of 0.13, 0.17, and 0.09, respectively. These levels of mutation do not particularly echo the frequencies of Rif-resistant bacterial colonies induced by each chimera (chimera F induced more mutations than chimera B and considerably more than chimera A, Fig. 2B), but they are similar to the level of mutation hA3F induced in viral transcripts in this experiment. From these data, it is therefore evident that levels of mutation detected in viral cDNAs do not correlate with viral infectivity (Fig. 2A).
TABLE 1.
Summary of mutations induced by hA3G/hA3F chimeras
| Sample | Viral infectivity (%) | No. of clones sequenced | Total bp sequenced | % Clones mutated | Total no. of mutationsa | Avg no. of mutationsa
|
|
|---|---|---|---|---|---|---|---|
| Per clone | Per 100 bp | ||||||
| No APOBEC | 100 | 30 | 19,500 | 3.3 | 1 | 0.03 | 0.01 |
| hA3G | 0.05 | 9 | 5,850 | 100.0 | 107 | 11.89 | 1.83 |
| hA3F | 13 | 33 | 21,450 | 36.4 | 23 | 0.70 | 0.11 |
| Chimera A | 40 | 37 | 24,050 | 37.8 | 32 | 0.89 | 0.13 |
| Chimera B | 67 | 35 | 22,750 | 34.3 | 38 | 1.09 | 0.17 |
| Chimera D | 52 | 32 | 20,800 | 15.6 | 5 | 0.16 | 0.02 |
| Chimera E | 83 | 34 | 22,100 | 8.8 | 3 | 0.09 | 0.01 |
| Chimera F | 54 | 39 | 25,350 | 35.9 | 23 | 0.59 | 0.09 |
G-to-A and C-to-T mutations only.
Previous studies have identified local consensus sites for APOBEC-mediated cytidine deamination (1, 8, 9, 16, 34). hA3G preferentially edits the 3′-C in CpC dinucleotides, whereas hA3F favors the C of TpC dinucleotides. Figure 4 shows the frequencies with which each base was found at the −2, −1, and +1 positions relative to the substrate C residue, for the three chimeras that edited HIV-1 cDNA. Chimera A has the C-terminal deaminase motif of hA3F and also edits cytidine residues in a TpC context. Chimeras B and F both contain the hA3G C-terminal deaminase motif and show as strong a preference for editing the 3′-C in CpC dinucleotides as hA3G. Thus, the mutations detected in viral reverse transcripts can be attributed to cytidine deamination by the hA3G/3F chimeric proteins, and the C-terminal deaminase domain is entirely responsible for the local consensus site preference.
FIG. 4.
Comparison of the preferred sequence context for dC deamination in HIV-1 by hA3G, hA3F, or chimeric proteins. All of the mutations in HIV-1 induced by either hA3G, hA3F, or chimeras A, B, or F were aligned with respect to the dC residue targeted for deamination (position zero). The frequency (as a percentage) with which each of the four bases is found at positions adjacent to this dC is indicated. The consensus sequence is shown at the bottom of each minitable.
APOBEC proteins reduce the levels of reverse transcripts.
It has been reported that HIV-1 reverse transcripts accumulate to lower levels in T cells infected with virions made in the presence of hA3G (14, 18, 25). We, and others, have speculated that uridine residues could serve as the recognition signal for the premature degradation of edited reverse transcripts by cellular DNA repair enzymes (8, 10, 18). Alternatively, it is possible that viral cDNA synthesis itself is impeded by APOBEC proteins (26, 31). Therefore, we used fluorescence-monitored quantitative PCR to measure the temporal accumulation of viral cDNA in cells infected with viruses produced in the presence of hA3G, hA3F, or the chimeric proteins. We found that for all infections the level of early reverse transcription products increased with time to a peak at around 6 h and then declined (Fig. 5A). The level of transcripts detected in cells infected with virus produced in the presence of hA3G was very low at all time points and only just detectable (Fig. 5, light green). The amount of viral cDNA from the equivalent hA3F virus was higher (Fig. 5, light blue) but still only ca. 25% of the level of reverse transcripts seen for control virus produced in the absence of any APOBEC protein at 6 h postinfection. Interestingly, greater amounts of viral cDNA were recovered from cells infected with viruses produced in the presence of the chimeras, and the relative levels of reverse transcripts appeared to correlate well with the infectivities of the same virus preparations (Fig. 5B). A corresponding trend in levels was seen when we measured a later product of reverse transcription (data not shown).
FIG. 5.
Effects of hA3G/hA3F chimeras on levels of viral reverse transcripts. (A) Quantitative real-time PCR analysis of acute HIV-1 infection in SupT1 cells. Equivalent amounts of vif-deficient virions produced from 293T cells expressing hA3G (light green), hA3F (light blue), chimera A (pink), chimera B (orange), chimera D (dark green), chimera E (dark blue), chimera F (purple), or no APOBEC (red) were added to SupT1 cells, and total DNA was harvested at the indicated times after infection. The relative levels of HIV-1 early reverse transcription products compared to standard samples are indicated. (B) Effects of hA3G/hA3F chimeras on HIV-1 infectivity. The viral preparations described above were used in parallel to challenge TZM-β-gal indicator cells, and productive infection was measured as the induction of β-galactosidase activity. Values are presented as the percent infectivity relative to virus produced in the absence of any APOBEC protein. Colors are as in panel A.
Correlation of viral infectivity with viral cDNA levels and mutational burden.
From the experiments described in the present study, it seemed that the antiviral phenotypes of the chimeric APOBEC proteins did not correlate with the cytidine deaminase potential of these proteins. Three chimeras were able to edit HIV-1 reverse transcripts to the same degree as hA3F (Table 1), and yet none were as antiviral (Fig. 2A). In addition, the mutational burden carried by these transcripts did not appear to tally with the levels of transcripts measured by quantitative PCR (Fig. 5A). However, the amount of transcripts detected did appear to match the degree of viral infectivity. In order to establish whether there were statistically significant correlations between any of these observations, the levels of mutation (in number of mutations per 100 bp), the percentage of sequenced clones with at least one mutation, or the level of reverse transcripts (as a percentage of the amount detected from virus produced in the absence of any APOBEC protein) were plotted against viral infectivity (as a percentage of infectivity of virus produced in the absence of any APOBEC protein) (Fig. 6). The Pearson r value is given for each graph, as an indication of correlation, and the probability of the correlation occurring by chance is given as the P value. Measurements from two independent experiments (experiments 1 and 4) were used to compile the graphs in panels A and B, and data from three independent experiments (experiments 2, 3, and 4) were used in panel C. Each experiment consisted of eight separate viral stocks, prepared either in the presence of wild-type or chimeric APOBEC proteins, or in the absence of any APOBEC protein. Multiple experiments were combined in this way so as to maximize the number of datum points and not bias the plot. As seen in Fig. 6A, there is clearly no correlation between the rate of mutation of reverse transcripts and viral infectivity, with an r value of only −0.51, and a P value of 0.052. Since hA3G is such a potent deaminase, it may skew the statistics, so we also recalculated the correlation with the hA3G values excluded: here, the r value drops to −0.13, with P increased to 0.65. To address the possibility that a single mutation is enough to reduce infectivity, we also plotted the percentage of reverse transcripts with at least one mutation against infectivity (Fig. 6B). This resulted in an r value of −0.58, with a P value of 0.025, still a very weak correlation, which is reduced further when the hA3G data are removed, giving an r value of −0.34 and a P value of 0.24.
FIG. 6.
Correlation of viral infectivity with frequency of mutations and amounts of reverse transcripts. Correlation plots of viral infectivity (as the percent infectivity relative to virus produced in the absence of any APOBEC protein) versus the average number of mutations per 100 bp of sequenced viral cDNA (A), the percentage of clones sequenced that had at least one mutation (B), and the relative amount of early reverse transcripts compared to the amount of cDNA in the absence of any APOBEC protein (C), as measured by quantitative PCR, were prepared. Each point represents an individual viral preparation, and different experiments are identified by different symbols: panels A and B show data from experiments 1 (⧫) and 4 (•), and panel C shows the results of experiments 2 (▪), 3 (▴), and 4. The Pearson r value gives a measure of the correlation between each pair of variables. P is the probability that this correlation occurs by chance. Statistics were calculated by using GraphPad Prism version 4.0c.
In sharp contrast to these findings, however, Fig. 6C shows a strong correlation between the levels of reverse transcripts accumulated in the target cell at 6 h postinfection and the infectivity of the same viral preparation. In this case, the r value is 0.89 (only dropping to 0.86 if the hA3G data are excluded) and the P value is <0.0001 (not altering if the hA3G data are excluded). These observations therefore indicate that the level of hypermutation the APOBEC proteins induce in HIV-1 cDNA is not correlative with antiviral activity of the APOBEC protein, but, rather, that the effect the APOBEC proteins have on the accumulation of reverse transcripts is more indicative of their antiviral function.
DISCUSSION
The human cytidine deaminases hA3G and hA3F both inhibit infection of vif-deficient HIV-1 and generate mutations in nascent viral transcripts. However, as shown here and elsewhere, hA3G is a more potent inhibitor than hA3F and induces a much greater frequency of mutation in viral cDNAs (1, 16, 34, 37). Because the two proteins share a high degree of sequence identity, we reasoned that chimeras between the two would retain both antiviral function and deaminase activity. One report has already published an active hA3G/hA3F chimera, although the reciprocal chimera was not antiviral (7). That publication demonstrated that the C-terminal deaminase domain of APOBEC proteins determined the local sequence preference for retroviral cDNA deamination. However, another study suggested that the N-terminal domain is also required for antiviral activity (20), and we have previously reported that mutations in the C-terminal catalytic domain can impair deaminase function without blocking antiviral activity (21). Accordingly, we wanted to use hA3G/hA3F chimeras to investigate whether the magnitude of the antiviral effect and enzymatic function are dictated by the same site(s) in these two proteins and then to map this region(s). To this end, we made a set of chimeras that had the N-terminal deaminase motif of one APOBEC and the C-terminal deaminase motif of the other. To our surprise, none of the chimeric proteins inhibited HIV-1 infection to the extent of hA3F (Fig. 2A), although all were expressed equally in producer cells, and all were packaged efficiently into virions produced by transient transfection (Fig. 3).
The reductions in antiviral function could not be explained by a lack of editing ability either, since not only did two of the chimeras significantly increase mutation frequency in bacterial DNA (Fig. 2B), severalfold more than hA3F in fact, but three chimeras induced mutations in viral reverse transcripts, with orders of magnitude similar to those of hA3F (Table 1). This not only confirms that the chimeric proteins are folded, active cytidine deaminases but also that they must be incorporated into HIV-1 virions. The chimeras that induced high levels of mutation in bacterial DNA, chimeras B and F, both encoded the C-terminal deaminase motif of hA3G. Interestingly, however, these two proteins did not induce the high levels of mutation in viral cDNAs characteristic of hA3G, implying that whereas in bacteria the C-terminal domain alone is responsible for catalytic function, the frequency of mutation of viral reverse transcripts is modulated by the N-terminal domain of the APOBEC protein. Moreover, since chimeras A and E did not edit viral cDNAs to the same extent as hA3G either (Table 1), the presence of the N-terminal deaminase motif of hA3G is not sufficient to confer high levels of mutation. Like Hache et al. (7), we also found that the dinucleotide preference for mutation is determined by the C-terminal deaminase domain of the APOBEC protein (Fig. 4).
Chimera B only differs from hA3G by 37 amino acids, covering the N-terminal deaminase motif. A total of 67% of the virions produced in the presence of chimera B are infectious compared to less than 1% of virions made in the presence of wild-type hA3G. This indicates that the N-terminal deaminase motif of hA3G is very important for enabling a potent antiviral phenotype. Since both chimera B and hA3G contain the conserved HXEX27-28PCX2C deaminase motif in their N-terminal domains and we have shown previously that this site does not mediate hypermutation (21), we suspect that the function(s) of the N-terminal deaminase motif is distinct from editing. We do not think that chimera B suffers from misfolding since it is quite active in bacteria (Fig. 2B), and it is efficiently packaged into virions when it is transiently expressed (Fig. 3). Notably, a similar chimera made by Hache et al. yielded comparable results in that it also lost antiviral activity but still edited bacterial DNA. Accordingly, we propose that the N-terminal deaminase motif of double domain APOBEC proteins may be required for binding to the substrate, for oligomerization of APOBEC proteins, or for another activity that prevents accumulation of reverse transcripts in the infected target cells (see below). Further mapping studies comparing hA3G to chimera B may be informative for defining the determinants of hA3G's particularly potent anti-HIV phenotype.
Since chimeras A and B induced slightly more mutations in viral cDNAs than hA3F (Table 1) but were not as antiviral (Fig. 2A), it is clear that deaminase function is not the sole determinant of antiviral function. Moreover, higher levels of mutation in the viral cDNAs did not correspond with lower amounts of reverse transcripts accumulating in target cells (Fig. 5A), as would be expected if deamination reliably led to degradation of the transcripts. For example, chimera B induced more mutations than hA3F and all of the other chimeras, and yet higher levels of cDNAs were detected in cells infected with virus stocks made in the presence of this chimera than with viruses produced in the presence of hA3F or the other chimeras. Degradation of reverse transcripts by cellular endonucleases that cleave abasic sites resulting from cytidine deamination, followed by uracil excision, has been proposed as a model to explain aspects of APOBEC-mediated antiviral function (8, 10). Several groups have reported decreases in the numbers of copies of reverse transcripts in target cells, if the virus had been produced in the presence of hA3G and the absence of Vif (14, 18, 25, 26, 31), although it is not possible to determine precisely whether this is due to decreases in cDNA synthesis or accelerated degradation. We have shown that hA3F also inhibits the accumulation of viral cDNA, although not as well as hA3G (Fig. 5A). This could, in part, account for the difference in antiviral potency between the two proteins, since the relative levels of reverse transcripts correlated well with viral infectivity, whereas the frequency of mutations in the transcripts did not (Fig. 6).
If heavily mutated transcripts are degraded, it is possible that the levels of mutations in transcripts that accumulate (and are measurable) do not represent the degree of editing in the products that are actually degraded. However, the frequency of mutation induced by the chimeras in the bacterial assay was not indicative of their antiviral activity either (Fig. 2). Whatever the case, the observable mutation frequency is evidently a poor marker of relative antiviral activity. Assuming the degree of mutation observed is the true level, it is difficult to understand how the same frequencies of mutation could induce markedly different degrees of cDNA degradation for infections carried out in the presence of the chimeras versus hA3F. In addition, a recent study revealed that hA3G inhibits HIV-1 infectivity independently of at least one cellular DNA repair enzyme, UNG2 (14). Therefore, it seems plausible that instead of hypermutation and degradation, a large part of the antiviral activity of APOBEC proteins could be due to the impeded synthesis of reverse transcripts. This could be independent of deamination and would explain why hypermutation-deficient hA3G mutants can still be antiviral. Any reverse transcripts that are synthesized, by avoiding this block, may acquire mutations in the presence of active cytidine deaminases, and this would be expected to amplify the antiviral effect, particularly in the case of an enzyme as potent as hA3G. Although seemingly improbable, it is perhaps also worth noting that hypermutation-deficient proteins, as well as wild-type APOBEC proteins, could potentially mediate a “rare” editing event that has (to date) gone undetected but underlies an important aspect of the antiviral effect.
Thus, we have shown that the antiviral activities of APOBEC proteins reflect the ability of the proteins to prevent accumulation of reverse transcripts in target cells, which is distinct from the degree of mutation each protein induces. Taken together with our previous report (21), we propose that cytidine deamination of viral reverse transcripts is neither necessary nor sufficient for APOBEC-induced antiviral activity.
Acknowledgments
This study was supported by the Royal Society, the United Kingdom Medical Research Council, and the Biotechnology and Biological Sciences Research Council. K.N.B. is a Royal Society Dorothy Hodgkin Research Fellow. M.H.M. is an Elizabeth Glaser Scientist supported by the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse. [DOI] [PubMed]
- 2.Bishop, K. N., R. K. Holmes, A. M. Sheehy, and M. H. Malim. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480-485. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hache, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280:10920-10924. [DOI] [PubMed] [Google Scholar]
- 8.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 9.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10:1247-1253. [DOI] [PubMed] [Google Scholar]
- 10.Harris, R. S., A. M. Sheehy, H. M. Craig, M. H. Malim, and M. S. Neuberger. 2003. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat. Immunol. 4:641-643. [DOI] [PubMed] [Google Scholar]
- 11.Huthoff, H., and M. H. Malim. 2005. Cytidine deamination and resistance to retroviral infection: toward a structural understanding of the APOBEC proteins. Virology 334:147-153. [DOI] [PubMed] [Google Scholar]
- 12.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser, S. M., and M. Emerman. 2006. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 80:875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieffer, T. L., P. Kwon, R. E. Nettles, Y. Han, S. C. Ray, and R. F. Siliciano. 2005. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 79:1975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 17.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defense by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 19.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 20.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 21.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 22.Noguchi, C., H. Ishino, M. Tsuge, Y. Fujimoto, M. Imamura, S. Takahashi, and K. Chayama. 2005. G to A hypermutation of hepatitis B virus. Hepatology 41:626-633. [DOI] [PubMed] [Google Scholar]
- 23.Rosler, C., J. Kock, M. Kann, M. H. Malim, H. E. Blum, T. F. Baumert, and F. von Weizsacker. 2005. APOBEC-mediated interference with hepadnavirus production. Hepatology 42:301-309. [DOI] [PubMed] [Google Scholar]
- 24.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 25.Simon, J. H., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sova, P., and D. J. Volsky. 1993. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 67:6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suspene, R., D. Guetard, M. Henry, P. Sommer, S. Wain-Hobson, and J.-P. Vartanian. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 102:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 29.Vartanian, J.-P., P. Sommer, and S. Wain-Hobson. 2003. Death and the retrovirus. Trends Mol. Med. 9:409-413. [DOI] [PubMed] [Google Scholar]
- 30.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G-A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H.-Y. Chung, E. Morita, H. E. Wang, T. Davis, G.-P. He, and D. M. Cimbora. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 33.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 36.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 37.Zennou, V., and P. D. Bieniasz. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 349:31-40. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]